Abstract
The Rev regulatory protein of human immunodeficiency virus (HIV) facilitates the nuclear export of unspliced and partially spliced HIV RNAs. Using a Rev:MS2 phage coat protein fusion that could be targeted to bind and activate the Rev-responsive element (RRE) RNA or heterologous MS2 phage operator RNA, we analyzed the role(s) of the arginine-rich RNA binding domain in RNA binding and transactivation. The arginine-rich domain could be functionally replaced by a stretch of nine arginines. However, polyarginine substitutions expanded the RNA binding specificity of the resultant mutant Rev protein. Polyarginine insertions in place of residues 24 to 60 that excised the RNA binding and oligomerization domains of Rev preserved the activation for MS2 RNA, but not for the RRE. A nine-arginine insertion outside of the natural context of the Rev nuclear localization signal domain was incompatible with activation of either RNA target. Insertions of fewer than eight arginines impaired RRE activation. Interrupted lysine clusters and disruption of the arginine stretch with lysine or neutral residues resulted in a similar phenotype. Some of these mutants with a null phenotype for RRE activated the heterologous MS2 RNA target. Under steady-state conditions, mutants that preserved the Rev response for RRE RNA localized to the nuclei; those with poor or no Rev response accumulated mostly in the cytoplasm. Many of the cytoplasmically resident derivatives became nuclear when leptomycin B (LMB) treatment inhibited nuclear export of nuclear export signal-containing proteins. Mutants that had a null activation potential for either RNA target were particularly resistant to LMB treatment. Abbreviated nuclear residence times and differences in RRE binding affinity may have compromised their activation potential for RRE. High-affinity binding to MS2 RNA through the intact coat protein was sufficient to overcome the short nuclear residence times and to facilitate MS2 activation by some derivatives.
The Rev regulatory protein of human immunodeficiency virus type 1 (HIV-1) is required for the expression of unspliced and partly spliced RNAs for the viral structural proteins (19, 31, 45). Rev modulates splicing, nuclear export, and cytoplasmic utilization of unspliced or partially spliced viral mRNAs (11, 13, 21, 22, 52, 54, 74) by binding to a highly structured Rev- responsive element (RRE) RNA sequence embedded in the env mRNA (17, 30, 33, 36, 52, 57, 72, 97). RRE RNA folds into four stem-loops designated A, C, D, and E or stem-loops I, III, IV, and V with a branched stem-loop structure (B, B1, or B2 [or stem-loop II A, B, or C]) linked by a central loop (36, 52, 57). Most of the RRE structure is dispensable for Rev activity, and a minimal structure composed of the B, B1, or B2 (or stem-loop II A, B, or C) subdomain was active both in vitro and in vivo (37, 41). Rev is a basic phosphoprotein that shuttles between the nucleus and the cytoplasm (24, 42, 61, 70, 95) but accumulates in the nucleus, concentrating in the nucleolus under steady-state conditions. Like many viral and cellular trans-activators, Rev contains separate peptide modules for RNA binding and activation functions. Within the N-terminal region of Rev, a basic sequence between positions 35 and 50, containing a total of nine arginine residues, mediates RNA binding (8, 32, 34, 44, 57, 76, 97, 98), nuclear targeting (nuclear localization signal [NLS]), and nucleolar localization (nucleolar localization signal [NOS]) functions (8, 15, 32, 39, 51, 88). This RNA binding and NLS domain of Rev has also been shown to interact with the importin-β subunit facilitating import of Rev into the nucleus (35, 66, 83). Rev protein oligomerization and optimal Rev function require sequences between positions 24 and 35 and positions 51 to 65 flanking the 35-to-50 NLS-NOS motif on the N and C termini, respectively (40, 46, 50, 53, 64, 98). Genetic studies have identified a leucine-rich motif near the C-terminal third of Rev between residues 75 and 93 as the effector domain (38, 56, 60, 88, 90). Substitution mutants with changes at selected leucine residues in this domain not only have a null phenotype but also dominantly interfere with the function of the wild-type Rev (5, 20, 51, 55). The effector domain comprises the nuclear export signal (NES) that interacts with the highly conserved CRM1 protein (also referred to as exportin 1) (6, 23, 25, 26, 65, 75) and nuclear eIF-5A (73). The interaction of importin-β with the NLS and the binding of CRM-1 to the NES results in shuttling of Rev between the nucleus and the cytoplasm.
In spite of the wealth of biochemical and biophysical data addressing the kinetics of Rev binding to RNA and the many elegant genetic and biochemical studies that have identified the cellular proteins recruited by the Rev effector domain to facilitate RNA export, several important questions remain to be answered. For instance, it is not established whether Rev contacts RNA in a sequence-specific manner or whether the interaction is mediated by the recognition of unusual bends and bubbles in the RNA secondary structure (2, 16, 36, 37, 58). Second, it is not clear whether Rev binds to a single site on the target RNA or whether there are hierarchical sites on the RNA for Rev binding (16, 34, 43, 44, 82). Some of the issues relevant to RNA binding kinetics and stoichiometry and to mapping of the core site on the RNA have been addressed in recent studies (16, 18, 34, 58, 86). However, the precise sequence requirements of the NLS domain in RNA binding and nuclear import and the role(s) of oligomerization domains in RNA binding specificity and nuclear transport have not been well defined. For instance, replacement of an arginine at position 35, 38, 39, or 44, the threonine at 34, or the asparagine at 40 in the context of 17-residue Rev peptide-spanning residues 34 to 50 abolished the in vitro RNA binding potential (63, 76, 98). However, alanine scanning mutagenesis of the same arginine-rich motif in the context of Rev demonstrated an inherent redundancy of arginines, since any of the eight arginines could be mutated individually without affecting RNA binding (32). A mutation that exchanged a tryptophan in the arginine-rich domain for a serine preserved RNA binding in vitro but impaired nuclear localization of Rev and therefore led to a null activation phenotype. However, mutants with a change at the same tryptophan in the context of a fusion with the hormone-responsive domain of glucocorticoid receptor retained nuclear targeting in response to hormone but were compromised for trans-activation, suggesting that RNA binding or some other function may have been compromised (39). A mutant in which an asparagine within the same domain was replaced by aspartic acid lost both RRE RNA binding and nuclear localization (32). Analysis of the putative multimerization domain(s) of Rev also led to somewhat ambiguous conclusions. Mutants with substitutions within the sequence flanking the 5′ end or the 3′ end of the NLS were defective for multimerization in vitro and lacked transactivation potential for the native RRE target. However, the same mutants were capable of in vivo interaction with the minimal RRE RNA target, SLIIB, and when introduced as a Tat-Rev fusion protein activated the HIV long terminal repeat (LTR)-containing promoter-proximal SLIIB RNA target (50, 53, 79, 80).
In this study, we have analyzed the basic domain of Rev with respect to RNA binding specificity, nuclear localization, and trans-activation of bound RNAs. Using Rev-MS2 coat protein fusions which could be targeted to the HIV-1 RRE or the unrelated MS2 phage operator RNA (59, 87), we show critical differences in the activation phenotype of Rev mutants for the various RRE derivatives and MS2 RNAs and discuss their mechanistic significance.
MATERIALS AND METHODS
gag expression plasmids.
The construction of the HIV-1 LTR-linked gag expression plasmids used in this study has been described elsewhere (36, 37, 87). Briefly, the recombinants were constructed by site-directed mutagenesis of RRE DNA using a commercial M13-based protocol (Mutagene kit; Bio-Rad Laboratories), and the respective mutant DNAs were exchanged for RRE in the HIV-1 LTR-linked gag expression vector containing wild-type (wt) RRE. The RREZ-MS that replaced the Rev-responsive stem-loop II sequences for the phage MS2 translational operator wt sequence, the mutant derivatives of MS2 operator, the bivalent chimeras containing various combinations of wt or mutant derivatives of RRE stem-loop, and MS2 have also been described (36, 37, 87). gag-TAR and gag-VAI were constructed by exchanging the RRE sequence in the HIV LTR-linked gag expression plasmid for the PCR-amplified HIV-1 TAR and adenovirus VAI DNAs, respectively (67). Human T-cell leukemia virus type 1 (HTLV-1) RexRe containing the HIV-1 gag expression plasmid was a gift from George Pavlakis, National Cancer Institute, Frederick, Md.).
Expression plasmids for Rev, Rev–MS-C fusion proteins, and other activators.
HIV-1 Rev was expressed either from the TAT-responsive HIV-1 LTR or from the constitutive Rous sarcoma virus (RSV) LTR (36). Rev–MS-C, denoting a tandem fusion of the Rev and MS-C open reading frames (ORFs), had been cloned into the RSV LTR-linked eukaryotic expression plasmid pRSV.5 (87). The various deletion and insertion mutants (including the polyarginine insertions, etc.) were constructed by one- or two-step site-directed mutagenesis of Rev–MS-C containing M13 mp18 phage DNA (87). Most of the Rev-MS chimeras were cloned at the XbaI site of an RSV LTR-linked eukaryotic expression plasmid, pRSV.5; others were cloned into a commercial RSV LTR expression plasmid, pRC (Invitrogen Corp). After we verifyied the DNA sequence of the individual Rev–MS-C, the fusion genes were PCR amplified with T7 phage RNA polymerase promoter and terminator sequence tags at the 5′ and 3′ ends, respectively. The resulting T7 templates were transcribed and translated in vitro using a coupled system (Promega Corp.) with [35S]methionine to label the proteins. The various Rev–MS-C derivatives yielded fusion proteins of the expected molecular mass that were immunoprecipitated with anti-Rev antiserum. HIV-1 LTR-linked or cytomegalovirus (CMV) promoter-linked Tat expression plasmids have been described before. RSV LTR-linked HTLV-1 Rex and HIV-1 LTR-linked linked Tev (incorporating the first exon of Tat, a small portion of env, and the second exon of Rev) were gifts from George Pavlakis.
Escherichia coli expression of Rev–MS-C fusion proteins.
The various Rev and Rev–MS-C fusion protein derivatives were expressed in E. coli as fusion proteins, linked to the C terminus of E. coli maltose-binding protein (MBP), from an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible β-galactosidase promoter using a commercial kit (New England Biolabs, Beverly, Mass.). Individual recombinant clones were transferred to the lone XbaI site in the MBP vector. MBP fused protein expression was induced by IPTG (1 mM) at an optical density at 600 nm (OD600) of 0.3, followed by a 4-h incubation at 30°C. Bacteria were disrupted using a French press at 10,000 lb/in2 in a buffer (10 ml per packed cell volume) containing 50 mM Tris-HCI (pH 7.8), 50 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol (DTT), 1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride (AEBSF), aprotinin (2 μg/ml), leupeptin (1 μg/ml), and pepstatin (2 μg/ml). The cell extracts were centrifuged at 25,000 × g for 15 min to collect the pellet containing inclusion bodies. MBP-tagged fusion proteins were purified by affinity chromatography on amylose resin as described by the manufacturer. Procedures pertinent to MBP tag excision and purification of the MBP-free proteins were done essentially as described by the manufacturer (New England Biolabs). Completion of Factor Xa digestion was monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting with rabbit anti-Rev antiserum. After removal of the residual maltose by hydroxylapatite chromatography, MBP and undigested MBP fusion proteins were removed by binding them to amylose resin. The respective fusion proteins in the flow-through fraction from the amylose chromatography were bound to carboxymethylcellulose, CM52, or phosphocellulose, P11 (Whatman Corp.), and batch eluted with NaCl. Protein concentrations were determined by Bio-Rad Bradford assay. Fractions were monitored for Rev by dot immunoblotting and PhosphorImager quantitation (Molecular Dynamics). The Rev protein eluates were pooled, concentrated, and dialyzed against phosphate-buffered saline (PBS) using Centricon 10 columns (Amicon Corp).
RNA synthesis and protein binding.
DNA templates, tagged at the 5′ end with the T7 promoter, were generated by PCR using primer pairs corresponding to the ends of the desired RNA. T7 RNA polymerase initiated transcription to generate [α-32P]UMP-labeled RNAs, and the RNA purification methods have been described elsewhere (36, 37, 87). RNA-protein binding was evaluated by electrophoretic mobility shift assay (EMSA). Reaction mixtures containing different protein samples, heparin (5 μg), and yeast tRNA (500 ng) in HEPES binding buffer (20 mM HEPES-KOH, pH 7.9; 62 mM KCl; 0.15 mM DTT; 6% glycerol) were preincubated at 30°C for 10 min. 32P-labeled RNA was added, and incubation continued for another 10 min at 30°C. Samples were electrophoresed at 30 mA and 4°C through a 5% native polyacrylamide gel in 0.5× Tris-borate-EDTA. Radioactivity was visualized by autoradiography of the dried gels. From preliminary titration experiments, at a protein/RNA ratio of about 5:1, approximately 90% of the RNA was converted into an RNA-protein complex. Rev was expressed in E. coli and purified to near homogeneity (92).
Transient-expression assay.
The general protocols were as described before (36, 37, 87). Approximately 2 × 106 HeLa or Cos-7 cells were electroporated at 300 V and 250 μF in a Bio-Rad electroporator with the individual gag plasmids (5 μg) and 2 μg of pHIV-TAT (pSV40 TAT for Cos cells) with or without the indicated Rev or the Rev-MS coat protein fusion protein plasmids. For expression of Rev, 2.5 or 5 μg of an HIV-1 LTR-linked or RSV LTR-linked plasmid was used. The Rev-MS fusion protein plasmids (5 μg per experiment) were expressed from the RSV LTR. HIV-1 LTR-linked chloramphenicol acetyltransferase (CAT) or luciferase (LUC) (2 μg) plasmid was added to normalize for constant LTR transcription (by CAT or LUC assay on aliquots) in transfections using HIV LTR-linked gag plasmids. CMV-CAT or RSV LTR-LUC were coexpressed for normalizing transfections not involving HIV LTR-based plasmids. Aliquots of transfected cells were removed at 48 h and used for immune detection of Rev and Rev-MS fusion proteins (see below). For immunofluorescence experiments, an aliquot of cells was plated on a coverslip immediately after electroporation and processed for Rev and Rev-MS fusion proteins. gag expression was quantified by p24 enzyme-linked immunosorbent assay (ELISA) of cell extracts (Coulter Diagnostics). p24 gag expression levels were normalized to constant values of CAT or LUC activity from HIV LTR-linked CAT. Each gag ELISA value from HeLa cell transfections represented a mean of five or six independent transfections using at least two preparations of plasmid DNA. CAT expression was quantified by using a commercial CAT ELISA kit (Boehringer Mannheim), and the LUC expression was quantified by using a commercial kit (Promega Corp.) designed for a luminometer.
Immunoblotting.
Immunoblotting for Rev and indirect immunofluorescence detection of Rev and Rev-containing proteins in transfected cells have been described in detail before (87). For immunoblotting, commercial (Intracel Corp., Issaquah, Wash.) rabbit polyclonal anti-Rev antiserum was used routinely to avoid the loss of detection of certain deletion mutants that may have lost immunoreactive epitopes. The filters were then incubated with the appropriate species-specific secondary antibody tagged with horseradish peroxidase. Finally, the immunoreactive bands were visualized by means of a commercial chemiluminescence protocol (Amersham Corp.) and quantified by scanning.
Immunofluorescence microscopy.
For indirect immunofluorescence microscopy, permeabilized and fixed cells on coverslips were treated with rabbit polyclonal anti-Rev antiserum (1:200 dilution) or commercial (Intracel Corp.) anti-Rev mouse monoclonal (epitope mapped to residues 96 to 110) antibody (1:500 dilution) in 0.2% bovine serum albumin (BSA) for 4 h at room temperature. This was followed by a 1-h reaction with fluorescein-labeled goat anti-rabbit Fab fragment (1:500) or donkey anti-mouse antiserum (1:200) in 0.2% BSA in PBS. The nucleocytoplasmic shuttling was investigated by leptomycin B (LMB; Sigma-Aldrich, St. Louis, Mo.) treatment. The LMB treatment conditions are described both in the text and in the figure legends. At the indicated times after transfection and drug treatment, the cells were processed for immunological staining as described elsewhere (87). Rabbit antiserum against Rev was used to maximize the recognition of Rev epitopes, some of which may have been lost in the Rev–MS-C mutants. Transfected HeLa cells were also counterstained with murine monoclonal antibodies against nuclear pore complex (Leinco Corp.), followed by Texas Red-conjugated goat anti-mouse immunoglobulin G (IgG). Cells were examined by confocal microscopy using a Leica confocal microscope, and the images were acquired by using Leica software. Transfections were done thrice for each expression plasmid in each cell type, and 12 fields were analyzed for each coverslip.
Protein cross-linking.
For cross-linking experiments, several bifunctional cross-linkers were tried. Each reagent was titrated against purified Rev in vitro using conditions recommended by the manufacturer (Pierce Chemicals). The reaction products were resolved by SDS-PAGE under nonreducing conditions and then screened for Rev bands by immunoblotting. Protein aggregation and solubility problems restricted the selection to DSS (disuccinimidyl suberate) and DTSSP [dithiobis-(sulfosuccinimidyl) propionate] for in vitro cross-linking experiments. Individual recombinant fusion proteins, purified after Factor Xa cleavage and MBP removal, were adjusted to constant Rev equivalents in 20 μl of PBS and reacted with 2 μl of freshly dissolved DSS (50 mg/ml in dimethyl sulfoxide [DMSO]) or DTSSP (50 mg/ml in DMSO) for 1 h at 0 to 4°C. Reactions were then terminated by addition of a 1/10 volume of a solution containing glycine (1 M) and ethanolamine (1 M), and the incubation was continued for another hour. Reaction products were denatured by the addition of SDS to 1% and then electrophoresed on SDS gels. After SDS-PAGE, the gel was blotted on a polyvinylidene difluoride (PVDF) membrane and processed for immunoblotting with rabbit anti-Rev antibody and chemiluminescence detection of the immunoreactive bands.
Cytotoxicity and permeability characteristics constrained the selection to one or two reagents for in vivo work. DTBP (dimethyl 3′,3′-dithiobispropionimidate) proved to be the most consistent for in vivo work. HeLa cell transfectants on 60-mm dishes were rinsed several times with ice-cold PBS, and the monolayers were dislodged by treatment with PBS containing 5 mM EDTA. The cells were collected by centrifugation, rinsed three times with ice-cold PBS, and suspended in 0.2 ml of PBS containing DTBP (1 mg/ml), diluted from a freshly dissolved stock solution (50 mg/ml in DMSO). The cell suspension was incubated at 4°C for 1 to 2 h, the excess DTBP was neutralized by addition of a 1/10 volume of PBS containing glycine (1 M) and ethanolamine (1 M), and incubation was continued for another hour. The cells were harvested by centrifugation and disrupted by two freeze-thaw cycles in 50 μl of a buffer containing 50 mM Tris HCl (pH 7.4), 0.5% NP-40, and 0.5% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}. The cell extracts were adjusted to 1 mM AEBSF and 1 μg of aprotinin, 1 μg of leupeptin, and 2 μg of pepstatin per ml and incubated at 37°C for 15 min with a mixture of pancreatic RNase A (2 μg/ml) and 10 U of RQ DNase (Promega Biotec). The digestion was stopped by the addition of an equal volume of a buffer containing 50 mM Tris HCl (pH 7.4), 4% SDS, and 12% (wt/vol) glycerol and heating at 95°C for 5 min. The samples were then subjected to SDS-PAGE under nonreducing conditions, followed by immunoblotting.
RESULTS
The putative RNA binding and NLS domain of Rev was functionally exchanged for a polyarginine insertion(s).
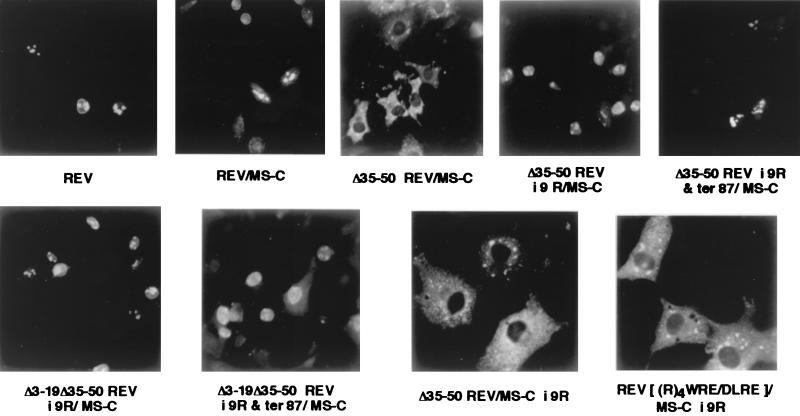
Initially, we assumed that the arginine-rich motif of Rev (residues 35 to 50) may be dispensable for activation of a heterologous RNA, such as MS2, if Rev were directed to this target as a Rev–MS-C fusion protein (59, 87). In the Rev–MS-C fusion protein and all other derivatives, the complete Rev ORF had been fused in frame to the MS2 phage coat protein ORF at the second codon of the latter, thus precluding internal initiation of the coat protein ORF. However, deletion (residues 35 to 50) and substitution (RRRRWRE to DLRE) mutations in the basic peptide motif of Rev resulted in the loss of nuclear accumulation and transactivation potential for both RRE and MS2 targets (data not shown). While the loss of response with RRE was expected since the RRE RNA binding domain had been disrupted, the failure of these mutant proteins to accumulate in the nucleus could have abolished activation of MS2 RNA. Mutational analysis of the arginine-rich RNA binding and NLS domain of Rev between residues 35 and 50 of Rev had suggested a functional redundancy of arginines. To determine whether all of the arginines in the basic domain of Rev are required, we exchanged the sequence between residues 35 and 50 for a string of nine Arg residues in the context of the Rev–MS-C (Δ35/50Rev-i-9R/MS-C) protein. Nine arginines were also inserted at the same locus of a double N-terminal deletion mutant (Δ3-19Δ35-50Rev-i-9R/MS-C). MS-C ORF translation was eliminated in the two nine-arginine (9R) mutants described above by forced termination at the 87th residue of Rev (Δ35-50Rev-i-9R & ter 87/MS-C; Δ3-19Δ35-50Rev-i-9R & ter 87/MS-C). Finally, nine arginines were also inserted at the 2nd codon of MS-C ORF in the 35-to-50 deletion to engineer Δ35/50Rev/MS-C-i-9R and in the DLRE substitution mutant to construct Rev[(R)4WRE/DLRE]/MS-C-i-9R.
The steady-state subcellular distribution of the various nine-arginine insertions visualized by indirect immunofluorescence is illustrated in Fig. 1. In this and other experiments of immunofluorescence microscopy, the steady-state subcellular distribution of various Rev derivatives was not different regardless of whether the Rev plasmids were transfected alone or in combination with the respective RRE- or MS2-containing gag reporter DNAs. The nine-arginine insertion into the 35-to-50 region of Rev in the Rev–MS-C fusion protein altered the predominantly cytoplasmic distribution of the 35-to-50 deletion mutant to that resembling the nuclear and nucleolar distribution of authentic Rev. A mutant in which Rev residues 3 to 19 in the above nine-Arg (9R) insertion were excised also behaved similarly. Forced termination of the above two 9R mutants at the 87th codon of Rev resulted in the Rev-like phenotype. Nine arginines were also inserted into the MS-C ORF, away from the natural context of the RNA binding and NLS domain of Rev in the deletion (residues 35 to 50) and substitution (RRRRWRE to DLRE). The resulting insertion mutants, Δ35/50Rev/MS-C-i-9R and Rev[(R)4WRE/DLRE]/MS-C-i-9R, displayed an almost exclusively cytoplasmic localization.
FIG. 1.
Indirect immunofluorescence detection of Rev, Rev–MS, and Rev–MS-C mutants with deletions or mutations in the RNA binding and/or NLS domain(s) of Rev or reciprocal insertions of nine arginines at the NLS of Rev or at the MS-C ORF. HeLa cells were transfected with the respective plasmids in the context of transient gag expression from an HIV-1 LTR-linked RREZ-MS (refer to Table 1) containing a gag expression plasmid. Following electroporation, 2 × 104 cells were plated on 8-mm coverslips in 24-well plates. At 48 to 72 h posttransfection, the monolayers were processed for immunofluorescence analysis using rabbit polyclonal anti-Rev antibody and FITC-conjugated goat F(ab)′ fragments against rabbit IgG. The cells were examined for immunofluorescence using a Zeiss Axiophot microscope.
Activation potential of the respective Rev–MS-C fusion proteins was determined by Rev-dependent gag expression from reporter plasmids containing RRE or MS2 RNA targets in transient transfections of HeLa or Cos-7 cells (Table 1). In this and other experiments, RREZ-MS was used as the target for MS2 coat protein binding. In RREZ-MS, the Rev-responsive core domain, SLIIB was exchanged for the MS2 translational operator RNA. Although the Rev–MS-C fusion protein was capable of activating the MS2 RNA when present as a concatenated tetramer (59, 87), RREZ–MS-C was far more efficient in this regard, presumably because the secondary structure of MS2 RNA is well preserved in this configuration. Neither the RREZ lacking the SLIIB motif nor the RREZ-MS chimera is bound or activated by Rev. To quantify the magnitude of Rev response, extracts of cells expressing wt Rev and gag RNA were serially diluted to obtain gag ELISA OD units of between 1 and 2. Under these conditions, the positive Rev response in different experiments ranged between 200 and 300 serial dilutions. The cutoff value (recommended by the manufacturer) for the negative response was at ELISA readings that fell below 10% of the control RRE and Rev values after the respective dilution(s) for the experiment. ELISA readouts of undiluted or serially diluted extracts of nontransfected cells and transfectants expressing RRE mutants nonresponsive to Rev fell below the 10% cutoff values of undiluted or serially diluted extracts of Rev or RRE cells. Values that ranged between 10 and 20% of that with RRE and Rev were scored as marginal responders. gag expression obtained with the Rev expression plasmid pRSV LTR Rev was arbitrarily set to 100. A 9R insertion between residues 35 and 50 of Rev (Δ35/50Rev-i-9R/MS-C) restored the transactivation of both RRE and MS2 RNA targets that was lost for the deletion mutant Δ35/50Rev/MS-C. A mutation (Δ3-19Δ35-50-Rev-i-9R/MS-C) deleting the N-terminal Rev sequence between positions 3 and 19 in the above nine-arginine insertion reduced slightly the magnitude of the Rev effect. When the arginine insertion mutants described above were truncated to the 87th residue of Rev by forced termination, the resulting mutants, Δ35-50Rev-i-9R & ter 87/MS-C and Δ3-19Δ35-50Rev-i-9R & ter-87/MS-C, which could not bind MS2 RNA, activated RRE and not MS2 target. These C-terminal truncated 9R mutants had reduced activation potential for RRE, reflecting their reduced RRE RNA binding affinity (Table 1). When nine arginines were placed outside of the deleted or substituted RNA binding and NLS domain of Rev (i.e., into the MS-C ORF), the resulting mutants, Δ35/50Rev/MS-C-i-9R and Rev[(R)4WRE/DLRE]/MS-C-i-9R, were devoid of activation for either RRE or MS2 RNA. Interestingly, these two MS-C insertions had markedly reduced binding affinity for RRE but not for MS2 RNA.
TABLE 1.
Homopolyarginine insertion rectifies the functional defect of a mutant deleting the RNA binding and NLS domain of Rev in a context-sensitive manner
| Rev–MS-C fusion protein | In vitro RNA bindinga
|
trans-Activationb (SD)
|
||
|---|---|---|---|---|
| RRE | MS2 | RRE | RREZ-MS2 | |
| Rev | +++++ | +++++ | 100 | 1.3 (0.7) |
| Rev–MS-C | +++++ | +++++ | 143 (9.7) | 114 (14.8) |
| Δ35/50Rev-i-9R/MS-C | +++++ | +++++ | 97 (14.5) | 122 (16.7) |
| Δ3-19Δ35-50Rev-i-9R/ MS-C | +++ | +++++ | 57 (7.5) | 89 (11.7) |
| Δ35-50Rev-i-9R & ter 87/MS-C | ++++ | +/− | 79 (11.5) | 0.6 (0.4) |
| Δ3-19Δ35-50Rev-i-9R & ter 87/MS-C | +++ | +/− | 38 (6.8) | 1.1 (0.3) |
| Δ35/50Rev/MS-C-i-9R | +/− | ++++ | 3.8 (1.7) | 2.2 (0.8) |
| Rev[(R)4WRE/DLRE]/ MS-C-i-9R | +/− | ++++ | 1.2 (0.8) | 4.7 (1.3) |
RNA binding was evaluated by EMSA at fixed levels for the respective RNAs but various levels of the individual proteins, expressed in E. coli as MBP fusion proteins. The binding potential of mutants is expressed as: ++++, 75%; +++, 50%; ++, 25%; +, 1%; and +/−, <0.5% of wt Rev or Rev–MS-C.
Mean GAG expression values from five experiments, normalized for transfection efficiency, are tabulated with the respective standard deviations. The positive values are denoted by boldface.
It was possible that mere insertion of nine arginines out of natural context may not have been sufficient to restore Rev function. Moreover, if the RNA binding and NLS or NOS domains of Rev were truly modular, a reciprocal insertion of these domains at a distant site, i.e., into the MS-C ORF, may have been better at compensating the defects of the corresponding deletion (residues 35 to 50) or substitution mutants (RRRRWRE to DLRE). However, insertion of an 8 (RRNRRRRW)-, 14 (residues 33 to 46)-, 16 (residues 35 to 50)-, 19 (residues 33 to 51)-, or 37 (residues 24 to 60)-residue Rev sequence near the N terminus of the MS-C ORF in the respective deletion (Δ35/50Rev/MS-C) or substitution (Rev[(R)4WRE/DLRE]/MS-C) mutants failed to restore the nuclear accumulation phenotype or the activation potential for either RRE or MS2 RNAs (data not shown).
Deletion of one or both of the Rev multimerization motifs in the context of the nine-arginine Rev–MS-C fusion protein results in a differential activation of RRE and MS2 targets.
Two motifs flanking the RNA binding and the NLS domain of Rev have been assigned a role in Rev protein oligomerization, a property that is essential for optimal trans-activation of RRE RNA. However, mutations in these domains do not affect activation of MS2 target by the Rev–MS-C protein (59). Since the MS2 coat protein has been known to form oligomers, especially within the phage particles (28, 62, 68, 85), it has been presumed that multimerization across the coat protein interface could substitute for the lack of Rev oligomerization patch. To address this point, we engineered mutations deleting one or both of the oligomerization motifs in the context of the wt Rev–MS-C or the 9R insertion mutant, Δ35/50Rev-i-9R/MS-C.
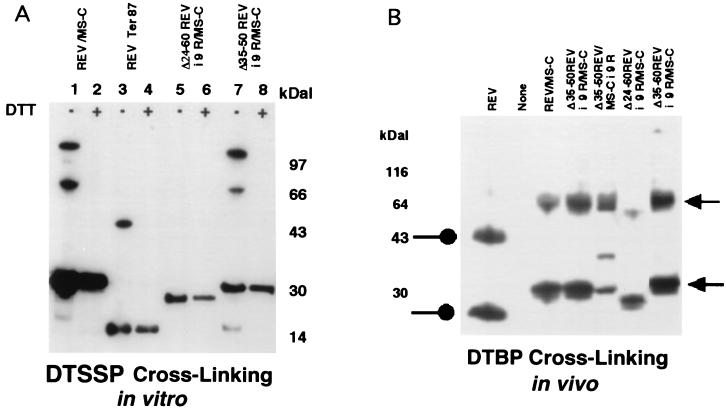
First, we examined the oligomerization properties of the various Rev or Rev–MS-C fusion proteins. The respective mutants were expressed in E. coli as fusion proteins tagged to MBP. MBP tags on selected Rev–MS-C mutant proteins were removed by Factor Xa proteolysis, and the respective Rev–MS-C fusion proteins were purified by negative selection on amylose resin, followed by batchwise elution from carboxymethyl cellulose (CM52) or phosphocellulose columns as described in Materials and Methods. Each protein was adjusted to a constant Rev equivalent before reacting it with the thiol-reversible bifunctional chemical cross-linking reagent DTSSP. Rev–MS-C protein reacted to yield two prominent bands of ca. 68 and 112 kDa, representing dimeric and tetrameric species (Fig. 2A, lane 1); both higher-molecular-mass species disappeared upon treatment with 1 M DTT (Fig. 2A, lane 1 versus lane 2). Under the same conditions, truncated Rev protein of 86 amino acids reacted to yield only a reversible dimeric species (Fig. 2A, lane 3 versus lane 4). With the 9R insertion at the NLS domain of Rev (Δ35/50Rev-i-9R/MS-C), the cross-linked dimers and tetramers were disrupted by DTT treatment (Fig. 2A, lane 7 versus lane 8). Excision of both the N- and the C-terminal multimerization motifs of Rev (Δ24/60Rev-i-9R/MS-C) abolished multimer formation (Fig. 2A, lanes 5 and 6).
FIG. 2.
Self-association of Rev and Rev–MS-C protein derivatives detected by chemical cross-linking in vitro and in vivo. (A) The indicated proteins were expressed in E. coli as MBP-tagged fusion proteins and purified by affinity chromatography. The MBP tag on each protein was excised by Factor Xa proteolysis, and the respective Rev and Rev–MS-C proteins were purified by negative selection on amylose resin, followed by cationic cellulose chromatography (see Materials and Methods). Purified proteins were adjusted to constant Rev equivalents and reacted with thiol-reversible (DTSSP) bifunctional cross-linker in solution as described in Materials and Methods. DTSSP-reacted products were treated with 1 M DTT (+) or left untreated (−) prior to electrophoresis. The protein bands were blotted on PVDF filters and detected by immunoblotting with polyclonal rabbit anti-Rev antiserum, followed by chemiluminescence. (B) In vivo Rev multimer formation analyzed by protein cross-linking. HeLa cells were electroporated with the respective Rev and Rev–MS-C derivatives. The transfectants were treated with the bifunctional chemical cross-linker DTBP and processed for SDS-PAGE and immunoblotting as described in Materials and Methods.
The fusion proteins were also evaluated for multimerization potential in vivo in HeLa cells. Transfectants were treated with the bifunctional cross-linker DTBP as described in Materials and Methods, and the fusion proteins were analyzed by immunoblotting cell extracts after SDS-PAGE under nonreducing conditions. Rev, Rev–MS-C, and Δ35/50Rev-i-9R/MS-C proteins were readily cross-linked to dimer species (Fig. 2B). Upon longer exposure, faint bands representing higher oligomers were also evident (data not shown). As expected from the in vitro experiments (not shown), Δ35/50Rev/MS-C-i-9R was not impaired for dimerization. Deletion of the C-terminal multimerization motif of Rev (Δ35/60Rev-i-9R/MS-C) did not interfere with multimer formation; deletion of the N-terminal motif (Δ24/50Rev-i-9R/MS-C) reduced the multimerization potential (not shown). Removal of the entire bipartite oligomerization motif (Δ24/60Rev-i-9R/MS-C) markedly reduced the intensity of the dimer band (Fig. 2B). Coexpression of Rev or Rev–MS-C-responsive gag-RRE or gag-RREZMS2 mRNAs did not materially alter the self-association properties of the respective Rev proteins (data not shown).
These results showed that excising the multimerization motifs of Rev severely impaired or abolished the multimer formation of the respective Rev–MS-C fusion protein despite the inherent potential of the coat protein moiety for oligomerization. The RNA binding properties of various Rev–MS-C derivatives (summarized in Table 2) supported these findings. With RRE or MS2 RNA, wt Rev–MS-C or the Δ35/50Rev-i-9R/MS-C mutant formed two or three slower-moving complexes in addition to a major retarded complex, representing monomer protein binding. Mutant proteins lacking the N-terminal (Δ24/50Rev-i-9R/MS-C) or both the N- and the C-terminal (Δ24/60Rev-i-9R/MS-C) multimerization motifs of Rev formed only the monomer complex.
TABLE 2.
Deletion of the multimerization domain(s) of Rev in the nine-arginine-substituted Rev–MS-C proteins has differential effects on RRE and MS2 targets
| Rev and Rev–MS-C protein | Oligomers in EMSAa
|
Subcellular locationb |
trans-Activationc (SD)
|
||
|---|---|---|---|---|---|
| RRE | MS2 | RRE | RREZ-MS2 | ||
| Rev | Yes | Yes | Nuc>>>Cyt | 100 | 1.3 (0.7) |
| Rev–MS-C | Yes | Yes | Nuc>>>Cyt | 148 (9.7) | 114 (14.8) |
| Δ35/50Rev-i-9R/MS-C | Yes | Yes | Nuc>>>Cyt | 97 (14.5) | 122 (16.7) |
| Δ35/60Rev-i-9R/MS-C | +/− | +/− | Nuc>>>Cyt | 3.7 (14.5) | 83 (17.1) |
| Δ24/50Rev-i-9R/MS-C | No | No | Nuc>>>Cyt | 3.3 (1.9) | 111 (18.3) |
| Δ24/60Rev-i-9R/MS-C | No | No | Nuc>>>Cyt | 3.7 (0.8) | 97 (12.1) |
| Δ24/60Rev-i-9R & ter 87/MS-C | ND | ND | Nuc>>>Cyt | 2.4 (1.4) | 3.3 (2.4) |
Oligomer formation was evaluated in EMSA using fixed amounts of 32P-labeled PPE or MS2 RNAs. +/−, occasional occurrence of dimer species; ND, not determined.
Subcellular localization data are from indirect immunofluorescence microscopy analyses. Nuc, nuclear; Cyt, cytoplasmic.
Mean GAG expression values from six experiments, normalized for transfection efficiency, are tabulated with the respective standard deviations. The positive values are denoted by boldface.
Activation potential for gag mRNAs containing RRE or MS2 targets by the multimerization-defective 9R mutants is shown in Table 2. Mutants deleting the N-terminal (Δ24/ 50Rev-i-9R/MS-C), C-terminal (Δ35/60Rev-i-9R/MS-C), or both the N- and the C-terminal (Δ24/60Rev-i-9R/MS-C) motifs of Rev failed to activate RRE RNA but retained wt potential for the MS-2 reporter. When Δ24/60Rev-i-9R/MS-C was truncated to the 87th residue of Rev by forced termination, the resulting mutant, Δ24/60Rev-i-9R & ter 87/MS-C, lost activation for both RRE and MS2 targets. All of the above mutants were mainly localized in the nuclei and were concentrated in the nucleoli like authentic Rev (Table 2). A nine-arginine insertion within the deletion from residues 24 to 60 in the context of Rev (Δ24/60Rev-i-9R) resulted in a similar null phenotype for both RNAs (not shown). Truncation to the 87th residue of Rev was chosen since residues 1 to 87 constitute the minimal functional Rev. When the nine arginines were removed from the Rev 24-to-60 deletion mutant, the resulting protein (Δ24/60Rev/MS-C) was cytoplasmic and was unable to activate either RNA (data not shown). Δ24/60Rev/MS-C bound MS2 but not RRE RNA in vitro and showed no evidence of multimer formation (41a).
Functionally competent polyarginine insertion mutants have a somewhat broadened specificity for RNA recognition in vivo.
We tested the polyarginine insertion mutants for fidelity of Rev function with different RNA targets. Toward this goal we assayed Rev-dependent gag expression from various gag-pol mRNAs carrying wt RRE, various RRE mutants, or heterologous targets such as MS-2, TAR, RexRe, and VAI RNAs. The results are summarized in Table 3. Many RRE mutants that were not responsive to Rev in the context of either wt Rev or the Rev–MS-C fusion protein were activated by the nine-Arg substitution mutant protein (denoted by italicized bold- face in Table 3). Mutations with base substitutions (such as AGC to ACG, target 4) that disrupted the secondary structure of RRE stem-loop II and mutants (targets 5 to 8) in which the topology of the individual stem-loops within stem-loop II has been rearranged have been shown to be nonresponsive to wt Rev (37). All of these mutants, except for target 8, were activated modestly by the 9R insertion mutant. The 9R mutant also activated mutant RREs with deletions of one or two of the three G's in the stem-loop II B (3G/2G, 3G/1G), substitutions for all three G's (3G/3A, 3G/3C, or 3G/3U), or changes of the GGG sequence to GUG. All of these mutants were nonresponsive to either Rev or Rev–MS-C. A few of the RRE mutants (target 4, AGC to ACG; target 10, 3G to 3A; target 15, 3G to 1G; and target 18, 3G to GUG [see Table 3]) were evaluated for Rev binding by filter-binding assays (by varying the protein concentration) with wt Rev and the 9R mutant. They were 100-fold less efficient than RRE at binding Rev, but they bound the 9R mutant 20 to 40% as efficiently as wt RRE (data not shown). Although this suggested that the 9R mutant may have reduced stringency for RNA binding, there was no evidence of promiscuous RNA binding. Heterologous RNA targets such as HIV-1 TAR, HTLV-1 RexRE, and adenovirus VAI RNAs displayed the null phenotype with the 9R mutant (Table 3). Under the same conditions HTLV-1 Rex and HIV-1 Tev protein induced excellent and modest expression, respectively, from the RexRE and TAR RNA containing gag mRNAs (data not shown). The 9R insertion mutant preserved the response for the MS2 RNA, since MS2 RNA binding was mediated by the unaltered MS-C moiety. To confirm that activation of RRE derivatives was mediated by binding to the nine arginines, two additional mutants were investigated. The 9R insertion mutant was truncated by introducing a termination codon at the 87th residue of Rev and thus lacked the MS-C ORF (Δ35-50Rev-i-9R & ter 87/MS-C). A second mutant (Δ35-50Rev-i-9R/MS-D) introduced a single nucleotide deletion at the seventh codon of MS-C ORF, causing a frameshift and early termination of the coat protein (87). Many of the null (in response to Rev or Rev–MS-C) RRE mutants, notably those at the bulged G's, such as 3G/3A, 3G/3U, 3G/2G, and 3G/1G, were activated by both truncation mutants, confirming that the binding to the respective RRE RNAs was mediated by the 9R sequence inserted in place of the NLS or NOS of Rev. As expected, both truncation mutants were devoid of activity with the MS2 targets.
TABLE 3.
Homo-poly-arginine substitution at the RNA binding and NLS domain of Rev in the Rev–MS-C fusion protein broadens the RRE specificity of the resulting protein
| RNA targets examined
|
trans-Activation by Rev and its derivativesa (SD)
|
|||||
|---|---|---|---|---|---|---|
| No. | Target | Rev | Rev–MS-C | Δ35/50Rev-i-9R/MS-C | Δ35/50Rev-i-9R/MS-D | Δ35/50Rev-i-9R & ter 87 |
| 1 | RRE | 100 | 148 (9.7) | 97 (14.5) | 87 (12.3) | 107 (16.1) |
| 2 | ERR (antisense) | 0.8 (0.2) | 14 (0.6) | 21 (1.2) | 0.6 (0.2) | 1.5 (1.2) |
| 3 | RRE-ΔSLIIB | 1.4 (0.4) | 0.8 (0.2) | 4.5 (1.7) | 1.5 (1.3) | 1.2 (0.9) |
| 4 | RRE (AGC to ACG, 62–64) | 7.1 (2.8) | 3.8 (1.4) | 21.3 (5.2) | 18.1 (6.4) | 28.1 (9.2) |
| 5 | RRE (SLIIAC′B) | 1.7 (0.2) | 0.2 (0.1) | 32.3 (5.7) | 22.8 (3.7) | 19.8 (2.8) |
| 6 | RRE (SLIIACB and AGC to ACG) | 2.7 (1.5) | 2.8 (1.8) | 27.3 (6.2) | ND | ND |
| 7 | RRE (SLIIAC′B and AGC, 62–64) | 3.5 (1.8) | 2.3 (0.7) | 35.4 (11.2) | ND | ND |
| 8 | RRE (SLIIA′B′) | 1.4 (0.7) | 3.1 (1.4) | 5.8 (2.4) | 5.8 (2.4) | 3.7 (1.8) |
| 9 | RRE (3G→3A) | 1.2 (0.5) | 0.8 (0.2) | 39.2 (7.8) | 29 (6.3) | 26 (7.3) |
| 10 | RRE (3G→3U) | 0.5 (0.3) | 0.8 (0.4) | 29.8 (5.5) | 36 (8.3) | 27 (6.7) |
| 11 | RRE (3G→3C) | 1.7 (0.7) | 1.7 (0.8) | 11.7 (3.8) | ND | ND |
| 12 | RRE (4G) | 47.8 (8.9) | 68.4 (12.7) | 84.5 (11.8) | 75 (13.8) | 59 (8.2) |
| 13 | RRE (AGGG) | 27.8 (4.4) | 33.2 (4.8) | 78.2 (14.7) | 58 (9.3) | 66 (10.1) |
| 14 | RRE (2G) | 17.8 (8.5) | 48.5 (12.4) | 81.7 (9.5) | 63 (10.1) | 71 (8.1) |
| 15 | RRE (1G) | 1.2 (0.7) | 1.7 (0.8) | 58.7 (12.3) | 37 (9.8) | 31 (6.6) |
| 16 | RRE (0G) | 0.7 (0.5) | 1.8 (0.8) | 8.8 (1.8) | 3.4 (0.9) | 2.6 (2.2) |
| 17 | RRE (3G→GCG) | 138 (17.7) | 88 (11.4) | 112 (15.7) | 72 (9.7) | 67 (8.8) |
| 18 | RRE (3G→GUG) | 5.8 (2.2) | 3.8 (2.2) | 36.7 (6.8) | 25 (5.8) | 41 (7.3) |
| 19 | RREZ-MS2 | 3.2 (1.4) | 114 (14.8) | 122 (16.7) | 1.8 (0.9) | 1.1 (0.6) |
| 20 | RREZ-MS2ΔA | 1.8 (0.8) | 22.8 (7.3) | 34.5 (8.1) | ND | ND |
| 21 | MS2 plus SLIIB | 37.8 (9.7) | 64 (10.7) | 55 (8.7) | 43 (7.6) | 48 (6.7) |
| 22 | MS2 plus SLIIB (3G/3A) | 1.7 (1.2) | 58 (7.8) | 66 (11.2) | ND | ND |
| 23 | MS2ΔA plus SLIIB | 44.3 (11.8) | 42.4 (9.2) | 71.2 (13.3) | ND | ND |
| 24 | MS2ΔA plus SLIIB (3G/3A) | 0.4 (0.4) | 1.3 (0.6) | 0.8 (0.6) | ND | ND |
| 25 | GAG-RexRE | 3.8 (1.4) | 2.4 (1.6) | 3.7 (1.6) | 1.7 (1.3) | 0.8 (0.6) |
| 26 | GAG-TAR | 1.1 (0.5) | 1.7 (0.9) | 3.3 (1.8) | 1.4 (0.8) | ND |
| 27 | GAG-VAI | 1.5 (0.7) | 0.8 (0.6) | 2.8 (1.4) | ND | ND |
Mean GAG expression values from five experiments, normalized for transfection efficiency, are tabulated with the respective standard deviations. The positive values are denoted by boldface, and positive values obtained with the 9R mutants are shown in boldface italics. Mutants 4 to 8 represent topological rearrangements of stem-loop II components; AGC to ACG refers to base substitution at positions 62 to 64 of RRE that forces the upstream three G's from a bulge to a base-paired configuration (36, 37). Mutants 9 to 18 denote base substitutions at the three-G bulge in the context full-length RRE (37). In mutants 19 and 20, RREZ refers to RRE RNA deleted for the Rev-responsive stem-loop II sequence and RREZ-MS2 is an RNA chimera that replaced RRE stem-loop II for the MS2 translational operator RNA. MS2ΔA denotes MS2 RNA mutant deleted for a critical A residue and severely compromised for MS2 coat protein binding. Constructs 21 to 24 represent various mutations in the bivalent chimera of stem-loop II and MS2 RNA (84). In mutants 25 to 27, a HTLV-1 RexRE, HIV-1 TAR, or adenovirus VAI sequence replaced RRE. ND, not done.
A minimum stretch of four or five uninterrupted arginines is required for trans-activation and steady-state nuclear accumulation.
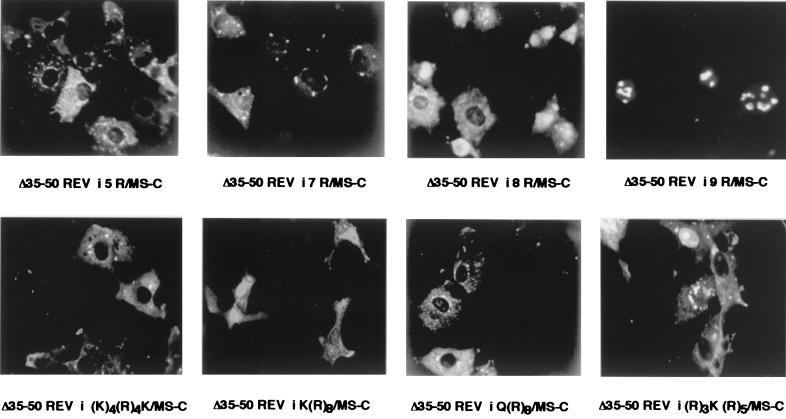
Arginine forks in the RNA binding motifs of Rev and Tat mediate binding to the respective cognate target RNAs, RRE and TAR. In vitro RNA binding and in vivo Tat assays have shown that a single arginine surrounded by three basic residues was sufficient for Tat binding to its cognate target, TAR (9, 10, 78). Similar studies using a 17-mer peptide corresponding to the 35-to-50 domain of Rev have shown that RNA binding occurs through two discrete arginine-rich domains (44, 76). Therefore, we investigated how many arginines were required for activation and whether an arginine cluster interrupted by other residues would be functional. gag expression from RRE or RREZ-MS2 plasmids was measured in the presence of mutants containing different numbers of arginines inserted in place of Rev residues 35 to 50. gag expression from RRE containing gag mRNA required a minimal presence of eight arginines. In contrast, an insertion mutant containing as few as three arginines was able to activate the RREZ-MS2 chimera (Table 4). Inserting more than nine arginines did not appreciably increase the magnitude of transactivation for either RNA target. A few of the above arginine mutants were expressed in E. coli as MBP fusion proteins, purified by affinity chromatography, and evaluated for RRE and MS2 RNA binding by EMSA. Fusion proteins with fewer than nine arginines had a somewhat reduced affinity for RRE RNA (a five-arginine mutant being fourfold less efficient than Rev). In contrast, their relative binding affinities for MS2 RNA were unaltered (Table 4). The differential response of the two RNA targets to Rev mutants containing fewer than seven arginines may have reflected the different affinities for RRE and MS2 RNAs. However, a different picture emerged when the steady-state subcellular distribution of the different arginine insertions was examined. Insertion mutants containing eight or fewer arginines were predominantly cytoplasmic under steady-state conditions (Fig. 3). Steady-state accumulation of mutants with nine or more insertions was predominantly, if not exclusively, nuclear with nucleolar concentration, resembling that of wt Rev.
TABLE 4.
Number of arginine insertions required for functional restoration of Rev–MS-C protein deleted for the RNA binding and NLS domain of Rev
| Arginine substitutions in Rev–MS-C | RNA bindinga
|
Subcellular locationb |
trans-Activationc (SD)
|
||
|---|---|---|---|---|---|
| RRE | MS2 | RRE | RREZ-MS2 | ||
| Δ35/50Rev-i-1R/MS-C | ND | ND | Cyt>>>Nuc | 0.8 (0.5) | 4.8 (1.8) |
| Δ35/50Rev-i-3R/MS-C | ND | ND | Cyt>>>Nuc | 1.8 (0.8) | 10.8 (3.2) |
| Δ35/50Rev-i-4R/MS-C | ND | ND | Cyt>>>Nuc | 3.1 (1.2) | 18.2 (4.8) |
| Δ35/50Rev-i-5R/MS-C | ++ | +++++ | Cyt>>>Nuc | 0.4 (0.3) | 27 (3.8) |
| Δ35/50Rev-i-7R/MS-C | ND | ND | Cyt>>Nuc | 0.8 (0.4) | 45 (8.9) |
| Δ35/50Rev-i-8R/MS-C | ++++ | +++++ | Cyt≈Nuc | 17 (4.5) | 68 (9.9) |
| Δ35/50Rev-i-9R/MS-C | +++++ | +++++ | Nuc>>>Cyt | 97 (14.5) | 122 (16.7) |
| Δ35/50Rev-i-10R/MS-C | ND | ND | Nuc>>>Cyt | 133 (17.5) | 111 (18.3) |
| Δ35/50Rev-i-11R/MS-C | +++++ | +++++ | Nuc>>>Cyt | 89 (13.8) | 156 (8.7) |
| Δ35/50Rev-i-12R/MS-C | ND | ND | Nuc>>>Cyt | 107 (8.8) | 108 (12.1) |
RNA binding was evaluated by EMSA at fixed levels of the respective RNAs but with various levels of the individual proteins, expressed in E. coli as MBP fusion proteins. The binding potentials of mutants are expressed as follows: ++++, 75%; +++, 50%; ++, 25%; +, 1%; and +/−, <0.5% of wt Rev or Rev–MS-C. ND, not determined.
Subcellular localization data are from indirect immunofluorescence microscopy analyses. Nuc, nuclear; Cyt, cytoplasmic.
Mean GAG expression values from five experiments, normalized for transfection efficiency, are tabulated with the respective standard deviations. The positive values are in boldface. wt Rev-induced GAG expression from RRE RNA is assigned a value of 100.
FIG. 3.
Immunofluorescence assay of selected Rev–MS-C fusion proteins with different arginine insertions (top row) or arginine clusters interrupted by other residues (bottom row) in place of the Rev NLS domain between residues 35 and 50.
Genetic analyses (32, 63, 76, 98) of the basic domain of Rev have not demonstrated a critical need for any one residue except for the Asn at position 40 and Try at position 45. Substitution of Asn at position 40 for Asp resulted in a protein that lacked RNA binding and nuclear accumulation. Depending on the context, substitution of Try at position 45 resulted in a loss of either RNA binding or nuclear targeting (32, 39). To evaluate the need for an uninterrupted Arg tract for activation of RRE target, we engineered several mutations that interrupted the Arg stretch at one or two places or exchanged a smaller cluster of arginines for lysines. Table 5 summarizes the in vivo activation results obtained with RRE or RREZ-MS2 targets with the different mutants. All of the substitution mutants except for Δ35/50Rev-i-(K)4(R)4K/MS-C activated the MS2 target to a reasonable extent (28 to 75% of the levels seen with wt Rev–MS-C fusion protein). However, many of these mutants exhibited a null activation phenotype for RRE, except for the Δ35/50Rev-i-Q(R)8/MS-C, Δ35/50Rev-i-(R)5Q(R)3/MS-C, and Δ35/50Rev-i-(R)6S(R)3/MS-C mutants. Even with these latter mutants, the magnitude of RRE activation was only about 10 to 20% of that with wt Rev. A few of these interrupted arginine cluster mutants were also evaluated for binding to RRE and MS2 RNAs (Table 5). Mutants [Δ35/50Rev-i-Q(R)8/MS-C and Δ35/50Rev-i-(R)3K(R)5/MS-C] that had a marginal activation potential for RRE bound RRE RNA about 25% as well as Rev. Δ35/50Rev-i-(K)4R(K)4/MS-C and Δ35/50Rev-i-(K)4(R)2(K)3/MS-C mutants that had a null activation phenotypes bound RRE RNA poorly in vitro. All of the above mutants bound MS2 RNA almost as well as the wt Rev–MS-C fusion protein. The substitution mutants had a predominantly cytoplasmic distribution under steady-state conditions, except for Δ35/50Rev-i-Q(R)8/MS-C, Δ35/50Rev-i-K(R)8/MS-C, and Δ35/50Rev-i-(R)3K(R)5/MS-C, which displayed occasional nuclear and nucleolar accumulation (Fig. 3). Activation of MS2 by mutants with fewer than eight arginines or interrupted arginines was due to the presence of intact MS-2 ORF. When seven arginines, (R)5Q(R)3, or (K)4(R)2(K)3 was inserted in place of residues 35 to 50 of Rev, the resulting Rev mutants were negative for MS2 activation, since they lacked the coat protein moiety. (R)5Q(R)3 was minimally active with RRE (ca. 10% of wt Rev levels); (K)4(R)2(K)3 and a seven-arginine insertion mutant were negative for RRE (data not shown).
TABLE 5.
trans-Activation potential of interrupted arginine tract insertions in the Rev–MS-C proteins deleted for the RNA binding and NLS domain of REV
| Arginine substitutions in Rev–MS-C | RNA bindinga
|
Subcellular locationb |
trans-Activationc (SD)
|
||
|---|---|---|---|---|---|
| RRE | MS2 | RRE | RREZ-MS2 | ||
| Δ35/50Rev-i-9R/MS-C | +++++ | +++++ | Nuc>>>Cyt | 97 (14.5) | 122 (16.7) |
| Δ35/50Rev-i-Q(R)8/MS-C | ++ | +++++ | Cyt>Nuc | 19.8 (2.8) | 88 (9.2) |
| Δ35/50Rev-i-K(R)8/MS-C | ND | ND | Cyt>Nuc | 6.4 (2.3) | 79 (10.8) |
| Δ35/50Rev-i-(R)3K(R)5/MS-C | ND | ND | Cyt>Nuc | 6.8 (2.4) | 65 (9.3) |
| Δ35/50Rev-i-(K)4R(K)4/MS-C | − | ++++ | Cyt>>>Nuc | 1.7 (0.5) | 6.8 (2.9) |
| Δ35/50Rev-i-(K)4(R)2(K)3/MS-C | +/− | ++++ | Cyt>>>Nuc | 0.8 (0.4) | 34 (6.9) |
| Δ35/50Rev-i-(R)5E(R)3/MS-C | ND | ND | Cyt>>>Nuc | 3.3 (1.5) | 81 (13.3) |
| Δ35/50Rev-i-(R)5Q(R)3/MS-C | ++ | ++++ | Cyt>Nuc | 14.8 (3.2) | 61 (9.2) |
| Δ35/50Rev-i-(R)2N(R)6/MS-C | ND | ND | Cyt>>Nuc | 7.8 (2.8) | 59 (7.1) |
| Δ35/50Rev-i-(R)6S(R)3/MS-C | ND | ND | Cyt>>Nuc | 16.4 (3.7) | 64 (9.3) |
RNA binding was evaluated by EMSA at fixed levels of the respective RNAs, but varying the individual proteins, expressed in E. coli as MBP fusion proteins. Binding potentials of mutants are expressed as follows: ++++, 75%; +++, 50%; ++, 25%; +, 1%; and +/−, <0.5% of wt Rev or Rev–MS-C. ND, not determined.
Subcellular localization data are from indirect immunofluorescence microscopy analyses. Nuc, nuclear; Cyt, cytoplasmic.
Mean GAG expression values from five experiments, normalized for transfection efficiency, are tabulated with the respective standard deviations. The positive values are in boldface. wt Rev-induced GAG expression from RRE RNA is assigned a value of 100.
Evaluation of nucleocytoplasmic shuttling potential of Rev and Rev–MS-C derivatives.
The different subcellular distributions of the Rev–MS-C derivatives may reflect different rates of nuclear import. All our derivatives had intact NES domains, and the export of NES domain-containing proteins is mediated by the nuclear export protein CRM1 in a saturable manner (23, 26, 65, 75). Inactivation of CRM1 by LMB (1, 23, 26, 47–49, 65, 96) would be expected to induce, in a dose-dependent manner, nuclear retention of even poorly imported proteins. The expression plasmids encoding Rev and Rev–MS-C derivatives used were individually transfected into HeLa or Cos-7 cells seeded on 8-mm coverslips. After 24 h, individual transfectants were treated with various amounts of LMB or were left untreated as described in Materials and Methods. Immunofluorescence microscopy results of selected transfections in HeLa cells are shown in Fig. 4. Fusion proteins were stained with fluorescein isothiocyanate (FITC)-conjugated antibodies against Rev. Texas Red-conjugated monoclonal antibody against the nuclear pore complex was used as a counterstain.
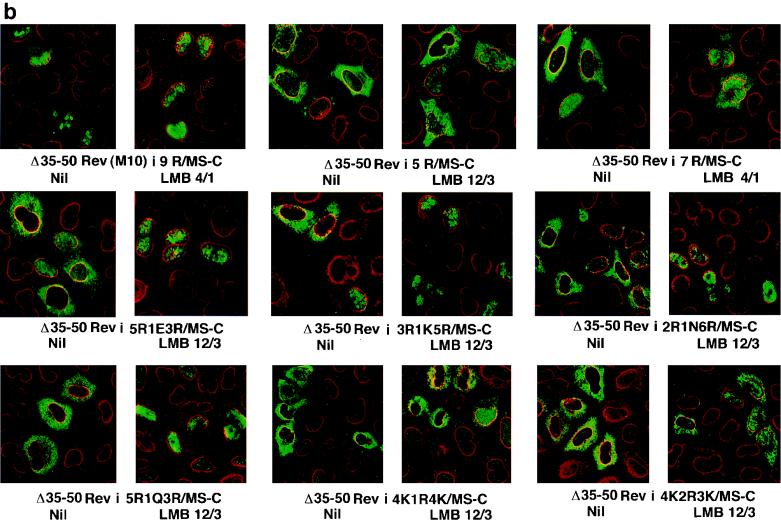
FIG. 4.
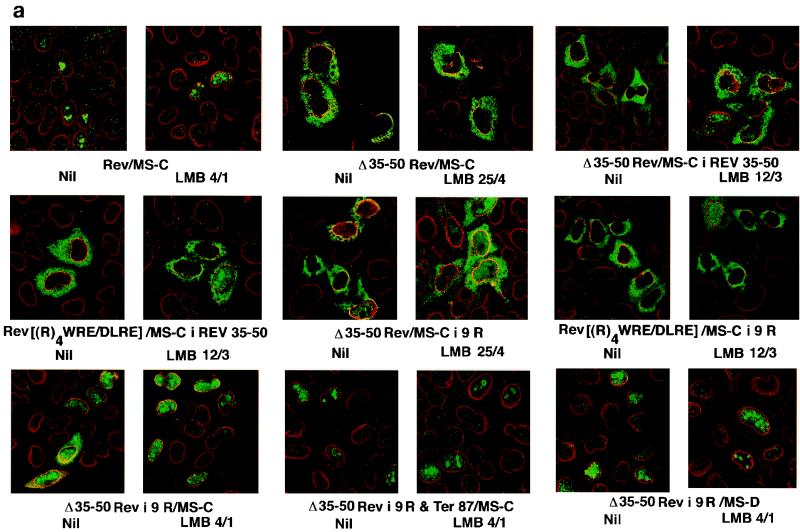
LMB-induced morphological changes in the subcellular localization of Rev and Rev–MS-C derivatives. (a) Changes in the distribution of mutants with deletions and insertions. (b) Changes in cells expressing proteins with multiple arginines or interrupted arginine or lysine strings. HeLa cells on 8-mm coverslips were transfected in quadruplicate with the indicated plasmids. Cells were rinsed and treated with LMB at 4 nM for 1 h (4/1), 12 nM for 3 h (12/3), or 25 nM for 4 h (25/4) or left untreated (Nil). At 24 to 36 h after transfection, transfectants were reacted with a mixture of rabbit anti-Rev antibodies and murine monoclonal antibodies against the nuclear pore complex. FITC-conjugated goat anti-rabbit IgG and Texas Red-tagged donkey anti-mouse IgG were used to label the fusion protein (green) and the nuclear pore complex (red). Images were visualized by using a ×63 objective lens on a Leica confocal microscope.
Rev–MS-C was localized exclusively in the nuclei of untreated or LMB-treated cells. This was similar to the behavior of wt Rev (not shown). Similar results were obtained in Cos-7 cells. The Δ35/50Rev/MS-C mutant that excised the arginine-rich NLS domain of Rev was retained exclusively in the cytoplasm even after treatment for 4 h with 25 nM LMB. A mutant substituting the four consecutive arginines in the Rev NLS domain for aspartate followed by leucine {Rev[(R)4WRE/DLRE]/MS-C-i-Rev} also behaved similarly. Mutants with reciprocal insertions of Rev NLS domain into the MS-C ORF of the above deletion (Δ35-50Rev/MS-C-i-Rev35-50) or substitution {Rev[(R)4WRE/DLRE]/MS-C-i-Rev35-50} mutants were exclusively cytoplasmic in untreated cells. LMB treatment induced these mutants to relocalize to nuclei in some but not in all cells (Fig. 4a). The magnitude of LMB-induced redistribution of these mutants was less pronounced in Cos-7 cells (not shown).
Δ35/50Rev-i-9R/MS-C mutant with nine arginines in place of the Rev NLS domain was distributed mostly in the nuclei of untreated HeLa cells. LMB treatment caused this mutant to have more enhanced nuclear localization. When the 9R fusion protein was truncated at the 87th residue of Rev (Δ35-50Rev-i-9R & ter 87/MS-C), it was exclusively nuclear, with nucleolar concentration in both LMB-treated and untreated cells. A similar situation prevailed with the fusion protein mutant (Δ35-50Rev-i-9R/MS-D) forced to terminate beyond the second codon of MS-C ORF by an in-frame deletion (Fig. 4a). When the 9R derivative was engineered to contain the M10 mutation in the NES domain of Rev, it was exclusively localized in the nuclei in LMB-treated and untreated cells (Fig. 4b). The mutant (Δ35-50Rev/MS-C-i-9R) with a 9R insertion within the MS-C ORF was exclusively cytoplasmic in untreated cells. LMB treatment redistributed this protein equally between nuclear and cytoplasmic compartments (Fig. 4b). In many cases where LMB induced the redistribution of (otherwise predominantly cytoplasmic) fusion protein(s) to nuclei, there was very little, if any, nucleolar concentration, in accord with an earlier report (49).
The five-arginine insertion mutant was mostly cytoplasmic in untreated cells and showed partial localization to the nuclei in response to LMB (Fig. 4b). LMB induced a more complete nuclear transfer of mutants with seven (Δ35/50Rev-i-7R/MS-C) or eight (data not shown) arginines. A similar LMB phenotype was observed with mutations interrupting nine arginines with a single lysine [Δ35/50Rev-i-(R)3K(R)5/MS-C], asparagine [Δ35/50Rev-i-(R)2N(R)6/MS-C], glutamate [Δ35/50Rev-i-(R)3E(R)5/MS-C], or glutamine [Δ35/50Rev-i-(R)5Q(R)3/MS-C] (Fig. 4b). Mutants with insertions of a lysine or glutamine followed by eight arginines [Δ35/50Rev-i-K(R)8/MS-C or Δ35/50Rev-i-Q(R)8/MS-C] showed modest nuclear accumulation in untreated cells and were redistributed exclusively into nuclei by LMB treatment (data not shown). A small fraction of cells expressing mutants with a polylysine stretch interrupted by one or two arginines [Δ35/50Rev-i-(K)4R(K)4/MS-C or Δ35/50Rev-i-(K)4(R)2(K)3/MS-C] displayed nuclear localization of respective fusion proteins after LMB treatment (Fig. 4b). All of the above arginine and lysine mutants displayed similar LMB-induced morphological changes in Cos-7 cells.
DISCUSSION
We have shown here that the entire basic domain of Rev could be functionally replaced by a string of nine arginines. The resulting protein, while retaining the inherent specificity of Rev for RRE RNA, displayed reduced stringency and recognized some RRE mutants that were not bound by wt Rev. Second, optimal in vitro RRE RNA binding and nuclear accumulation required the presence of at least eight uninterrupted arginines. Finally, deletion of oligomerization domains of Rev that flank the NLS domain markedly reduced or abolished multimer formation of the resulting 9R mutant, Δ24/60Rev-i-9R/MS-C, despite the inherent potential of the MS2 coat protein to form multimers.
Polyarginine insertions expand the RNA binding specificity of Rev.
Using nine-arginine substituted Rev–MS-C mutants forced to express a truncated nine-arginine substituted Rev of 87 residues or a frame-shifted Rev–MS-C protein forced to terminate beyond the 2nd residue of MS-C, we established that the activation phenotype for RRE derivatives was mediated by the nine-arginine motif. In vitro RRE RNA binding studies with a 17-mer Rev basic domain peptide have shown that initial RNA binding occurs through interaction of an arginine fork with the adjacent phosphates of a pUpGGG sequence at the junction between stem-loop IIA and stem-loop IIB of the RRE secondary structure. A second arginine fork mediated interaction with a pGpCU sequence in SLIIB, and additional base-specific interactions with the major groove of SLIIB RNA in the A form helix are also critical binding events (43, 44). Although our nine-arginine insertion mutant was not active with unrelated RNAs such as TAR, VAI, and RexRe, its reactivity with RRE mutants and variants was somewhat less stringent (Table 3). Most notably, changes at the GGG sequence that may have disrupted the initial contact of the 17-mer peptide to RNA through arginine forks were recognized by the nine-arginine mutant. Other RRE mutations capable of introducing subtle changes in the secondary (loss of noncanonical GG base pairing with the AGC-ACG at 62-to-64 mutant), and tertiary (SLIIAC′B, SLIIACB & AGC-to-ACG, and SLIIAC′B & AGC 62-to-64 mutants) structure were also modestly responsive with the nine-arginine mutant. However, more drastic changes, such as the excision of the entire SLIIB or inverted complement of SLIIB, were nonresponsive. In vitro binding potential of the nine-arginine protein for the respective mutant RRE RNAs correlated with their in vivo response (not shown). Our analysis suggests that if there are other sites on RRE RNA for Rev binding, these sites must be contained within stem-loop II, since RRE lacking SLIIB had a null activation phenotype. Therefore, to a first approximation, our data suggest that the binding of the homopolymeric arginines to any one of the multiple RRE loci is equivalent to that of Rev binding to all the potential sites in RRE. We cannot formally exclude, however, the possibility that the 9R mutant may bind to another, undetected RRE subsequently.
It was of interest that the 9R mutant was unable to activate TAR-containing mRNAs that responded positively to the HIV-1 Tev protein. Previous work has shown that a nine-arginine substitution for the arginine-rich RNA binding motif of HIV Tat preserved activation for both HIV-1 and HIV-2 LTR (14). In vitro, a nine-arginine-substituted Rev peptide of residues 22 to 85 bound TAR RNA with a reduced affinity compared to RRE RNA. However, the nine-arginine-substituted Rev–MS-C fusion protein was poor at binding TAR RNA (41a). Whereas interactions of Tev or the nine-arginine-substituted Tat protein may be reinforced in vivo by other protein-protein interactions with the activation domain of Tat (viz. cyclin T1) or by Tat oligomerization (27, 69, 89), sequences flanking the arginines in our 9R Rev–MS-C mutant did not facilitate such enhancement of TAR RNA binding.
When nine arginines were inserted away from the N terminus of Rev as in Δ35/50 Rev/MS-C-i-9R, RRE RNA binding was impaired (Table 1). It is likely that, at the distant locus, the nine-arginine string lacks the conformational integrity provided by the N-terminus Rev that is required for optimal RNA binding affinity and discrimination of RRE binding specificity (18). Although this mutant bound MS2 RNA almost as well as had wt Rev–MS-C, its negative phenotype for activation of MS2 RNA was probably due to its mostly cytoplasmic residence. Mutants inserting five or fewer arginines in place of NLS of Rev displayed a two- to fivefold-reduced affinity for RRE RNA compared with the binding potential of Rev, Rev–MS-C, or a 9R mutant (data not shown). Therefore, it is possible that optimal binding to wt RRE can occur with a minimal number of arginine forks. Unlike the case of Tat, where a single arginine flanked by Lys provides an optimal arginine fork for TAR RNA binding, the arginine forks of Rev may have to have to be presented as an uninterrupted arginine cluster for RRE binding (10, 76, 78). Interruption of the nine-arginine stretch by a single lysine, (R)3K(R)5, or asparagine, (R)3N(R)5, also resulted in reduced RRE RNA binding (data not shown). A lysine cluster interrupted by two tandem arginines, (K)4(R)2(K)3, was also impaired for RRE binding. Some of these substituted arginine and lysine mutants were somewhat impaired for RRE activation but not for MS2 RNA. This was expected since they all had intact MS-C ORF and bound MS2 RNA as well as wt Rev–MS-C. Their activation potential for MS2 RNA was lost when they were truncated to express only the substituted Rev ORF.
Requirements of arginine-rich motif for nuclear localization.
Genetic analyses of the arginine-rich domain of Rev have suggested an inherent functional redundancy of arginines (32). Apart from a requirement for α helicity (3, 4, 76, 77), changes at the individual arginines are tolerated with no loss of function. Changes at other residues in this motif were also tolerated except for the asparate at position 38 and tryptophan at position 45 (32, 39, 76, 77). We found that the entire basic domain of Rev could be exchanged for a string of nine arginines with no loss of activation potential for RRE, and the MS2 RNA and the resulting protein accumulated in the nucleus and nucleoli under steady-state conditions. Optimal trans-activation of RRE required the presence of at least eight arginines. However, insertions of four to seven arginines were competent for activation of MS2 RNA, although these mutants were mostly cytoplasmic. The high-affinity binding of the MS2 RNA to the coat protein may compensate for the poor nuclear accumulation of the sparse arginine insertions at the Rev NLS. In general, there was a correlation between the steady-state nuclear accumulation and the RRE activation potential of the various mutants.
If the RNA binding and NLS or NOS domains of Rev were truly modular, reciprocal insertion of the deleted sequence at a distant site, i.e., within the MS-C ORF, should have compensated for the defects of the corresponding deletion mutants. However, inserting Rev sequences of 8 (RRNRRRRW), 14 (residues 33 to 46), 16 (residues 35 to 50), 19 (residues 33 to 51), or 37 (residues 24 to 60) residues into the deletion and substitution mutants failed to restore the activation potential for either the RRE or the MS2 RNAs (data not shown). Under steady-state conditions, these mutants were mostly, if not exclusively, cytoplasmic. They bound RRE RNA less efficiently in vitro than wt Rev (not shown), implying that optimal RRE RNA binding requires positional integrity of the Rev NLS domain and may be stabilized by higher-order interactions with the neighboring Rev domain(s).
Nucleocytoplasmic shuttling.
Since the Rev–MS-C mutants that were impaired for nuclear accumulation had intact CRM1 binding NES domains, LMB treatment (96) would be expected to block their nuclear egress. Δ35/50Rev/MS-C lacking the NLS domain was probably not imported into the nuclei and remained cytoplasmic even with maximal LMB treatment. A similar phenotype was observed with a mutant that exchanged four sequential arginines followed by a tryptophan in the Rev NLS for asparate followed by leucine [(R)4WRE/DLRE]. Inserting the 16-residue Rev sequence (positions 16 to 50) into the deletion and substitution mutants failed to restore nuclear accumulation of the mutants. LMB treatment induced partial relocalization of these insertion mutants into the nuclei, suggesting that these mutants are imported into the nucleus, albeit inefficiently. A nine-arginine insertion at the MS-C ORF (Δ35/50Rev/MS-C-i-9R) with a defective phenotype for both RRE and MS2 RNAs was also partially relocalized to the nuclei after LMB treatment. All of these insertion mutants had an intact MS-C ORF protein and bound MS2 RNA as well as wt Rev–MS-C in vitro. The failure of these (out-of-context) insertion mutants to activate MS2 RNA may have been due to the inability of the insertion mutants to recover completely the lost nuclear targeting of the NLS deletion and/or substitution mutants.
Mutants with arginine strings interrupted or flanked by some other residue or mutants with short arginine strings were somewhat impaired for nuclear accumulation, resembling the RevM6 or RevN40D mutants (7, 53, 80) in this regard. LMB treatment relocalized them to the nuclei almost completely. RRE binding by these mutant proteins was, in general, less efficient than that by wt Rev. Therefore, abbreviated nuclear residence time and/or reduced RRE binding affinity may underlie the poor activation of RRE by these mutants. Mutants in which interrupted lysine strings were inserted instead of nine arginines were mostly, if not exclusively, cytoplasmic. LMB induced a partial redistribution to nuclei. Both lysine cluster mutants were poor RRE binders and, as expected, failed to activate RRE. One lysine cluster mutant (4K2R3K) was active for MS2, while the other one (4K1R4K) was not, suggesting that in vivo MS2 RNA binding may have been compromised for the latter.
Arginine-rich NLS of the type found in HIV Rev and Tat or HTLV-1 Rex represent a novel class of motifs that depend on direct interaction with importin-β for nuclear import (35, 83). In general, such domains contain three or four sequential arginines (29, 66, 81, 83, 84, 91). Since insertion of three to four arginines in place of the Rev NLS allowed activation of MS2 RNA (Table 4), intranuclear levels of this mutant may have been at the threshold level for activation of this target. In light of these observations, a motif composed of three arginines may be sufficient for importin-β-mediated nuclear import of our Rev-MS-C fusion proteins.
Target-dependent oligomerization requirements for Rev function.
From a purely kinetic and biochemical standpoint, it has not been possible to determine whether Rev binds to RRE as a monomer, followed by the in situ assembly of additional Rev molecules, or whether Rev multimerization is a prerequisite for RNA binding (86). There is evidence that Rev multimerization may reinforce monomer binding. Although the minimally responsive SLIIB RRE RNA can only bind a Rev monomer (16, 82, 86), it was not activated by the multimerization-defective Rev, suggesting that additional Rev molecules have to be recruited by protein-protein interactions to stabilize the SLIIB RNA-Rev complex. Protein multimerization may then lock the productive RNA-protein complex with fast koff rates. Enhanced RNA binding affinity can bypass the requirement for protein oligomerization to stabilize the initial RNP complex. MS2 phage coat protein has been demonstrated to exist as dimers and tetramers in the phage particle (28, 62, 68, 85). However, the calculated affinity of MS2 coat protein for its target is greater than the corresponding affinity of Rev for RRE by an order of magnitude (12, 71, 87, 93, 94). Interaction between the coat protein and MS2 RNA may thus bypass the need for protein multimerization. By extrapolation, gain-of-function Rev mutants with enhanced affinity for RRE RNA may function in vivo without the need for oligomerization.
ACKNOWLEDGMENTS
We thank Alicia Buckler-White of NIAID for oligonucleotide synthesis. We are grateful to Paul Wingfield for the purified Rev protein expressed in E. coli. We thank Barbara Felber of NCI/FCRDC and Marie Lou Hammerskjold of SUNY, Buffalo, for anti-Rev antisera. We thank John Hanover and Dona Love of NIDDK for advice concerning LMB treatment. We thank Kuan Teh-Jeang, Eric Freed, and Jonathan Silver of NIAID for critical reviews and comments.
REFERENCES
- 1.Askjaer P, Jensen T H, Nilsson J, Englmeier L, Kjems J. The specificity of the CRM1-Rev nuclear export signal interaction is mediated by RanGTP. J Biol Chem. 1998;273:33414–33422. doi: 10.1074/jbc.273.50.33414. [DOI] [PubMed] [Google Scholar]
- 2.Bartel D, Zapp M, Green M, Szostak J. HIV-1 Rev regulation involves recognition of non-Watson-Crick base pairs in viral RNA. Cell. 1991;67:529–536. doi: 10.1016/0092-8674(91)90527-6. [DOI] [PubMed] [Google Scholar]
- 3.Battiste J L, Mao H, Rao N S, Tan R, Muhandiram D R, Kay L E, Frankel A D, Williamson J R. Alpha helix-RNA major groove recognition in an HIV-1 rev peptide-RRE RNA complex. Science. 1996;273:1547–1551. doi: 10.1126/science.273.5281.1547. [DOI] [PubMed] [Google Scholar]
- 4.Battiste J L, Tan R, Frankel A D, Williamson J R. Binding of an HIV Rev peptide to Rev responsive element RNA induces formation of purine-purine base pairs. Biochemistry. 1994;33:2741–2747. doi: 10.1021/bi00176a001. [DOI] [PubMed] [Google Scholar]
- 5.Bevec D, Dobrovnik M, Hauber J, Böhnlein E. Inhibition of human immunodeficiency virus type 1 replication in human T cells by retroviral-mediated gene transfer of a dominant-negative Rev trans-activator. Proc Natl Acad Sci USA. 1992;89:9870–9874. doi: 10.1073/pnas.89.20.9870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bogerd H P, Fridell R A, Madore S, Cullen B R. Identification of a novel cellular cofactor for the Rev/Rex class of retroviral regulatory proteins. Cell. 1995;82:485–494. doi: 10.1016/0092-8674(95)90437-9. [DOI] [PubMed] [Google Scholar]
- 7.Böhnlein E, Berger J, Hauber J. Functional mapping of the human immunodeficiency virus type 1 Rev RNA binding domain: new insights into the domain structure of Rev and Rex. J Virol. 1991;65:7051–7055. doi: 10.1128/jvi.65.12.7051-7055.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Böhnlein S, Pirker F P, Hofer L, Zimmermann K, Bachmayer H, Böhnlein E, Hauber J. Transdominant repressors for human T-cell leukemia virus type I Rex and human immunodeficiency virus type 1 Rev function. J Virol. 1991;65:81–88. doi: 10.1128/jvi.65.1.81-88.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calnan B J, Biancalana S, Hudson D, Frankel A D. Analysis of arginine-rich peptides from the HIV Tat protein reveals unusual features of RNA-protein recognition. Genes Dev. 1991;5:201–210. doi: 10.1101/gad.5.2.201. [DOI] [PubMed] [Google Scholar]
- 10.Calnan B J, Tidor B, Biancalana S, Hudson D, Frankel A D. Arginine-mediated RNA recognition: the arginine fork. Science. 1991;252:1167–1171. doi: 10.1126/science.252.5009.1167. [DOI] [PubMed] [Google Scholar]
- 11.Campbell L H, Borg K T, Haines J K, Moon R T, Schoenberg D R, Arrigo S J. Human immunodeficiency virus type 1 Rev is required in vivo for binding of poly(A)-binding protein to Rev-dependent RNAs. J Virol. 1994;68:5433–5438. doi: 10.1128/jvi.68.9.5433-5438.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carey J, Cameron V, de Hasseth P, Uhlenbeck O. Sequence specific interaction of R17 coat protein with its ribonucleic acid binding site. Biochemistry. 1983;22:2601–2610. doi: 10.1021/bi00280a002. [DOI] [PubMed] [Google Scholar]
- 13.Chang D D, Sharp P A. Regulation by HIV Rev depends upon recognition of splice sites. Cell. 1989;59:789–795. doi: 10.1016/0092-8674(89)90602-8. [DOI] [PubMed] [Google Scholar]
- 14.Chang Y N, Jeang K T. The basic RNA-binding domain of HIV-2 Tat contributes to preferential trans-activation of a TAR2-containing LTR. Nucleic Acids Res. 1992;20:5465–5472. doi: 10.1093/nar/20.20.5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cochrane A, Perkins A, Rosen C. Identification of sequences important in the nucleolar localization of human immunodeficiency virus Rev: relevance of nucleolar localization to function. J Virol. 1990;64:881–885. doi: 10.1128/jvi.64.2.881-885.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cook K S, Fisk G J, Hauber J, Usman N, Daly T J, Rusche J R. Characterization of HIV-1 REV protein: binding stoichiometry and minimal RNA substrate. Nucleic Acids Res. 1991;19:1577–1583. doi: 10.1093/nar/19.7.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daly T, Cook K, Gray G, Maione T, Rusche J R. Specific binding of HIV-1 recombinant Rev protein to the Rev-responsive element in vitro. Nature. 1989;342:816–819. doi: 10.1038/342816a0. [DOI] [PubMed] [Google Scholar]
- 18.Daly T J, Doten R C, Rusche J R, Auer M. The amino terminal domain of HIV-1 Rev is required for discrimination of the RRE from nonspecific RNA. J Mol Biol. 1995;253:243–258. doi: 10.1006/jmbi.1995.0549. [DOI] [PubMed] [Google Scholar]
- 19.Emerman M, Vazeux R, Peden K. The rev gene product of the human immunodeficiency virus affects envelope-specific RNA localization. Cell. 1989;57:1155–1165. doi: 10.1016/0092-8674(89)90053-6. [DOI] [PubMed] [Google Scholar]
- 20.Escaich S, Kalfoglou C, Plavec I, Kaushal S, Mosca J D, Bohnlein E. RevM10-mediated inhibition of HIV-1 replication in chronically infected T cells. Hum Gene Ther. 1995;6:625–634. doi: 10.1089/hum.1995.6.5-625. [DOI] [PubMed] [Google Scholar]
- 21.Feinberg M, Jarrett R, Aldovini A, Gallo R, Wong-Staal F. HTLV III expression and production involve complex regulation at the levels of splicing and translation of viral RNA. Cell. 1986;46:807–817. doi: 10.1016/0092-8674(86)90062-0. [DOI] [PubMed] [Google Scholar]
- 22.Felber B, Hadzopoulu-Cladaras M, Cladaras C, Copeland T, Tse A, Pavlakis G. Rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA. Proc Natl Acad Sci USA. 1989;86:1495–1499. doi: 10.1073/pnas.86.5.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 24.Fouts D E, True H L, Cengel K A, Celander D W. Site-specific phosphorylation of the human immunodeficiency virus type-1 Rev protein accelerates formation of an efficient RNA-binding conformation. Biochemistry. 1997;36:13256–13262. doi: 10.1021/bi971551d. [DOI] [PubMed] [Google Scholar]
- 25.Fritz C C, Zapp M L, Green M R. A human nucleoporin-like protein that specifically interacts with HIV Rev. Nature. 1995;376:530–533. doi: 10.1038/376530a0. [DOI] [PubMed] [Google Scholar]
- 26.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 27.Garber M E, Wei P, KewalRamani V N, Mayall T P, Herrmann C H, Rice A P, Littman D R, Jones K A. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 1998;12:3512–3527. doi: 10.1101/gad.12.22.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Golmohammadi R, Valegard K, Fridborg K, Liljas L. The refined structure of bacteriophage MS2 at 2.8 Å resolution. J Mol Biol. 1993;234:620–639. doi: 10.1006/jmbi.1993.1616. [DOI] [PubMed] [Google Scholar]
- 29.Gorlich D, Henklein P, Laskey R A, Hartmann E. A 41 amino acid motif in importin-alpha confers binding to importin-beta and hence transit into the nucleus. EMBO J. 1996;15:1810–1817. [PMC free article] [PubMed] [Google Scholar]
- 30.Hadzoupoulu-Cladaras M, Felber B, Cladaras C, Athanasopoulos A, Tse A, Pavlakis G. The Rev (Trs/Art) protein of human immunodeficiency virus type I affects viral mRNA and protein expression via cis-acting sequences in the env region. J Virol. 1989;63:1265–1274. doi: 10.1128/jvi.63.3.1265-1274.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hammarskjold M L, Heimer J, Hammarskjold B, Sangwan I, Albert L, Rekosh D. Regulation of human immunodeficiency virus env expression by the rev gene product. J Virol. 1989;63:1959–1966. doi: 10.1128/jvi.63.5.1959-1966.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hammerschmid M, Palmeri D, Ruhl M, Jaksche H, Weichselbraun I, Bohnlein E, Malim M H, Hauber J. Scanning mutagenesis of the arginine-rich region of the human immunodeficiency virus type 1 Rev trans-activator. J Virol. 1994;68:7329–7335. doi: 10.1128/jvi.68.11.7329-7335.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heaphy S, Dingwall C, Ernberg M, Gait M, Green S, Karn J, Lowe A, Singh M, Skinner M. HIV-1 regulator of virion expression (Rev) binds to an RNA stem-loop structure located in the Rev response element region. Cell. 1990;60:685–693. doi: 10.1016/0092-8674(90)90671-z. [DOI] [PubMed] [Google Scholar]
- 34.Heaphy S, Finch J T, Gait M J, Karn J, Singh M. Human immunodeficiency virus type 1 regulator of virion expression, rev, forms nucleoprotein filaments after binding to a purine-rich “bubble” located within the rev-responsive region of viral mRNAs. Proc Natl Acad Sci USA. 1991;88:7366–7370. doi: 10.1073/pnas.88.16.7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henderson B R, Percipalle P. Interactions between HIV Rev and nuclear import and export factors: the Rev nuclear localisation signal mediates specific binding to human importin-beta. J Mol Biol. 1997;274:693–707. doi: 10.1006/jmbi.1997.1420. [DOI] [PubMed] [Google Scholar]
- 36.Holland S M, Ahmad N, Maitra R K, Wingfield P, Venkatesan S. Human immunodeficiency virus Rev protein recognizes a target sequence in Rev-responsive element RNA within the context of RNA secondary structure. J Virol. 1990;64:5966–5975. doi: 10.1128/jvi.64.12.5966-5975.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holland S M, Chavez M, Gerstberger S, Venkatesan S. A specific sequence with a bulged guanosine residue(s) in a stem-bulge-stem structure of Rev-responsive element RNA is required for trans activation by human immunodeficiency virus type 1 Rev. J Virol. 1992;66:3699–3706. doi: 10.1128/jvi.66.6.3699-3706.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hope T J, Bond B L, McDonald D, Klein N P, Parslow T G. Effector domains of human immunodeficiency virus type 1 Rev and human T-cell leukemia virus type I Rex are functionally interchangeable and share an essential peptide motif. J Virol. 1991;65:6001–6007. doi: 10.1128/jvi.65.11.6001-6007.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hope T J, Huang X J, McDonald D, Parslow T G. Steroid-receptor fusion of the human immunodeficiency virus type 1 Rev transactivator: mapping cryptic functions of the arginine-rich motif. Proc Natl Acad Sci USA. 1990;87:7787–7791. doi: 10.1073/pnas.87.19.7787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hope T J, Klein N P, Elder M E, Parslow T G. trans-Dominant inhibition of human immunodeficiency virus type 1 Rev occurs through formation of inactive protein complexes. J Virol. 1992;66:1849–1855. doi: 10.1128/jvi.66.4.1849-1855.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang X, Hope T, Bond B, McDonald D, Grahl K, Parslow T. Minimal Rev-responsive element for type 1 human immunodeficiency virus. J Virol. 1991;65:2131–2134. doi: 10.1128/jvi.65.4.2131-2134.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41a.Jeong K S, Nam Y S, Venkatesan S. Deletions near the N-terminus of HIV-1 Rev reduce RNA binding affinity and dominantly interfere with Rev function irrespective of the RNA target. Arch Virol. 2000;145:2443–2467. doi: 10.1007/s007050070001. [DOI] [PubMed] [Google Scholar]
- 42.Kalland K H, Szilvay A M, Brokstad K A, Saetrevik W, Haukenes G. The human immunodeficiency virus type 1 Rev protein shuttles between the cytoplasm and nuclear compartments. Mol Cell Biol. 1994;14:7436–7444. doi: 10.1128/mcb.14.11.7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kjems J, Brown M, Chang D D, Sharp P A. Structural analysis of the interaction between the human immunodeficiency virus Rev protein and the Rev response element. Proc Natl Acad Sci USA. 1991;88:683–687. doi: 10.1073/pnas.88.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kjems J, Calnan B J, Frankel A D, Sharp P A. Specific binding of a basic peptide from HIV-1 Rev. EMBO J. 1992;11:1119–1129. doi: 10.1002/j.1460-2075.1992.tb05152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Knight D M, Flomerfelt F A, Ghrayeb J. Expression of the art/trs protein of HIV and study of its role in viral envelope synthesis. Science. 1987;236:837–840. doi: 10.1126/science.3033827. [DOI] [PubMed] [Google Scholar]
- 46.Kubota S, Furuta R, Maki M, Hatanaka M. Inhibition of human immunodeficiency virus type 1 Rev function by a Rev mutant which interferes with nuclear/nucleolar localization of Rev. J Virol. 1992;66:2510–2513. doi: 10.1128/jvi.66.4.2510-2513.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner E P, Wolff B, Yoshida M, Horinouchi S. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc Natl Acad Sci USA. 1999;96:9112–9117. doi: 10.1073/pnas.96.16.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kudo N, Wolff B, Sekimoto T, Schreiner E P, Yoneda Y, Yanagida M, Horinouchi S, Yoshida M. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp Cell Res. 1998;242:540–547. doi: 10.1006/excr.1998.4136. [DOI] [PubMed] [Google Scholar]
- 49.Love D C, Sweitzer T D, Hanover J A. Reconstitution of HIV-1 rev nuclear export: independent requirements for nuclear import and export. Proc Natl Acad Sci USA. 1998;95:10608–10613. doi: 10.1073/pnas.95.18.10608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Madore S J, Tiley L S, Malim M H, Cullen B R. Sequence requirements for Rev multimerization in vivo. Virology. 1994;202:186–194. doi: 10.1006/viro.1994.1334. [DOI] [PubMed] [Google Scholar]
- 51.Malim M, Bohnlein S, Fenrick R, Le S, Maizel J, Cullen B. Functional dissection of the HIV-1 Rev trans-activator-derivation of a trans-dominant repressor of Rev function. Cell. 1989;58:205–214. doi: 10.1016/0092-8674(89)90416-9. [DOI] [PubMed] [Google Scholar]
- 52.Malim M, Hauber J, Le S, Maizel J, Cullen B. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989;338:254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 53.Malim M H, Cullen B R. HIV-1 structural gene expression requires the binding of multiple Rev monomers to the viral RRE: implications for HIV-1 latency. Cell. 1991;65:241–248. doi: 10.1016/0092-8674(91)90158-u. [DOI] [PubMed] [Google Scholar]
- 54.Malim M H, Cullen B R. Rev and the fate of pre-mRNA in the nucleus: implications for the regulation of RNA processing in eukayotes. Mol Cell Biol. 1993;13:6180–6189. doi: 10.1128/mcb.13.10.6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Malim M H, Freimuth W W, Liu J, Boyle T J, Lyerly H K, Cullen B R, Nabel G J. Stable expression of transdominant Rev protein in human T cells inhibits human immunodeficiency virus replication. J Exp Med. 1992;176:1197–1201. doi: 10.1084/jem.176.4.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Malim M H, McCarn D F, Tiley L S, Cullen B R. Mutational definition of the human immunodeficiency virus type 1 Rev activation domain. J Virol. 1991;65:4248–4254. doi: 10.1128/jvi.65.8.4248-4254.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Malim M H, Tiley L S, McCarn D F, Rusche J R, Hauber J, Cullen B R. HIV-1 structural gene expression requires binding of the Rev trans-activator to its RNA target sequence. Cell. 1990;60:675–683. doi: 10.1016/0092-8674(90)90670-a. [DOI] [PubMed] [Google Scholar]
- 58.Mann D A, Mikaelian I, Zemmel R W, Green S M, Lowe A D, Kimura T, Singh M, Butler P J, Gait M J, Karn J. A molecular rheostat. Co-operative rev binding to stem I of the rev-response element modulates human immunodeficiency virus type-1 late gene expression. J Mol Biol. 1994;241:193–207. doi: 10.1006/jmbi.1994.1488. [DOI] [PubMed] [Google Scholar]
- 59.McDonald D, Hope T J, Parslow T G. Posttranscriptional regulation by the human immunodeficiency virus type 1 Rev and human T-cell leukemia virus type I Rex proteins through a heterologous RNA binding site. J Virol. 1992;66:7232–7238. doi: 10.1128/jvi.66.12.7232-7238.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mermer B, Felber B K, Campbell M, Pavlakis G N. Identification of trans-dominant HIV-1 rev protein mutants by direct transfer of bacterially produced proteins into human cells. Nucleic Acids Res. 1990;18:2037–2044. doi: 10.1093/nar/18.8.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meyer B E, Malim M H. The HIV-1 Rev trans-activator shuttles between the nucleus and the cytoplasm. Genes Dev. 1994;8:1538–1547. doi: 10.1101/gad.8.13.1538. [DOI] [PubMed] [Google Scholar]
- 62.Ni C Z, Syed R, Kodandapani R, Wickersham J, Peabody D S, Ely K R. Crystal structure of the MS2 coat protein dimer: implications for RNA binding and virus assembly. Structure. 1995;3:255–263. doi: 10.1016/S0969-2126(01)00156-3. [DOI] [PubMed] [Google Scholar]
- 63.Olsen H S, Beidas S, Dillon P, Rosen C A, Cochrane A W. Mutational analysis of the HIV-1 Rev protein and its target sequence, the Rev responsive element. J Acquir Immune Defic Syndr. 1991;4:558–567. [PubMed] [Google Scholar]
- 64.Olsen H S, Cochrane A W, Dillon P J, Nalin C M, Rosen C A. Interaction of the human immunodeficiency virus type 1 Rev protein with a structured region in env mRNA is dependent on multimer formation mediated through a basic stretch of amino acids. Genes Dev. 1990;4:1357–1364. doi: 10.1101/gad.4.8.1357. [DOI] [PubMed] [Google Scholar]
- 65.Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 66.Palmeri D, Malim M H. Importin beta can mediate the nuclear import of an arginine-rich nuclear localization signal in the absence of importin alpha. Mol Cell Biol. 1999;19:1218–1225. doi: 10.1128/mcb.19.2.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Park H, Davies M V, Langland J O, Chang H W, Nam Y S, Tartaglia J, Paoletti E, Jacobs B L, Kaufman R J, Venkatesan S. TAR RNA-binding protein is an inhibitor of the interferon-induced protein kinase PKR. Proc Natl Acad Sci USA. 1994;91:4713–4717. doi: 10.1073/pnas.91.11.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Peabody D S. The RNA binding site of bacteriophage MS2 coat protein. EMBO J. 1993;12:595–600. doi: 10.1002/j.1460-2075.1993.tb05691.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rana T M, Jeang K T. Biochemical and functional interactions between HIV-1 Tat protein and TAR RNA. Arch Biochem Biophys. 1999;365:175–185. doi: 10.1006/abbi.1999.1206. [DOI] [PubMed] [Google Scholar]
- 70.Richard N, Iacampo S, Cochrane A. HIV-1 Rev is capable of shuttling between the nucleus and cytoplasm. Virology. 1994;204:123–131. doi: 10.1006/viro.1994.1516. [DOI] [PubMed] [Google Scholar]
- 71.Romaniuk P, Lowary P, Wu H, Stormo G, Uhlenbeck O. RNA binding site of R17 coat protein. Biochemistry. 1987;26:1563–1568. doi: 10.1021/bi00380a011. [DOI] [PubMed] [Google Scholar]
- 72.Rosen C A, Terwilliger E, Dayton A, Sodroski J G, Haseltine W A. Intragenic cis-acting art gene-responsive sequences of the human immunodeficiency virus. Proc Natl Acad Sci USA. 1988;85:2071–2075. doi: 10.1073/pnas.85.7.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ruhl M, Himmelspach M, Bahr G M, Hammerschmid F, Jaksche H, Wolff B, Aschauer H, Farrington G K, Probst H, Bevec D, et al. Eukaryotic initiation factor 5A is a cellular target of the human immunodeficiency virus type 1 Rev activation domain mediating trans-activation. J Cell Biol. 1993;123:1309–1320. doi: 10.1083/jcb.123.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sodroski J, Goh W C, Rosen C, Dayton A, Terwilliger E, Haseltine W. A second post-transcriptional trans-activator gene required for HTLV-III replication. Nature. 1986;321:412–417. doi: 10.1038/321412a0. [DOI] [PubMed] [Google Scholar]
- 75.Stade K, Ford C S, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 76.Tan R, Chen L, Buettner J A, Hudson D, Frankel A D. RNA recognition by an isolated alpha helix. Cell. 1993;73:1031–1040. doi: 10.1016/0092-8674(93)90280-4. [DOI] [PubMed] [Google Scholar]
- 77.Tan R, Frankel A D. Costabilization of peptide and RNA structure in an HIV Rev peptide-RRE complex. Biochemistry. 1994;33:14579–14585. doi: 10.1021/bi00252a025. [DOI] [PubMed] [Google Scholar]
- 78.Tao J, Frankel A D. Specific binding of arginine to TAR RNA. Proc Natl Acad Sci USA. 1992;89:2723–2726. doi: 10.1073/pnas.89.7.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Thomas S L, Hauber J, Casari G. Probing the structure of the HIV-1 Rev trans-activator protein by functional analysis. Protein Eng. 1997;10:103–107. doi: 10.1093/protein/10.2.103. [DOI] [PubMed] [Google Scholar]
- 80.Thomas S L, Oft M, Jaksche H, Casari G, Heger P, Dobrovnik M, Bevec D, Hauber J. Functional analysis of the human immunodeficiency virus type 1 Rev protein oligomerization interface. J Virol. 1998;72:2935–2944. doi: 10.1128/jvi.72.4.2935-2944.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tiganis T, Flint A J, Adam S A, Tonks N K. Association of the T-cell protein tyrosine phosphatase with nuclear import factor p97. J Biol Chem. 1997;272:21548–21557. doi: 10.1074/jbc.272.34.21548. [DOI] [PubMed] [Google Scholar]
- 82.Tiley L S, Malim M H, Tewary H K, Stockley P G, Cullen B R. Identification of a high-affinity RNA-binding site for the human immunodeficiency virus type 1 Rev protein. Proc Natl Acad Sci USA. 1992;89:758–762. doi: 10.1073/pnas.89.2.758. . (Erratum, 89:1997.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Truant R, Cullen B R. The arginine-rich domains present in human immunodeficiency virus type 1 Tat and Rev function as direct importin beta-dependent nuclear localization signals. Mol Cell Biol. 1999;19:1210–1217. doi: 10.1128/mcb.19.2.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Truant R, Fridell R A, Benson R E, Bogerd H, Cullen B R. Identification and functional characterization of a novel nuclear localization signal present in the yeast Nab2 poly(A)+ RNA binding protein. Mol Cell Biol. 1998;18:1449–1458. doi: 10.1128/mcb.18.3.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Valegard K, Liljas L, Fridborg K, Unge T. The three-dimensional structure of the bacterial virus MS2. Nature. 1990;345:36–41. doi: 10.1038/345036a0. [DOI] [PubMed] [Google Scholar]
- 86.Van Ryk D I, Venkatesan S. Real-time kinetics of HIV-1 Rev-Rev response element interactions. Definition of minimal binding sites on RNA and protein and stoichiometric analysis. J Biol Chem. 1999;274:17452–17463. doi: 10.1074/jbc.274.25.17452. [DOI] [PubMed] [Google Scholar]
- 87.Venkatesan S, Gerstberger S M, Park H, Holland S M, Nam Y. Human immunodeficiency virus type 1 Rev activation can be achieved without Rev-responsive element RNA if Rev is directed to the target as a Rev/MS2 fusion protein which tethers the MS2 operator RNA. J Virol. 1992;66:7469–7480. doi: 10.1128/jvi.66.12.7469-7480.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Venkatesh L K, Chinnadurai G. Mutants in a conserved region near the carboxy-terminus of HIV-1 Rev identify functionally important residues and exhibit a dominant negative phenotype. Virology. 1990;178:327–330. doi: 10.1016/0042-6822(90)90414-m. [DOI] [PubMed] [Google Scholar]
- 89.Wei P, Garber M E, Fang S M, Fischer W H, Jones K A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 90.Weichselbraun I, Farrington G K, Rusche J R, Böhnlein E, Hauber J. Definition of the human immunodeficiency virus type 1 Rev and human T-cell leukemia virus type I Rex protein activation domain by functional exchange. J Virol. 1992;66:2583–2587. doi: 10.1128/jvi.66.4.2583-2587.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Weis K, Ryder U, Lamond A I. The conserved amino-terminal domain of hSRP1 alpha is essential for nuclear protein import. EMBO J. 1996;15:1818–1825. [PMC free article] [PubMed] [Google Scholar]
- 92.Wingfield P T, Stahl S J, Payton M A, Venkatesan S, Misra M, Steven A C. HIV-1 Rev expressed in recombinant Escherichia coli: purification, polymerization, and conformational properties. Biochemistry. 1991;30:7527–7534. doi: 10.1021/bi00244a023. [DOI] [PubMed] [Google Scholar]
- 93.Witherell G W, Gott J M, Uhlenbeck O C. Specific interaction between RNA phage coat proteins and RNA. Prog Nucleic Acid Res Mol Biol. 1991;40:185–220. doi: 10.1016/s0079-6603(08)60842-9. [DOI] [PubMed] [Google Scholar]
- 94.Witherell G W, Wu H N, Uhlenbeck O C. Cooperative binding of R17 coat protein to RNA. Biochemistry. 1990;29:11051–11057. doi: 10.1021/bi00502a006. [DOI] [PubMed] [Google Scholar]
- 95.Wolff B, Cohen G, Hauber J, Meshcheryakova D, Rebeck C. Nucleocytoplasmic transport of the Rev protein of human immunodeficiency virus type 1 is dependent on the activation domain of the protein. Exp Cell Res. 1995;217:31–41. doi: 10.1006/excr.1995.1060. [DOI] [PubMed] [Google Scholar]
- 96.Wolff B, Sanglier J J, Wang Y. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem Biol. 1997;4:139–147. doi: 10.1016/s1074-5521(97)90257-x. [DOI] [PubMed] [Google Scholar]
- 97.Zapp M, Green M. Sequence-specific RNA binding by the HIV-1 Rev protein. Nature. 1989;342:714–716. doi: 10.1038/342714a0. [DOI] [PubMed] [Google Scholar]
- 98.Zapp M L, Hope T J, Parslow T G, Green M R. Oligomerization and RNA binding domains of the type 1 human immunodeficiency virus Rev protein: a dual function for an arginine-rich binding motif. Proc Natl Acad Sci USA. 1991;88:7734–7738. doi: 10.1073/pnas.88.17.7734. [DOI] [PMC free article] [PubMed] [Google Scholar]