Abstract
Short amino acid sequences in the cytosolic domains of transmembrane proteins are recognized by specialized adapter proteins which are part of coated vesicles utilized to transport membrane proteins between the trans-Golgi network (TGN) and the plasma membrane (forward and backward). Previously, we and others reported that the membrane-proximal tyrosine residues Y712 (human immunodeficiency virus [HIV]) and Y721 (simian immunodeficiency virus [SIV]) in the envelope glycoprotein (Env) of the primate lentiviruses are crucial for the association of Env with clathrin-associated adapter complex AP-2. The same tyrosine-based endocytosis motifs in the cytosolic domains (EnvCD) of transmembrane gp41 of HIV type 1 (HIV-1) and SIV, respectively, were also shown to modulate the interaction with TGN- and endosome-based clathrin-associated complex AP-1. Our findings suggested that EnvCD binding to AP-1, unlike the association of EnvCD with AP-2, is dependent largely on residues other than Y712 and Y721. Here, we tested if motifs downstream of Y712 affect HIV-1 EnvCD–AP-1 binding and Env trafficking. Mutational analysis revealed that the C-terminal leucine-based motif in Env was crucial for the recruitment of AP-1 in vitro and in Env-expressing cells. In addition to affecting Env–AP-1 association, mutations at the C terminus of Env also altered the subcellular localization of Env, suggesting that proper post-Golgi routing of Env depends on its recruitment of AP-1. Finally, the C-terminal dileucine was shown to assist the membrane-proximal Y712 motif in restricting the cell surface expression of Env.
The envelope glycoprotein (Env) is an essential component of retroviruses because it mediates the selective attachment of virus to its target cell (19). Env is not necessary for the formation and the release of retroviral particles, but Env of lentiviruses, like the glycoproteins of other enveloped RNA viruses, has been implicated in the spatial restriction of virus production (2, 5, 51). In addition, Env contributes to controlling the rate with which virions exit the host cell (44, 50). Env's position in the viral replication cycle is thus pivotal not only because it controls viral entry but also because it regulates when and where exactly virus will be released during the late phase of the viral life cycle.
Polarization of lentiviral release is observed not only in epithelial cells but also in lymphocytes and probably in neurons (8, 24, 41, 55). Such a directed release of virus may be particularly important for an efficient spread of virus in the crowded environment of lymph nodes, where infection is propagated locally, but polarized secretion is probably also important during systemic dissemination, when the virus crosses tissue barriers.
How Env targets virus release to certain areas of the cell surface is not known. However, viral exit at distinct sites coincides with Env localization in these areas. It seems likely, therefore, that Env accumulation at distinct sites is a prerequisite for the polarized release of infectious particles. Consequently, signals which direct Env from the trans-Golgi network (TGN) to these sites are crucial for directional virus secretion (see, e.g., references 8 and 31).
To elucidate how Env targets virus release to particular areas of the cell surface, we analyzed its trafficking during the late phase of the viral replication cycle. As part of these investigations we characterized the interaction of Env's cytosolic domain (EnvCD) with proteins of the cellular sorting machinery. The targeting of membrane proteins from the Golgi apparatus to the plasma membrane or to compartments of the endosomal system is mediated by specialized clathrin-coated vesicles (CCVs) (22, 26, 32, 35, 45). Endocytosis of membrane proteins gives rise to the formation of similar transport vesicles at the plasma membrane. Selection of the membrane proteins into CCVs largely depends on the interaction of signals in the cytosolic domains of these proteins with specific adapter proteins (APs). These APs are part of the vesicle coats forming at the cytosolic leaflet of the lipid bilayer. The existence of four different APs (AP-1, AP-2, AP-3, and AP-4) in mammals has been described so far (4). All APs are heterotetrameric complexes which are composed of two large (∼100-kDa) subunits, one medium (∼50-kDa) subunit, and one small (∼20-kDa) subunit. Membrane proteins are recruited into specific CCVs because the medium subunit, often referred to as medium chain (μ1, μ2, μ3, or μ4), or one of the large chains (β1) or both recognize sorting signals in the cytosolic domains of membrane proteins. The existence of isoforms for two of the medium chains (μ1 and μ3) was revealed recently. It is thought that the selectivity with which CCVs transport their cargo to its final destination in a particular cell type is determined at least partly by the cell-specific compositions of the APs (11, 37, 39).
Unless inhibited by the internal structural proteins of the virus, newly synthesized human immunodeficiency virus type 1 (HIV-1) Env and simian immunodeficiency virus (SIV) Env undergo endocytosis after their arrival at the cell surface (10, 46, 47; S. Wyss and M. Thali, unpublished data). For both viruses it was shown that Env internalization is mediated by an interaction of the AP-2 clathrin adapter with a membrane-proximal tyrosine-based endocytosis signal (1, 3, 6, 38). AP-2 most likely associates with EnvCD via the AP-2 medium chain μ2. When added to EnvCD independent of the other AP-2 subunits, μ2 bound to it with the same specificity as that for the intact AP-2 complex (3). In those studies HIV-1 and SIV Env were also shown to associate with the medium chain of AP-1 (μ1) (38) and with the whole AP-1 complex (1, 6). Unlike AP-2, which appears to localize exclusively at the plasma membrane, AP-1 recruits membrane proteins into CCVs budding at the TGN and from a yet-undefined endosomal subcompartment (for a review, see reference 16). In our previous studies we noticed that Y712, which is critical for HIV-1 Env–AP-2 association, also influenced the binding of μ1 to Env, but we found that this tyrosine motif was far less important for the interaction of Env with the intact AP-1 complex than it was for Env–AP-2 association (M. Boge and M. Thali, unpublished observation). Furthermore, a comparison of the previous studies on Env-adapter interactions led us to conclude that Env–AP-1 binding may be dependent on multiple motifs in EnvCD.
We thus set out to test if signals downstream of the membrane-proximal tyrosine-based motif mediate Env–AP-1 interaction. Our results reveal the importance of a C-terminal dileucine for the association of HIV-1 Env with the AP-1 adapter. We also provide evidence for an involvement of the same C-terminal signal in the proper subcellular localization of Env. Furthermore, this study demonstrates that the AP-1-binding site in Env, in conjunction with the membrane-proximal Y712, is implicated in controlling how much Env is expressed at the surfaces of infected cells though it does not act as an endocytosis signal.
MATERIALS AND METHODS
Cell culture and transfections.
HeLa cells were grown in Dulbecco's modified Eagle's medium (DMEM; Life Technologies) supplemented with 10% fetal calf serum (FCS; Life Technologies), 100 U of penicillin (Life Technologies)/ml, and 100 μg of streptomycin (Life Technologies)/ml. Human T-cell line Jurkat was grown in RPMI 1640 medium supplemented with 10% FCS and 100 U of penicillin/ml and 100 μg of streptomycin/ml.
Transfections were performed using standard calcium phosphate precipitation techniques or by electroporation. Analysis of the transfected cells was performed 48 h after transfection.
Binding assays. (i) Recombinant proteins.
All constructs used in the in vitro binding assays were made by ligation of PCR (Pwo DNA polymerase; Boehringer)-amplified DNA fragments into pGEX-3X (Pharmacia) as described previously (3). Site-directed mutagenesis was performed by applying the Quick Change system (Stratagene). The following primers were used: Y712A, 5′GAGTTAGGCAGGGAGCTTCTCCATTATCG-3′; Y768A, 5′GTGCCTCTTCGCTGCCACCGCTTGAG-3′, and LL855/856AA, 5′GGAAAGGATTGCGGCCTAGGATGGGTGG-3′. Mutations were confirmed by DNA sequencing.
(ii) Expression of fusion proteins for the binding assays.
Glutathione S-transferase (GST) fusion proteins were produced in Escherichia coli strain BL21 and purified on glutathione-Sepharose 4B beads (Amersham Pharmacia Biotech). Precultures (4 ml of Luria-Bertani [LB] medium supplemented with 0.5 M d-sorbitol and 2.5 mM betaine) were grown overnight at 37°C. They were transferred to 400 ml of LB medium supplemented with 1 M d-sorbitol and 2.5 mM betaine and grown for 8 h at 37°C. Cultures were shifted to 25°C and induced overnight with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The purification on glutathione-Sepharose 4B beads was performed according to the manufacturer's instructions, including a heat shock step with 50 mM Tris-HCl (pH 7.4)–10 mM MgSO4–2 mM ATP for 10 min at 37°C. The concentration and quality of the fusion proteins were monitored on a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel after coloration by Coomassie blue (Sigma Chemicals) staining.
(iii) Binding assay I: Adapter precipitations in T-cell lysates.
T-cell lysate was prepared from Jurkat cells essentially as described previously (9). Briefly, the cells were washed three times with cytosol buffer (25 mM HEPES [pH 7.0], 125 mM CH3COOK, 2.5 mM [CH3COO]2Mg, 1.0 mM dithiothreitol, 1 mg of glucose/ml), and the resulting pellet was resuspended in an equal volume of cytosol buffer containing 1 mM phenylmethylsulfonyl fluoride. The cell suspension was frozen in liquid nitrogen, thawed on ice, and drawn five times through a 21-gauge syringe. After centrifugation for 30 min at 20,000 × g and 4°C, the supernatant was transferred to new tubes in 100-μl aliquots and stored at −80°C. Aliquots were precleared in 900 μl of phosphate-buffered saline (PBS) containing 1.5 mg of GST proteins on Sepharose 4B beads at 35°C for 30 min. Three hundred microliters of precleared lysate was incubated with 100 μg of fusion protein on Sepharose 4B beads at 35°C for 2 h. Beads were washed three times with PBS and boiled in gel sample buffer. Proteins were transferred to nitrocellulose membranes and probed with Sigma 100/2 (anti-α-adaptin) or 100/1 (anti-β-adaptin) antibodies. Before immunostaining, staining with Ponceau S (Sigma) was used to verify that equal amounts of the different GST fusion proteins had been precipitated and transferred to the filters. Bound proteins were visualized using specific antibodies followed by binding of secondary antibodies and enhanced chemiluminescence (Pierce).
(iv) Binding assay II: SPR analysis.
Association of EnvCD with cellular adapters AP-1 and AP-2 was analyzed in real time by surface plasmon resonance (SPR) using a BIAcore AB model 3000 biosensor. In brief, an anti-GST monoclonal antibody (BIAcore AB) was immobilized on all four flow cells of a CM5 sensor chip at equal densities according to the manufacturer's instructions. Subsequently the chip was used to capture GST-EnvCD fusion proteins. All interaction experiments were performed with buffer A (20 mM HEPES-NaOH [pH 7.0], 150 mM NaCl, 10 mM KCl, 2 mM MgCl2, 0.2 mM dithiothreitol) at a flow rate of 20 μl/min. Association for 2 min was followed by dissociation for 2 min, during which buffer A was perfused. A short-pulse injection (15 s) of 20 mM NaOH–0.5% SDS was used to regenerate the sensor chip surface after each experimental cycle. The GST-EnvCD-derived sensor chip remained stable and retained its specific binding capacity for all experimental cycles of association and dissociation and regeneration. Pure AP-1 and AP-2 were prepared from pig brain as described previously (17, 18) and used at final protein concentrations ranging from 25 to 750 nM. To exclude distortions due to injection and mixing, segments of the sensograms recorded 15 to 20 s after switching from buffer flow to adapter solution or 5 to 10 s after switching back to running buffer were used for the calculations of the association and dissociation rates, respectively.
Kinetic parameters and equilibrium dissociation constants were determined from sensograms recorded at different adapter concentrations. The association constant, Ka, the dissociation constant, kd, and the equilibrium constant, KD (=kd/ka) were determined using the BIAcore kinetic evaluation software, assuming pseudo-first-order kinetics (A = B = AB). The model calculates ka and the steady-state response level, Req, by fitting data to the equation R = Req (1 − e−(kaCn + kd)[t − t (0)]), where t is the time in seconds and C is the molar concentration of adapters in the injection solution. The steric interference factor, n, which describes the valency of the interaction between adapters and the GST-EnvCD fusion protein, was set to 1. kd was determined by fitting the data to the equation R = R0e−kd[t − t(0)], where R0 is the response level at the beginning of the dissociation phase. This model has recently been applied to describe the interactions of adapters with cytosolic domains (14, 18) and is described in more detail elsewhere (21).
Indirect immunofluorescence: subcellular localization of Env.
Transfected cells were redistributed into fresh plates containing sterile glass coverslips for reattachment. After transfer of the coverslips into 24-well plates cells were incubated for 5 min in 500 μl of 0.5% bovine serum albumin (BSA)-DMEM at room temperature (RT). Thereafter, cells were fixed and permeabilized in 200 μl of ice-cold methanol and 200 μl of ice-cold acetone for 5 min each. The cells were carefully washed three times with PBS and incubated with anti-Env monoclonal antibodies (b12; dilution 1:1,000; a kind gift from P. Parren and D. Burton) or with serum from an infected individual in 0.5% BSA-DMEM in a final volume of 20 μl per coverslip for 60 min at 37°C. Cells were than washed three times with PBS, and nonspecific secondary antibody binding was blocked using PBS with 10% FCS for 20 min at RT before the cells were incubated with the fluorophore-conjugated secondary antibody (1:100 dilution in PBS, in a final volume of 20 μl per coverslip) for 30 min at RT in the dark. Coverslips were washed two times in PBS and one time in water to remove all traces of PBS and mounted on slides using Mowiol 4-88 (Calbiochem). Cells were analyzed with a Zeiss Axiophot microscope using a 63× oil immersion lens.
Generation and transfection of CD8-EnvCD chimeras.
A DNA fragment encoding EnvCD of HIV-1 LAI Env (amino acid residues 707 to 856) was obtained by PCR and cloned in-frame with the extracellular and transmembrane (TM) domains of human CD8 alpha chain (residues 1 to 211) into the pJ.CN vector (48) to generate the pCD8-HIV construct. The HIV-1 LAI Env dileucine motifs at positions 814 and 815 and 855 and 856 were mutated into alanine residues to create pCD8-LL814/815AA and pCD8-LL855/856AA, respectively, by PCR-directed mutagenesis using appropriate primers. The mutation of L at positions 855 and 856 to A (LL855/856AA mutation) was also introduced into pCD8-HIV-Y712A (1) in which the essential tyrosine residue of the tyrosine-based motif was changed into an alanine residue (pCD8-Y712A-LL855/856AA). Mutations were verified by DNA sequencing using the Sanger dideoxy termination method adapted to the ABI 373A automated sequencer. The pJ.CNstop vector, containing a stop codon downstream of the coding sequence for the transmembrane domain of CD8, was used as a control for CD8 cell surface expression (48). The pRSV-GFP vector, containing the open reading frame for the green fluorescent protein (GFP) under the control of the Rous sarcoma virus long terminal repeat promoter, was provided by T. Bordet (ICGM, Paris, France).
HeLa cells for experiments involving CD8-EnvCD chimeras were grown in DMEM (Gibco BRL) supplemented with glutamine, antibiotics, and 10% FCS. HeLa cells (8 × 106 cells per point) were suspended in 200 μl of DMEM–10% FCS–10 mM HEPES and mixed with 50 μl of 200 mM NaCl containing 25 μg of the appropriate plasmids. Electroporation was performed at 200 V and 960 μF using 4-mm-wide cuvettes in a Bio-Rad gene pulser. Cells were collected for analysis 48 h later.
Flow cytometry.
Expression of the CD8-EnvCD hybrid at the cell surface was monitored by flow cytometry. HeLa cells (106) subjected to electroporation with 4 μg of pRSV-GFP and 8 μg of the various pCD8-EnvCD vectors were washed twice with PBS and incubated for 1 h at 4°C with CD8-RD1 antibody diluted in 1% BSA-PBS (Coulter Coultronics). They were then washed three times with 1% BSA-PBS and fixed in 1% formaldehyde-PBS. Flow cytometry analysis was performed on a Coultronics Epics Elite instrument. Values shown in Table 1 are the averages of three independent experiments. The results for the CD8-EnvCD hybrids were compared to those for the wild type by means of Student's t test for unpaired samples (Stat View software package). Values are given as means ± standard errors of means (SEM).
TABLE 1.
AP-1 recruitment: quantification of the AP-1 signal in HeLa cells expressing different CD8 chimeras
| Vector pCD8 | Fluorescence intensity of γ-adaptin signal (mean of arbitrary units ± SEM)a (n) | Pb | γ-Adaptin signal (%)c |
|---|---|---|---|
| NTd | 85.3 ± 3.3 (48) | 100 | |
| EnvCD wt | 108.3 ± 7.5 (53) | <0.05 | 127 |
| LL814/815AA | 107.6 ± 9.1 (49) | <0.05 | 126 |
| LL855/856AA | 80.1 ± 5.8 (51) | >0.05 | 93 |
Values are the averages of two independent experiments. Transfected HeLa cells expressing different chimeras were processed for fluorescence analysis. The AP-1 recruitment induced by the cytosolic domain of HIV TM gp41 was determined by confocal microscopy and image analysis as described in Materials and Methods.
The results for the mutant CD8-EnvCD hybrids were compared to those for the nontransfected cells by Student's t test.
Intensity of γ-adaptin signal in the perinuclear area obtained as a percentage of the fluorescence intensity of γ-adaptin measured in nontransfected cells.
NT, the pRSV-GFP vector alone was used for cell transfection.
The overall expression of the CD8-EnvCD hybrids in transfected cells was monitored by Western blotting with anti-CD8 H160 polyclonal antibodies (Santa Cruz Biotechnology) as described by Berlioz-Torrent et al. (1) (not shown).
Indirect immunofluorescence and quantitation of AP-1 recruitment.
HeLa cells (8 × 104) underwent electroporation with 10 μg of pCD8-EnvCD vectors and were spread on glass coverslips in 24-well plates. The cells on glass coverslips were washed twice with PBS and fixed for 20 min in 4% paraformaldehyde-PBS at room temperature. They were then quenched by incubation for 10 min in PBS–0.1 M glycine and permeabilized for 30 min in 0.05% saponin–0.2% BSA-PBS at room temperature.
(i) AP-1 staining.
The cells were incubated for 1 h at room temperature with a γ-adaptin antibody (Sigma) diluted in 0.05% saponin-PBS, washed four times in 0.05% saponin-PBS, and stained with an anti-mouse Cy3 antibody (Jackson ImmunoResearch). Cells were then incubated for 20 min with 10% mouse serum-PBS, washed two times, and stained with CD8-fluorescein isothiocyanate (FITC) antibody (Coulter Coultronics) for 45 min at RT. Cells were then diluted in 0.05% saponin–0.2% BSA-PBS, washed three times in 0.05% saponin–0.2% BSA-PBS, and mounted with 5 μl of Moviol (Calbiochem) on microsope slides. Confocal microscopy was performed with a Bio-Rad MRC1024 instrument. Optical sections were mounted using the Adobe Photoshop software package.
(ii) Quantitation of AP-1 recruitment.
The fluorescence associated with AP-1 was quantitated at the single-cell level by confocal laser scanning microscopy and immunofluorescence analysis using a Bio-Rad MRC1024 confocal microscope. Simultaneous double-fluorescence acquisition was performed using the 488- and 568-nm laser lines to excite FITC (CD8 signal) and Cy3 (γ-adaptin) dyes. In each experiment the laser beam and the photomultipliers were first adjusted to the AP-1 signal of the transfected cells in order to avoid saturation in recording the signal of γ-adaptin binding in a 250-Gy color scale. For each sample medial optical slices of 45 to 55 different cells were recorded. A focal plane representative of the perinuclear staining by γ-adaptin was then chosen, and the intensities of fluorescence (mean intensities of fluorescence per pixel) of the γ-adaptin and the CD8 staining were calculated within this area using a 0- to 250-Gy color scale. All data were saved in different series, and statistical analysis (mean intensities and standard deviations) was performed. The confidence limits of the results obtained were assessed by Student's t test.
Endocytosis assays.
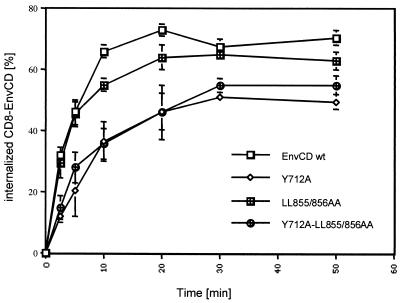
HeLa cells (8 × 106) were subjected to electroporation with 4 μg of pRSV-GFP and 8 μg of CD8-EnvCD chimera vectors. Forty-eight hours later, cells (107) were washed twice with PBS and incubated for 1 h at 4°C with CD8 Leu/2a antibody diluted in 1% BSA-PBS (Becton Dickinson). Background staining was measured by fluorescence-activated cell sorting by transferring 106 cells into a fivefold excess of PBS before staining with CD8 Leu/2a antibody. Following three washes with 1% BSA-PBS, the cells were incubated at 37°C. Aliquots were removed at various times and placed in fivefold excess of cold PBS solution. Following three washes, all fractions were incubated with anti-mouse phycoerythrin antibody for 1 h at 4°C and washed three times with cold 1% BSA-PBS. Cells were then fixed in 1% formaldehyde-PBS. Flow cytometry analysis was performed on a Coultronics Epics Elite instrument. The percentage of CD8 chimeras internalized was calculated by subtracting the initial mean of fluorescence of GFP+ CD8+ cells at 4°C from that obtained at each incubation at 37°C and then dividing by the initial mean of fluorescence of GFP+ CD8+ cells at 4°C. Values shown in Fig. 6 are the averages of three independent experiments. Values are given as means ± standard errors of means (SEM).
FIG. 6.
Internalization kinetics of CD8-EnvCD hybrids. HeLa cells transiently expressing either of the indicated CD8-EnvCD hybrids were incubated at 37°C. Aliquots were removed at various times, and flow cytometry was performed to measure the amount of CD8-EnvCD remaining at the cell surface (see Materials and Methods for details). Data are from three independent experiments.
RESULTS
Binding of clathrin-associated complex AP-1 to HIV-1 Env depends on a leucine-based signal at the C terminus of EnvCD.
To characterize the signals required for the association of HIV-1 EnvCD with adapter complexes responsible for the transport of membrane proteins, we analyzed the binding of Env to adapters in cell extracts and in live cells. Previously we and others have shown that residue Y712 in HIV-1 EnvCD and the corresponding Y721 in SIV EnvCD are implicated in the recognition of clathrin-associated adapter complexes AP-1 and AP-2 and their respective medium chains, μ1 and μ2 (1, 3, 38). However, in our studies we also noticed that Y712- and Y721-to-alanine changes affected Env binding to AP-1 much less if situated in the context of the full-length EnvCD than if introduced in a truncated version of EnvCD (1). These findings suggested that signals downstream of Y712 and Y721 are important for efficient Env–AP-1 interaction and thus probably also for correct post-Golgi sorting of Env.
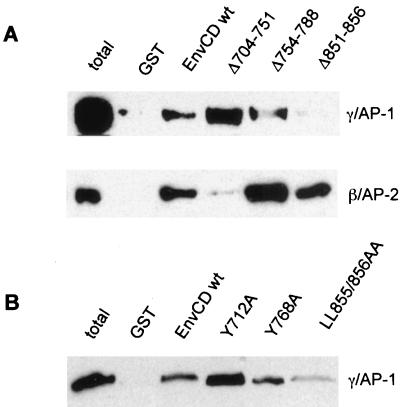
To map the regions in HIV-1 Env required for the recruitment of AP-1, we tested a panel of EnvCD mutants lacking either the membrane-proximal Y712-based motif, the more-distal Y768 motif, or sequences toward the C terminus of EnvCD. Figure 1 shows a multiple sequence alignment for the intracellular domain of HIV-1 Env. Like the extracellular part, EnvCD contains multiple short but highly conserved areas (Fig. 1), some of which overlap with potential tyrosine-based or leucine-based sorting signals. To measure EnvCD–AP-1 association, we utilized GST-EnvCD fusion proteins for the precipitation of adapter complexes, as previously described (3). As shown in Fig. 2A, while the deletion of the membrane-proximal region of HIV EnvCD (residues 704 to 751 [Δ704–751]), which contains the Y712-based sorting motif, abolished binding of AP-2, this deletion did not negatively affect Env–AP-1 interaction. The only Env deletion mutants whose association with AP-1 was reduced were those with truncated C termini (e.g., a mutant with a deletion of residues 851 to 856 [Δ851–856]). Deletion of the last six amino acids of Env substantially reduced Env–AP-1 interaction while not affecting the association of Env with AP-2. Similar reductions of EnvCD–AP-1 interactions were observed if more (10 or 16) amino acids or as few as 3 amino acids were deleted at the C terminus (data not shown), strongly suggesting that the C terminus of HIV-1 Env is crucial for its association with the AP-1 adapter.
FIG. 1.
Sequence comparison for EnvCDs of the different HIV-1 subgroups. The amino acid sequences of representative isolates of different clades of the HIV-1 groups M, N, and O were aligned, and the sequence of SIV MM239 is also shown for comparison. The amino acids are numbered as described by Korber et al. (23). Amino acids conserved in all HIV-1 isolates and in SIV CPZ are shaded. Dileucines and YxxL motifs are in boldface. With respect to these two motifs, the sequences of the isolates listed here (A, U455; B, HXB2R; C, ETH2220; D, ELI; AE, 92TH011A; F1, 93BR020.1; GH, AG_VI525A2; H, 90CR056; O, ANT70; CPZ, CPZGAB; SMM, 239) match the consensus sequences of the different clades (28).
FIG. 2.
Interaction of HIV-1 EnvCD with clathrin-associated protein complexes AP-1 and AP-2 from T-lymphocyte lysates. Equal amounts of the various immobilized GST-EnvCD fusion proteins were incubated with T-cell cytosol, and bound material was separated by SDS-PAGE. (A) The effect on AP-1 binding (top) and AP-2 binding (bottom) of deletions in the cytosolic domain of Env was assessed by immunoblotting using an antibody against either one of the large chains of AP-1 (γ-adaptin) or an antibody against the large-chain β2 of AP-2. For comparison, an aliquot of the T-cell extract was loaded in the first lane. (B) As in panel A, immobilized GST-EnvCD fusion proteins were incubated with T-cell cytosol and bound material was separated by SDS-PAGE. The effect on AP-1 recruitment of mutations in the Env tail was revealed by Western blotting using an antibody against the large chain γ of AP-1.
The C terminus of HIV-1 Env contains at its very end two leucine residues (L855 and L856). Like tyrosine-based motifs, dileucine motifs can serve as recognition sequences for AP complexes (for a recent review, see reference 15). In Fig. 1 tyrosine-based as well as dileucine motifs are numbered irrespective of whether or not they have been shown to function as sorting signals. The C-terminal dileucine, unlike the other three dileucines in EnvCD, is preceded by a nearby acidic amino acid (at position 852). Charged and polar residues upstream of dileucines are thought to help in exposing this motif, thus allowing it to be recognized by adapters (42, 43). As becomes also apparent in the alignment in Fig. 1, the C-terminal dileucine but neither of the other dileucines is conserved also in the cytosolic domain of SIV MN239.
To test the hypothesis that the C-terminal dileucine in EnvCD is part of the recognition signal for the AP-1 complex, we analyzed the binding to AP-1 of EnvCD in which we replaced the dileucine with a pair of alanines. As shown in Fig. 2B, the C-terminal dileucine is indeed critical for EnvCD–AP-1 interaction. The binding of AP-1 to the LL855/856AA mutant was significantly reduced compared with its binding to wild-type EnvCD (EnvCD wt). The same mutation did not affect EnvCD–AP-2 binding at all (not shown). Interestingly, the Y712A mutant bound the AP-1 adapter more efficiently than wild-type Env. This finding is reminiscent of the result presented in Fig. 2A, where the mutant lacking the membrane-proximal region encompassing Y712 (Δ704–751) displayed a relatively enhanced interaction with AP-1. In conclusion, the results of these binding assays demonstrate that Env–AP-1 interactions in the context of the full-length EnvCD depend on sequences at the C terminus including the conserved dileucine. The same dileucine, in contrast, is not critical for Env–AP-2 binding.
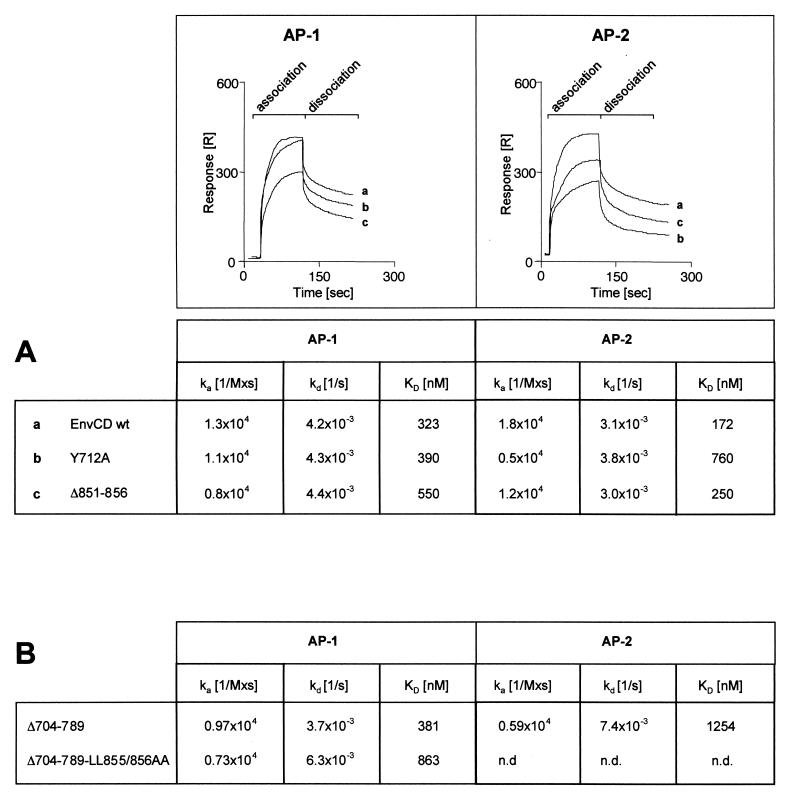
To confirm and also to quantify the negative effect of C-terminal mutations on Env–AP-1 association by an independent method, we measured the affinity of the binding of AP-1 and AP-2 to EnvCD mutants by SPR spectroscopy (14, 17, 18, 21). In SPR spectroscopy the binding of a ligand to, and its dissociation from, its receptor, which is immobilized on a sensor chip, can be analyzed with polarized light in real time. This technique measures changes in the refractive index in the vicinity of the receptor, which occur upon ligand binding. We coupled our GST-EnvCD fusion proteins to the sensor chip surface via an immobilized anti-GST antibody. Different concentrations of purified AP-1 and AP-2 complexes were added to these chips, and the association and dissociation curves served as basis for the calculation of kinetic rate constants, which are summarized in Fig. 3. This analysis again revealed the importance of the C terminus for EnvCD–AP-1 interaction because the Δ851–856 mutant yielded a KD of only 550 nM compared to 323 nM calculated for EnvCD wt. In contrast, the binding of AP-2 to EnvCD was found to be only marginally affected if the last six amino acids were deleted but was reduced to near background level (down to 760 nM from 172 nM measured for wild-type EnvCD) when the membrane-proximal Y712 was mutated (Y712A). The results obtained by the SPR analysis thus exactly paralleled what we observed in the in vitro GST pull-down binding assay, where we used lymphocyte lysate as a source of AP-1 (compare Fig. 2 and 3). In the latter assay we had also noticed that elimination of the region containing Y712 or mutation of Y712 can lead to an increase in AP-1 recruitment. We thus decided to test the effect of the LL855/856AA mutation on the interaction with AP-1 in the context of a truncated version of EnvCD. This truncated version contains only the C-terminal half of EnvCD (residues 789 to 856). The results shown in Fig. 3B demonstrate, however, that simply destroying the AP-2 binding site by either deletion or targeted mutation is not sufficient to enhance EnvCD–AP-1 binding because the affinity of AP-1 for the mutant with residues 704 to 789 deleted (Δ704–789 wt) is slightly lower than the affinity of AP-1 for the full-length tail (381 versus 323 nM). Importantly however, the results shown in Fig. 3B for the LL855/856AA mutant (Δ704–789-LL855/856AA) confirm that a replacement of the very C-terminal dileucine by a pair of alanines is sufficient to negatively affect EnvCD–AP-1 association (reducing the KD from 381 nM as measured for Δ704–789 wt to 863 nM, calculated for the dileucine mutant).
FIG. 3.
Binding of purified adapters to EnvCD analyzed by SPR spectroscopy. GST-EnvCD fusion proteins were coupled to SRP sensor chips via an anti-GST antibody at equal densities. Purified adapter complexes were passed over the EnvCD-derived chip surfaces for 2 min (association) before the chip surface was washed for 2 min (dissociation). GST alone did not show any binding of adapters. Sensograms of EnvCD-adapter interaction recorded in real time (A, top) were used to calculate ka (on rate), kd (off rate), KD (ka/kd) as described in Materials and Methods. The increase or decrease in resonance units (RU) per second and thus the slope of the curve carries the relevant information for the calculation of on and off rates, respectively, while the absolute RU values at the end points (after association or dissociation) are negligible. Note that the differences in KD are due mainly to changes in the on rate, while no gross difference between the rates of dissociation of the adapters from the fusion proteins was detectable. (A) Analysis of adapter (AP-1 and AP-2) binding to full-length EnvCD fusion proteins. (B) A deleted version (lacking the N-terminal half of EnvCD) served as the backbone for affinity measurements of wild-type and LL855/856AA mutant Env.
Subcellular localization of Env correlates with its ability to bind AP-1.
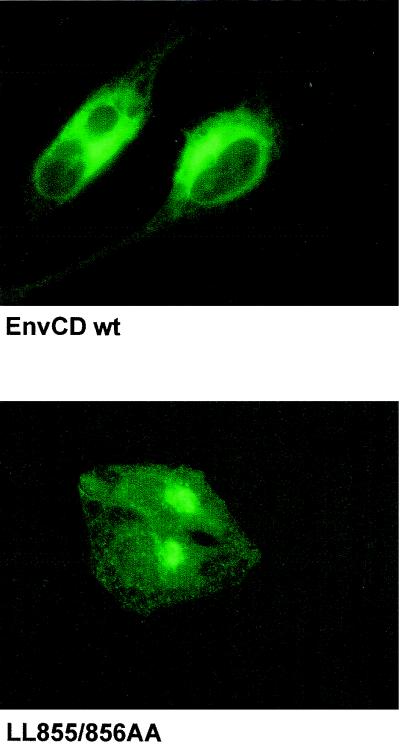
To assess the consequences for intracellular Env routing of mutations at the C terminus of EnvCD, we performed immunofluorescence analysis of cells expressing wild-type Env or Env mutants either alone or in the context of the whole virus. HIV-1 Env was visualized either with patient serum or with a monoclonal antibody against the SU part (Env gp120) as the primary antibody. Untransfected cells revealed virtually no background antibody reactivity (not shown). As documented in Fig. 4, an important fraction of wild-type Env at steady state localized to the perinuclear region. In addition, Env was found in more peripheral zones of the cytoplasm and at the plasma membrane. When analyzing Env with a mutated C terminus we noticed differences in the intracellular distribution of the protein. While both the wild-type and the mutant Env still resided prominently in the perinuclear area, the staining of the mutant protein in the periphery of the cytoplasm seemed reduced. Such a change of the intracellular distribution was observed in cells expressing the Env mutants alone, as is shown in Fig. 4, and also in cells expressing Env mutants in the context of the whole virus (not shown). Furthermore, the localization in the cytoplasm seemed restricted to the same degree as documented here for the LL855/856AA mutant if 3, 10, or 16 amino acids were deleted at the C terminus of EnvCD. The observed difference in the intracellular distribution of Env was not due to differences in the kinetics of Env synthesis or the overall stability because the synthesis rate and the steady-state level of the LL855/856AA mutant did not differ from the corresponding values measured for wild-type Env (not shown).
FIG. 4.
Localization of Env in HeLa cells. HeLa cells transiently expressing wild-type Env or the LL855/856AA mutant were fixed, permeabilized, and stained with an anti-Env antibody (b12) and with an FITC-conjugated antihuman secondary antibody.
The C-terminal dileucine is important for AP-1 recruitment in Env-expressing cells.
To analyze more precisely what changes in the intracellular distribution of Env are provoked by mutating the C-terminal dileucine motif, we performed colocalization studies by laser confocal microscopy using the previously described CD8–HIV-1 EnvCD chimeras (1). In these CD8–HIV-1 EnvCD chimeras the extracellular/luminal part and the membrane-spanning domain of the CD8 alpha-chain antigen have been joined to residues 707 to 856 of the HIV-1 EnvCD. Colocalization of these chimeras and full-length HIV-1 Env had been confirmed previously by confocal microscopy (not shown). As with full-length Env we did pulse-chase experiments and established that mutations at the C terminus did not affect the synthesis rate, the steady-state level of the protein, or the half-life of the chimera (not shown). Also, CD8–HIV-1 EnvCD chimeras are incorporated faithfully into virus-like particles (G. Blot, C. Berlioz-Torrent, and R. Benarous, unpublished results), thus further validating the use of these chimeras to study sorting signals in EnvCD.
The analysis of EnvCD–AP-1 binding together with the examination of the steady-state intracellular distribution of C-terminal Env mutants suggested a C terminus-dependent recruitment of AP-1 during the post-Golgi sorting of this membrane protein. HeLa cells were thus transfected with wild-type or with mutant CD8-EnvCD chimeras, and transfected cells were doubly stained with an antibody against CD8 and an antibody against AP-1. Like Env expressed as a full-length protein (Fig. 4, top), the CD8-EnvCD chimeras were found to be concentrated in a perinuclear region and in peripheral dots in the cytoplasm (Fig. 5). Some more-diffuse staining throughout the cytosol, which is clearly Env specific because it is absent from those neighboring cells which are not transfected, was also easily recognizable. CD8 signals were barely detectable at the cell surface, however.
FIG. 5.
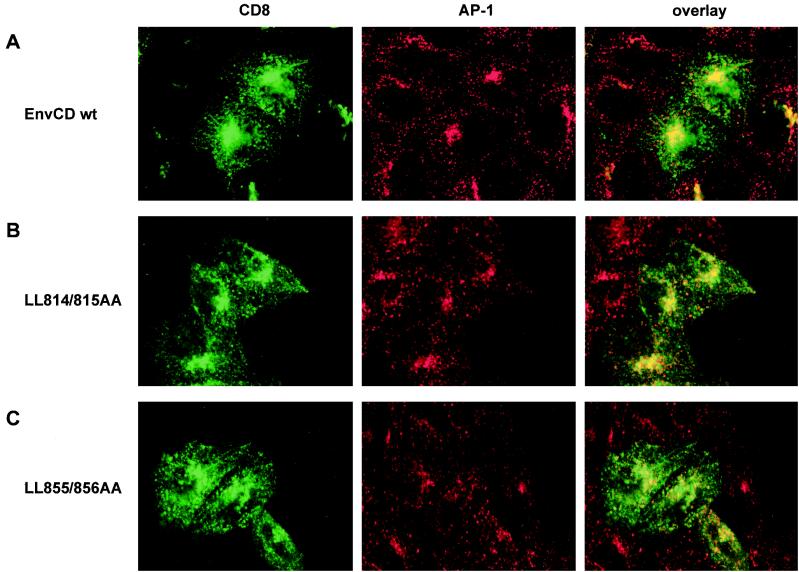
Colocalization of CD8-EnvCD chimeras and AP-1 in HeLa cells. Cells were transfected with pCD8-HIV (EnvCD wt) (top), pCD8-LL814/815AA (middle), or pCD8-LL855/856AA (bottom). Cells were fixed and permeabilized and doubly stained with an anti-CD8-FITC-conjugated antibody (left column) and an anti-γ-adaptin antibody and with a Texas red-conjugated antimouse secondary antibody (middle column). Colocalization of CD8-EnvCD and AP-1, or lack thereof, was determined by confocal microscopy. Superpositions (yellow) of the two stainings are shown for representative medial sections (right column).
The expression of CD8-EnvCD wt chimera provoked a concentration of AP-1 adapters in the perinuclear region of transfected cells as visualized by the staining with anti-γ-adaptin antibody (Fig. 5A, middle). Similarly, a considerable fraction of AP-1 was recruited to the perinuclear compartment in cells transfected with the LL814/815AA mutant (Fig. 5B, middle). In both cases colocalization of CD8-EnvCD and AP-1 was observed in the perinuclear region, where AP-1 was recruited by Env expression, while such colocalization was not detectable in the periphery (Fig. 5A and B, right). The γ-adaptin staining of nontransfected cells differed considerably from what was observed in cells expressing EnvCD wt or the LL814/815AA mutant. AP-1 vesicles were found dispersed more equally throughout the cells and much less AP-1 was recruited to membranes in the perinuclear region of the nontransfected cells (compare the γ-adaptin staining in nontransfected and Env-expressing cells in Fig. 5A, middle). Strikingly, a lack of AP-1 recruitment to perinuclear membranes was also obvious in cells expressing the LL855/856AA mutant, as visualized in Fig. 5C, middle. Furthermore, colocalization of AP-1 and CD8-LL855/856AA was considerably reduced compared to what was observed with EnvCD wt and the LL814/815AA chimeras (compare right panels of Fig. 5).
A quantitative analysis of the γ-adaptin staining in the perinuclear region is summarized in Table 1. Whereas the expression of both CD8-EnvCD wt and the LL814/815AA mutant led to an increased recruitment onto perinuclear membranes of AP-1, the level of such recruitment in cells expressing CD8-LL855/856AA was comparable to that observed in mock-transfected cells. Reduced association between LL855/856AA mutant Env and adapter AP-1 in our in vitro binding assays (Fig. 2 and 3) thus correlates with failure of mutant Env to couple AP-1 to perinuclear membranes in cells.
In summary, the results presented in Fig. 5 and Table 1 demonstrate that the mutation of the C-terminal dileucine altered HIV-1 Env's capacity to mobilize AP-1 to perinuclear membranes and led to an reduction in Env–AP-1 colocalization.
Mutation of the C-terminal dileucine in combination with a Y712A change leads to increased cell surface expression of Env.
To assess if the alterations of intracellular routing described above resulted in modified levels of Env cell surface expression, we analyzed the effects of the LL855/856AA mutation on the localization at the plasma membranes of CD8-EnvCD chimeras. The overall expression of the chimeras in a defined number of transfected cells was monitored by analyzing identical amounts of cell extracts. Levels of expression of the wild type and the LL855/856 mutant chimera were found to be similar in all our experiments (data not shown). Mutation of LL855/856 did not increase significantly the cell surface expression of Env as determined by flow cytometry analysis of the CD8-EnvCD chimera (Table 2). However, evidence for a role of the C-terminal dileucine in modulation of cell surface expression was gained in experiments where we analyzed the effects of combining mutations in this dileucine motif with the tyrosine-to-alanine substitution in the membrane-proximal tyrosine-based motif (GYSLP; Fig. 1), the well-characterized and potent internalization signal in HIV-1 EnvCD (1, 3, 10, 38, 46). As shown in Table 2, replacement by alanines of the dileucine 855/856 in the context of the mutated juxtamembrane endocytosis signal substantially enhanced the percentage of GFP+ CD8+ cells as well as the mean CD8 fluorescence intensity of these cells. This result reveals that the C-terminal dileucine together with Y712 contributes to restricting the surface expression of HIV-1 Env.
TABLE 2.
Cell surface expression of CD8-EnvCD hybrids in HeLa cellsa
| Vector pCD8 | % GFP+ CD8+ cellsb | Mean CD8 fluorescence intensityc | Pd |
|---|---|---|---|
| NTe | 0.4 ± 0.1 | 1.4 ± 0.1 | |
| EnvCD wt | 41.7 ± 4.1 | 4.5 ± 0.6 | |
| LL855/856AA | 49.4 ± 6.5 | 5.3 ± 0.7 | >0.05 |
| Y712A | 60.2 ± 2.1 | 6.1 ± 0.3 | <0.05 |
| Y712A-LL855/856AA | 68.4 ± 1.0 | 8.4 ± 0.3 | <0.05 |
Values are the averages of three independent experiments. After 48 h, surface expression of CD8 was analyzed in GFP+ cells by flow cytometry.
Percentage of GFP+ cells expressing CD8 hybrids at the cell surface. Values are means ± SEM.
Means of CD8 fluorescence intensity of GFP+ cells expressing the CD8 hybrid at the cell surface ± SEM.
The results for the mutant CD8-EnvCD hybrids were compared to those for the wild type by means of Student's t test for unpaired samples. Data obtained with the double mutant (tyrosine and dileucine) were compared to those obtained with the CD8-Y712A chimera.
NT, the pRSV-GFP vector alone was used for cell transfection.
LL855/856 do not function as an endocytosis signal.
To distinguish, finally, if the C-terminal dileucine assists in the restriction of Env cell surface expression because it functions as an endocytosis signal or if it does so by controlling the transport of Env to the plasma membrane, we measured the internalization kinetics for the different CD8-EnvCD chimeras. The results are summarized in Fig. 6. The internalization rate, as determined during the first 5 min of the experiment and thus before part of the internalization Env recycles back to the cell surface (which gives raise to the flattening of the internalization curves) for the LL855/856AA mutant was the same as that for the EnvCD wt chimera. While mutations of dileucine 855/856 slightly decreased the amount of internalized Env after 10 min of incubation of cells at 37°C, the contribution of this motif to the overall endocytosis of Env must clearly be minor: when comparing the internalization kinetics of the Y712A-LL855/856AA double mutant with the kinetics of the mutant which has the Y712-to-A change only, we found that dileucine 855/856 did not enhance the rate with which Env was retrieved from the cell surface, because the double mutant did not show an endocytosis rate lower than that of the Y712A mutant. In conclusion, dileucine 855/856 does not assist in the restriction of Env cell surface expression by enhancing the retrieval of Env from the plasma membrane but by controlling how much Env is transported to the cell surface.
DISCUSSION
Trafficking and cell surface expression of the lentiviral glycoprotein in infected cells are strictly regulated, and evidence that such regulation participates in controlling where at the cell surface virus is secreted is accumulating. In this study we identified the C terminus of the cytosolic domain of HIV-1 transmembrane TM gp41 to be important for proper subcellular localization of Env. Env mutants whose C-terminal sequences have been changed showed an altered intracellular distribution. These mutants failed to interact efficiently with clathrin-associated protein complex AP-1 in vitro and in live cells, suggesting an involvement of AP-1 in the correct routing of Env.
In our previous studies (1, 3), we noticed that the membrane-proximal tyrosine residue (Y712) in HIV-1 EnvCD, which mediates Env endocytosis and AP-2 binding, also influenced the binding of Env to the medium chain μ1 of the AP-1 adapter and to the intact (heterotetrameric) AP-1 complex, respectively, albeit to a much lesser extent than it affected Env–AP-2 binding. The finding that the Env–AP-1 interaction was only reduced significantly in the context of a truncated Env tail, not in the context of the complete cytosolic domain of Env, suggested that signals downstream of Y712 may be important for efficient Env–AP-1 interaction. Using two different methods we now find that a dileucine-containing sequence at the highly conserved C terminus of EnvCD is important for Env–AP-1 interaction (Fig. 2 and 3). Dileucine motifs were shown previously to participate to the interaction of AP-1 with various proteins, including CD4, CD3γ of the T-cell receptor αβ–CD3 receptor complex, FcRII-B2, and the lentiviral Nef (7, 13, 15). However, it has become apparent in many studies that the presence of a putative adapter binding motif does not allow a prediction of whether the motif is functional. Also, some leucine-based motifs involved in AP-1 binding require an acidic amino acid at position −4 and/or −5 relative to the first leucine of the doublet in order to be functional (29, 42), while the C-terminal dileucine in HIV-1 Env has a glutamic acid at position −3. Nevertheless, the present analysis clearly establishes that leucines L855 and L856 form a dileucine motif that is critical for the binding of AP-1.
Interactions between AP-1 and the cytosolic domains of membrane proteins often depend on the cooperation of multiple signals localized in their cytosolic tails (20, 49, 52, 53, 54). Amino acids in the N-terminal half of EnvCD may contribute to AP-1 binding, as evidenced by our finding that Env–AP-1 association varied for the different backbones used in our experiments (Fig. 2 and 3). Regions upstream of LL855/856 could contribute directly to AP-1 binding or they could act indirectly via influencing EnvCD conformation or the status of EnvCD oligomerization (27). In addition, residues in the direct vicinity of LL855/856 may also support Env–AP-1 binding because the deletion of the last six amino acids affected Env–AP-1 binding more severely than mutation of leucines LL855/856 (compare Fig. 2A and B).
The results of our binding assays suggest that Y712 in the context of a full-length HIV-1 EnvCD is not critical for EnvCD–AP-1 interaction. These findings differ from results presented in one of our previous studies (1) and in a recent study on SIV Env-adapter binding (6). In both studies it was found that mutations of the critical tyrosine in short peptides centered either on Y712 of HIV-1 Env or on Y721 of SIV Env reduced not only AP-2 binding but also the association with the AP-1 complex. Adapter binding to the motif per se, that is, when isolated from the rest of the cytosolic tail, thus clearly differs from adapter binding to the whole EnvCD. Our results now reveal a domain localized downstream of the proximal tyrosine-based motif in the long cytosolic tail of primate lentiviruses, which is important for Env–AP-1 association.
In this study we also show that EnvCD induced the recruitment of AP-1 to the perinuclear area (Fig. 5). Mutation of LL855/856 severely affected this function and also the colocalization of Env and AP-1 complexes (Fig. 5 and Table 1), suggesting that efficient Env–AP-1 binding may be critical for the recruitment of AP-1 to juxtanuclear membranes. The nature of the perinuclear compartment in which interactions between Env and AP-1 take place is currently not clear. AP-1 has been found in CCVs budding from the TGN and from the endosomal compartment (25; for a recent overview, see reference 36). This adapter participates, e.g., in the transport of cellular membrane proteins such as mannose-6-phosphate receptors from the TGN to the endosomal compartment, but AP-1 was also implicated in the transferrin receptor recruitment at the endosomal compartment of polarized epithelial cells (12). While it remains to be established where the interaction between Env and AP-1 takes place, the lack of AP-1 recruitment is likely to result in rerouting of Env and in an altered subcellular localization.
This study gives further support to the notion that the presence of viral Env at the cell surface is tightly regulated (33, 34). The results summarized in Table 2 demonstrate that the mutation of the C-terminal dileucine essentially doubled the enhancing effect on Env cell surface expression that was observed when the membrane-proximal, Y712-based endocytosis signal is destroyed. It is not manifest immediately that LL855/856 contribute to restraining the cell surface expression of Env because a mutation of the C-terminal dileucine alone, unlike a mutation of Y712 (46, 47), was not sufficient to alter the presence of Env at the cell surface significantly. The C-terminal dileucine-based signal thus needs to cooperate with Y712 in order to retain Env inside the cell.
Enhancement of Env cell surface expression could result from a default in endocytosis. Results shown in Fig. 6, however, demonstrate that the C-terminal dileucine does not act as an endocytosis signal, either alone or in conjuction with the tyrosine signal. The dileucine thus appears to affect some step in the forward transport of Env, e.g., it could have a role in recycling Env to the cell surface. Our results do not support that hypothesis. Indeed, efficient recycling requires an optimal rate of endocytosis. However, in our experiments the enhancing effect of the LL855/856AA mutation on CD8-EnvCD cell surface expression was higher in the context of a mutated Y712-based signal, the major endocytosis signal in Env, than it was in the context of wild-type HIV EnvCD (Table 2 and Fig. 6).
Consistent with data shown in Fig. 5 we currently favor the hypothesis that Env–AP-1 interactions are implicated in the correct sorting of Env from the TGN to the cell surface. We thus postulate that the dileucine mutation changes the routing of HIV-1 Env at the TGN and that such an altered transit to the plasma membrane, in conjuction with a defect in the Y712-based motif, results in an augmented cell surface expression of Env. In all, our results support the notion raised by us and others (1, 6) that additional signals at the C termini of the cytosolic domains of HIV-1 Env and SIV Env cooperate with the membrane-proximal tyrosine-based motif to maintain low levels of Env at the cell surface.
Control of the level of forward transport as demonstrated in this study may be coupled to directing Env to the appropriate domains at the cell surface upon its exit from the Golgi. Evidence for an involvement of AP complexes in the polarized delivery of membrane proteins is accumulating: two recent reports have raised the possibility that AP-1-containing vesicles budding from either the TGN or the endosomal compartment mediate basolateral sorting in MDCK cells (12, 40). Furthermore, the presence of a cell-specific medium chain in the adapter complex AP-1 may be important for the selection of some proteins into basolateral vesicles (11). The C terminus of EnvCD was shown to restrict HIV-1 release to the basolateral membrane in polarized epithelial MDCK cells (30). In view of all these reports, our finding that changes in the C terminus of EnvCD affect the recruitment of AP-1 as well as the subcellular localization of Env points to a potential involvement of AP-1 in localizing virus release. Such an overlap of signals controlling the level of Env cell surface expression and polarization of virus release would not be without precedent. The membrane-proximal tyrosine residue which is crucial for Env endocytosis has been demonstrated to act as polarity signal for HIV-1 and SIV particle release in T lymphocytes (10, 23, 30, 47).
In summary, we have shown that the efficient interaction of HIV-1 EnvCD with clathrin-associated adapter AP-1 depends on the intactness of a C-terminal dileucine. The same motif provides a signal necessary for proper subcellular localization of Env and, together with the membrane-proximal endocytosis signal, is also implicated in regulating how much Env is present at the surface of virus-producing cells. It is currently unknown if Env is sorted directly from the TGN to the plasma membrane or if it reaches the cell surface via the endosomal compartment. The answer to this question will shed further light on how exactly lentiviruses achieve control over the levels and the localization of Env at the cell surface.
ACKNOWLEDGMENTS
S.W. and C.B.-T. are co-first authors.
This work was supported by grants from the Swiss National Science Foundation (SNF), as well as from the ANRS and SIDACTION (France), and by fellowships from FRM (C.B.-T.) and the French Ministry of Research and Technology (G.B.). C.B.-T. is a SIDACTION fellow, and M.T. is the recipient of a career award from the SNF.
We acknowledge the technical assistance of Isabelle Bouchaert and Manuel Favre. Thanks to Dennis Burton, Paul Parren, and Walter Hunziker for providing anti-Env antibody b12 and for critical reading of the manuscript and discussions, respectively.
REFERENCES
- 1.Berlioz-Torrent C, Shacklett B L, Erdtmann L, Delamarre L, Bouchaert I, Sonigo P, Dokhelar M C, Benarous R. Interactions of the cytoplasmic domains of human and simian retroviral transmembrane proteins with components of the clathrin adapter complexes modulate intracellular and cell surface expression of envelope glycoproteins. J Virol. 1999;73:1350–1361. doi: 10.1128/jvi.73.2.1350-1361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blau D M, Compans R W. Polarization of viral entry and release in epithelial cells. Semin Virol. 1996;7:245–253. [Google Scholar]
- 3.Boge M, Wyss S, Bonifacino J S, Thali M. A membrane-proximal tyrosine-based signal mediates internalization of the HIV-1 envelope glycoprotein via interaction with the AP-2 clathrin adaptor. J Biol Chem. 1998;273:15773–15778. doi: 10.1074/jbc.273.25.15773. [DOI] [PubMed] [Google Scholar]
- 4.Bonifacino J S, Dell'Angelica E C. Molecular bases for the recognition of tyrosine-based sorting signals. J Cell Biol. 1999;145:923–926. doi: 10.1083/jcb.145.5.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boulan E R, Pendergast M. Polarized distribution of viral envelope proteins in the plasma membrane of infected epithelial cells. Cell. 1980;20:45–54. doi: 10.1016/0092-8674(80)90233-0. [DOI] [PubMed] [Google Scholar]
- 6.Bowers K, Pelchen-Matthews A, Höning S, Vance P J, Creary L, Haggarty B S, Romano J, Ballensiefen W, Hoxie J A, Marsh M. The simian immunodeficiency virus envelope glycoprotein contains multiple signals that regulate its cell surface expression and endocytosis. Traffic. 2000;1:661–674. doi: 10.1034/j.1600-0854.2000.010810.x. [DOI] [PubMed] [Google Scholar]
- 7.Bresnahan P A, Yonemoto W, Ferrell S, Williams-Herman D, Geleziunas R, Greene W C. A dileucine motif in HIV-1 Nef acts as an internalization signal for CD4 downregulation and binds the AP-1 clathrin adaptor. Curr Biol. 1998;8:1235–1238. doi: 10.1016/s0960-9822(07)00517-9. [DOI] [PubMed] [Google Scholar]
- 8.Deschambeault J, Lalonde J P, Cervantes-Acosta G, Lodge R, Cohen E A, Lemay G. Polarized human immunodeficiency virus budding in lymphocytes involves a tyrosine-based signal and favors cell-to-cell viral transmission. J Virol. 1999;73:5010–5017. doi: 10.1128/jvi.73.6.5010-5017.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dietrich J, Kastrup J, Nielsen B L, Odum N, Geisler C. Regulation and function of the CD3gamma DxxxLL motif: a binding site for adaptor protein-1 and adaptor protein-2 in vitro. J Cell Biol. 1997;138:271–278. doi: 10.1083/jcb.138.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egan M A, Carruth L M, Rowell J F, Yu X, Siliciano R F. Human immunodeficiency virus type 1 envelope protein endocytosis mediated by a highly conserved intrinsic internalization signal in the cytoplasmic domain of gp41 is suppressed in the presence of the Pr55gag precursor protein. J Virol. 1996;70:6547–6556. doi: 10.1128/jvi.70.10.6547-6556.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Folsch H, Ohno H, Bonifacino J S, Mellman I. A novel clathrin adaptor complex mediates basolateral targeting in polarized epithelial cells. Cell. 1999;99:189–198. doi: 10.1016/s0092-8674(00)81650-5. [DOI] [PubMed] [Google Scholar]
- 12.Futter C E, Gibson A, Allchin E H, Maxwell S, Ruddock L J, Odorizzi G, Domingo D, Trowbridge I S, Hopkins C R. In polarized MDCK cells basolateral vesicles arise from clathrin-gamma-adaptin-coated domains on endosomal tubules. J Cell Biol. 1998;141:611–623. doi: 10.1083/jcb.141.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greenberg M, DeTulleo L, Rapoport I, Skowronski J, Kirchhausen T. A dileucine motif in HIV-1 Nef is essential for sorting into clathrin-coated pits and for downregulation of CD4. Curr Biol. 1998;8:1239–1242. doi: 10.1016/s0960-9822(07)00518-0. [DOI] [PubMed] [Google Scholar]
- 14.Heilker R, Manningkrieg U, Zuber J F, Spiess M. In vitro binding of clathrin adaptors to sorting signals correlates with endocytosis and basolateral sorting. EMBO J. 1996;15:2893–2899. [PMC free article] [PubMed] [Google Scholar]
- 15.Heilker R, Spiess M, Crottet P. Recognition of sorting signals by clathrin adaptors. Bioessays. 1999;21:558–567. doi: 10.1002/(SICI)1521-1878(199907)21:7<558::AID-BIES4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 16.Hirst J, Robinson M S. Clathrin and adaptors. Biochim Biophys Acta. 1998;1404:173–193. doi: 10.1016/s0167-4889(98)00056-1. [DOI] [PubMed] [Google Scholar]
- 17.Honing S, Griffith J, Geuze H J, Hunziker W. The tyrosine-based lysosomal targeting signal in lamp-1 mediates sorting into golgi-derived clathrin-coated vesicles. EMBO J. 1996;15:5230–5239. [PMC free article] [PubMed] [Google Scholar]
- 18.Honing S, Sandoval I V, von Figura K. A dileucine-based motif in the cytoplasmic tail of LIMP-II and tyrosinase mediates selective binding of AP-3. EMBO J. 1998;17:1304–1314. doi: 10.1093/emboj/17.5.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunter E. Viral entry and receptors. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 71–120. [PubMed] [Google Scholar]
- 20.Jones B G, Thomas L, Molloy S S, Thulin C D, Fry M D, Walsh K A, Thomas G. Intracellular trafficking of furin is modulated by the phosphorylation state of a casein kinase II site in its cytoplasmic tail. EMBO J. 1995;14:5869–5883. doi: 10.1002/j.1460-2075.1995.tb00275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jonsson U, Fagerstam L, Ivarsson B, Johnsson B, Karlsson R, Lundh K, Lofas S, Persson B, Roos H, Ronnberg I. Real-time biospecific interaction analysis using surface plasmon resonance and a sensor chip technology. BioTechniques. 1991;11:620–627. [PubMed] [Google Scholar]
- 22.Kirchhausen T, Bonifacino J S, Riezman H. Linking cargo to vesicle formation: receptor tail interactions with coat proteins. Curr Opin Cell Biol. 1997;9:488–495. doi: 10.1016/s0955-0674(97)80024-5. [DOI] [PubMed] [Google Scholar]
- 23.Korber B T, Foley B T, Kuiken C L, Satish K, Pillai K, Sodroski J G. Numbering positions in HIV relative to HXB2CG. In: Korber B, Kuiken C L, Foley B, Hahn B, McCutchan F, Mellors J W, Sodroski J, editors. Human retroviruses and AIDS 1998: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Los Alamos National Laboratory; 1998. [Google Scholar]
- 24.LaBranche C C, Sauter M M, Haggarty B S, Vance P J, Romano J, Hart T K, Bugelski P J, Marsh M, Hoxie J A. A single amino acid change in the cytoplasmic domain of the simian immunodeficiency virus transmembrane molecule increases envelope glycoprotein expression on infected cells. J Virol. 1995;69:5217–5227. doi: 10.1128/jvi.69.9.5217-5227.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le Borgne R, Hoflack B. Mannose 6-phosphate receptors regulate the formation of clathrin-coated vesicles in the TGN. J Cell Biol. 1996;37:335–345. doi: 10.1083/jcb.137.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le Borgne R, Hoflack B. Mechanisms of protein sorting and coat assembly: insights from the clathrin-coated vesicle pathway. Curr Opin Cell Biol. 1998;10:499–503. doi: 10.1016/s0955-0674(98)80065-3. [DOI] [PubMed] [Google Scholar]
- 27.Lee S, Wang C, Liang J Y P, Hong S, Huang C, Chen S S L. Multimerization potential of the cytoplasmic domain of the human immunodeficiency virus type 1 transmembrane glycoprotein gp41. J Biol Chem. 2000;275:15809–15819. doi: 10.1074/jbc.M000601200. [DOI] [PubMed] [Google Scholar]
- 28.Leitner T, Korber B, Robertson D, Gao F, Hahn B. Updated proposal of reference sequences of HIV-1 genetic subtypes. In: Korber B, Hahn B, Foley B, Mellors J W, Leitner T, Myers G, McCutchan F, Kuiken C L, editors. Human retroviruses and AIDS 1997: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Los Alamos National Laboratory; 1997. [Google Scholar]
- 29.Letourneur F, Klausner R D. A novel di-leucine motif and a tyrosine-based motif independently mediate lysosomal targeting and endocytosis of CD3 chains. Cell. 1992;69:1143–1157. doi: 10.1016/0092-8674(92)90636-q. [DOI] [PubMed] [Google Scholar]
- 30.Lodge R, Gottlinger H, Gabuzda D, Cohen E A, Lemay G. The intracytoplasmic domain of gp41 mediates polarized budding of human immunodeficiency virus type 1 in MDCK cells. J Virol. 1994;68:4857–4861. doi: 10.1128/jvi.68.8.4857-4861.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lodge R, Lalonde J P, Lemay G, Cohen E A. The membrane-proximal intracytoplasmic tyrosine residue of HIV-1 envelope glycoprotein is critical for basolateral targeting of viral budding in MDCK cells. EMBO J. 1997;16:695–705. doi: 10.1093/emboj/16.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marks M S, Ohno H, Kirchhausen T, Bonifacino J S. Protein sorting by tyrosine-based signals: adapting to the Ys and wherefores. Trends Cell Biol. 1997;7:124–128. doi: 10.1016/S0962-8924(96)10057-X. [DOI] [PubMed] [Google Scholar]
- 33.Marsh M, Pelchen-Matthews A, Hoxie J A. Roles for endocytosis in lentiviral replication. Trends Cell Biol. 1997;7:1–4. doi: 10.1016/S0962-8924(97)20038-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marsh M, Pelchen-Matthews A. Endocytosis in viral replication. Traffic. 2000;1:525–532. doi: 10.1034/j.1600-0854.2000.010701.x. [DOI] [PubMed] [Google Scholar]
- 35.Mellman I. Endocytosis and molecular sorting. Annu Rev Cell Dev Biol. 1996;12:575–625. doi: 10.1146/annurev.cellbio.12.1.575. [DOI] [PubMed] [Google Scholar]
- 36.Meyer C, Zizioli D, Lausmann S, Eskelinen E L, Hamann J, Saftig P, von Figura K, Schu P. Mu1A-adaptin-deficient mice: lethality, loss of AP-1 binding and rerouting of mannose 6-phosphate receptors. EMBO J. 2000;19:2193–2203. doi: 10.1093/emboj/19.10.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mostov K, ter Beest M B, Chapin S J. Catch the mu1B train to the basolateral surface. Cell. 1999;99:121–122. doi: 10.1016/s0092-8674(00)81643-8. [DOI] [PubMed] [Google Scholar]
- 38.Ohno H, Aguilar R C, Fournier M C, Hennecke S, Cosson P, Bonifacino J S. Interaction of endocytic signals from the HIV-1 envelope glycoprotein complex with members of the adaptor medium chain family. Virology. 1997;238:305–315. doi: 10.1006/viro.1997.8839. [DOI] [PubMed] [Google Scholar]
- 39.Ohno H, Tomemori T, Nakatsu F, Okazaki Y, Aguilar R C, Foelsch H, Mellman I, Saito T, Shirasawa T, Bonifacino J S. Mu1B, a novel adaptor medium chain expressed in polarized epithelial cells. FEBS Lett. 1999;449:215–220. doi: 10.1016/s0014-5793(99)00432-9. [DOI] [PubMed] [Google Scholar]
- 40.Orzech E, Schlessinger K, Weiss A, Okamoto C T, Aroeti B. Interactions of the AP-1 Golgi adaptor with the polymeric immunoglobulin receptor and their possible role in mediating brefeldin A-sensitive basolateral targeting from the trans-Golgi network. J Biol Chem. 1999;274:2201–2215. doi: 10.1074/jbc.274.4.2201. [DOI] [PubMed] [Google Scholar]
- 41.Owens R J, Dubay J W, Hunter E, Compans R W. Human immunodeficiency virus envelope protein determines the site of virus release in polarized epithelial cells. Proc Natl Acad Sci USA. 1991;88:3987–3991. doi: 10.1073/pnas.88.9.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pond L, Kuhn L A, Teyton L, Schutze M P, Tainer J A, Jackson M R, Peterson P A. A role for acidic residues in di-leucine motif-based targeting to the endocytic pathway. J Biol Chem. 1995;270:19989–19997. doi: 10.1074/jbc.270.34.19989. [DOI] [PubMed] [Google Scholar]
- 43.Rapoport I, Chen Y C, Cupers P, Shoelson S F, Kirchhausen T. Dileucine-based sorting signals bind to the beta chain of AP-1 at a site distinct and regulated differently from the tyrosine-based motif-binding site. EMBO J. 1998;17:2148–2155. doi: 10.1093/emboj/17.8.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ritter G D, Jr, Yamshchikoy G, Cohen S J, Mulligan M J. Human immunodeficiency virus type 2 glycoprotein enhancement of particle budding: role of the cytoplasmic domain. J Virol. 1996;70:2669–2673. doi: 10.1128/jvi.70.4.2669-2673.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robinson M S. Coats and vesicle budding. Trends Cell Biol. 1997;7:99–102. doi: 10.1016/S0962-8924(96)10048-9. [DOI] [PubMed] [Google Scholar]
- 46.Rowell J F, Stanhope P E, Siliciano R F. Endocytosis of endogenously synthesized HIV-1 envelope protein. Mechanism and role in processing for association with class II MHC. J Immunol. 1995;155:473–488. [PubMed] [Google Scholar]
- 47.Sauter M M, Pelchen-Matthews A, Bron R, Marsh M, LaBranche C C, Vance P J, Romano J, Haggarty B S, Hart T K, Lee W M, Hoxie J A. An internalization signal in the simian immunodeficiency virus transmembrane protein cytoplasmic domain modulates expression of envelope glycoproteins on the cell surface. J Cell Biol. 1996;132:795–781. doi: 10.1083/jcb.132.5.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sawai E T, Baur A, Struble H, Peterlin B M, Levy J A, Cheng-Mayer C. Human immunodeficiency virus type 1 Nef associates with a cellular serine kinase in T lymphocytes. Proc Natl Acad Sci USA. 1994;91:1539–1543. doi: 10.1073/pnas.91.4.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schafer W, Stroh A, Berghofer S, Seiler J, Vey M, Kruse M L, Kern H F, Klenk H D, Garten W. Two independent targeting signals in the cytoplasmic domain determine trans-Golgi network localization and endosomal trafficking of the proprotein convertase furin. EMBO J. 1995;14:2424–2435. doi: 10.1002/j.1460-2075.1995.tb07240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schubert U, Bour S, Willey R L, Strebel K. Regulation of virus release by the macrophage-tropic human immunodeficiency virus type 1 AD8 isolate is redundant and can be controlled by either Vpu or Env. J Virol. 1999;73:887–896. doi: 10.1128/jvi.73.2.887-896.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swanstrom R, Wills J W. Synthesis, assembly, and processing of viral proteins. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 263–334. [PubMed] [Google Scholar]
- 52.Takahashi S, Nakagawa T, Banno T, Watanabe T, Murakami K, Nakayama K. Localization of furin to the trans-Golgi network and recycling from the cell surface involves Ser and Tyr residues within the cytoplasmic domain. J Biol Chem. 1995;270:28397–28401. doi: 10.1074/jbc.270.47.28397. [DOI] [PubMed] [Google Scholar]
- 53.Teuchert M, Schafer W, Berghofer S, Hoflack B, Klenk H D, Garten W. Sorting of furin at the trans-Golgi network. Interaction of the cytoplasmic tail sorting signals with AP-1 Golgi-specific assembly proteins. J Biol Chem. 1999;274:8199–8207. doi: 10.1074/jbc.274.12.8199. [DOI] [PubMed] [Google Scholar]
- 54.Voorhees P, Deignan E, van Donselaar E, Humphrey J, Marks M S, Peters P J, Bonifacino J S. An acidic sequence within the cytoplasmic domain of furin functions as a determinant of trans-Golgi network localization and internalization from the cell surface. EMBO J. 1995;14:4961–4975. doi: 10.1002/j.1460-2075.1995.tb00179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weclewicz K, Ekstrom M, Kristensson K, Garoff H. Specific interactions between retrovirus Env and Gag proteins in rat neurons. J Virol. 1998;72:2832–2845. doi: 10.1128/jvi.72.4.2832-2845.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]