Abstract
Epstein-Barr virus (EBV) is a strict human pathogen for which no small animal models exist. Plasmids that contain the EBV plasmid origin of replication, oriP, and express EBV nuclear antigen 1 (EBNA1) are stably maintained extrachromosomally in human cells, whereas these plasmids replicate poorly in rodent cells. However, the ability of oriP and EBNA1 to maintain the entire EBV episome in proliferating rodent cells has not been determined. Expression of the two human B-cell receptors for EBV on the surfaces of murine B cells allows efficient viral entry that leads to the establishment of latent EBV infection and long-term persistence of the viral genome. Latent gene expression in these cells resembles the latency II profile in that EBNA1 and LMP1 can be detected whereas EBNA2 and the EBNA3s are not expressed.
Epstein-Barr virus (EBV), also designated human herpesvirus 4, establishes latent infection in human hosts and is associated with a wide variety of human malignancies (29, 41). Although humans are the exclusive natural host for EBV, several other Old World primates are infected with closely related herpesviruses of the same subgroup, lymphocryptovirus (3, 14, 23, 37). Lymphocryptovirus genomes are colinear and homologous with the EBV genome (15), and the structural and nonstructural proteins are frequently well conserved (11, 25, 39). Despite these similarities, differences in disease progression and the complications arising from working on primates hinder the use of these viruses as a model for EBV infection (32). The ability to genetically manipulate and study mice in the laboratory makes a murine model for EBV infection preferable. Murine gammaherpesvirus 68 (MHV-68) establishes latent infection in lymphoid tissues, but the disease pattern caused by MHV-68 differs from that of EBV (43, 45, 46). In addition, MHV-68 does not encode the complement of EBV latency-associated and/or transforming proteins, including the nuclear antigens (EBNAs) and latent membrane proteins LMP1 and LMP2A/B, indicating fundamental differences between it and EBV (2, 48). To determine if mice may able to serve as a suitable model for EBV infection it is necessary to determine the viral and/or host restrictions that uniquely direct EBV susceptibility to humans.
Murine cells lines are typically resistant to infection by EBV, but reports have indicated that introducing the expression of human CD21 (hCD21), the cellular receptor involved in binding the EBV envelope glycoprotein gp350 (36, 47), promotes EBV infection of murine L cells (1, 6). However, this may be a cell type-dependent phenomenon, since the requirements for viral entry into epithelial and fibroblast cell lines appear to be quite different than those for entry into B cells (16, 27, 33, 54). In addition to the binding of gp350 with CD21, EBV entry into human B cells requires additional interaction with a second cellular receptor, HLA class II, with the ternary gH-gL-gp42 complex (26, 27, 50). Moreover, studies indicate that the expression of CD21 on human lymphocytes is not sufficient for EBV entry and that lymphoid cell lines expressing CD21 in the absence of HLA class II are either resistant to entry or undergo an abortive infection (12, 26). Murine B cells express homologues of both of EBV receptors, murine CD21 (mCD21) and two isotypes of major histocompatibility complex (MHC) class II, I-A and I-E. mCD21 is not able to mediate EBV infection in the absence of MHC class II (34), but its ability to mediate entry in the presence of MHC class II has not been determined.
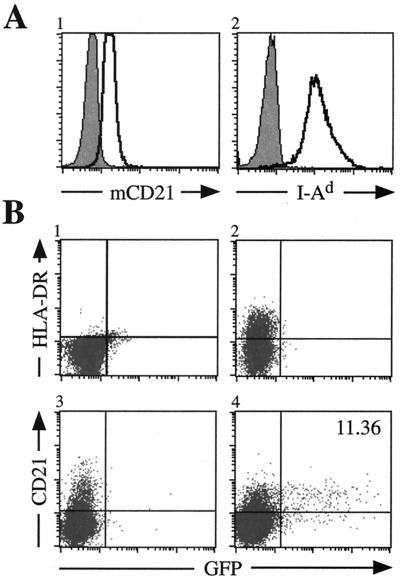
Analysis of mCD21 and I-Ad expression on the murine B-cell line M12 (19) by flow cytometry indicates moderate expression of mCD21 and abundant I-Ad expression (Fig. 1A). Similar results using the same antibodies were observed for a different murine B-cell line, A20 (American Type Culture Collection). However, both of these cells are resistant to infection by a recombinant EBV encoding the enhanced green fluorescence protein (EBfaV-GFP) (Fig. 1B, panel 1, and data not shown) (44). To determine whether either murine molecule could mediate entry if complemented by the human homologue, hCD21 and HLA-DR were singly transfected into M12 cells. Neither expression of HLA-DR nor that of hCD21 on the surface resulted in EBV entry (Fig. 1B, panels 2 and 3). This indicates that neither mCD21 nor MHC class II I-Ad can perform the entry-mediating function of its human homologue. Coexpression of hCD21 and HLA-DR resulted in efficient entry of EBV into M12 cells, as 11.36% of M12 cells expressing hCD21 and HLA-DR are infected by EBfaV-GFP (Fig. 1B, panel 4). This rate of infection resembles that observed when CD21-positive human lymphocyte cell lines are transiently transfected with HLA class II and infected with EBfaV-GFP (12, 13). Similar results were also observed for A20 cells transfected with hCD21 and HLA-DR (data not shown), indicating that EBV entry into murine B cells requires the expression of both hCD21 and HLA class II.
FIG. 1.
EBV infection of M12 cells is dependent upon the dual expression of hCD21 and HLA class II. (A) Surface expression of mCD21 and I-Ad (clear histograms) on M12 cells analyzed by flow cytometry. mCD21 expression was determined using an anti-mCD21 antibody directly conjugated to fluorescein isothiocyanate (Pharmingen). I-Ad was observed by staining cells with a biotin-conjugated anti-I-Ad antibody (Pharmingen) detected by streptavidin-conjugated allophycocyanin (APC) (Pharmingen). Shaded histograms represent cells stained with an isotype-matched control antibody. (B) Susceptibility of M12 cells to EBfaV-GFP infection. pSG5 (Stratagene)-transfected cells do not express HLA-DR (panel 1) or hCD21 (data not shown) and are resistant to EBV infection. M12 cells transiently expressing HLA-DR (panel 2) or hCD21 (panel 3) are inefficiently infected with EBV. However, M12 cells expressing both hCD21 and HLA-DR (panel 4) are efficiently infected by EBfaV-GFP. A20 and M12 cells were infected by EBfaV-GFP as previously described 24 h after transfection (12, 13). Analysis of infected cells occurred 24 h after infection. HLA-DR expression was observed using an anti-HLA-DR antibody directly conjugated to APC (Becton Dickinson); hCD21 expression was recognized by a biotin-conjugated anti-hCD21 antibody (Ancell) detected using streptavidin-conjugated APC.
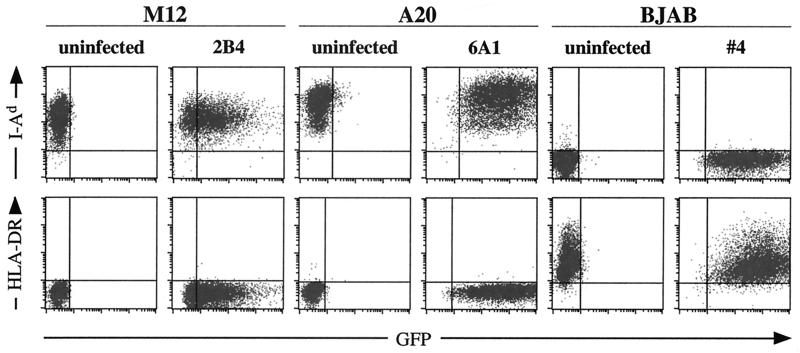
To determine if the EBV genome can persist as an episome in murine B cells, M12 and A20 cells transiently transfected with HLA-DR and hCD21 were infected with EBfaV-GFP and plated in 96-well dishes under G418 selection, and individual clones were selected and expanded. Cells of the BJAB line, a human EBV-negative Burkitt's lymphoma cell line (20), were infected in parallel and served as a control. Approximately 1% of GFP-positive cells from each cell line resulted in colony formation (data not shown). Figure 2 shows representative data from the parental and one infected clonal cell line for A20, M12, and BJAB stained for mouse- and human-specific cell markers. Both the parental and EBV-infected M12 and A20 cells stained positive for the murine specific I-Ad, whereas the human BJAB cells did not (Fig. 2). Conversely, the parental and infected BJAB cells stained positive for HLA-DR in contrast to both murine cell lines, which were negative (Fig. 2). In addition, all of the EBV-infected cell lines were positive for GFP expression, whereas each parental cell line was not. All cell lines were also monitored for CD18 expression using mouse- and human-specific antibodies that resulted in the expected staining pattern, reflecting the species origin of each cell line (data not shown). These results indicate that EBV can infect murine cells when the appropriate human receptors are expressed and that the EBV genome can persist when grown under selection.
FIG. 2.
Verification of cell populations using species-specific cell surface markers. M12, A20, and BJAB cells were stained identically for expression of I-Ad and HLA-DR. As expected, murine cell lines, regardless of EBV infection, are recognized by the anti-I-Ad antibody but not by the anti-HLA-DR antibody. The converse is true for BJAB cells.
Two months after passage of infected cell lines under selection, all clonal lines maintained green fluorescence, exhibited blastoid morphology, and grew in clumps, indicative of latent EBV infection (data not shown). In addition, in the absence of drug selection during the same time period, many GFP-negative cells were readily observed, indicating that the genome may be lost from these infected clones (data not shown). The stable maintenance of the EBV genome in cell lines under drug selection was surprising, since small plasmids containing oriP and EBNA1 are not stably replicated as episomes in rodent cells (21, 53). Thus, to verify EBV extrachromosomal genome maintenance, the presence of EBV episomes within these cell lines was analyzed. Of 28 EBV-infected A20 cell lines, 22 demonstrated episomal maintenance by Gardella gel analysis (data not shown). Similarly, 13 of 17 M12 lines infected with EBV also had readily detected EBV episomes (data not shown). These data suggest that in EBV-infected murine B-cell lines, approximately 75 to 80% (78.6% for A20 and 76.5% for M12) of cells support appropriate episomal maintenance. To determine how the murine frequency compares to the maintenance of EBV in a human B-cell line, 11 infected BJAB lines were also analyzed. EBV episomes were detected in 9 of the 11, or approximately 81.8% (data not shown), which corresponds with previous results (31). These observations suggest that murine B-cell lines have an ability to maintain EBV episomes similar to that of human B cells.
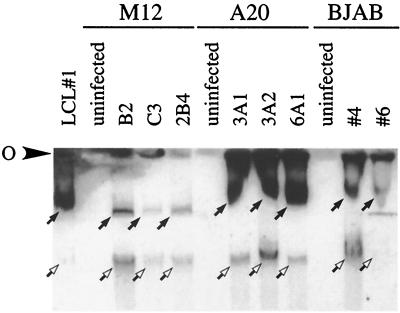
Selected cell lines that demonstrated the ability to maintain EBV episome 2 months after infection were cultured for an additional 4 months to determine the longevity of episomal maintenance. As seen with Gardella gel analysis with cell lines cultured for 2 months, M12 cells appeared to have fewer copies of the EBV genome than A20 and BJAB EBV-infected cell lines (Fig. 3). BJAB cells seem to harbor EBV at intermediate levels, whereas A20 cells sustain the genome at relatively high copy numbers (Fig. 3). In addition, linear forms of the EBV genome were detected in both of the human and murine cell lines (Fig. 3). It is also interesting that GFP fluorescence loosely correlates with copy number in that M12 cells have relatively low GFP expression and appear to harbor a low number of episomes, as indicated by Gardella analysis (Fig. 2 and 3). Comparatively, A20 and BJAB have a much higher copy number and fluoresce more strongly with GFP (Fig. 2 and 3).
FIG. 3.
Long-term persistence of the EBV genome in murine B-cell lines. Equal numbers of viable cells from each clonal cell line were harvested and analyzed by agarose gel electrophoresis. Horizontal Gardella gel analysis was carried out as previously described (10), with the addition of 15 μg of chloroquine/ml to the stacking gel. Southern blotting was carried out as previously described (5), and blots were probed using 32P-radiolabeled BamA fragments. O, origin of electrophoresis. Detection of the episomal and linear versions of the EBV genomes is indicated by closed and open arrows, respectively.
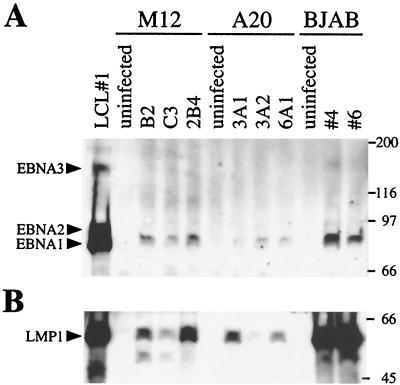
To further characterize EBV infection of murine B cells, latent EBV gene expression was also determined using human sera reactive with latently expressed EBV proteins. Western analysis of equal numbers of M12, A20, and BJAB cells shows that though EBNA1 is produced in these cells, EBNA2 and the EBNA3s are not (Fig. 4A). A Western blot for LMP1 indicates that LMP1 is also expressed (Fig. 4B). LMP2 detection was not investigated because EBfaV-GFP contains a deletion of LMP2A/B. This latent gene expression resembles latency II, which is characterized by nasopharyngeal carcinoma and EBV-positive Hodgkin's and T cell lymphomas (7). Despite lower levels of protein, the M12 and A20 expression profile is comparable to that of BJAB cells infected with EBfaV-GFP (Fig. 4). The pattern of latent gene expression in EBV positive Burkitt's lymphoma cell lines grown in culture can be quite variable (30, 31, 35, 42, 49). The EBV-positive M12 and A20 cell lines, with the more restricted type II latency, resemble BJAB cells infected in the present study as well as EBV-positive BJAB cells described in previous studies (31, 49). This indicates that EBV can establish a latent infection in murine B-cell lines resembling latent infection of at least some human B-cell lines infected in vitro.
FIG. 4.
Detection of latent protein expression in latently EBV-infected cell lines. Equal numbers of viable cells from each clonal cell line was harvested and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. (A) Western blot analysis using human antiserum that recognizes EBNA1, EBNA2, and the EBNA3s. LCL#1 cells express all three EBNAs, whereas the murine cells express EBNA1 alone. (B) Western blot analysis using polyclonal rabbit sera to detect LMP1. Molecular weights (in thousands) are on the right.
Clonal EBV-infected M12 and A20 cell lines demonstrate the ability to maintain the EBV episome over long periods of time (Fig. 3). For EBV to persist in latently infected cells, it must properly replicate and partition its genome during mitosis. To accomplish this, EBV uses a single viral element in cis, oriP, and a single viral protein in trans, EBNA1 (52). oriP consists of two clusters that contain binding sites for EBNA1; the family of repeats (FR) and the region of dyad symmetry (DS), both of which are required for stable replication of oriP plasmids in human cells (40). Biochemical studies indicate that DNA synthesis initiates from the DS for oriP plasmids in human cells that express EBNA1 (9). Several reports have indicated that while oriP plasmids can replicate and be properly partitioned in primate, bovine, feline, and canine cells, they fail to properly replicate in rodent cells (21, 53). This failure has been attributed to the inability of the DS to function properly as an origin of DNA synthesis in rodent cells (21).
While the DS does not function as an origin of DNA synthesis in rodent cells, reports indicate a possible mechanism for EBV genome replication during latency in rodent cells. Experiments have demonstrated that the latent EBV genomes present in Raji cells do not use the DS as an origin of replication; instead, a replication origin elsewhere in the genome is predominantly used (28). Consistent with this observation, other studies have shown that EBV genomes with the DS deleted are not impaired in their ability to infect, establish latency in, or replicate episomally in primary human B cells and human B-cell lines (24, 38). These DS deletion-containing genomes must possess an alternative origin of DNA synthesis elsewhere in the viral genome that operates during latent infection, possibly the same origin detected in Raji cells. These studies indicate that a similar alternate origin of replication also functions in M12 and A20 cells latently infected with EBV.
Unlike the elements in the DS, there is evidence that in rodent cells EBNA1 properly facilitates plasmid maintenance and partitioning through the elements contained within the FR. First, EBNA1 can bind the FR in rodent cells and activate transcription (51), a property that has been attributed to the prolonged maintenance of transcriptional reporter plasmids within transfected cells. Second, hybrid plasmids that contain the FR and human chromosomal DNA sequences do replicate stably in rodent cells that express EBNA1 (21). It is proposed that for these plasmids EBNA1 and the FR provide maintenance and partitioning functions while the human chromosomal DNA sequences provide a replication origin that operates in rodent cells. Finally, when rodent fibroblasts are fused with human cells containing either the entire EBV genome carrying a selectable marker or oriP plasmids containing large human genomic inserts, the EBV genome and oriP plasmids are maintained efficiently (18). Thus, it is likely that the cis and trans plasmid maintenance and partitioning functions of EBV do function in rodent cells, consistent with the observations reported here.
Previous studies have indicated that expression of hCD21 on murine L cells results in susceptibility of these cells to EBV infection (1, 6). However, the expression of CD21 on human lymphocytes is in itself not sufficient to promote efficient EBV entry and productive infection (12, 26). The necessity of other factors for EBV entry into mouse cells is further borne out by the observation that transgenic mice expressing hCD21 are not susceptible to EBV infection (17). Although lymphocytes from these mice are able to bind EBV particles, no entry or EBV gene products were detected (17). To ascertain the requirements for EBV infection of murine cells, hCD21 and HLA-DR were transfected into murine B-cell lines. Expression of either molecule alone does not allow EBV entry, yet coexpression of both resulted in efficient infection (Fig. 1). These data suggest that one of the blocks to EBV infection of murine cells resides in the expression of cellular receptors. To some extent the finding that I-Ad cannot serve as a coreceptor for EBV entry is surprising, considering the high degree of similarity between HLA and MHC class II molecules (4, 8). In particular, although the glutamic acid at position 46 of the beta chain in HLA class II, which is essential for EBV entry (13), is found in the corresponding region of I-Ad, I-Ad does not mediate entry, as shown in this study.
The ability of EBV to enter murine B-cell lines transfected with hCD21 and HLA-DR and to sustain long-term episomal replication indicates that transgenic mice expressing these receptors might provide a suitable small animal model of EBV infection. Nonetheless, in order for such a system to mimic human infection, other aspects of the EBV life cycle must also take place. In particular, the ability of the EBV gene products to promote cellular proliferation and transformation must also be assessed. Transgenic mice expressing either LMP1 or LMP2A display alterations of normal B-cell function that might be expected given the known functions of these viral proteins in EBV infection (5, 22). This suggests that EBV infection resulting in the production of these latent gene products might indeed promote the proliferation and transformation associated with EBV infection in humans. In light of this and the data presented here, transgenic mice bearing hCD21 and HLA-DR may offer a novel system to study aspects of EBV infection and disease that have previously been difficult to investigate due to the restriction of EBV infection to humans.
Acknowledgments
We thank the people in the laboratories of R. Longnecker, A. Aiyar, and P. Spear for providing advice and help. We also thank G. Kansas for the gift of antibodies to the human and murine CD18.
K.M.H. is supported by the training program on the Cellular and Molecular Basis of Disease (T32 GM08061) of the National Institutes of Health. A.A. was a Special Fellow of the Leukemia and Lymphoma Society of America when these studies were initiated. He is currently supported by the Leukemia Research Foundation and grant CA82177 from the National Cancer Institute. R.L. is a Scholar of the Leukemia and Lymphoma Society of America and is supported by Public Health Service grants CA62234 and CA73507 from the National Cancer Institute and grant DE13127 from the National Institute of Dental and Craniofacial Research.
REFERENCES
- 1.Ahearn J M, Hayward S D, Hickey J C, Fearon D T. Epstein-Barr virus (EBV) infection of murine L cells expressing recombinant human EBV/C3d receptor. Proc Natl Acad Sci USA. 1988;85:9307–9311. doi: 10.1073/pnas.85.23.9307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baer R, Bankier A T, Biggin M D, Deininger P L, Farrell P J, Gibson T J, Hatfull G, Hudson G S, Satchwell S C, Seguin C, et al. DNA sequence and expression of the B95–8 Epstein-Barr virus genome. Nature. 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 3.Bocker J F, Tiedemann K H, Bornkamm G W, zur Hausen H. Characterization of an EBV-like virus from African green monkey lymphoblasts. Virology. 1980;101:291–295. doi: 10.1016/0042-6822(80)90506-1. [DOI] [PubMed] [Google Scholar]
- 4.Brown J H, Jardetzky T S, Gorga J C, Stern L J, Urban R G, Strominger J L, Wiley D C. Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature. 1993;364:33–39. doi: 10.1038/364033a0. [DOI] [PubMed] [Google Scholar]
- 5.Caldwell R G, Wilson J B, Anderson S J, Longnecker R. Epstein-Barr virus LMP2A drives B cell development and survival in the absence of normal B cell receptor signals. Immunity. 1998;9:405–411. doi: 10.1016/s1074-7613(00)80623-8. [DOI] [PubMed] [Google Scholar]
- 6.Carel J C, Frazier B, Ley T J, Holers V M. Analysis of epitope expression and the functional repertoire of recombinant complement receptor 2 (CR2/CD21) in mouse and human cells. J Immunol. 1989;143:923–930. [PubMed] [Google Scholar]
- 7.Cohen J I. Epstein-Barr virus infection. N Engl J Med. 2000;343:481–492. doi: 10.1056/NEJM200008173430707. [DOI] [PubMed] [Google Scholar]
- 8.Fremont D H, Hendrickson W A, Marrack P, Kappler J. Structures of an MHC class II molecule with covalently bound single peptides. Science. 1996;272:1001–1004. doi: 10.1126/science.272.5264.1001. [DOI] [PubMed] [Google Scholar]
- 9.Gahn T A, Schildkraut C L. The Epstein-Barr virus origin of plasmid replication, oriP, contains both the initiation and termination sites of DNA replication. Cell. 1989;58:527–535. doi: 10.1016/0092-8674(89)90433-9. [DOI] [PubMed] [Google Scholar]
- 10.Gardella T, Medveczky P, Sairenji T, Mulder C. Detection of circular and linear herpesvirus DNA molecules in mammalian cells by gel electrophoresis. J Virol. 1984;50:248–254. doi: 10.1128/jvi.50.1.248-254.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerber P, Pritchett R F, Kieff E D. Antigens and DNA of a chimpanzee agent related to Epstein-Barr virus. J Virol. 1976;19:1090–1099. doi: 10.1128/jvi.19.3.1090-1099.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haan K, Kwok W, Longnecker R, Speck P. Epstein-Barr Virus entry utilizing HLA-DP or DQ as a coreceptor. J Virol. 2000;74:2451–2454. doi: 10.1128/jvi.74.5.2451-2454.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haan K M, Longnecker R. Coreceptor restriction within the HLA-DQ locus for Epstein-Barr virus infection. Proc Natl Acad Sci USA. 2000;97:9252–9257. doi: 10.1073/pnas.160171697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heller M, Gerber P, Kieff E. DNA of herpesvirus pan, a third member of the Epstein-Barr virus-herpesvirus papio group. J Virol. 1982;41:931–939. doi: 10.1128/jvi.41.3.931-939.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heller M, Gerber P, Kieff E. Herpesvirus papio DNA is similar in organization to Epstein-Barr virus DNA. J Virol. 1981;37:698–709. doi: 10.1128/jvi.37.2.698-709.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janz A, Oezel M, Kurzeder C, Mautner J, Pich D, Kost M, Hammerschmidt W, Delecluse H J. Infectious Epstein-Barr virus lacking major glycoprotein BLLF1 (gp350/220) demonstrates the existence of additional viral ligands. J Virol. 2000;74:10142–10152. doi: 10.1128/jvi.74.21.10142-10152.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kearns-Jonker M, Monell-Torrens E, Abbasi F, Holers V M, Notkins A L, Sigounas G. EBV binds to lymphocytes of transgenic mice that express the human CR2 gene. Virus Res. 1997;50:85–94. doi: 10.1016/s0168-1702(97)00052-x. [DOI] [PubMed] [Google Scholar]
- 18.Kelleher Z T, Fu H, Livanos E, Wendelburg B, Gulino S, Vos J M. Epstein-Barr-based episomal chromosomes shuttle 100 kb of self-replicating circular human DNA in mouse cells. Nat Biotechnol. 1998;16:762–768. doi: 10.1038/nbt0898-762. [DOI] [PubMed] [Google Scholar]
- 19.Kim K J, Kanellopoulos-Langevin C, Merwin R M, Sachs D H, Asofsky R. Establishment and characterization of BALB/c lymphoma lines with B cell properties. J Immunol. 1979;122:549–554. [PubMed] [Google Scholar]
- 20.Klein G, Lindahl T, Jondal M, Leibold W, Menezes J, Nilsson K, Sundstrom C. Continuous lymphoid cell lines with characteristics of B cells (bone-marrow-derived), lacking the Epstein-Barr virus genome and derived from three human lymphomas. Proc Natl Acad Sci USA. 1974;71:3283–3286. doi: 10.1073/pnas.71.8.3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krysan P J, Calos M P. Epstein-Barr virus-based vectors that replicate in rodent cells. Gene. 1993;136:137–143. doi: 10.1016/0378-1119(93)90457-e. [DOI] [PubMed] [Google Scholar]
- 22.Kulwichit W, Edwards R H, Davenport E M, Baskar J F, Godfrey V, Raab-Traub N. Expression of the Epstein-Barr virus latent membrane protein 1 induces B cell lymphoma in transgenic mice. Proc Natl Acad Sci USA. 1998;95:11963–11968. doi: 10.1073/pnas.95.20.11963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landon J C, Ellis L B, Zeve V H, Fabrizio D P. Herpes-type virus in cultured leukocytes from chimpanzees. J Natl Cancer Inst. 1968;40:181–192. [PubMed] [Google Scholar]
- 24.Lee A M, Diamond M E, Yates J L. Genetic evidence that EBNA-1 is needed for efficient, stable latent infection by Epstein-Barr virus. J Virol. 1999;73:2974–2982. doi: 10.1128/jvi.73.4.2974-2982.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levy J A, Levy S B, Hirshaut Y, Kafuko G, Prince A. Presence of EBV antibodies in sera from wild chimpanzees. Nature. 1971;233:559–560. doi: 10.1038/233559a0. [DOI] [PubMed] [Google Scholar]
- 26.Li Q, Spriggs M K, Kovats S, Turk S M, Comeau M R, Nepom B, Hutt-Fletcher L M. Epstein-Barr virus uses HLA class II as a cofactor for infection of B lymphocytes. J Virol. 1997;71:4657–4662. doi: 10.1128/jvi.71.6.4657-4662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Q, Turk S M, Hutt Fletcher L M. The Epstein-Barr virus (EBV) BZLF2 gene product associates with the gH and gL homologs of EBV and carries an epitope critical to infection of B cells but not of epithelial cells. J Virol. 1995;69:3987–3994. doi: 10.1128/jvi.69.7.3987-3994.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Little R D, Schildkraut C L. Initiation of latent DNA replication in the Epstein-Barr virus genome can occur at sites other than the genetically defined origin. Mol Cell Biol. 1995;15:2893–2903. doi: 10.1128/mcb.15.5.2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Longnecker R. Molecular biology of Epstein-Barr virus. In: McCance D, editor. Human tumor viruses. Washington, D.C.: ASM Press; 1998. pp. 133–172. [Google Scholar]
- 30.Marchini A, Kieff E, Longnecker R. Marker rescue of a transformation-negative Epstein-Barr virus recombinant from an infected Burkitt lymphoma cell line: a method useful for analysis of genes essential for transformation. J Virol. 1993;67:606–609. doi: 10.1128/jvi.67.1.606-609.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marchini A, Longnecker R, Kieff E. Epstein-Barr virus (EBV)-negative B-lymphoma cell lines for clonal isolation and replication of EBV recombinants. J Virol. 1992;66:4972–4981. doi: 10.1128/jvi.66.8.4972-4981.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moghaddam A, Rosenzweig M, Lee-Parritz D, Annis B, Johnson R P, Wang F. An animal model for acute and persistent Epstein-Barr virus infection. Science. 1997;276:2030–2033. doi: 10.1126/science.276.5321.2030. [DOI] [PubMed] [Google Scholar]
- 33.Molesworth S J, Lake C M, Borza C M, Turk S M, Hutt Fletcher L M. Epstein-Barr Virus gH is essential for penetration of B cells but also plays a role in attachment of virus to epithelial cells. J Virol. 2000;74:6324–6332. doi: 10.1128/jvi.74.14.6324-6332.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Molina H, Brenner C, Jacobi S, Gorka J, Carel J C, Kinoshita T, Holers V M. Analysis of Epstein-Barr virus-binding sites on complement receptor 2 (CR2/CD21) using human-mouse chimeras and peptides. At least two distinct sites are necessary for ligand-receptor interaction. J Biol Chem. 1991;266:12173–12179. [PubMed] [Google Scholar]
- 35.Murray R J, Young L S, Calender A, Gregory C D, Rowe M, Lenoir G M, Rickinson A B. Different patterns of Epstein-Barr virus gene expression and of cytotoxic T-cell recognition in B-cell lines infected with transforming (B95.8) or nontransforming (P3HR1) virus strains. J Virol. 1988;62:894–901. doi: 10.1128/jvi.62.3.894-901.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nemerow G R, Houghten R A, Moore M D, Cooper N R. Identification of an epitope in the major envelope protein of Epstein-Barr virus that mediates viral binding to the B lymphocyte EBV receptor (CR2) Cell. 1989;56:369–377. doi: 10.1016/0092-8674(89)90240-7. [DOI] [PubMed] [Google Scholar]
- 37.Neubauer R H, Rabin H, Strnad B C, Nonoyama M, Nelson-Rees W A. Establishment of a lymphoblastoid cell line and isolation of an Epstein-Barr-related virus of gorilla origin. J Virol. 1979;31:845–848. doi: 10.1128/jvi.31.3.845-848.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paola N, Schildkraut C L, Yates J L. Initiation of DNA replication within oriP is dispensable for stable replication of the latent Epstein-Barr virus chromosome after infection of established cell lines. J Virol. 2000;74:8563–8574. doi: 10.1128/jvi.74.18.8563-8574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rabin H, Strnad B C, Neubauer R H, Brown A M, Hopkins III R F, Mazur R A. Comparisons of nuclear antigens of Epstein-Barr virus (EBV) and EBV-like simian viruses. J Gen Virol. 1980;48:265–272. doi: 10.1099/0022-1317-48-2-265. [DOI] [PubMed] [Google Scholar]
- 40.Reisman D, Yates J, Sugden B. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol Cell Biol. 1985;5:1822–1832. doi: 10.1128/mcb.5.8.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rickinson A, Kieff E. Epstein-Barr virus. In: Fields B, Knipe D, Howley P, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2397–2446. [Google Scholar]
- 42.Rowe M, Rowe D T, Gregory C D, Young L S, Farrell P J, Rupani H, Rickinson A B. Differences in B cell growth phenotype reflect novel patterns of Epstein-Barr virus latent gene expression in Burkitt's lymphoma cells. EMBO J. 1987;6:2743–2751. doi: 10.1002/j.1460-2075.1987.tb02568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simas J P, Efstathiou S. Murine gammaherpesvirus 68: a model for the study of gammaherpesvirus pathogenesis. Trends Microbiol. 1998;6:276–282. doi: 10.1016/s0966-842x(98)01306-7. [DOI] [PubMed] [Google Scholar]
- 44.Speck P, Kline K A, Cheresh P, Longnecker R. Epstein-Barr virus lacking latent membrane protein 2 immortalizes B cells with efficiency indistinguishable from wild-type virus. J Gen Virol. 1999;80:2193–2203. doi: 10.1099/0022-1317-80-8-2193. [DOI] [PubMed] [Google Scholar]
- 45.Sunil-Chandra N P, Arno J, Fazakerley J, Nash A A. Lymphoproliferative disease in mice infected with murine gammaherpesvirus 68. Am J Pathol. 1994;145:818–826. [PMC free article] [PubMed] [Google Scholar]
- 46.Sunil-Chandra N P, Efstathiou S, Arno J, Nash A A. Virological and pathological features of mice infected with murine gamma-herpesvirus 68. J Gen Virol. 1992;73:2347–2356. doi: 10.1099/0022-1317-73-9-2347. [DOI] [PubMed] [Google Scholar]
- 47.Tanner J, Weis J, Fearon D, Whang Y, Kieff E. Epstein-Barr virus gp350/220 binding to the B lymphocyte C3d receptor mediates adsorption, capping, and endocytosis. Cell. 1987;50:203–213. doi: 10.1016/0092-8674(87)90216-9. [DOI] [PubMed] [Google Scholar]
- 48.Virgin H W, IV, Latreille P, Wamsley P, Hallsworth K, Weck K E, Dal Canto A J, Speck S H. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J Virol. 1997;71:5894–5904. doi: 10.1128/jvi.71.8.5894-5904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang F, Marchini A, Kieff E. Epstein-Barr virus (EBV) recombinants: use of positive selection markers to rescue mutants in EBV-negative B-lymphoma cells. J Virol. 1991;65:1701–1709. doi: 10.1128/jvi.65.4.1701-1709.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang X, Hutt-Fletcher L M. Epstein-Barr virus lacking glycoprotein gp42 can bind to B cells but is not able to infect. J Virol. 1998;72:158–163. doi: 10.1128/jvi.72.1.158-163.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wysokenski D A, Yates J L. Multiple EBNA1-binding sites are required to form an EBNA1-dependent enhancer and to activate a minimal replicative origin within oriP of Epstein-Barr virus. J Virol. 1989;63:2657–2666. doi: 10.1128/jvi.63.6.2657-2666.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yates J, Warren N, Reisman D, Sugden B. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc Natl Acad Sci USA. 1984;81:3806–3810. doi: 10.1073/pnas.81.12.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yates J L, Warren N, Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature. 1985;313:812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]
- 54.Yoshiyama H, Imai S, Shimizu N, Takada K. Epstein-Barr virus infection of human gastric carcinoma cells: implication of the existence of a new virus receptor different from CD21. J Virol. 1997;71:5688–5691. doi: 10.1128/jvi.71.7.5688-5691.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]