Abstract
Background: O-6-methylguanine-DNA methyltransferase is responsible for the direct repair of O6-methylguanine lesions induced by alkylating agents, including temozolomide. O-6-methylguanine-DNA methyltransferase promoter hypermethylation is a well-established biomarker for temozolomide response in glioblastoma patients, also correlated with therapeutic response in colorectal cancer. Objectives: The ARETHUSA clinical trial aims to stratify colorectal cancer patients based on their mismatch repair status. Mismatch repair-deficient patients are eligible for treatment with immune checkpoint inhibitors (anti-PDL-1), whereas mismatch repair-proficient samples are screened for O-6-methylguanine-DNA methyltransferase promoter methylation to identify those suitable for temozolomide treatment. Methods: In this context, a subset of ARETHUSA metastatic colorectal cancer samples was used to compare two different techniques for assessing O-6-methylguanine-DNA methyltransferase hypermethylation: Methyl-BEAMing, a highly sensitive digital PCR approach that combines emulsion PCR and flow cytometry, and droplet digital PCR, a more automated procedure that enables the rapid, operator-independent analysis of a large number of samples. Results: Our study clearly demonstrates that the results obtained using Methyl-BEAMing and droplet digital PCR are comparable, with both techniques showing similar accuracy, sensitivity, and reproducibility. Conclusions: Digital droplet PCR proved to be an efficient method for detecting gene promoter methylation. However, the Methyl-BEAMing method has proved more sensitive for detecting low quantities of DNA.
Keywords: MGMT, DNAmethylation, Methyl-BEAMing, digital PCR, metastatic colorectal cancer
1. Introduction
Temozolomide (TMZ) exerts its therapeutic effect by inducing DNA methylation at the N-7 and O6 positions of guanine and the N-3 position of adenine. O6-methylguanine (O6-meG) pairs with thymine instead of cytosine during DNA replication, thereby activating the post-replication mismatch repair (MMR) system, which leads to cell cycle arrest and apoptosis. The O6-methylguanine-DNA methyltransferase (MGMT) enzyme recognizes the O6-meG mismatch and repairs the lesion by transferring the alkyl group from guanine to a cysteine residue in its active site [1,2]. Epigenetic regulation through the methylation of the MGMT gene promoter prevents protein synthesis in cancer cells, consequently increasing their sensitivity to alkylating agents. Indeed, in glioblastoma patients, MGMT promoter hypermethylation is an established predictive biomarker for TMZ response and is approved for use in this cancer type [3,4]. Furthermore, MGMT promoter methylation is present in 30–40% of metastatic colorectal cancer (mCRC) cases and is directly correlated with increased DNA damage. Several phase 2 clinical studies have utilized MGMT methylation as a predictive and prognostic marker for TMZ response in chemotherapy-refractory mCRC [5]. Patients with high levels of promoter methylation exhibit longer disease-free survival upon TMZ treatment compared to those with reduced methylation levels and increased MGMT protein expression [6,7]. In general, the failure of the MMR system leads to the accumulation of DNA defects, and specifically in colorectal cancer (CRC), MMR dysfunction occurs in approximately 5% of total mCRC cases [7]. Based on MMR status, CRC patients are categorized as mismatch repair-proficient (MMRp) or mismatch repair-deficient (MMRd) [8,9]. A subset of MMRd CRC exhibits a microsatellite instability (MSI) phenotype, characterized by the accumulation of mutations in short, repetitive DNA regions known as microsatellites [10]. Patients with MSI-positive tumors have a better prognosis compared to those with microsatellite-stable (MSS) tumors and thus exhibit distinct histological features, which make them responsive to treatment with immune checkpoint inhibitors such as the anti-PD1 antibody pembrolizumab [11,12,13]. Based on previous studies, we contributed to a two-step clinical trial called ARETHUSA [14]. The trial begins with an initial screening in which MMRd mCRC patients are treated with pembrolizumab, while MMRp patients are assessed for MGMT protein expression and methylation status. Patients lacking MGMT protein and displaying promoter hypermethylation are selected for the first (priming) phase of the trial and treated with TMZ to induce a hypermutant status. Those patients who develop a tumor mutational burden (TMB) of ≥20 mutations per megabase (mut/Mb) upon progression after TMZ treatment proceed to the second phase, where they are treated with pembrolizumab as monotherapy [14]. Since MGMT promoter methylation status plays a critical role during the priming phase of this trial, it is essential to select and validate a sensitive and specific method for methylation analysis in order to minimize issues related to sampling and tumor heterogeneity. Traditionally, MGMT status has been assessed qualitatively using methylation-specific polymerase chain reaction (MSP) and bisulfite pyrosequencing. To improve the assessment of MGMT methylation, herein, we firstly employed a quantitative technique called Methylation Beads, Emulsion, Amplification, and Magnetic technology (Methyl-BEAMing) [15,16]. Methyl-BEAMing is considered one of the most reliable techniques for quantitatively evaluating MGMT promoter methylation [15]. In this study, we aimed to compare Methyl-BEAMing analysis with droplet digital PCR (ddPCR), a fully automated technology that may offer greater protection against operator-dependent errors, thus allowing the simultaneous analysis of many DNA samples in a shorter time period [17]. Our study demonstrates that both Methyl-BEAMing and ddPCR provide similar reproducibility, accuracy, and specificity for the quantitative assessment of MGMT methylation in clinical samples. However, Methyl-BEAMing showed higher sensitivity for detecting low quantities of DNA.
2. Material and Methods
2.1. DNA Extraction
FFPE tissues were sectioned using a microtome (Leica Biosystem, Nussloch, Germany) to obtain five 10 µm thick slices. Serial sections were cut, and IHC scoring was performed in a semi-quantitative fashion, taking into account both the extension and intensity of staining. Positive MGMT staining was defined as the staining intensity of the majority of tumor cells, as previously described [7]. To ensure an unbiased evaluation, at least two independent histologists scored the FFPE tissues and provided the cellularity values for each sample. Bulk DNA for molecular analysis was extracted from these tissues using the QIAamp DNA FFPE tissue kit (Qiagen, Hilden, Germany), following the manufacturer’s instructions. The protocol consists of six steps: the removal of paraffin, lysis using proteinase K, and heating at 90 °C to break the formalin-induced crosslinks. Subsequent steps included DNA binding, washing, and elution from the tissue.
2.2. DNA Quantification
DNA quantification was carried out using two methods: Nanodrop (DeNovix, Wilmington, DE, USA) and Qubit 4 (ThermoFisher, Waltham, MA, USA). Nanodrop quantifies nucleic acids via spectrophotometry by measuring the UV light absorption of nucleic acids; DNA concentration is directly proportional to the absorbed light. In contrast, Qubit is a highly sensitive fluorometer that selectively binds to nucleic acids, minimizing non-specific binding. For double-stranded DNA (dsDNA) quantification, we used two reagent types: the broad-range kit (2–1000 ng) and the high-sensitivity kit (0.2–100 ng) for dsDNA.
2.3. Treatment with Bisulfite
Bisulfite conversion was performed using the EZ DNA Methylation Gold kit (Zymo Research, Orange, CA, USA) according to the manufacturer’s protocol. Sodium bisulfite (HSO3−) from the CT Conversion Reagent was added to the DNA samples to distinguish between methylated and unmethylated cytosines. Upon incubation at high temperatures, unmethylated cytosines were converted to sulfonated cytosines and sulfonated uracils through deamination. In the final step, cytosines were irreversibly converted into uracils. DNA samples were then eluted from the columns using the elution buffer provided by the kit. Unmethylated cytosines were converted to uracils, whereas methylated cytosines remained unchanged due to the presence of a CH3 methyl group at the 5-carbon position.
2.4. Methyl-BEAMing Assay
The Methyl-BEAMing technique involves two rounds of DNA amplification: the first step uses specific primers for methylated and unmethylated DNA, while, in the second step, primers covalently bound to magnetic beads are compartmentalized into microdroplets. In Methyl-BEAMing, methylated beads are generated after the PCR amplification of individual DNA molecules in aqueous nanocompartments suspended in a continuous oil phase. The status of these beads is then detected by flow cytometry using fluorescent probes that specifically hybridize to methylated or unmethylated sequences. In contrast, ddPCR is a fully automated technique that uses nanodroplet sample partitioning, offering high sensitivity without the need for a standard curve. Bisulfite-converted DNA was analyzed using the Methyl-BEAMing technique to detect the methylation status of the MGMT gene promoter. The protocol begins with the initial polymerase chain reaction (PCR) amplification of the purified DNA using tagged primers to enrich the target locus. Amplicons were then diluted (1/16,000) and reamplified using tag-coated beads to enhance sensitivity. The second round of amplification was conducted in an emulsion, enabling the physical separation and independent amplification of different templates. PCR mixtures were prepared according to previously established conditions [15]. After amplification, the emulsion was broken using isopropanol/butanol, and the amplicons were hybridized with fluorescent probes specific to methylated or unmethylated bisulfite-converted templates. Flow cytometric analysis was performed using the Accuri C6 (BD Biosciences, Franklin Lakes, NJ, USA). The percentage of methylation was calculated by dividing the number of methylated events by the total number of specific events. A minimum of 200 cumulative events (methylated + unmethylated) were required to validate the results. All analyses were conducted in duplicate; final results were obtained by averaging the values, which were then normalized according to the percentage of tumor cells in the sample. A Positive Predictive Value of 0.67 and Negative Predictive Value of 0.98 had been determined previously with ROC analysis [15]. To check the quality of the assay, in each run, internal controls corresponding to 100%, 50%, and 0% methylation were used.
2.5. Droplet Digital PCR
Droplet digital PCR is a digital PCR method in which DNA samples are split into thousands of separate reaction droplets. PCR occurs in each droplet, which contains DNA molecules and fluorescent probes. A mixture of DNA, primers, and probes was prepared in a final volume of 20 µL. DNA samples and oil were loaded into the wells of the droplet generator. A vacuum system ensured that both the sample and oil passed through microfluidic circuits, forming a uniform dispersion of droplets. Each well containing 20 µL of mixture was divided into highly uniform compartments. The droplets were transferred to a 96-well PCR plate, and amplification was carried out in a conventional thermal cycler. After amplification, the plate was loaded into a reader where droplets passed through a two-color detector. Droplets were classified as positive or negative based on fluorescence amplitude, with the following binary threshold: 1 = positive, 0 = negative. For the ddPCR reaction, 5–10 µL of DNA template was mixed with 10 µL of ddPCR Supermix for Probes (Bio-Rad, Hercules, CA, USA) and 5 µL of the primer and probe mix (fluorophores FAM and HEX). The sample and mix were added to a cartridge along with 60 µL of oil required for droplet generation (Auto-DG, Bio-Rad). The droplets were transferred to a 96-well plate and subjected to temperature-dependent amplification in a thermal cycler. Finally, fluorescence was evaluated using the QX200 Droplet Reader (Bio-Rad).
2.6. Methylation Assay Controls
Ultramer oligomers of 250 bp (representing fully methylated or unmethylated bisulfite-converted templates) were used as positive controls. The specificity and sensitivity of ddPCR were verified using a synthetic methylation scale (ranging from 0% to 100% MGMT methylation) by mixing the two controls. Each amplification batch included two positive controls (methylated and unmethylated) and one negative control (no template).
2.7. Statistical Analyses
Statistical analyses, including correlation, linear regression, and Kappa statistics, were conducted using Prism 7.00 for Windows (GraphPad Software, La Jolla, MA, USA). Fisher’s exact test was performed to calculate the probability of the data for the 2 × 2.
3. Results
3.1. Methyl-BEAMing Analysis of MGMT Methylation
A subset of 342 samples from mCRC patients enrolled in the ARETHUSA trial was analyzed (Ethical Committee: Comitato Etico Milano Area 3 Italy; approval date 6 August 2018, IFOM-CPT002/2018/PO001; EudraCT number, 2018-001441-14). Formalin-fixed, paraffin-embedded (FFPE) tissue samples were previously evaluated for MGMT protein expression through immunohistochemistry (IHC). Samples with low-to-absent MGMT protein expression were processed to assess MGMT gene hypermethylation. This analysis was initially performed using Methyl-BEAMing, which involved two rounds of DNA amplification, thereby increasing the sensitivity of the protocol and enabling the detection of a small number of target molecules. Specific fluorescent probes for methylated or unmethylated DNA sequences were then used for flow cytometry analysis.
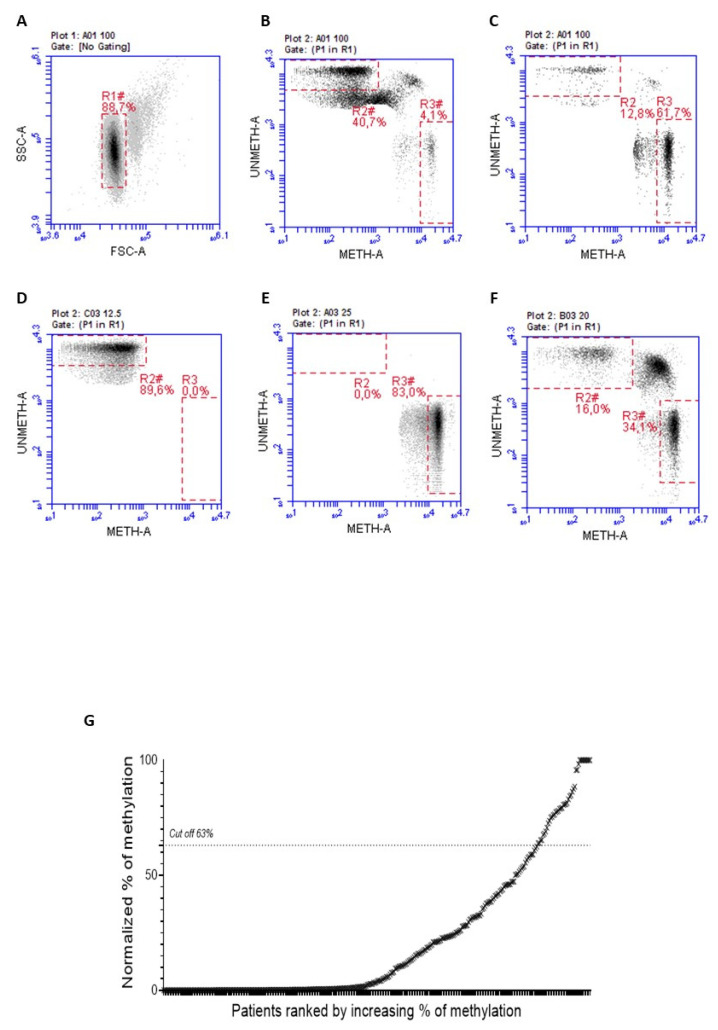
MGMT-negative FFPE mCRC samples were first subjected to Methyl-BEAMing to determine the methylation levels of the MGMT gene promoter. A previous ROC analysis determined the cutoff value to distinguish between hypermethylated and unmethylated samples [7]. A threshold of 63% was set to identify positive samples and quantitatively predict the therapeutic response. After two rounds of PCR amplification, the promoter methylation status was detected by flow cytometry, using probes specific to methylated and unmethylated sequences. As shown in Figure 1A–F, gate R2 represents unmethylated events, while R3 contains methylated events; R2 and R3 values are normalized according to the percentage of cancer cells obtained from IHC analysis. The final graph from this analysis allowed us to classify samples as positive or negative based on the percentage of events in gates R2 and R3. Using the Methyl-BEAMing assay, we found that the MGMT promoter was methylated in 7.78% of cases (Figure 1G).
Figure 1.
Representative plots of the gate strategy used for the flow cytometric analysis. (A) ARETHUSA samples analyzed through the Methyl-BEAMing technique were first grouped based on their complexity events. (B) Gate R2 (46.6%) includes unmethylated events, whereas R3 (4.1%) contains methylated ones. (C) The value of 61.7% referred to gate R3 is representative of a methylated sample. (D) Unmethylated sample control (R3 0%). (E) Methylated sample control (R3 83%). (F) The 50% control, obtained by mixing the unmethylated control with the methylated one (R3 34.1%) (G) Methylation profiling of CRC tissue samples assessed by Methyl-BEAMing.
3.2. Droplet Digital PCR Analysis
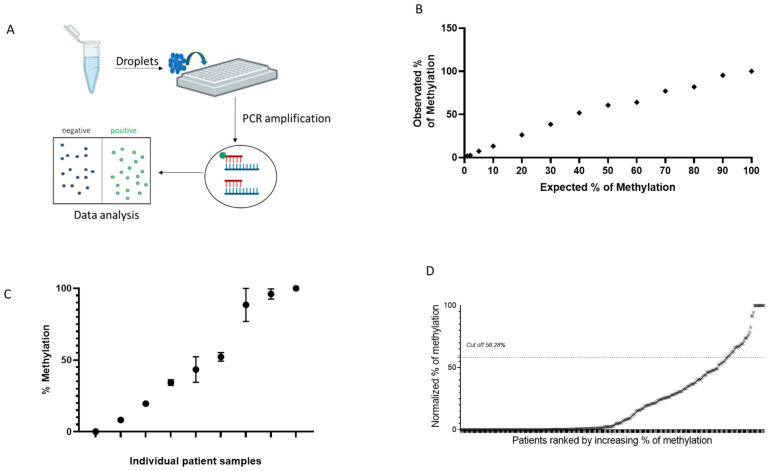
MGMT methylation analysis obtained by ddPCR is reported in Figure 2. Samples were processed through a two-color plate reader, where droplets were classified as positive or negative based on the emitted fluorescence (Figure 2A). ddPCR is one of the most precise and sensitive techniques, allowing the absolute quantification of DNA molecules in each sample. To assess the specificity and sensitivity of this method, a calibration curve was performed using an ultramer oligonucleotide mixture (Figure 2B). As shown by the linear curve, ddPCR analysis was highly reproducible and accurate. Reproducibility was further confirmed by performing three independent bisulfite treatments on separate mCRC samples (Figure 2C).
Figure 2.
Assessment of MGMT gene promoter hypermethylation using the ddPCR technique. (A) Scheme of ddPCR: DNA amplification occurs independently in thousands of drops through a water-in-oil emulsion. Droplets are classified as positive or negative based on the emitted fluorescence wavelength. (B) Linearity of quantification of ultramer oligonucleotide mixture assessed by ddPCR. (C) Methylation analysis by ddPCR using three independent bisulfite treatments in nine different mCRC samples. (D) MGMT methylation profiling of CRC tissue samples obtained by ddPCR.
To evaluate the capability of ddPCR for MGMT methylation assessment, the same CRC samples previously analyzed using Methyl-BEAMing were tested with ddPCR. First, a new threshold value for distinguishing positive from negative samples was determined using the linear regression equation y = a × x + b, where the independent variable (x) corresponded to the cutoff used in the Methyl-BEAMing analysis. With the calculated cutoff value of 58.28%, we found that 10.28% of samples displayed MGMT gene methylation (Figure 2D). Notably, 17 samples were excluded from ddPCR analysis due to insufficient DNA, which could not be amplified, rendering them undetectable.
3.3. Methyl-BEAming and Droplet Digital PCR Comparison
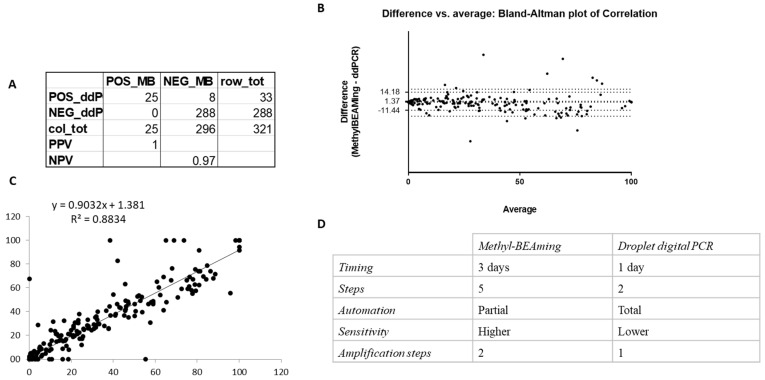
To determine whether the number of hypermethylated samples detected by both Methyl-BEAMing and ddPCR was comparable (Figure 3), we created a contingency table and found that the specificity of both methods was very similar (Figure 3A). Notably, due to the inability of ddPCR to detect low DNA amounts, the comparison was conducted on only 321 samples. To assess the concordance between the two techniques for methylation detection, we constructed a Bland–Altman plot, which showed that the results obtained using Methyl-BEAMing and ddPCR were comparable, though Methyl-BEAMing was more sensitive for samples with a low DNA content (Figure 3B). Furthermore, linear regression analysis confirmed these findings, with a coefficient of determination (r2 = 0.8834, p < 0.0001), indicating that the independent variable (BEAMing data) could predict the dependent variable (ddPCR data) (Figure 3C). Despite the high concordance between the methods, differences between them were noted and are summarized in Figure 3D.
Figure 3.
Evaluation of the concordance between results obtained using Methyl-BEAMing and those obtained using ddPCR. (A) Contingency table was carried out to qualitatively compare the number of methylated samples evaluated by either Methyl-BEAMing or ddPCR analysis. (B) Bland–Altman plot shows the agreement between the results of both techniques used. (C) Correlation and linear regression between Methyl-BEAMing and droplet digital PCR results. (D) The table reports differences between Methyl-BEAMing and ddPCR, including the time of execution, number of amplification steps, automation grade, and sensitivity.
3.4. MGMT Methylation Is Increased in Aged Women with mCRC
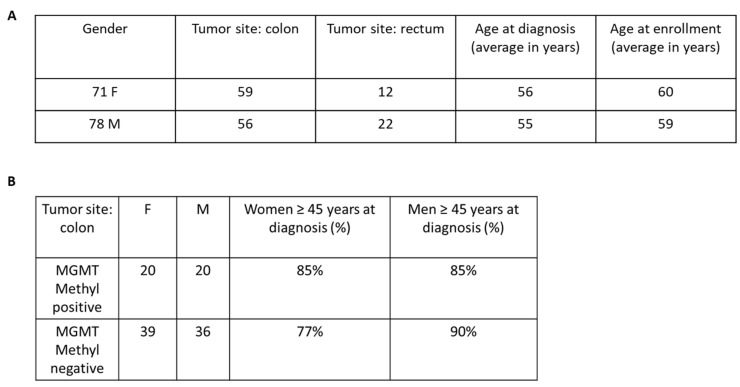
To investigate whether MGMT methylation status had predictive value in tumor-bearing patients, we conducted a small-scale epidemiologic study using clinical data from the ARETHUSA trial (Figure 4). Unfortunately, for several patients, the Electronic Case Report Forms were not properly collected. Therefore, this analysis was performed on a smaller cohort of 148 patients for whom we had complete medical information (Figure 4). Based on previous studies suggesting a correlation between MGMT methylation and advanced age in colon cancer patients [18], we specifically analyzed this subgroup. Furthermore, we considered tumor location and subdivided the data into patients with either rectal or colon cancer. Although our analysis did not reach statistical significance, we observed an interesting trend showing that younger women had lower methylation frequencies compared to women over 45 years old (Figure 4B). Notably, this difference was not observed in male patients, further supporting a correlation between age, gender, tumor location, and increased MGMT methylation.
Figure 4.
Analysis of the incidence of MGMT gene promoter methylation in a cohort of mCRC patients. (A) Clinical features collected for 148 mCRC patients. (B) This Table shows a non-statistically significant trend of increased MGMT methylation frequency in older women compared to younger ones (Fisher’s exact probability test one-tailed p-value = 0.4963).
4. Discussion
Chemoresistance to standard therapy remains a significant challenge in the treatment of metastatic colorectal cancer (mCRC) patients. In this context, identifying new therapeutic targets and predictive biomarkers for pharmacological treatment remains an unmet clinical need. The hypermethylation of the MGMT gene promoter and the resulting inactivation of protein expression play a key role in the early stages of colorectal cancer (CRC) development, as it is associated with an increase in G>A point mutations in other cancer-associated genes, such as KRAS and TP53 [4]. Moreover, MGMT promoter hypermethylation may increase sensitivity to alkylating agents like Dacarbazine and Temozolomide, as observed in patients with glioblastoma, advanced melanoma, and neuroendocrine tumors [6,19,20]. However, the efficacy of Temozolomide in mCRC remains controversial. Several phase II clinical trials enrolling MGMT-deficient mCRC patients did not show a clear improvement in progression-free survival or overall survival [21,22]. Notably, our data align with previous findings showing that older age and female gender are generally associated with higher levels of MGMT and/or p16 gene methylation. Despite the analysis being conducted on a limited cohort of mCRC patients, the observed trend supports the idea that MGMT hypermethylation may play an important role in colorectal cancer. In recent years, clinical studies on glioblastoma and mCRC patients have used methylation-specific PCR (MSP) to assess MGMT gene methylation [23]. However, it has been demonstrated that while MSP-based selection is necessary, it is not sufficient for the optimal stratification of patients who may benefit from alkylating agent therapy. Thus, using MSP alone for MGMT molecular analysis may be inadequate and requires further investigation. Recent studies have applied novel digital methods for patient stratification, providing greater prognostic and predictive significance for TMZ treatment. Among these, Methyl-BEAMing technology has proven more effective in assessing MGMT promoter methylation in glioblastoma samples compared to MSP or pyrosequencing [19]. Similarly, a retrospective study on FFPE mCRC samples using both standard and digital methods demonstrated the efficacy of Methyl-BEAMing in predicting prognosis and therapeutic response. To date, the highest prediction accuracy (87%) for treatment response has been achieved by combining multiple techniques to assess MGMT gene status, including Methyl-BEAMing, mass spectrometry, and RNAseq [17]. Additionally, MGMT protein expression detection via immunohistochemistry (IHC) has been evaluated as an exploratory biomarker in several studies, which have shown a high concordance between MGMT protein expression and quantitative MGMT methylation assessed using Methyl-BEAMing. These findings suggest that combining different methods is crucial in achieving the most accurate prediction. Furthermore, MMR evaluation proved crucial in predicting the TMZ treatment response in mCRC patients. In the ARETHUSA study, MGMT protein expression and promoter hypermethylation were assessed in MSS/MMRp patients prior to TMZ treatment, which was used as a noncanonical strategy to convert immunologically “cold tumors” into “hot tumors”. In this regard, MGMT hypermethylation assessment was found to be a crucial factor in patient selection. Our study directly compares Methyl-BEAMing and droplet digital PCR (ddPCR) for the assessment of MGMT promoter methylation. Both methods share a similar mechanism of action: they are based on molecular compartmentalization to perform large-scale amplification and can detect very low amounts of the target sequence. The results were normalized to tumor cell counts (as estimated by a histopathologist), and statistical analyses were performed to assess assay concordance. The analysis demonstrated a good correlation between the two techniques, further confirmed by linear regression analysis and sample distribution on a Bland–Altman plot. The sensitivity and specificity values indicate that both methods effectively identify false-positive and false-negative samples. Specifically, ddPCR yields optimal results when sufficient initial DNA is present, while Methyl-BEAMing is better suited for evaluating MGMT methylation in samples with limited DNA. The two-phase amplification process in Methyl-BEAMing enhances protocol sensitivity, but it also increases the error rate due to the dilution of the amplification product. This issue can be mitigated by using the fully automated ddPCR technique.
Overall, our study clearly demonstrates that combining IHC with either Methyl-BEAMing or ddPCR offers an accurate stratification of mCRC patients, leading to better prognostic and predictive outcomes for Temozolomide treatment.
Acknowledgments
We thank all the staff involved in the Candiolo Cancer Institute FPO-IRCCS facilities for their job and support.
Author Contributions
Methodology and validation: M.M.; Methodology and Data curation: V.P., N.C. and L.L.; Formal analysis: S.E.B., F.I. and A.C.; Resource, Conceptualization of the clinical trial and Supervision: F.P., A.R., E.G., L.G., D.C., N.P., A.A., E.F.B. and K.B.B.; Resource, Conceptualization of the clinical trial, and Project administration: M.G.Z., G.M., A.S., S.S. and A.S.-B.; Resource, Conceptualization of the clinical trial, and Investigation: E.B., M.C.A. and E.V.; Validation and Supervision: G.S. and S.M.; Methodology and Investigation: L.B.; Investigation Supervision, Project administration and Founding acquisition: F.D.N.; Data curation, Writing—review & editing, and supervision: F.M. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board of Comitato Etico Milano Area 3 Italy (Approval code: IFOM-CPT002/2018/PO001; EudraCT number 2018-001441-14; Approval date: 6 August 2018).
Data Availability Statement
The data that support the findings of this study are available upon request from the authors.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
Work in the authors’ laboratories has been supported by funding from AIRC under 5 per Mille 2018-ID. 21091 program (with F.D.N. as Group Leader); AIRC under IG 2018-ID. 21407 project to F.D.N.F. Pietrantonio reported receiving institutional research grants from BMS, Incyte, Agenus, Amgen, Lilly, and AstraZeneca and personal fees from BMS, MSD, Amgen, Merck-Serono, Pierre-Fabre, Servier, Bayer, Takeda, Astellas, Johnson & Johnson, Rottapharm, Ipsen, AstraZeneca, GSK, Daiichi-Sankyo, Seagen/Pfizer, and Beigene.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Oldrini B., Vaquero-Siguero N., Mu Q., Kroon P., Zhang Y., Galán-Ganga M., Bao Z., Wang Z., Liu H., Sa J.K., et al. MGMT genomic rearrangements contribute to chemotherapy resistance in gliomas. Nat. Commun. 2020;11:3883. doi: 10.1038/s41467-020-17717-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao J., Yang S., Cui X., Wang Q., Yang E., Tong F., Hong B., Xiao M., Xin L., Xu C., et al. A novel compound EPIC-0412 reverses temozolomide resistance via inhibiting DNA repair/MGMT in glioblastoma. Neuro Oncol. 2023;25:857–870. doi: 10.1093/neuonc/noac242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orzan F., De Bacco F., Crisafulli G., Pellegatta S., Mussolin B., Siravegna G., D’Ambrosio A., Comoglio P.M., Finocchiaro G., Boccaccio C. Genetic Evolution of Glioblastoma Stem-Like Cells from Primary to Recurrent Tumor. Stem Cells. 2017;35:2218–2228. doi: 10.1002/stem.2703. [DOI] [PubMed] [Google Scholar]
- 4.Wick W., Gorlia T., Bady P., Platten M., van den Bent M.J., Taphoorn M.J., Steuve J., Brandes A.A., Hamou M.F., Wick A., et al. Phase II Study of Radiotherapy and Temsirolimus versus Radiochemotherapy with Temozolomide in Patients with Newly Diagnosed Glioblastoma without MGMT Promoter Hypermethylation (EORTC 26082) Clin. Cancer Res. 2016;22:4797–4806. doi: 10.1158/1078-0432.CCR-15-3153. [DOI] [PubMed] [Google Scholar]
- 5.Pietrantonio F., Perrone F., de Braud F., Castano A., Maggi C., Bossi I., Gevorgyan A., Biondani P., Pacifici M., Busico A., et al. Activity of temozolomide in patients with advanced chemorefractory colorectal cancer and MGMT promoter methylation. Ann. Oncol. 2014;25:404–408. doi: 10.1093/annonc/mdt547. [DOI] [PubMed] [Google Scholar]
- 6.Trillo Aliaga P., Spada F., Peveri G., Bagnardi V., Fumagalli C., Laffi A., Rubino M., Gervaso L., Guerini Rocco E., Pisa E., et al. Should temozolomide be used on the basis of O6-methylguanine DNA methyltransferase status in patients with advanced neuroendocrine tumors? A systematic review and meta-analysis. Cancer Treat. Rev. 2021;99:102261. doi: 10.1016/j.ctrv.2021.102261. [DOI] [PubMed] [Google Scholar]
- 7.Sartore-Bianchi A., Pietrantonio F., Amatu A., Milione M., Cassingena A., Ghezzi S., Caporale M., Berenato R., Falcomata C., Pellegrinelli A., et al. Digital PCR assessment of MGMT promoter methylation coupled with reduced protein expression optimises prediction of response to alkylating agents in metastatic colorectal cancer patients. Eur. J. Cancer. 2017;71:43–50. doi: 10.1016/j.ejca.2016.10.032. [DOI] [PubMed] [Google Scholar]
- 8.Kopetz S., Grothey A., Yaeger R., Van Cutsem E., Desai J., Yoshino T., Wasan H., Ciardiello F., Loupakis F., Hong Y.S., et al. Encorafenib, binimetinib, and cetuximab. N. Engl. J. Med. 2019;381:1632–1643. doi: 10.1056/NEJMoa1908075. [DOI] [PubMed] [Google Scholar]
- 9.Sinicrope F.A., Sargent D.J. Molecular pathways: Microsatellite instability in colorectal cancer: Prognostic, predictive, and therapeutic implications. Clin. Cancer Res. 2012;18:1506–1512. doi: 10.1158/1078-0432.CCR-11-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiricny J. The multifaceted mismatch-repair system. Nat. Rev. Mol. Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 11.Boland C.R., Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073–2087.e2073. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Smedt L., Lemahieu J., Palmans S., Govaere O., Tousseyn T., Van Cutsem E., Prenen H., Tejpar S., Spaepen M., Matthijs G., et al. Microsatellite instable vs. stable colon carcinomas: Analysis of tumour heterogeneity, inflammation and angiogenesis. Br. J. Cancer. 2015;113:500–509. doi: 10.1038/bjc.2015.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le D.T., Uram J.N., Wang H., Bartlett B.R., Kemberling H., Eyring A.D., Skora A.D., Luber B.S., Azad N.S., Laheru D., et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N. Engl. J. Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeinalian M., Hashemzadeh-Chaleshtori M., Salehi R., Emami M.H. Clinical Aspects of Microsatellite Instability Testing in Colorectal Cancer. Adv. Biomed. Res. 2018;7:28. doi: 10.4103/abr.abr_185_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crisafulli G., Sartore-Bianchi A., Lazzari L., Pietrantonio F., Amatu A., Macagno M., Barault L., Cassingena A., Bartolini A., Luraghi P., et al. Temozolomide Treatment Alters Mismatch Repair and Boosts Mutational Burden in Tumor and Blood of Colorectal Cancer Patients. Cancer Discov. 2022;12:1656–1675. doi: 10.1158/2159-8290.CD-21-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barault L., Amatu A., Bleeker F.E., Moutinho C., Falcomata C., Fiano V., Cassingena A., Siravegna G., Milione M., Cassoni P., et al. Digital PCR quantification of MGMT methylation refines prediction of clinical benefit from alkylating agents in glioblastoma and metastatic colorectal cancer. Ann. Oncol. 2015;26:1994–1999. doi: 10.1093/annonc/mdv272. [DOI] [PubMed] [Google Scholar]
- 17.Schwartz S., Szeto C., Tian Y., Cecchi F., Corallo S., Calegari M.A., Di Bartolomeo M., Morano F., Raimondi A., Fuca G., et al. Refining the selection of patients with metastatic colorectal cancer for treatment with temozolomide using proteomic analysis of O6-methylguanine-DNA-methyltransferase. Eur. J. Cancer. 2019;107:164–174. doi: 10.1016/j.ejca.2018.11.016. [DOI] [PubMed] [Google Scholar]
- 18.Blanc-Durand F., Tang R., Pommier M., Nashvi M., Cotteret S., Genestie C., Le Formal A., Pautier P., Michels J., Kfoury M., et al. Clinical Relevance of BRCA1 Promoter Methylation Testing in Patients with Ovarian Cancer. Clin. Cancer Res. 2023;29:3124–3129. doi: 10.1158/1078-0432.CCR-22-3328. [DOI] [PubMed] [Google Scholar]
- 19.Stupp R., Mason W.P., van den Bent M.J., Weller M., Fisher B., Taphoorn M.J.B., Belanger K., Brandes A.A., Marosi C., Bogdahn U., et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 20.Amatu A., Sartore-Bianchi A., Moutinho C., Belotti A., Bencardino K., Chirico G., Cassingena A., Rusconi F., Esposito A., Nichelatti M., et al. Promoter CpG island hypermethylation of the DNA repair enzyme MGMT predicts clinical response to dacarbazine in a phase II study for metastatic colorectal cancer. Clin. Cancer Res. 2013;19:2265–2272. doi: 10.1158/1078-0432.CCR-12-3518. [DOI] [PubMed] [Google Scholar]
- 21.Krakowczyk L., Strzelczyk J.K., Adamek B., Zalewska-Ziob M., Arendt J., Półtorak S., Maciejewski B., Wiczkowski A. Methylation of the MGMT and p16 genes in sporadic colorectal carcinoma and corresponding normal colonic mucosa. Med. Sci. Monit. 2008;14:BR219-25. [PubMed] [Google Scholar]
- 22.Esteller M., Toyota M., Sanchez-Cespedes M., Capella G., Peinado M.A., Watkins D.N., Issa J.P., Sidransky D., Baylin S.B., Herman J.G. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is associated with G to A mutations in K-ras in colorectal tumorigenesis. Cancer Res. 2000;60:2368–2371. [PubMed] [Google Scholar]
- 23.Middleton M.R., Grob J.J., Aaronson N., Fierlbeck G., Tilgen W., Seiter S., Gore M., Aamdal S., Cebon J., Coates A., et al. Randomized phase III study of temozolomide versus dacarbazine in the treatment of patients with advanced metastatic malignant melanoma. J. Clin. Oncol. 2000;18:158–166. doi: 10.1200/JCO.2000.18.1.158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available upon request from the authors.