Abstract
Analysis of genomic and expression data allows both identification and characterization of novel retroviruses. We describe a recombinant type C murine retrovirus, similar to the Mus dunni endogenous retrovirus, with VL30-like long terminal repeats and murine leukemia virus-like coding sequences. This virus is present in multiple copies in the mouse genome and expressed in a range of mouse tissues.
Data mining genomic databases not only allows the discovery of new genes; it can also lead to the discovery and characterization of novel retroviruses and transposable elements and will prove invaluable in exploring the role of retroelements in host genome evolution. Here we combine analysis of both genomic and expression data to characterize a novel murine type C retrovirus from DNA sequence databases. This retrovirus is present in multiple copies in the Mus musculus genome, representing multiple recent insertions, and is transcribed in a range of mouse tissues. The virus is recombinant, with murine leukemia virus (MLV)-like coding sequences and long terminal repeats (LTRs) derived from a VL30 retroelement. Analysis of expression data suggests that a 70-bp repeat region of the LTR containing the polyadenylation signal can be tandemly duplicated and may have properties that enhance its cooption into the host genome.
Complete simple type C retroviruses (accession no. AF151794 and X94150) were used as query sequences for searching the nonredundant high-throughput (HTG), genome survey sequences (GSS), and nucleic acid databases supplied by the Australian National Genomic Information Service (http://www.angis.org.au). The databases were searched using BlastN (1). The results were manually screened for multiple high scoring pairs (HSP) that spanned more than 2,000 continuous nucleotides of the subject sequence. These matches (plus 5,000 bp upstream and downstream) were run through FRAMES (Wisconsin Package, version 8.1-UNIX, August 1995) to look for the open reading frame (ORF) pattern typical of simple retroviruses. For the expression analysis, we used all Mus musculus expressed sequence tags (ESTs) in GenBank 117. We have developed a number of Perl scripts to parse the Blast output and semiautomate the process of filtering and sewing the blast matches (http://www.bit.uq.edu.au/retrovirus/).
The reference proviral genome sequence is 8,665 nucleotides (112341 to 121005) from an M. musculus PAC clone (12). Similar sequences (with 3% average difference across coding sequence) can be found in a number of other clones on several different chromosomes (2, 4, 6). The provirus has open reading frames for gag, pro-pol, and env (101 bp of overlapping reading frames [+1 bp] between pol and env). Upstream of the gag ATG start codon, a CTG start codon defines the start of 99 bp of glycosylated Gag (which has been implicated in pathogenicity in murine leukemia viruses [3]). We defined the LTRs by imperfect inverted 9-bp repeats (11) and identified the TATA box, primer-binding sites, polyadenylation and polypurine signals, and splice sites (see Fig. 2; also see http://www.bit.uq.edu.au/retrovirus/). The untranslated region downstream of U5 contains two copies of a 35-bp repeat (which is present in up to 10 copies in VL30-like LTRs). This virus, here referred to as M. musculus retrovirus (MmERV), is most closely related to Mus dunni endogenous retrovirus (MDEV) but is clearly a distinct variant, differing at 10% of sites in the protein-coding sequences (Fig. 1).
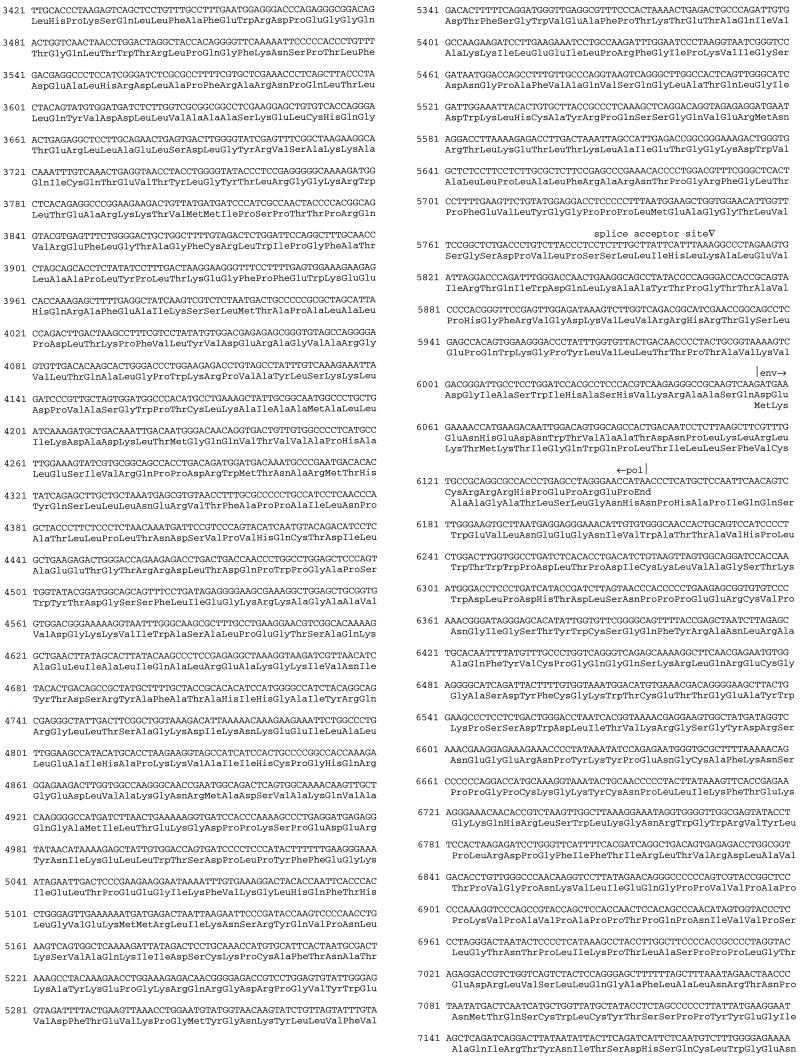
FIG. 2.
Nucleotide sequence of the proviral genome of a novel type C retrovirus (MmERV) from positions 112341 to 121005 of M. musculus PAC clone 657p21 (accession no. AC005743) has a total length of 8,665 bp. The 9-bp imperfect inverted repeats that define the LTRs have been underlined. Note that 2 bp are deleted from the terminal copies of these repeats upon insertion. The CAT box is not clearly identifiable (13), though a candidate sequence appears upstream of the TATA box on the opposite strand. The repeat region spanning U3 and R is marked by bold horizontal bars, and the following features are identified: flanking 9-bp repeats (double underline), pol-like motif (bold type), 12-bp inverse palindrome (boxed), and polyadenylation signal (boxed). This region containts a putative ORF (213 bp, positions 378 to 590: MSFDPRALVYVLSCCCFIKSCLQHFEFGLSVFLGPRLSRGLSEGLPSGVFHLGARPGSVRPPRDPRPTLR). Upstream of the gag ATG start codon, a CTG start codon defines the start of 99 bp of glycosylated gag. The untranslated region between U5 and the start of glycosylated gag contains a variable number (two to six) of perfect 35-bp repeats.
FIG. 1.
Maximum-likelihood phylogeny of an alignment of conserved regions of coding sequences from available whole genome sequences of mammalian type C leukemia retroviruses. Porcine endogenous retrovirus (PERV, accession AF038600), M. dunni endogenous retrovirus MDEV (AF053745), koala endogenous retrovirus (7) (KoRV, AF151794), Gibbon ape leukemia virus (GALV, M26927), gibbon ape leukemia virus strain X (GALVX, GLU60065), feline leukemia virus (FELV, M18247 and M19392), murine leukemia virus (MLV, MLU13766), Friend murine leukemia virus (FMLV, M93134 and M81185), rat leukemia virus (RLV, MLVGAG), Moloney murine leukemia virus (MMLV, REMLM). Sequences were aligned by eye; positions too variable to be confidently aligned were excluded from the analysis, leaving a final alignment of 4,779 bp. We assumed an HKY+Γ model, with transition-transversion ratio (ti/tv), (ti/tv) base composition, and gamma shape parameter (α) estimated from the data) (ti/tv = 3.1, α = 0.4). Scale in substitutions per site.
EST data for mice demonstrate that MmERV is transcribed in a range of mouse tissues, including heart, skin, and T cells (with complete EST hits for all genes and LTRs, including ESTs that overlap gene boundaries). The presence of MmERV in a variety of mouse tissues, presumably from a number of different laboratory stocks of M. musculus, may indicate that it is endogenous in the mouse genome. However, the LTRs are perfect copies of each other, and the coding sequences of the different MmERV clones are very similar, implying relatively recent insertions of the MmERV provirus into the mouse genome (8), so MmERV may also exist as an exogenous virus of M. musculus. The recombinant nature of MmERV might confer some of the expression properties of VL30 elements. These retroelements do not have functional coding sequences, but strong promoters and enhancers and promiscuous packaging signals allow them to transpose and to cross-infect cells (including human cells) by efficiently copackaging with MLV-type virions (5, 8, 13). The closest relative of MmERV, the M. dunni retrovirus MDEV, also has VL30-like LTRs and can be induced to produce virions (13). It therefore seems possible that MmERV is replication competent.
Expression data reveal that a 70-bp region of the MmERV LTR which spans the R-U5 boundary and contains the polyadenylation signal (nucleotides 383 to 452; Fig. 2) is conserved among diverse VL30-like LTRs and is tandemly duplicated in some LTRs (possibly representing a case of multimerization of regulatory elements to increase viral replication rates [14] or to alter expression patterns [8]). This region appears to provide the polyadenylation signal for a range of ESTs. Furthermore, the region occurs in EST transcripts that apparently are not transcribed from proviruses or VL30 elements (for example, ESTs AA572611, AW261579, and AV343291). These observations suggest that this repeat region is not only important in the transcription of viral (and retroelement) sequences but may be active in the expression of nonviral sequences. This observation is consistent with previous examples of mammalian genes adopting polyadenylation signals from retroviruses and transposable elements (8–10).
A closer examination of this repeat region reveals that it has a number of curious features. It is flanked by 9-bp imperfect repeats and contains a 12-bp inverted palindrome (CCAGAGCTCTGG). This palindrome coincides with a sequence that closely matches a highly conserved motif of pol (Fig. 2). This sequence lies within a small ORF (213 bp, positions 378 to 590). The significance of this ORF is unknown; it is not well preserved between isolates, and the potential product has no significant matches to sequences in the current databases. An additional upstream copy of the 9-bp flanking repeat (308 to 316) defines a 78-bp region which contains the TATA box; this region also receives many hits to ESTs that are apparently not viral in origin. These features, and the observed duplication of the poly(A) addition region and its apparent cooption in the mouse genome, may indicate some form of transpositional mobility, past or present. The possibility that these sequences represent “cassettes” containing strong viral promoters and enhancers that may be duplicated or coopted into expression of both host and viral genes warrants further exploration.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers W M, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Birren B, et al. Mus musculus clones RP23–113D4 ( AC021768), RP23–191A4 ( AC012147), CT7–378P20 ( AC015886). Boston, Mass: Whitehead Institute/MIT Center for Genome Research; 2000. [Google Scholar]
- 3.Corbin A, Prats A C, Darlix J L, Sitbon M. A nonstructural gag-encoded glycoprotein precursor is necessary for efficient spreading and pathogenesis of murine leukemia viruses. J Virol. 1994;68:3857–3867. doi: 10.1128/jvi.68.6.3857-3867.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Department of Energy Joint Genome Institute. 2000. Mus musculus clones RP23–369C16 (accession AC079538), RP23–205N19 (AC079494), RP23–435B12 (AC079555), RP23–33B2 (AC073758), RP23–261I4 (AC073736), RP23–13703 (AC073686).
- 5.French N S, Norton J D. Structure and functional properties of mouse VL30 retrotransposons. Biochim Biophys Acta. 1997;1352:33–47. doi: 10.1016/s0167-4781(97)00009-2. [DOI] [PubMed] [Google Scholar]
- 6.Han J, et al. Mus musculus chromosome 2 clone RP23–390N5: accession AC074337. New York, N.Y: Department of Molecular Genetics, Albert Einstein College of Medicine Genome Center; 2000. [Google Scholar]
- 7.Hanger J J, Bromham L D, McKee J J, O'Brien T M, Robinson W F. The nucleotide sequence of koala (Phascolarctos cinereus) retrovirus: a novel type C endogenous virus related to gibbon ape leukemia virus. J Virol. 2000;74:4264–4272. doi: 10.1128/jvi.74.9.4264-4272.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keshet E, Schiff R, Itin A. Mouse retrotransposons: a cellular reservoir for long terminal repeat (LTR) elements with diverse transcriptional specificities. Adv Cancer Res. 1991;56:215–251. doi: 10.1016/s0065-230x(08)60482-0. [DOI] [PubMed] [Google Scholar]
- 9.Mager D L, Hunter D G, Schertzer M, Freeman J D. Endogenous retroviruses provide the primary polyadenylation signal for two new human genes (HHLA2 and HHLA3) Genomics. 1999;59:255–263. doi: 10.1006/geno.1999.5877. [DOI] [PubMed] [Google Scholar]
- 10.Murnane J P, Morales J F. Use of mammalian interspersed repetitive (MIR) element in the coding and processing sequences of mammalian genes. Nucleic Acids Res. 1995;23:2837–2839. doi: 10.1093/nar/23.15.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parent I, Qin Y, Vandebroucke A-T, Walon C, Delferriere N, Godfroid E, Burtonboy G. Characterization of a C-type retrovirus isolated from an infected cell line: complete nucleotide sequence. Arch Virol. 1998;143:1077–1092. doi: 10.1007/s007050050357. [DOI] [PubMed] [Google Scholar]
- 12.Wang Q, Fu Y, Pan H, Dumanski J, Roe B A. Mus musculus chromosome unknown clone rp21–657p21 strain 129S6/SvEvTac: accession AC005743. Norman: Department of Chemistry and Biochemistry, University of Oklahoma; 2000. [Google Scholar]
- 13.Wolgamot G, Bonham L, Miller A D. Sequence analysis of Mus dunni endogenous virus reveals a hybrid VL30/gibbon ape leukemia virus-like structure and a distinct envelope. J Virol. 1998;72:7459–7466. doi: 10.1128/jvi.72.9.7459-7466.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolgamot G, Miller A D. Replication of Mus dunni endogenous retrovirus depends on promoter activation followed by enhancer multimerization. J Virol. 1999;73:9803–9809. doi: 10.1128/jvi.73.12.9803-9809.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]