Abstract
E3 ubiquitin ligases known as plant U-box (PUB) proteins regulate a variety of aspects of plant growth, development, and stress response. However, the functions and characteristics of the PUB gene family in alfalfa remain unclear. This work involved a genome-wide examination of the alfalfa U-box E3 ubiquitin ligase gene. In total, 210 members were identified and divided into five categories according to their homology with the members of the U-box gene family in Arabidopsis thaliana. The phylogenetic analysis, conserved motifs, chromosomal localization, promoters, and regulatory networks of this gene were investigated. Chromosomal localization and covariance analyses indicated that the MsPUB genes expanded MsPUB gene family members through gene duplication events during evolution. MsPUB genes may be involved in the light response, phytohormone response, growth, and development of several biological activities, according to cis-acting element analysis of promoters. In addition, transcriptome analysis and expression analysis by qRT-PCR indicated that most MsPUB genes were significantly upregulated under cold stress, drought stress, and salt stress treatments. Among them, MsPUBS106 and MsPUBS185 were significantly and positively correlated with cold resistance in alfalfa. MsPUBS110, MsPUBS067, MsPUBS111 and MsPUB155 were comprehensively involved in drought stress, low temperature, and salt stress resistance. All things considered, these discoveries offer fresh perspectives on the composition, development, and roles of the PUB gene family in alfalfa. They also provide theoretical guidance for further investigations into the mechanisms regulating the development, evolution, and stress tolerance of MsPUB.
Keywords: alfalfa, PUB gene family, abiotic stress, expression pattern, qRT-PCR
1. Introduction
During the process of development, plants are frequently impacted by many different kinds of abiotic factors, including low temperatures, salinity, and salt. For maintaining the course of ordinary development and growth, plants have developed a series of regulatory processes [1]. Ubiquitination represents a post-translational modification and is often implicated in the growth, development, and stress processes in plants induced by abiotic factors [2]. The ubiquitin–proteasome system (UPS) is a complex system whose core members include ubiquitin (Ub), deubiquitylating enzyme (DUB), ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), ubiquitin ligase enzyme (E3), and 26S proteasome [3]. In order to add ubiquitin to particular target proteins, ubiquitin is first activated by E1, and then it moves to E2 and lastly the E3 ligase [4]. The 26S proteasome typically breaks down target proteins that bind polyubiquitin chains, although monoubiquitinated proteins have the ability to control protein function. E3 ubiquitin ligases are the primary factor that determines the specificity of protein ubiquitination. They work by recognizing and binding to substrates [5]. Depending on how many subunits they have, E3 ubiquitin ligases can be divided into two groups: monomeric and multisubunit. Three types of monomeric E3 ubiquitin ligases can be distinguished: U-box E3 ubiquitin ligases, really interesting new gene (RING), and homology to E6-AP C-terminus (HECT) [6]. In contrast, anaphase-promoting complex/cyclosome (APC/C) E3 ubiquitin ligase complexes and cullin-RING ligase (CRL) are examples of multisubunit E3 ubiquitin ligases [7].
U-box type E3 ubiquitin ligases are widespread in eukaryotes as members of the monomeric group [8]. The 75 amino acid U-box domain of U-box E3 ligase was first found in yeast [9]. In recent years, a growing quantity of plants has been found to contain U-box (PUB) proteins. For example, the PUB gene members are 59 in sorghum [10], 62 in tomato [11], 62 in white pear [12], 65 in cabbage [13], 77 in rice [14], 91 in banana [15], 99 in kale [16], and 121 in moso bamboo [17]. To date, a large number of studies have been conducted to explore the various functions of the U-box gene. In Arabidopsis, AtPUB18 and AtPUB19 mutants are more salt-sensitive than the wild type during seed germination, while AtPUB18 and AtPUB19 negatively inhibited the ABA-mediated drought stress response [18]. In the apple, the U-box E3 ubiquitin ligase MdPUB23 reduces cold stress tolerance by degrading the cold stress regulatory protein MdICE1 [19]. Additionally, it was demonstrated that MdPUB24 is ethylene-activated and promotes the breakdown of apple chlorophyll [20]. In the strawberry, FaU-box98 was comprehensively involved in the resistance to ABA, low temperatures, and salt stress. FaU-box98 and FaU-box136 were significantly and positively correlated with fruit ripening, while FaU-box52 was negatively correlated [21]. Furthermore, it has been shown that the peach PUB genes are connected to anxin and ABA [22]. These results suggest that PUB genes play a critical role in various biological processes, such as fruit ripening and stress responses [23]. However, studies on PUB genes in alfalfa are still limited and require further investigation.
Worldwide, alfalfa (Medicago sativa L.) is a highly valued feed crop that is abundantly grown. Although it has ecological and economic importance, a variety of factors frequently affect its yield and quality. Therefore, understanding the genetic traits of drought, cold, and salt tolerance in alfalfa is crucial for agricultural development. This study conducted a comprehensive genome-wide analysis of the PUB gene family in alfalfa, including phylogenetic analysis, conserved motifs, chromosomal localization, cis-acting elements, and protein–protein interactions. Additionally, the study explored the responses of alfalfa PUB genes under different abiotic stress conditions. These findings offer valuable insights into the role of PUB genes in enhancing alfalfa’s stress resilience and provide a foundation for further functional studies of U-box E3 ubiquitin ligase genes in alfalfa.
2. Results
2.1. Genomic Characterization of Members of the PUB Gene Family in Alfalfa
From the alfalfa genome, 210 MsPUB genes were successfully extracted in accordance with the amino acid sequence of the alfalfa PUB gene family. Based on where their chromosomes were located, they were given the names MsPUBS001 through MsPUBS210. Table S1 displays the genomic details of these MsPUB genes, such as their names, chromosomal locations, intron counts, and protein lengths (amino acid residues). The longest proteins among these genes are encoded by MsPUBS003, MsPUBS004, MsPUBS012, and MsPUBS013; the shortest proteins are encoded by MsPUBS123, MsPUBS126, and MsPUBS208, with 228 amino acid residues. In addition, MsPUBS206 and MsPUBS207 had the greatest number of introns (17). Interestingly, they are both members of group VI.
2.2. Phylogenetic Analysis of the MsPUBs in Alfalfa
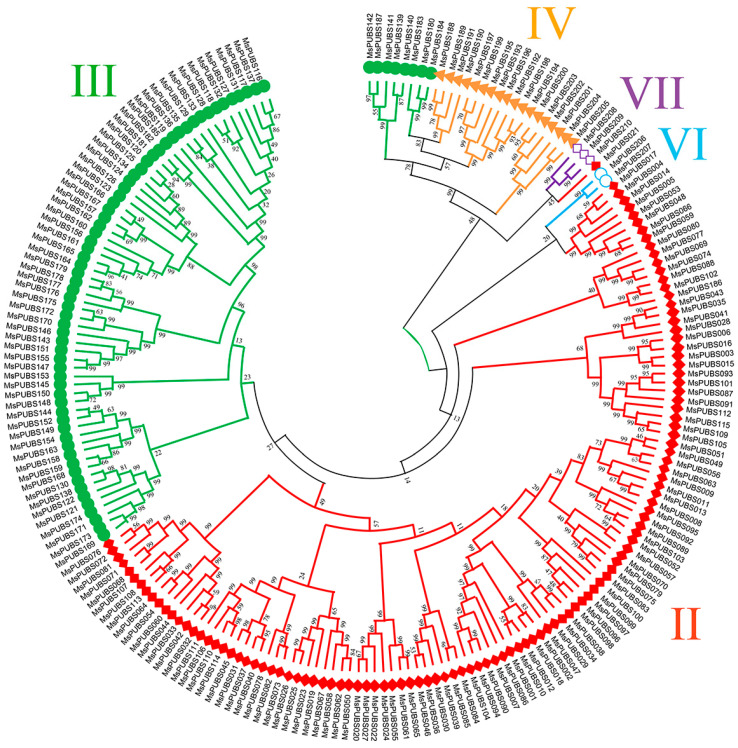
A phylogenetic tree was constructed using the neighbor-joining (NJ) method to reveal the evolutionary relationships among MsPUB genes (Figure 1). Based on the tree topology and consistency with the AtPUB classification, the 210 MsPUB gene family members in alfalfa were divided into five major groups: group II, group III, group IV, group VI, and group VII, as shown in Figure 1. Specifically, group II contained the largest number of members, with 115 members, followed by groups III and IV with 72 and 18 members. In addition, group VI has the smallest number of members, with only two members.
Figure 1.
Phylogenetic relationships between MsPUB genes. Sequence comparison of 210 MsPUB proteins. Using 1000 bootstrap iterations, the neighbor-joining (NJ) method was used to construct the phylogenetic tree. The five groups of members of the MsPUB gene family are II, III, IV, VI, and VII. Five distinct colors are used to point out the genes in each group.
2.3. Conserved Motif Analysis of the MsPUB Gene Family
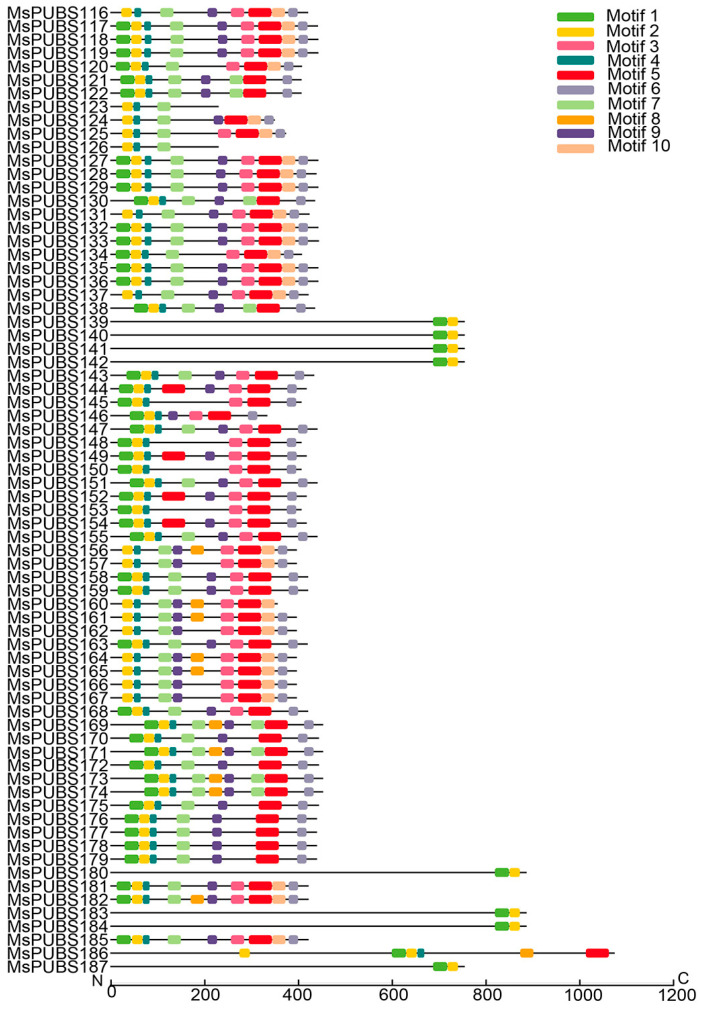
Using the MEME tool (Multiple EM for motif elicitation, version 4.8.1), the conserved motifs and gene structure of the alfalfa PUB genes were examined. A total of 10 individual motifs were found in different regions of MsPUBs. The findings demonstrated that there were similar conserved motifs in different subfamilies (Figure S1). Detailed information of the 10 conserved motifs can be found in Figure S2. All members of the MsPUB genes in group III contain motif 2, and most contain motifs 1, 4, 5, 6, 7 and 9 (Figure 2). Most of the members in group IV contain three motifs, and the members in group VI contain motif 2 and motif 9; the MsPUB genes in group VII all contain motif 1. The composition of the conserved motifs in MsPUB genes showed that there were obvious motif differences among the five groups, which supported the results of the phylogenetic tree analysis.
Figure 2.
Distribution of conserved motifs in group III of the MsPUB genes in alfalfa.
2.4. Chromosomal Localization Analysis of the MsPUB Genes
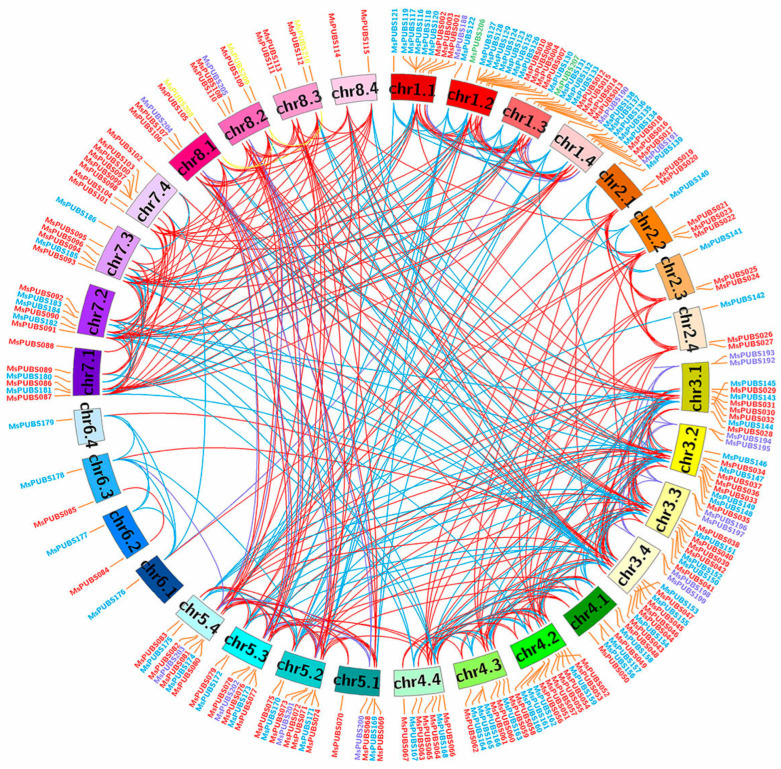
The localization map of MsPUB genes on chromosomes was constructed using the MCScanX (version python) and Circos software (version 0.69), as shown in Figure 3. Out of the 210 MsPUB genes, 209 were successfully mapped onto 32 chromosomes of alfalfa, displaying an uneven distribution. With 18 genes altogether, chr1.2 had the greatest number of MsPUB genes. Chr6.1 and chr6.4 had the lowest number of MsPUB genes, each with only 1 MsPUB gene. The MCScanX program discovered 466 gene duplication occurrences in the alfalfa PUB gene family based on the BLAST results. The findings show that the increase in the MsPUB gene family in the alfalfa genome is largely due to these gene duplication events. The MsPUB gene family has obtained an increased number of members owing to the gene duplication event.
Figure 3.
Distribution and replication of MsPUB genes. The 210 MsPUB genes are located in a ring formed by the 32 chromosomes (chr1.1–chr8.4) of alfalfa. Gene interactions are represented by the colors inside the circles.
2.5. Cis-Acting Elements Analysis of the MsPUB Promoter
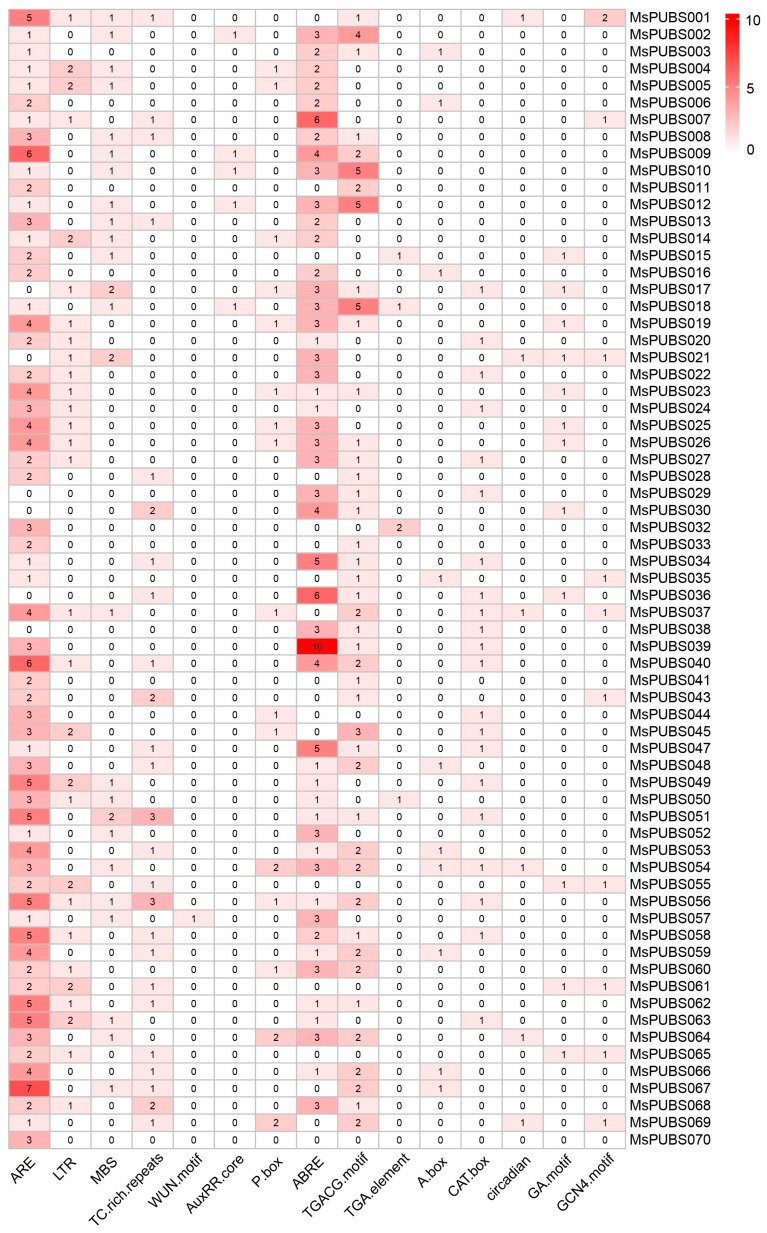
Using the PlantCARE database, cis-acting elements in the promoter region of MsPUB genes were investigated in order to learn more about their function. After identifying 83 different types of cis-acting elements, based on the correlation between cis-elements and known regulatory functions, including plant growth and development, hormone response, and abiotic and biotic stress, 15 cis-acting elements were selected for further analysis [24]. The cis-acting elements of all members of the MsPUB gene family are shown in Figure S3. The cis-acting elements in the promoter regions of the first 70 Ms PUB genes are shown in Figure 4. In the category of growth and developmental elements, we identified the light-responsive regulatory element A-box, the circadian regulatory element, the meristematic tissue expression element CAT-box, the endosperm expression element GCN4 motif, and the light-responsive element involved in GA motif. Furthermore, the plant response to biotic stress is mediated by ARE, TC-rich repeats, and the WUN motif, which are found in a total of 482,104 and 11 cis-acting elements, respectively. There were 100 low-temperature-responsive cis-element LTRs and 123 drought-induced element MBSs among the abiotic stress-response elements. Among the hormone-response elements, there were 393 abscisic acid-related elements (ABRE) and 508 MeJA-related elements (255 TGACG motifs, 253 CGTCA motifs), as well as 76 gibberellin-responsive P-box motifs, 32 growth hormone-related AuxRR-core elements, and 14 TGA elements. The results presented reveal that MsPUB genes might be actively involved in regulating the responses of phytohormones and different stressors.
Figure 4.
Analysis of cis-acting elements in the promoter regions of the top 70 MsPUB genes. The promoter region of MsPUB genes contains fifteen cis-acting elements; the more cis-acting elements, the darker the color. The ARE, LTR, MBS, TC-rich repeats, and the WUN motif are related to biotic and abiotic stresses; phytohormone responsiveness is associated with the AuxRR-core, P-box, ABRE, TGACG motif, and TGA element. Plant growth and development are associated with the A-box, CAT-box, circadian, GA motif, and GCN4 motif.
2.6. Genetic Regulatory Network Analysis of MsPUB Genes
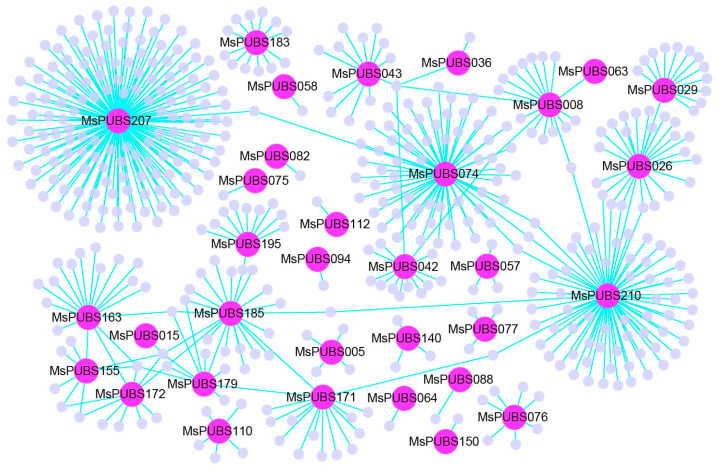
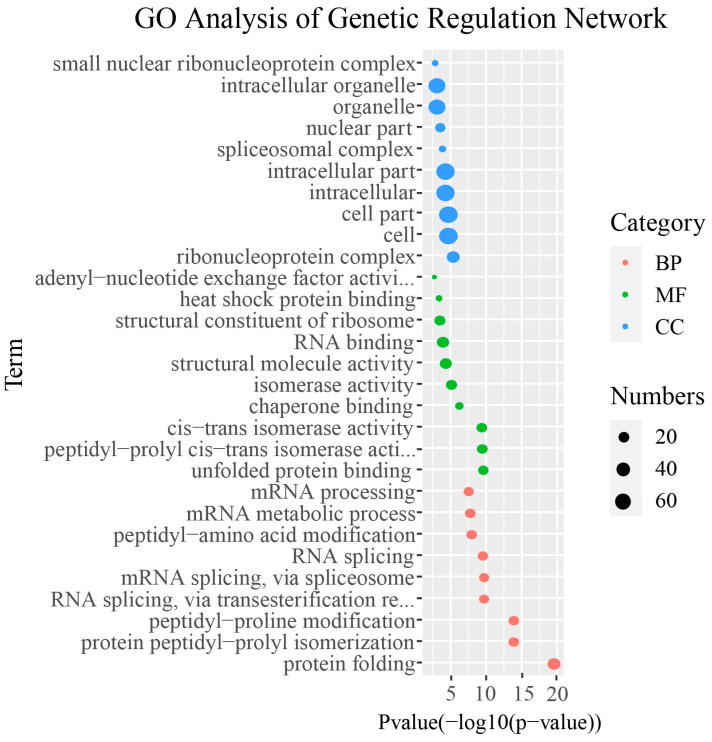
The gene regulatory network (GRN) is a complex network system composed of a series of genes and their regulatory factors. As shown in Figure 5, the GRNs of MsPUBs and their interacting genes contain a total of 493 genes and 498 interactions. From the GRN, the majority of MsPUBs were discovered to interact with many functional genes. For example, MsPUBS207 interacts with 149 genes, MsPUBS210 interacts with 72 genes, MsPUBS074 interacts with 65 genes, and MsPUBS026 interacts with 24 genes. GO enrichment analysis was performed using R software (version 4.3.3) with the topGO program package (version 2.38.31). In Figure 6, a large number of MsPUB genes are distributed within cells and organelles. In addition, these functional genes are mainly focused on regulating ribosomal protein assembly and ribosomal RNA splicing.
Figure 5.
Analysis of the interactions between MsPUB genes in alfalfa using gene regulatory networks. The gene regulatory network (GRN) of MsPUB genes and their interactions was built using Cytoscape and was based on relationships between genes in Arabidopsis thaliana. The cyan line shows the interaction of alfalfa, the pink nodes relate to MsPUB genes, and the purple nodes correspond to genes interacting with MsPUB genes.
Figure 6.
A review of the genes that interact with MsPUB genes and their gene ontology enrichment. The GO terms for molecular functions (MFs), cellular components (CCs), and biological processes (BPs) are shown as red, green, and blue dots, respectively. The GO term is displayed on the Y-axis, while the X-axis displays the p-value of the topGO enrichment analysis with −log 10 transformation or −log 10 (p). The number of genes involved in the GO keywords is reflected in the size of the circles.
2.7. Expression Analysis of Alfalfa MsPUBs Under Abiotic Stresses
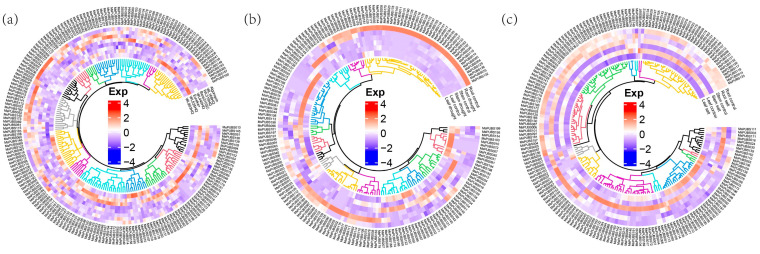
To learn more about how MsPUB responds to abiotic stressors through engagement, the RNA-seq of alfalfa under different stresses was downloaded for further expression analysis. In Figure 7a, under cold stress treatment, the expression levels of MsPUBS023, MsPUBS113, MsPUBS185, and MsPUBS205 were upregulated. MsPUB147 exhibited the highest expression levels, indicating a stronger response to cold stress compared to other genes. In Figure 7b, MsPUB gene expression was upregulated in the tissues of the roots, stems, and leaves when the drought stress treatment was applied; the expression level in the roots was remarkably higher. For instance, this was the case in the MsPUBS022, MsPUBS036, and MsPUBS090 genes. In addition, MsPUBS172 showed the highest expression under salt stress, suggesting it is more responsive to salt stress than the other genes. In Figure 7c, when the salt stress treatment was applied to the root, stem, and leaves, the majority of MsPUB genes had elevated expression levels, such as MsPUBS032, MsPUBS059, and so on. Furthermore, MsPUB183 also had the highest expression under salt stress, indicating its greater responsiveness to salt stress relative to the other genes. These results demonstrated the important contribution of the MsPUB gene family to abiotic stress response.
Figure 7.
Expression profiles of MsPUB under stress conditions. (a) Eight varieties of alfalfa were analyzed under cold stress. (b) Under drought stress, the performance of Wilson and PI467895 alfalfa was studied in three tissues of the root, stem, and leaf. (c) Under salt stress, the performance of Wilson and PI467895 alfalfa in three tissues of rhizome leaves was investigated. The R platform was used to present the mean expression levels (FPKM values), which were measured using Salmon software (version 0.12.0).
2.8. qRT-PCR Verification of MsPUB Gene Expression Under Abiotic Stress
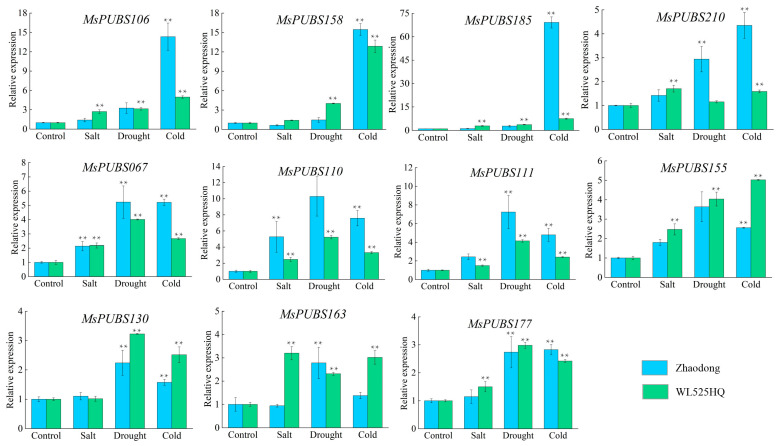
Eleven MsPUB genes were chosen for quantitative reverse transcription–polymerase chain reaction (qRT-PCR) analyses in order to confirm the genes’ quick sensitivity to abiotic stress. GAPDH was used as an internal reference, and each experiment was repeated three times (Table S2). The results showed that the expression levels of 11 MsPUB genes were significantly changed under cold stress, drought stress, and salt stress, as shown in Figure 8. Among them, the transcript levels of MsPUBS106, MsPUBS158, MsPUBS185, and MsPUBS210 showed a significant upward trend after cold stress treatments and were more obvious in the Zhaodong variety. In addition, MsPUBS177 and MsPUBS130 were significantly upregulated in expression under drought stress treatment, compared with other abiotic stresses and the control. It is significant that in conditions of cold, drought, and salt stress, MsPUBS155, MsPUBS110, MsPUBS111, and MsPUBS067 were considerably upregulated. These four MsPUB genes have a broad spectrum in the abiotic stress response of alfalfa. Overall, MsPUB genes were clearly engaged in alfalfa’s response to abiotic stressors, as shown by the generally similar RNA-seq and qRT-PCR data analysis results.
Figure 8.
qRT-PCR analysis of MsPUBs under abiotic stress. qRT-PCR analysis of MsPUBs under salt, drought, and cold stresses. Eleven abiotic stress-responsive genes were selected for qRT-PCR experiments. The X-axis represents control, salt treatment, drought treatment, and cold treatment. The Y-axis represents the relative expression level of MsPUB genes. The relative expression was calculated using the 2−ΔΔCT method, with the expression level of the “control” set to 1. GAPDH was used as the internal control. The blue color represents Zhaodong and the green color represents WL525HQ. Statistical significance was determined using the t-test (** p < 0.01). Asterisks indicate significant differences.
3. Discussion
Within the E3 ubiquitin ligase family, plants include a substantial amount of U-box genes [25]. So far, extensive research has been carried out on the traits and roles of the PUB gene family for several plant species, such as cabbage, rice, and white pears [12,13,14]. In wheat, TaPUB1 increases the plant’s resilience to salt and drought, whereas TaPUB4 influences the anther’s metabolism of sucrose and starch, which in turn controls the formation of pollen [26,27]. Alfalfa, as an important perennial herbaceous plant, has both economic and ecological values [28]. In this investigation, we discovered 210 PUB genes in the genome of alfalfa (Table S1). The phylogenetic analysis of MsPUB genes was conducted using the Arabidopsis PUB gene family as a reference. It was visible from the phylogenetic tree analysis that the 210 MsPUBs were divided into five categories (Figure 1). The group I PUB genes present in the Arabidopsis U-box gene family are missing in alfalfa, which may be due to the fact that Arabidopsis only has one member in the group I PUB gene family. During the evolutionary divergence of the PUB gene families between Arabidopsis and alfalfa, this specific gene group may have been lost or altered in alfalfa. Alfalfa may have compensated for this loss through other PUB genes or alternative mechanisms, potentially resulting in different stress regulation pathways. Furthermore, the development of this family inside the alfalfa genome has been shown by the significantly higher number of PUB genes in the genome compared to the banana [15] and pear [12], indicating the expansion of this family in the alfalfa genome. The MsPUB gene family contained 466 repeated occurrences, according to the results of the collinearity analysis (Figure 3). This duplication appears to have resulted from a recent genome-wide duplication event in the alfalfa genome, revealing a non-random conserved duplication pattern that is comparatively frequent among species [29]. We conducted further analysis on the types of gene duplication and found that duplication events in the PUB gene family in alfalfa are primarily segmental duplications. Segmental duplications typically involve multiple genes, which can significantly increase the number of gene family members, consistent with our observation of 210 MsPUB gene family members (Table S1). Because these segmental duplication gene copies are distributed across different genomic regions, they are likely influenced by distinct regulatory elements, gradually leading to functional diversification and adaptation to various environmental pressures [30]. Therefore, we propose that this duplication pattern may have driven the adaptive evolution of the PUB gene family in alfalfa, providing an important genetic foundation for environmental resilience and enhanced survival.
Many U-box genes influence drought tolerance in plants through diverse mechanisms, according to previous studies. For instance, OsPUB67, AtPUB11, AtPUB18, and AtPUB19 are ABA-dependently engaged in drought tolerance [18,31,32]. GmPUB6 overexpressing plants showed significantly upregulated drought-responsive genes, suggesting that GmPUB6 mediates ABA signaling pathways and osmotic stress to improve plant drought tolerance [33]. We also found a large number of abiotic response elements in the alfalfa PUB promoter region, particularly those associated with abiotic stress, ABA, JA, and GA responses (Figure 4). In the identification of hormone-related elements, the ABRE element is associated with the abscisic acid (ABA) signaling pathway, the P-box element is linked to the gibberellin (GA) signaling pathway, and the AuxRR-core element is involved in the auxin signaling pathway [34]. These three elements primarily regulate plant defense responses, especially in response to abiotic stresses such as water stress, drought, and salt stress. This suggests that MsPUB genes may respond to abiotic stresses by modulating hormonal signaling pathways. We speculate that the PUB genes may induce alfalfa hormone signal transduction in response to abiotic stress. Additionally, GO analysis shows that MsPUB genes are primarily associated with the regulation of ribosomal protein assembly and ribosomal RNA splicing. These functions may be linked to the ability of U-box type E3 ubiquitin ligases to respond to abiotic stress through protein degradation pathways mediated by ubiquitination (Figure 6).
Abiotic stressors like cold, salinity, and drought are harmful for plant growth and development, which reduces global food yield significantly [12]. In order to survive and keep developing, plants have developed complex systems to react quickly to changes in their surroundings. While AtPUB25 and AtPUB26 are involved in the reaction of plants to low temperatures, AtPUB22 and AtPUB23 are negative regulators that mediate the response to drought [35]. As a consequence of the study’s findings, the majority of genes, including MsPUBS023, MsPUBS113, MsPUBS185, and MsPUBS205, had their expression levels elevated under cold stress. Certain MsPUB genes exhibit stronger expression under specific stress conditions. For instance, MsPUBS147 showed the highest expression under cold stress, suggesting that it may play a key role in cold stress response. Similarly, MsPUBS172 exhibited higher expression under salt stress, while MsPUBS183 showed elevated expression under drought stress, indicating that these genes may be more sensitive to salt or drought stress compared to others. These differential expression patterns suggest that these MsPUB genes could serve as potential markers for specific stress responses, such as cold or salt tolerance, and may contribute to the development of stress-resistant varieties. However, further functional studies are needed to confirm their roles in stress tolerance mechanisms (Figure 7). At the same time, a cis-acting element LTR implicated in low temperature responsiveness exists in the promoter region of the MsPUB genes (Figure 4). In addition, the expression levels of MsPUBS022, MsPUBS036, and MsPUBS090 were significantly higher in roots compared to stems and leaves, suggesting that these genes have a tissue-specific function, likely tied to processes that are particularly important in root development or response to environmental stress. Such tissue-specific expression patterns are common for genes involved in stress resistance, nutrient uptake, or developmental pathways, with roots playing a key role in sensing and adapting to soil conditions like drought, salinity, or pathogen presence. It also aligns with the earlier outcomes. During the early developmental stages of banana fruit tissues, the MaU-box gene family was the most expressed in roots [12]. Similarly, in foxtail millet, the expression of SiPUB genes varied significantly across different tissues, with the majority showing high expression levels in the roots. These observations suggest that PUB genes play a crucial role in root development as well as other growth and developmental processes in crops [36]. The high responsiveness of the PUB genes to abiotic stress in alfalfa was further verified by qRT-PCR (Figure 8). In conclusion, our systematic identification of the alfalfa PUB gene family laid the groundwork for next studies aimed at functionally validating the PUB gene family.
4. Materials and Methods
4.1. Genome Identifcation of MsPUBs in Alfalfa
The Pfam database was used to retrieve the Hidden Markov Model (HMM) file (PF04564) corresponding to the U-box domain so as to identify the members of the alfalfa PUB gene family [17]. The members of the alfalfa genome with U-box domains were found using HMMER (version 3.1) The expected (e) cutoff of 0.01 was used to identify and validate the domains of the PUB proteins. The alfalfa genome was searched using the Arabidopsis U-box protein (PUB) sequence as a BLAST query sequence, with an E-value threshold of 1 × 10−5 and coverage set at 80%. The PUB sequence was downloaded from the TAIR database (https://www.arabidopsis.org) (accessed on 15 January 2024) [37]. All possible PUB genes were identified by their genomic location, length of protein, and number of introns, which were retrieved from the alfalfa genome. Subsequently, based on how similar these MsPUB genes were to Arabidopsis PUB genes, they were divided into a number of categories [9].
4.2. Phylogenetic Analysis of MsPUBs in Alfalfa
For the evolutionary analysis of MsPUB genes, a number of alignments of putative alfalfa PUB protein sequences were performed using MUSCLE (version 5.1.0)with default settings [38]. Phylogenetic tree construction was conducted using the neighbor-joining (NJ) approach with 1000 bootstrap repetitions and MEGA 11.0 software [39].
4.3. Analysis of the Distribution of Motif Composition in MsPUB Proteins in Alfalfa
The MEME program was used to search for conserved motifs in the sequences of alfalfa PUB proteins, and the following settings were used to look for conserved motifs in MsPUB protein sequencing: up to ten motifs; arbitrary number of repetitions of a single motif; and minimum and maximum widths of six and fifty [40]. TBtools (version 2.102) is used to present all results [41].
4.4. Chromosomal Localization Analysis of MsPUBs in Alfalfa
Using BLASTP software (version 2.9.0+), all alfalfa proteins were compared to one another. To learn more about the specific locations of the chromosomes that make up the PUB gene family, the genome file’s alfalfa annotation was utilized. Based on this information gene, duplicates were identified and characterized through MCSanX software (Version python) using default parameters [42]. The chromosomal localization of PUB genes in alfalfa was mapped by Circos and the TBtools software (version 2.102) and revealed the syntenic relationships of MsPUB genes [43].
4.5. Cis-Acting Element Analysis of MsPUBs in Alfalfa
The 2000 bp genomic sequence upstream of the start codon of the MsPUB genes was taken from the genomic sequence in order to study cis-acting elements in the promoter of the MsPUB genes. Utilizing the PlantCARE database, cis-acting elements were analyzed [44].
4.6. Analysis of the Gene Regulatory Network of the MsPUB Gene Family in Alfalfa
The AraNet database (V2) provided information on the Arabidopsis gene regulatory network (GRN) [45]. All proteins of alfalfa and Arabidopsis were retrieved by BLAST to establish homologous relationships. The homologs of Arabidopsis or alfalfa genes were identified among the hits with the highest scores, which were obtained by performing these searches at an E-value threshold of 1 × 10−5. Based on the two BLAST results, the procedure produced homologous gene pairs. After that, an alfalfa-like GRN was created utilizing the Arabidopsis GRN and the data from these homologous gene pairs. Using Cytoscape software (Version 3.9.1), the MsPUB gene-containing alfalfa subnetwork was located and investigated and the results were displayed [46]. Using topGO (Version 2.38.31), gene ontology (GO) analysis of enrichment was carried out on the subnetwork, with an acceptable threshold of 0.05, which identified extremely rich phrases using GRN (from the AraNet database (V2) functions and showed representations of the most significant terms, as the software protocol specifies.
4.7. Analysis of MsPUBs Gene Expression Using RNA-Seq Data
The NCBI database provided the alfalfa RNA-seq datasets. RNA-seq data were used to examine the responses of eight alfalfa cultivars to cold stress (accession number: PRJNA780579) [47] and three different tissues of Wilson and PI467895 under drought stress and salt stress response (accession number: PRJNA667169) [48]. Salmon software (version 0.12.0) was utilized to compute the expression levels of every gene, and the transcript sequences of the alfalfa genome were compared with the RNA-seq data [49]. R software (version 4.3.3) was used to cluster and display the data [47].
4.8. Plant Materials and Stress Treatments
Alfalfa seeds from Zhaodong and WL525HQ were germinated and then transplanted into a mixture of perlite and sand in a 3:1 volume ratio [50]. The growth temperature was 24 °C, the illumination time was 16 h, and the darkness was 8 h. Irrigation with Hoagland nutrient solution was performed every 2 days. After 45 days of growth, all seedlings were divided into four groups. For both the salt and drought treatments, two groups of seedlings received irrigations using 20% PEG-6000 and 100 mM NaCl nutritional solution. The other group of seedlings was cooled in a refrigerator at 4 °C. The final group of seedlings received regular nutrient solution irrigation without any additional substances, serving as the control. For all four groups, alfalfa leaves were collected after treatments under normal light conditions for 1 h, 3 h, and 4 h. Leaves at three different time points were collected and mixed. After being instantly frozen in liquid nitrogen, every sample was kept at −80 °C in order to facilitate further RNA extraction.
4.9. QRT-PCR Analysis
Total RNA was extracted from alfalfa leaves subjected to control, low-temperature stress (4 °C), drought stress, and salt stress using the RNApure Plant Kit (Tiangen, Beijing, China) according to the manufacturer’s instructions. DNase I from the kit was used to remove genomic DNA during RNA extraction. The integrity of RNA was confirmed by gel electrophoresis, and the RNA was stored at −80 °C until further use. For cDNA synthesis, 1 µg of total RNA was reverse-transcribed using the PrimeScript RT Kit (Toyobo, Shanghai, China) according to the manufacturer’s protocol. The synthesized cDNA was stored at −20 °C for subsequent quantitative real-time PCR (qRT-PCR) analysis. According to the nucleotide sequence of PUB family genes, the Primer 5.0 software was used to design 11 pairs of particular primers (Table S2). SYBR PreMix Ex TaqTMII (TOYOBO, Shanghai, China) was used for qRT-PCR. The qRT-PCR program was set as follows: 95 °C for 2 min, followed by 40 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min. Each experiment was performed in triplicate. A melting curve analysis was conducted at the end of each run to verify the specificity of the amplification. The corresponding expression level of MsPUBs was computed using the 2−∆∆CT technique [51].
5. Conclusions
This study found 210 PUB genes in alfalfa and performed several analyses on them, including GO annotations, qRT-PCR assays, cis-acting elements, chromosomal localization, gene expression analyses, and phylogenetic and motif composition. We discovered that there were gene duplication events across the evolution of the PUB gene family. Meanwhile, ABRE, P-box, TGACG motif elements, MBS, LTR, and other abiotic stress and parts of the hormone response are abundantly present in the promoter regions of MsPUBs. In alfalfa, MsPUB genes showed a strong reaction to abiotic stress, suggesting that MsPUB genes play a vital role in regulating how plants respond to various kinds of abiotic challenges. The findings of this work serve as a foundation for more research into the role of the PUB gene family in alfalfa under abiotic stressors.
Acknowledgments
We are grateful to the high-performance computing center of Harbin Normal University for the support with our analysis works. We are also grateful to Manman Li and Tianxiang Zhang for their technical assistance in this research.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms252212324/s1.
Author Contributions
Y.S. and W.H. conceived the study and revised the manuscript; S.L., X.C. and M.G. conducted the experiments; Y.S., S.L., X.C., X.Z., W.H., M.G. and C.G. analyzed the data; Y.S., S.L. and X.C. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
None of the plant materials used in this study included any wild species at risk of extinction. No specific permits were required for sample collection in this study. We complied with relevant institutional, national, and international guidelines and legislation on plant study.
Data Availability Statement
The datasets presented in this study can be found in Supplementary Materials.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This research was funded by the Natural Science Foundation of Heilongjiang Province (grant number LH2022C050), the Open Fund of Yunnan Province Flower Breeding Key Laboratory (FKL-202203); the Agriculture Joint Special Project of Science and Technology Plan Project of Yunnan Science and Technology Department (202301BD070001-208), the Green Food Brand Build a Special Project (Floriculture) supported by the Yunnan Provincial Finance Department (530000210000000013742), and the Natural and Science Foundation of China (grant number U21A20182).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Lim C.J., Ali A., Park J., Shen M., Park K.S., Baek D., Yun D.-J. HOS15-PWR chromatin remodeling complex positively regulates cold stress in Arabidopsis. Plant Signal. Behav. 2021;16:1893978. doi: 10.1080/15592324.2021.1893978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Saharin R., Hellmann H., Mooney S. Plant E3 Ligases and Their Role in Abiotic Stress Response. Cells. 2022;11:890. doi: 10.3390/cells11050890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fu X., Tang X., Liu W., Ghimire S., Zhang H., Zhang N., Si H. Ubiquitination in plant biotic and abiotic stress. Plant Growth Regul. 2023;103:33–50. doi: 10.1007/s10725-023-01095-w. [DOI] [Google Scholar]
- 4.Wang S., Lv X., Zhang J., Chen D., Chen S., Fan G., Ma C., Wang Y. Roles of E3 Ubiquitin Ligases in Plant Responses to Abiotic Stresses. Int. J. Mol. Sci. 2022;23:2308. doi: 10.3390/ijms23042308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coll-Martínez B., Crosas B. How the 26S Proteasome Degrades Ubiquitinated Proteins in the Cell. Biomolecules. 2019;9:395. doi: 10.3390/biom9090395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smalle J., Vierstra R.D. The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 2004;55:555–590. doi: 10.1146/annurev.arplant.55.031903.141801. [DOI] [PubMed] [Google Scholar]
- 7.Toma-Fukai S., Shimizu T. Structural Diversity of Ubiquitin E3 Ligase. Molecules. 2021;26:6682. doi: 10.3390/molecules26216682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shu K., Yang W. E3 Ubiquitin Ligases: Ubiquitous Actors in Plant Development and Abiotic Stress Responses. Plant Cell Physiol. 2017;58:1461–1476. doi: 10.1093/pcp/pcx071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Azevedo C., Santos-Rosa M.J., Shirasu K. The U-box protein family in plants. Trends Plant Sci. 2001;6:354–358. doi: 10.1016/S1360-1385(01)01960-4. [DOI] [PubMed] [Google Scholar]
- 10.Cui J., Ren G., Bai Y., Gao Y., Yang P., Chang J. Genome-wide identification and expression analysis of the U-box E3 ubiquitin ligase gene family related to salt tolerance in sorghum (Sorghum bicolor L.) Front. Plant Sci. 2023;14:1141617. doi: 10.3389/fpls.2023.1141617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma B., Taganna J. Genome-wide analysis of the U-box E3 ubiquitin ligase enzyme gene family in tomato. Sci. Rep. 2020;10:9581. doi: 10.1038/s41598-020-66553-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang C., Song B., Dai Y., Zhang S., Huang X. Genome-wide identification and functional analysis of U-box E3 ubiquitin ligases gene family related to drought stress response in Chinese white pear (Pyrus bretschneideri) BMC Plant Biol. 2021;21:235. doi: 10.1186/s12870-021-03024-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang P., Zhu L., Li Z., Cheng M., Chen X., Wang A., Wang C., Zhang X. Genome-Wide Identification of the U-Box E3 Ubiquitin Ligase Gene Family in Cabbage (Brassica oleracea var. capitata) and Its Expression Analysis in Response to Cold Stress and Pathogen Infection. Plants. 2023;12:1437. doi: 10.3390/plants12071437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim M.-S., Kang K.-K., Cho Y.-G. Molecular and Functional Analysis of U-box E3 Ubiquitin Ligase Gene Family in Rice (Oryza sativa) Int. J. Mol. Sci. 2021;22:12088. doi: 10.3390/ijms222112088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu H., Dong C., Sun D., Hu Y., Xie J. Genome-Wide Identification and Analysis of U-Box E3 Ubiquitin–Protein Ligase Gene Family in Banana. Int. J. Mol. Sci. 2018;19:3874. doi: 10.3390/ijms19123874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu D., Xie Q., Liu Q., Zuo T., Zhang H., Zhang Y., Lian X., Zhu L. Genome-Wide Distribution, Expression and Function Analysis of the U-Box Gene Family in Brassica oleracea L. Genes. 2019;10:1000. doi: 10.3390/genes10121000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou J., Hu Y., Li J., Yu Z., Guo Q. Genome-Wide Identification and Expression Analysis of the Plant U-Box Protein Gene Family in Phyllostachys edulis. Front. Genet. 2021;12:710113. doi: 10.3389/fgene.2021.710113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bergler J., Hoth S. Plant U-box armadillo repeat proteins AtPUB18 and AtPUB19 are involved in salt inhibition of germination in Arabidopsis. Plant Biol. 2011;13:725–730. doi: 10.1111/j.1438-8677.2010.00431.x. [DOI] [PubMed] [Google Scholar]
- 19.Wang D.-R., Zhang X.-W., Xu R.-R., Wang G.-L., You C.-X., An J.-P. Apple U-box-type E3 ubiquitin ligase MdPUB23 reduces cold-stress tolerance by degrading the cold-stress regulatory protein MdICE1. Hortic. Res. 2022;9:uhac171. doi: 10.1093/hr/uhac171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei Y., Jin J., Xu Y., Liu W., Yang G., Bu H., Li T., Wang A. Ethylene-activated MdPUB24 mediates ubiquitination of MdBEL7 to promote chlorophyll degradation in apple fruit. Plant J. 2021;108:169–182. doi: 10.1111/tpj.15432. [DOI] [PubMed] [Google Scholar]
- 21.Jiang L., Lin Y., Wang L., Peng Y., Yang M., Jiang Y., Hou G., Liu X., Li M., Zhang Y., et al. Genome-wide identification and expression profiling reveal the regulatory role of U-box E3 ubiquitin ligase genes in strawberry fruit ripening and abiotic stresses resistance. Front. Plant Sci. 2023;14:1171056. doi: 10.3389/fpls.2023.1171056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan B., Lian X., Cheng J., Zeng W., Zheng X., Wang W., Ye X., Li J., Li Z., Zhang L., et al. Genome-wide identification and transcriptome profiling reveal that E3 ubiquitin ligase genes relevant to ethylene, auxin and abscisic acid are differentially expressed in the fruits of melting flesh and stony hard peach varieties. BMC Genom. 2019;20:892. doi: 10.1186/s12864-019-6258-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye Q., Wang H., Su T., Wu W.H., Chen Y.F. The Ubiquitin E3 Ligase PRU1 Regulates WRKY6 Degradation to Modulate Phosphate Homeostasis in Response to Low-Pi Stress in Arabidopsis. Plant Cell. 2018;30:1062–1076. doi: 10.1105/tpc.17.00845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marand A.P., Eveland A.L., Kaufmann K., Springer N.M. cis-Regulatory Elements in Plant Development, Adaptation, and Evolution. Annu. Rev. Plant Biol. 2023;74:111–137. doi: 10.1146/annurev-arplant-070122-030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su Y., Ngea G.L.N., Wang K., Lu Y., Godana E.A., Ackah M., Yang Q., Zhang H. Deciphering the mechanism of E3 ubiquitin ligases in plant responses to abiotic and biotic stresses and perspectives on PROTACs for crop resistance. Plant Biotechnol. J. 2024;22:2811–2843. doi: 10.1111/pbi.14407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang W., Wang W., Wu Y., Li Q., Zhang G., Shi R., Yang J., Wang Y., Wang W. The involvement of wheat U-box E3 ubiquitin ligase TaPUB1 in salt stress tolerance. J. Integr. Plant Biol. 2020;62:631–651. doi: 10.1111/jipb.12842. [DOI] [PubMed] [Google Scholar]
- 27.Kim J.H., Kim M.S., Seo Y.W. Overexpression of a plant U-box gene TaPUB4 confers drought stress tolerance in Arabidopsis thaliana. Plant Physiol. Biochem. 2023;196:596–607. doi: 10.1016/j.plaphy.2023.02.001. [DOI] [PubMed] [Google Scholar]
- 28.Wang D., Khurshid M., Sun Z.M., Tang Y.X., Zhou M.L., Wu Y.M. Genetic Engineering of Alfalfa (Medicago sativa L.) Protein Pept. Lett. 2016;23:495–502. doi: 10.2174/0929866523666160314152618. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y., Wang X., Tang H., Tan X., Ficklin S.P., Feltus F.A., Paterson A.H. Modes of gene duplication contribute differently to genetic novelty and redundancy, but show parallels across divergent angiosperms. PLoS ONE. 2011;6:e28150. doi: 10.1371/journal.pone.0028150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qiao X., Li Q., Yin H., Qi K., Li L., Wang R., Zhang S., Paterson A.H. Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants. Genome Biol. 2019;20:38. doi: 10.1186/s13059-019-1650-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qin Q., Wang Y., Huang L., Du F., Zhao X., Li Z., Wang W., Fu B. A U-box E3 ubiquitin ligase OsPUB67 is positively involved in drought tolerance in rice. Plant Mol. Biol. 2020;102:89–107. doi: 10.1007/s11103-019-00933-8. [DOI] [PubMed] [Google Scholar]
- 32.Cho S.K., Ryu M.Y., Song C., Kwak J.M., Kim W.T. ArabidopsisPUB22 and PUB23 Are Homologous U-Box E3 Ubiquitin Ligases That Play Combinatory Roles in Response to Drought Stress. Plant Cell. 2008;20:1899–1914. doi: 10.1105/tpc.108.060699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang N., Liu Y., Cai Y., Tang J., Li Y., Gai J. The soybean U-box gene GmPUB6 regulates drought tolerance in Arabidopsis thaliana. Plant Physiol. Biochem. 2020;155:284–296. doi: 10.1016/j.plaphy.2020.07.016. [DOI] [PubMed] [Google Scholar]
- 34.Danquah A., de Zelicourt A., Colcombet J., Hirt H. The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol. Adv. 2014;32:40–52. doi: 10.1016/j.biotechadv.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 35.Seo D.H., Ryu M.Y., Jammes F., Hwang J.H., Turek M., Kang B.G., Kwak J.M., Kim W.T. Roles of four Arabidopsis U-box E3 ubiquitin ligases in negative regulation of abscisic acid-mediated drought stress responses. Plant Physiol. 2012;160:556–568. doi: 10.1104/pp.112.202143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou X., Li Y., Wang J., Zhao Y., Wang H., Han Y., Lin X. Genome-wide identification of U-box gene family and expression analysis in response to saline-alkali stress in foxtail millet (Setaria italica L. Beauv) Front. Genet. 2024;15:1356807. doi: 10.3389/fgene.2024.1356807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edgar R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tamura K., Stecher G., Kumar S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021;38:3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen C., Chen H., Zhang Y., Thomas H.R., Frank M.H., He Y., Xia R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y., Tang H., Debarry J.D., Tan X., Li J., Wang X., Lee T.H., Jin H., Marler B., Guo H., et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lescot M., Déhais P., Thijs G., Marchal K., Moreau Y., Van de Peer Y., Rouzé P., Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee T., Yang S., Kim E., Ko Y., Hwang S., Shin J., Shim J.E., Shim H., Kim H., Kim C., et al. AraNet v2: An improved database of co-functional gene networks for the study of Arabidopsis thaliana and 27 other nonmodel plant species. Nucleic Acids Res. 2015;43:D996–D1002. doi: 10.1093/nar/gku1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang X., Yang H., Li M., Bai Y., Chen C., Guo D., Guo C., Shu Y. A Pan-Transcriptome Analysis Indicates Efficient Downregulation of the FIB Genes Plays a Critical Role in the Response of Alfalfa to Cold Stress. Plants. 2022;11:3148. doi: 10.3390/plants11223148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Medina C.A., Samac D.A., Yu L.X. Pan-transcriptome identifying master genes and regulation network in response to drought and salt stresses in Alfalfa (Medicago sativa L.) Sci. Rep. 2021;11:17203. doi: 10.1038/s41598-021-96712-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang X., Yang H., Li M., Chen C., Bai Y., Guo D., Guo C., Shu Y. Time-course RNA-seq analysis provides an improved understanding of genetic regulation in response to cold stress from white clover (Trifolium repens L.) Biotechnol. Biotechnol. Equip. 2022;36:745–752. doi: 10.1080/13102818.2022.2108339. [DOI] [Google Scholar]
- 51.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in Supplementary Materials.