Abstract
Enterococcus faecalis was tested for the ability to persist in mouse peritoneal macrophages in two separate studies. In the first study, the intracellular survival of serum-passaged E. faecalis 418 and two isogenic mutants [cytolytic strain FA2-2(pAM714) and non-cytolytic strain FA2-2(pAM771)] was compared with that of Escherichia coli DH5α by infecting BALB/c mice intraperitoneally and then monitoring the survival of the bacteria within lavaged peritoneal macrophages over a 72-h period. All E. faecalis isolates were serum passaged to enhance the production of cytolysin. E. faecalis 418, FA2-2(pAM714), and FA2-2(pAM771) survived at a significantly higher level (P = 0.0001) than did E. coli DH5α at 24, 48, and 72 h. Internalized E. faecalis 418, FA2-2(pAM714), and FA2-2(pAM771) decreased 10-, 55-, and 31-fold, respectively, over the 72-h infection period, while internalized E. coli DH5α decreased 20,542-fold. The difference in the rate of survival of E. faecalis strains and E. coli DH5α was most prominent between 6 and 48 h postinfection (P = 0.0001); however, no significant difference in killing was observed between 48 and 72 h postinfection. In the second study, additional E. faecalis strains from clinical sources, including DS16C2, MGH-2, OG1X, and the cytolytic strain FA2-2(pAM714), were compared with the nonpathogenic gram-positive bacterium, Lactococcus lactis K1, for the ability to survive in mouse peritoneal macrophages. In these experiments, the E. faecalis strains and L. lactis K1 were grown in brain heart infusion (BHI) broth to ensure that there were equal quantities of injected bacteria. E. faecalis FA2-2(pAM714), DS16C2, MGH-2, and OG1X survived significantly better (P < 0.0001) than did L. lactis K1 at each time point. L. lactis K1 was rapidly destroyed by the macrophages, and by 24 h postinfection, viable L. lactis could not be recovered. E. faecalis FA2-2(pAM714), DS16C2, MGH-2, and OG1X declined at an equivalent rate over the 72-h infection period, and there was no significant difference in survival or rate of decline among the strains. E. faecalis FA2-2(pAM714), MGH-2, DS16C2, and OG1X exhibited an overall decrease of 25-, 55-, 186-, and 129-fold respectively, between 6 and 72 h postinfection. The overall reduction by 1.3 to 2.27 log units is slightly higher than that seen for serum-passaged E. faecalis strains and may be attributable to the higher level of uptake of serum-passaged E. faecalis than of E. faecalis grown in BHI broth. Electron microscopy of infected macrophages revealed that E. faecalis 418 was present within an intact phagocytic vacuole at 6 h postinfection but that by 24 h the infected macrophages were disorganized, the vacuolar membrane was degraded, and the bacterial cells had entered the cytoplasm. Macrophage destruction occurred by 48 h, and the bacteria were released. In conclusion, the results of these experiments indicate that E. faecalis can persist for an extended period in mouse peritoneal macrophages.
Enterococcus faecalis is a gram-positive, facultatively anaerobic, coccal bacterium that causes a variety of community- and hospital-acquired infections in humans (for reviews, see references 9, 28, 33, and 39), including infections of the blood, endocardium, genitourinary tract, abdomen, wounds, and skin and soft tissue (e.g., burns, decubitus ulcers, and diabetic foot ulcers). The two most life-threatening infections caused by E. faecalis are bacteremia and endocarditis. E. faecalis causes 5 to 8% of all cases of bacteremia and 5 to 20% of all cases of endocarditis (∼55% in intravenous drug users [9, 17, 18, 33, 39, 40, 47]). Bacteremia due to E. faecalis can lead to septicemia, septic shock, and death or, alternatively, to the formation of acute or subacute endocarditis (17). Enterococcal endocarditis is a serious consequence of E. faecalis infection, with a mortality rate of 17 to 46% (37).
E. faecalis is part of the normal flora of the oral cavity and may also be found in the gastrointestinal tract, male urethra, and female vaginal tract of humans (9, 17, 28, 39). Under certain circumstances (in patients with indwelling catheters, intravenous lines, previous antibiotic use, or abscesses), E. faecalis breaches the host defenses by contamination of instrumentation or by direct extension to the bloodstream to cause bacteremia and/or endocarditis (9, 37, 38, 40). In some 42% of nosocomial bloodstream infections, there is no obvious explanation of how E. faecalis gained entry to the blood. To successfully cause infection, the bacterium must overcome the clearance functions of the host immune system. E. faecalis produces several virulence factors, including cytolysin (hemolysin/bacteriocin) (16, 25, 27), aggregation substance (10, 22, 24, 31, 41), gelatinase (protease) (4), and superoxide dismutase (6, 44), which could potentially modify the effectiveness of host defenses. There have been a limited number of studies which have addressed the interaction of E. faecalis or its products with cellular host defenses.
The interaction of E. faecalis with primary macrophages or macrophage-like cell lines has received limited attention (3, 19). Human monocytes respond to E. faecalis lipoteichoic acid by simultaneously synthesizing the inflammatory cytokines tumor necrosis factor alpha, interleukin-6, and interleukin-1β (3). Despite this inflammatory response, the ability of macrophages to eliminate E. faecalis may be inhibited or delayed in vivo. Wells et al. first hypothesized that intestinal bacteria such as E. faecalis can utilize macrophages as a vehicle for translocation across the intestinal epithelial cells to the mesenteric lymph nodes, where the bacteria could be released to proliferate and spread hematogenously to other sites (53). Animal studies support this hypothesis, since antibiotic-induced E. faecalis overgrowth in the intestines of mice led to the subsequent recovery of the bacterium from the mesenteric lymph nodes and livers of infected animals (52).
Wells et al. examined the survival of E. faecalis in mouse peritoneal macrophages infected in vivo following intraperitoneal injection of E. faecalis into mice (51). Their studies revealed that E. faecalis survived within mouse peritoneal macrophages for 2 h and that intracellular survival of E. faecalis over the infection period was comparable to that of the facultative intracellular pathogen Listeria monocytogenes. However, their studies were focused on determining the oral infectivity of various enteric bacteria rather than on the actual ability of E. faecalis to survive within macrophages for an extended period. Therefore, it is difficult to form a conclusion about the susceptibility of E. faecalis to killing by macrophages.
Since survival and sequestration within macrophages may contribute to the pathogenesis of E. faecalis infections and, furthermore, may hinder the efficacy of antimicrobial therapy, this study focused on determining whether E. faecalis isolates survive within mouse peritoneal macrophages for an extended period. Studies in this laboratory indicated that at least six E. faecalis isolates survived for 72 h in mouse peritoneal macrophages and that cytolysin or gelatinase had no effect on intracellular survival in an in vivo-in vitro macrophage infection model.
MATERIALS AND METHODS
Bacterial strains.
E. faecalis 418 was isolated from a culture of Fusobacterium necrophorum ATCC 27852 after infection of a mixed F. necrophorum-E. faecalis culture into mouse peritoneal macrophages resulted in rapid destruction of the F. necrophorum and recovery of the E. faecalis isolate 6 h postinfection. F. necrophorum ATCC 27852 was originally recovered from a sheep with foot rot, and E. faecalis 418 presumably was a fecal contaminant present in the foot rot. E. faecalis 418 was repeatedly recovered from separate vials of F. necrophorum ATCC 27852. E. faecalis FA2-2(pAM714) (24), a cytolysin-positive strain, E. faecalis FA2-2(pAM771) (24), a noncytolytic isogenic mutant, and E. faecalis OG1X (26), a gelatinase mutant, were kindly supplied by Mike Gilmore (University of Oklahoma Health Sciences Center, Oklahoma City, Okla.). Strains FA2-2(pAM714) and FA2-2(pAM771) were assayed for the production of cytolysin by previously published methods (25). Don Clewell (University of Michigan School of Dentistry, Ann Arbor, Mich.) kindly provided E. faecalis DS16C2, a derivative of clinical strain DS16 that contains cytolysin but lacks plasmid pAD2 (15). E. faecalis MGH-2 (clinical isolate, mouse virulent) was kindly supplied by Michael Cohen (Parke-Davis Pharmaceutical Research, Division of Warner-Lamber Co., Ann Arbor, Mich.) (11). These E. faecalis strains were chosen for study since they were human clinical isolates (or derivatives of clinical isolates) and were sensitive to vancomycin (1 to 2 μg/ml) and gentamicin (16 μg/ml) when tested by the broth dilution method as specified by National Committee for Clinical Laboratory Standards guidelines. Lactococcus lactis K1, a nonpathogenic, gram-positive bacterium, served as a negative control and was kindly provided by John Thompson (Oral Infection and Immunity Branch, National Institute of Dental and Craniofacial Research, National Institutes of Health, Bethesda, Md.) (49). Escherichia coli DH5α [F− endA1 hsdR17 (rk−mk+) supE44 λ− recA1 gyrA96 relA1 Δ(argF-lacZYA)U169 φ80dlacZΔM15] was purchased from GIBCO/BRL, Life Technologies, Gaithersburg, Md.), and served as the negative control in these experiments.
Reagents and mice.
Dulbecco’s modified Eagle’s medium (DMEM), penicillin G, vancomycin, Sarkosyl, and DNase I were obtained from Sigma Chemical Co., St. Louis, Mo. Gentamicin, HEPES buffer solution, MEM nonessential amino acids solution, and glutamine were obtained from Life Technologies. Fetal bovine serum was obtained from Summit Biotechnology, Fort Collins, Colo. Nonhemolyzed rabbit serum was supplied by Pel-Freez Biologicals, Rogers, Ark. The rabbit serum was heat inactivated at 56°C for 1 h prior to use. Brain heart infusion (BHI) broth and agar were purchased from Difco, Detroit, Mich. BALB/c mice (10-week-old males) were purchased from Harlan Sprague-Dawley, Indianapolis, Ind.
Assay for survival in mouse peritoneal macrophages.
Experimental methods were based on those established for studying the survival and multiplication of Salmonella typhimurium in macrophages (5, 21, 34). E. faecalis 418, FA2-2(pAM771), and FA2-2(pAM714) were passaged twice by being grown in sterile, nonhemolyzed rabbit serum (heat inactivated at 56°C for 1 h) and E. coli DH5α was grown aerobically at 37°C in Luria-Bertani (LB) broth to an identical density prior to injection into mice. In separate experiments, E. faecalis FA2-2(pAM714), DS16C2, OG1X, and MGH-2 were grown aerobically at 37°C in BHI broth for 16 h. After overnight growth, the bacteria were pelleted in a microcentrifuge at 13,800 χ g for 2 min and the cell pellet was resuspended in an equivalent volume of phosphate-buffered saline (PBS) for injection. E. faecalis strains and E. coli DH5α (107 to 108 CFU) were injected intraperitoneally into 10-week-old BALB/c mice, and following an infection period of 4 h, peritoneal macrophages were harvested by peritoneal lavage (two applications of 5 ml of PBS [pH 7.2] [lacking Ca2+ and Mg2+]). The infected peritoneal macrophages were centrifuged for 10 min at 900 × g (at room temperature) and suspended in DMEM containing 10 mM HEPES, 2 mM glutamine, 10% fetal bovine serum, and 1× nonessential amino acids (this combination is designated “DMEM complete medium” throughout this paper), supplemented with vancomycin (10 μg/ml) and gentamicin (150 μg/ml). The cell suspension was dispensed into 24-well tissue culture plates and incubated at 37°C under 8% CO2. After exposure to antibiotics for 2 h (i.e., 6 h postinfection) at 37°C under 8% CO2 to kill extracellular bacteria, the infected macrophages were washed three times with DMEM containing 10 mM HEPES buffer, and duplicate wells of infected macrophages were lysed with 0.5% Sarkosyl containing 2 μg of DNase per ml. Dilutions of lysates were made in BHI broth and plated on BHI agar to quantitate viable intracellular bacteria. The remaining wells of infected macrophages were maintained in DMEM complete medium containing vancomycin (2 μg/ml) and gentamicin (10 μg/ml) for the duration of the experiment. At 6, 24, 48, and 72 h postinfection, supernatant fluids from each well were removed and extracellular bacteria were quantitated by plating the fluids on BHI agar. Duplicate wells of infected macrophages were lysed with detergent at 24, 48, and 72 h postinfection, and lysates were plated as described above to recover viable bacteria.
Transmission electron microscopy.
Mice were injected intraperitoneally with E. faecalis 418 and E. coli DH5α, and peritoneal macrophages were harvested, plated into 24-well tissue culture plates, and exposed to antibiotics as described above. At 6, 24, 48, and 72 h postinfection, tissue culture medium was removed and cells were overlaid with 1 ml of a fixative solution consisting of 4% formaldehyde, 1% glutaraldehyde, and 0.1% sodium cacodylate (supplied by Paragon Biotech Inc., Baltimore, Md.). The fixed cells contained in the 24-well tissue culture plates were immediately transported to Paragon Biotech for processing and for transmission electron microscopy.
Assessment of macrophage viability.
Infected and noninfected mouse peritoneal macrophages were quantitated at 6, 24, 48, and 72 h to determine whether infection resulted in death of the infected macrophages. The macrophages were detached from tissue culture wells (2 wells) with cell scrapers and mixed with trypan blue dye, and viable macrophages were visualized under an inverted microscope and counted with a hemacytometer.
Statistical analysis of infection data.
All infection experiments described were performed three times and subjected to statistical analysis. The results of these experiments were analyzed by Sean Mahabir, Phillip Chapman, and Jill Smith, Colorado State University Statistics Department, with SAS computer software. Regression lines were fit for each strain for each time segment, and Bonferroni intervals were used to perform pairwise differences between slopes for strains at each time interval. A mixed-model analysis of variance was also performed to test for significance of strain-time interaction. A SAS-PROC MIXED program was used to perform a mixed-model analysis of variance.
RESULTS
Comparative survival of E. faecalis strains, E. coli DH5α, and L. lactis K1 in mouse peritoneal macrophages.
In initial experiments, the intracellular survival of E. faecalis 418, FA2-2(pAM714), and FA2-2(pAM771) was compared with that of E. coli DH5α by infecting mice intraperitoneally, recovering infected macrophages 4 h later, and then monitoring the survival of intracellular bacteria over a period of 72 h within peritoneal macrophages maintained in vitro. In these initial studies, all the E. faecalis isolates were passaged in nonhemolyzed rabbit serum prior to injection, since it was previously reported that growth of E. faecalis in rabbit serum is required for optimum expression of cytolysin. E. coli DH5α was used as a negative control, since this laboratory strain was found to be susceptible to killing by mouse peritoneal macrophages. By necessity, E. coli DH5α was grown in LB broth since it grew poorly in rabbit serum. In preliminary experiments, both E. coli DH5α and E. coli LE392 served as negative controls; however, there was no difference between the survival of the two E. coli strains and therefore only E. coli DH5α was included as the negative control in subsequent studies.
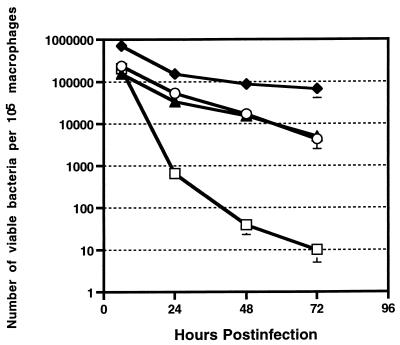
All serum-passaged E. faecalis strains were equal in their ability to survive inside macrophages and exhibited a similar rate of decline over the 72-h period (Fig. 1). There was no significant difference in the levels of the E. faecalis strains and E. coli DH5α at the 6-h time point. However, all E. faecalis strains were recovered at significantly higher levels (P < 0.0001) than E. coli DH5α at the 24-, 48-, and 72-h time points. By the 72-h time point, the E. faecalis strains exhibited a definite superiority in their ability to survive intracellularly, since numbers of internalized E. faecalis 418, FA2-2(pAM714), and FA2-2(pAM771) organisms decreased only 10-, 55-, and 31-fold, respectively, between 6 and 72 h postinfection while the number of E. coli DH5α organisms decreased 20,542-fold. The number of intracellular E. faecalis organisms slowly declined at a rate of 1 to 1.5 log units over the 72-h period. In contrast, the number of viable E. coli DH5α organisms declined more rapidly, with an initial reduction of 2 log units between 6 and 24 h postinfection and a similar reduction between 24 and 48 h postinfection, followed by a rate of decline of approximately 0.5 log unit between 48 and 72 h postinfection. Statistical analysis of the rate of reduction of E. coli DH5α and E. faecalis over the infection period revealed that E. coli DH5α was reduced at a significantly greater (P = 0.0001) rate than were the E. faecalis strains between 6 and 48 h postinfection but that the differences in the rates diminished between 48 and 72 h postinfection. Another common laboratory strains of E. coli, LE392, declined at a rate similar to that of E. coli DH5α (data not shown). The viability of both infected and uninfected mouse peritoneal macrophages decreased approximately 10-fold over the course of the experiment.
FIG. 1.
Recovery of viable E. faecalis and E. coli DH5α from infected murine peritoneal macrophages at 6, 24, 48, and 72 h postinfection. E. faecalis strains were passaged through serum, while E. coli DH5α was grown in LB broth prior to infection. The results represent the mean and standard error of three experiments. In some cases, the standard error is not indicated, because it is too small to be visible on the graph. Symbols: ○, E. faecalis FA2-2(pAM714); ▴, E. faecalis FA2-2(pAM771); □, E. coli DH5α; ⧫, E. faecalis 418.
These initial experiments were extended to include E. faecalis strains from different clinical sources and to include a more relevant negative control—the closely related, innocuous, gram-positive bacterium, L. lactis K1. These experiments were performed by growing E. faecalis FA2-2(pAM714), MGH-2, DS16C2, and OG1X and L. lactis K1 overnight in BHI broth, since L. lactis K1 did not grow at a rate similar to the E. faecalis strains in serum. The bacteria were collected by centrifugation, and a bacterial suspension was prepared in PBS for injection into mice. Macrophage survival was monitored as described above by determining the number of viable intracellular bacteria over the 72-h time course.
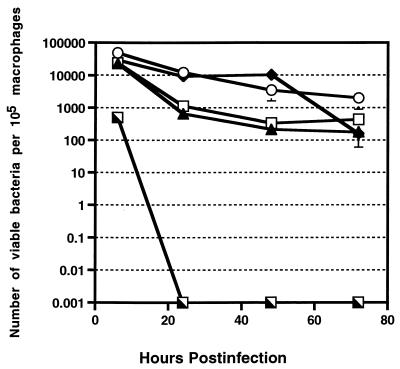
The E. faecalis strains grown in BHI broth [FA2-2(pAM714), MGH-2, DS16C2, and OG1X] showed no significant difference in their ability to survive in mouse peritoneal macrophages and exhibited similar rates of decline over the 72-h infection period (Fig. 2). However, it is interesting that the number of BHI-grown E. faecalis organisms recovered at 6 h postinfection was consistently 10-fold smaller than the number of serum-passaged E. faecalis organisms. E. faecalis FA2-2(pAM714), MGH-2, DS16C2, and OG1X (grown in BHI broth) showed an overall decrease of 25-, 55-, 186-, and 129-fold, respectively, between 6 and 72 h postinfection. The overall reduction of 1.3 to 2.27 log units is slightly higher than that seen for serum-passaged E. faecalis strains. This difference may be attributable to the fact that serum-passaged E. faecalis strains were phagocytosed at a higher rate than were BHI broth-grown strains and that the initial burden to the macrophage was higher with the serum-passaged E. faecalis strains. However, the overall reduction of the BHI broth-grown E. faecalis strains is still significantly smaller than the 4- to 4.5-log-unit reduction in E. coli DH5α over the 72-h infection period described above.
FIG. 2.
Recovery of viable E. faecalis and L. lactis K1 from infected murine peritoneal macrophages at 6, 24, 48, and 72 h postinfection. All E. faecalis strains and L. lactis K1 were grown in BHI broth prior to infection. The results represent the mean and standard error of two experiments. In some cases, the standard error is not indicated, because it is too small to be visible on the graph. Symbols: ○, E. faecalis FA2-2(pAM714); ▴, E. faecalis OG1X; □, E. faecalis MGH-2; ⧫, E. faecalis DS16C2; ┐, L. lactis K1.
All E. faecalis strains were markedly superior to the negative control, L. lactis K1, in their ability to survive in the macrophages. By 6 h postinfection, the level of L. lactis K1 was drastically reduced in the mouse peritoneal macrophages compared with those of the E. faecalis strains (P = 0.0001); it recovered at a 63-fold lower level than the E. faecalis strains. L. lactis K1 succumbed rapidly to the killing effects of the macrophage, and by 24 h postinfection no viable L. lactis K1 was recovered from the infected macrophages.
Electron micrographs of E. faecalis-infected mouse peritoneal macrophages.
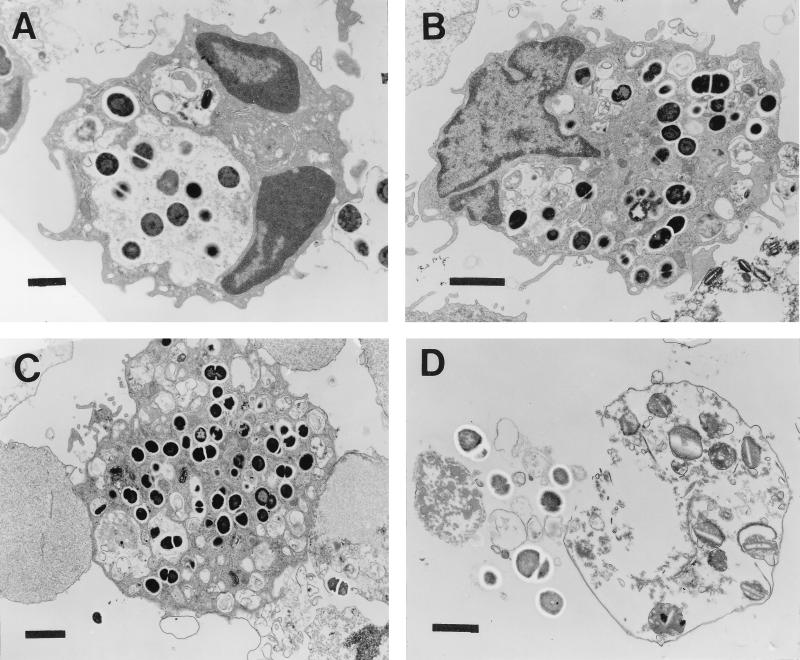
E. faecalis 418-infected macrophages were subjected to transmission electron microscopy at 6, 24, 48, and 72 h postinfection to confirm that intact bacterial cells were present within the macrophage and to determine whether the bacteria were present in a membrane-bound vacuole or in the cytoplasm. Representative electron micrographs of E. faecalis 418-infected macrophages at the different time points are shown in Fig. 3. By 6 h postinfection, E. faecalis bacteria were phagocytosed by the macrophage and were found intact within a phagocytic vacuole. At all time points postinfection, it appeared that dividing, intact enterococci were present within the macrophage, suggesting that some of the phagocytosed bacteria were capable of replication. At 24 h postinfection, the infected macrophages were disorganized, the vacuolar membrane appeared to be degraded in areas, and the bacterial cells were present in the cytoplasm. By 48 h postinfection, macrophages that had ingested numerous E. faecalis bacteria appeared to be disintegrating, whereas the majority of the intracellular E. faecalis isolates appeared to be intact and were surrounded by an electron-transparent halo. At the 72-h time point, infected macrophages were destroyed and both intact and degraded bacteria were present inside the remnants of the macrophages.
FIG. 3.
Electron micrographs of mouse peritoneal macrophages infected with E. faecalis 418. Infected macrophages were processed for transmission electron microscopy (see Materials and Methods) at 6 h (A), 24 h (B), 48 h (C), and 72 h (D) postinfection. Bars, 1 μm (A and D) or 2 μm (B and C).
DISCUSSION
E. faecalis is a significant cause of nosocomial infection, and therefore it must possess the ability to compete at some level with host cellular defense mechanisms in order to survive in tissue and blood. The experiments described above demonstrate that E. faecalis is superior to a closely related bacterium, L. lactis, and a nonpathogenic strain of E. coli in its ability to survive in mouse peritoneal macrophages for 72 h. This finding allows speculation that survival in macrophages contributes to the pathogenicity of this bacterium. It is of interest that virulent strains of Streptococcus bovis, a bacterium that causes septicemia in pigeons, multiply within pigeon peritoneal macrophages whereas avirulent strains are killed by macrophages (13).
Preliminary experiments in this laboratory revealed that E. faecalis 418, a sheep foot rot isolate, possessed the ability to survive in mouse peritoneal macrophages. These experiments were repeated in this study, and the role of cytolysin in macrophage survival was examined since animal studies by other investigators showed that noncytolytic mutants are 10-fold less virulent than wild-type, cytolytic E. faecalis isolates when tested in a mouse peritoneal infection model (27). Mouse peritoneal macrophages were infected in vivo with the cytolytic strain E. faecalis FA2-2(pAM714) and the noncytolytic, isogenic mutant, E. faecalis FA2-2(pAM771). Quantitation of viable, intracellular bacteria in infected macrophages maintained in vitro over 72 h revealed that there was no significant difference between the cytolytic and noncytolytic strains, suggesting that cytolysin does not play a role in intracellular survival. It was initially surprising that E. faecalis cytolysin is not necessary for macrophage survival, since hemolysins of other intracellular pathogens, including Listeria monocytogenes (43, 48) and Shigella flexneri (46), are essential for release of the bacteria into the cytoplasm and intracellular survival. However, it is possible that the E. faecalis cytolysin is different from other latter hemolysins since it is a member of the lantibiotic family, a group of bacteriocins which are active against other gram-positive bacteria (16). Therefore, the E. faecalis cytolysin may not be important in macrophage survival but, as proposed by previous investigators (16), may contribute to the local ecology of infections by eliminating competing bacteria and allowing overgrowth of cytolysin-producing E. faecalis strains. Alternatively, the cytolysin may contribute to pathogenicity by causing localized tissue damage. Another explanation for the lack of an effect of cytolysin in macrophage survival is that cytolysin may not be produced under the conditions of the assay. Studies in this laboratory indicated that cytolysin is not produced by E. faecalis grown in tissue culture media under a carbon dioxide atmosphere. Further experiments involving reverse transcriptase PCR for detection of cytolysin mRNA are necessary to determine whether cytolysin is produced by bacteria residing in the macrophage.
The initial infection studies in mouse peritoneal macrophages were extended to include additional E. faecalis strains from clinical sources [i.e., DS16C2, MGH-2, OG1X, and FA2-2(pAM714)] and a more relevant negative control strain, L. lactis K1. L. lactis K1 was included as a control strain since it is closely related to E. faecalis and has not been associated with human disease. L. lactis K1 failed to grow at the same rate as E. faecalis in rabbit serum; therefore, in subsequent experiments all E. faecalis strains and the L. lactis control was grown in BHI broth prior to infection. The results of macrophage infection with the additional E. faecalis strains essentially mimicked the results of the initial experiments—all the E. faecalis strains survived within the macrophages for the duration of the experiment, and E. faecalis greatly exceeded the ability of the negative control, L. lactis K1, to survive intracellularly. These experiments indicated that gelatinase and cytolysin play no role in macrophage survival under these test conditions. E. faecalis strains grown in BHI broth exhibited a slightly increased rate of decline during intracellular growth compared to E. faecalis strains grown in serum. This difference is presumably due to an increased uptake of serum-passaged E. faecalis by macrophages, resulting in increased macrophage burden and reduced killing.
Examination of E. faecalis 418-infected mouse peritoneal macrophages by transmission electron microscopy revealed that intact diplococci were present intracellularly at all times postinfection, suggesting either that the bacteria replicated intracellularly or that they remained in a viable, resting state. Furthermore, electron micrographs of infected macrophages revealed that E. faecalis could be found lying in the cytoplasm of the macrophage, an observation that was previously reported for S. bovis within splenic macrophages (13).
Quantitation of viable intracellular E. faecalis 418 over the course of the infection of the mouse peritoneal macrophages indicated that the number of bacteria did not increase, as has been reported for intracellular pathogens such as Brucella abortus (2, 30), Listeria monocytogenes (32, 35, 43), and Salmonella typhimurium (8, 34), but decreased slightly (approximately 10- to 55-fold) over the 72-h period in primary mouse macrophages. The slight reduction in the number of viable intracellular bacteria may be explained in three ways. First, there may be two populations of bacteria inside the macrophage, one which survives and multiplies and another which is killed intracellularly, an observation that has been made for macrophages infected with Salmonella typhimurium (1) and Listeria monocytogenes (12). Alternatively, E. faecalis may not replicate in the macrophage but may remain quiescent and undergo delayed death due to intrinsic resistance of the bacterium to the internal environment. Since E. faecalis is an opportunistic pathogen and has not previously been reported to survive intracellularly for extended periods, one might speculate that this bacterium lacks the mechanisms used by overt pathogens to multiply within the macrophage but may resist macrophage killing due to the production of enzymes that inactivate reactive oxygen intermediates generated by the oxidative burst. E. faecalis produces superoxide dismutase and NADH peroxidase, enzymes which may serve to reduce the detrimental effects of superoxide anion and hydrogen peroxide, respectively, in the macrophage (6, 42, 44). E. faecalis may simply persist in the macrophage, a property which might allow the bacterium to resist killing by antibiotics or to remain viable while translocating across the intestinal wall and which might also facilitate entry into the mesenteric lymph nodes and into the blood. The third possibility is that the decrease in the number of viable intracellular bacteria may reflect the slow uptake of gentamicin and vancomycin (or penicillin), antibiotics which are effective for killing E. faecalis. It is difficult experimentally to eliminate this last possibility, since for long-term survival assays it is necessary to include these antibiotics in the tissue culture media to prevent overgrowth of extracellular E. faecalis. The results of studies on whether various antibiotics enter the macrophage at a level sufficient to kill intracellular bacteria have been conflicting (7, 14, 23, 29, 36, 50). However, vancomycin and aminoglycosides are taken up slowly, and aminoglycosides localize primarily in lysosomes, where they are partially inactivated by the acidic pH (for a review, see reference 36).
The results reported in this study indicate that E. faecalis is capable of surviving for a prolonged period in mouse peritoneal macrophages. Knowledge of the detailed mechanisms used by E. faecalis to evade the bactericidal effects of the macrophage will require further studies of the bacterial products produced within the macrophage.
ACKNOWLEDGMENTS
We thank Paul Gulig and Ian Orme for helpful discussions during the conduct of this research.
This work was supported in part by a College Research Council Award and a Career Enhancement Award from Colorado State University.
REFERENCES
- 1.Abshire K Z, Neidhardt F C. Growth rate paradox of Salmonella typhimurium within host macrophages. J Bacteriol. 1993;175:3744–3748. doi: 10.1128/jb.175.12.3744-3748.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldwin C L, Winter A J. Macrophages and Brucella. In: Swilling B S, Eisenstein T K, editors. Macrophage-pathogen interactions. New York, N.Y: Marcel Dekker, Inc.; 1994. pp. 363–380. [Google Scholar]
- 3.Bhakdi S, Klonisch T, Nuber P, Fischer W. Stimulation of monokine production by lipoteichoic acids. Infect Immun. 1991;59:4614–4620. doi: 10.1128/iai.59.12.4614-4620.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bleiwess A S, Zimmerman L N. Properties of proteinase from Streptococcus faecalis var. liquefaciens. J Bacteriol. 1964;88:653–659. doi: 10.1128/jb.88.3.653-659.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowe F, Heffron F. Isolation of Salmonella mutants defective for intracellular survival. Methods Enzymol. 1994;236:509–526. doi: 10.1016/0076-6879(94)36039-1. [DOI] [PubMed] [Google Scholar]
- 6.Britton L, Malinowski D P, Fridovich I. Superoxide dismutase and oxygen metabolism in Streptococcus faecalis—comparisons with other organisms. J Bacteriol. 1978;134:229–236. doi: 10.1128/jb.134.1.229-236.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown K N, Percival A. Penetration of antimicrobials into tissue culture cells and leukocytes. Scand J Infect Dis Suppl. 1978;14:251–260. [PubMed] [Google Scholar]
- 8.Buchmeier N A, Heffron F. Intracellular survival of wild-type Salmonella typhimurium and macrophage-sensitive mutants in diverse populations of macrophages. Infect Immun. 1989;57:1–7. doi: 10.1128/iai.57.1.1-7.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chenoweth C, Schaberg C. The epidemiology of enterococci. Eur J Clin Microbiol Infect Dis. 1990;9:80–89. doi: 10.1007/BF01963631. [DOI] [PubMed] [Google Scholar]
- 10.Clewell D B. Bacterial sex pheromone-induced plasmid transfer. Cell. 1993;73:9–12. doi: 10.1016/0092-8674(93)90153-h. [DOI] [PubMed] [Google Scholar]
- 11.Cohen M, Yoder S L, Huband M D, Roland G E, Courtney C L. In vitro and in vivo activities of clinafloxacin, CI-900 (PD 131112), and PD 138312 versus enterococci. Antimicrob Agents Chemother. 1995;39:2123–2127. doi: 10.1128/aac.39.9.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Chastellier C, Berche P. Fate of Listeria monocytogenes in murine macrophages: evidence for simultaneous killing and survival of intracellular bacteria. Infect Immun. 1994;62:543–553. doi: 10.1128/iai.62.2.543-553.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Herdt P, Haesebrouch F, Charlier G, Ducatelle R, Devriese L A. Intracellular survival and multiplication of virulent and less virulent strains of Streptococcus bovis in pigeon macrophages. Vet Microbiol. 1995;45:157–169. doi: 10.1016/0378-1135(95)00035-9. [DOI] [PubMed] [Google Scholar]
- 14.Drevets D A, Canono B P, Leenen P J M, Campbell P A. Gentamicin kills intracellular Listeria monocytogenes. Infect Immun. 1994;62:2222–2228. doi: 10.1128/iai.62.6.2222-2228.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franke A E, Clewell D B. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of “conjugal” transfer in the absence of conjugative plasmid. J Bacteriol. 1981;145:494–502. doi: 10.1128/jb.145.1.494-502.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilmore M S, Segarra R A, Booth M C, Bogie C P, Hall L R, Clewell D B. Genetic structure of the Enterococcus faecalis plasmid pAD1-encoded cytolytic toxin system and its relationship to lantibiotic determinants. J Bacteriol. 1994;176:7335–7344. doi: 10.1128/jb.176.23.7335-7344.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graninger W, Raguette R. Nosocomial bacteremia due to Enterococcus faecalis without endocarditis. Clin Infect Dis. 1992;15:49–57. doi: 10.1093/clinids/15.1.49. [DOI] [PubMed] [Google Scholar]
- 18.Gray J, Marsh P J, Stewart D, Pedler S J. Enterococcal bacteraemia: a prospective study of 125 episodes. J Hosp Infect. 1994;27:179–186. doi: 10.1016/0195-6701(94)90125-2. [DOI] [PubMed] [Google Scholar]
- 19.Greenberg J W, Fischer W, Joiner K A. Influence of lipoteichoic acid structure on recognition by the macrophage scavenger receptor. Infect Immun. 1996;64:3318–3325. doi: 10.1128/iai.64.8.3318-3325.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gross A, Spiesser S, Terraza A, Rouot B, Caron E, Dornand J. Expression and bactericidal activity of nitric oxide synthase in Brucella suis-infected murine macrophages. Infect Immun. 1998;66:1309–1316. doi: 10.1128/iai.66.4.1309-1316.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gulig P A, Curtiss R., III Plasmid-associated virulence of Salmonella typhimurium. Infect Immun. 1987;55:2891–2901. doi: 10.1128/iai.55.12.2891-2901.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guzman C A, Pruzzo C, Lipira G, Calegari L. Role of adherence in pathogenesis of Enterococcus faecalis urinary tract infection and endocarditis. Infect Immun. 1989;57:1834–1838. doi: 10.1128/iai.57.6.1834-1838.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hand W L, Corwin R W, Steinberg T H, Grossman G D. Uptake of antibiotics by human alveolar macrophages. Am Rev Respir Dis. 1984;129:933–937. doi: 10.1164/arrd.1984.129.6.933. [DOI] [PubMed] [Google Scholar]
- 24.Ike Y, Clewell D B. Genetic analysis of the pAD1 pheromone response in Streptococcus faecalis, using transposon Tn917 as an insertional mutagen. J Bacteriol. 1984;158:777–783. doi: 10.1128/jb.158.3.777-783.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ike Y, Clewell D B, Segarra R A, Gilmore M S. Genetic analysis of the pAD1 hemolysin/bacteriocin determinant in Enterococcus faecalis: Tn917 insertional mutagenesis and cloning. J Bacteriol. 1990;172:155–163. doi: 10.1128/jb.172.1.155-163.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ike Y, Craig R A, White B A, Yagi Y, Clewell D B. Modification of Streptococcus faecalis sex pheromones after acquisition of plasmid DNA. Proc Natl Acad Sci USA. 1983;80:5369–5373. doi: 10.1073/pnas.80.17.5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ike Y, Hashimoto H, Clewell D B. Hemolysin of Streptococcus faecalis subspecies zymogenes contributes to virulence in mice. Infect Immun. 1984;45:528–530. doi: 10.1128/iai.45.2.528-530.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jett B D, Huycke M M, Gilmore M S. Virulence of enterococci. Clin Microbiol Rev. 1994;7:462–478. doi: 10.1128/cmr.7.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson J D, Hand W L, Francis J B, King-Thompson N, Corwin R W. Antibiotic uptake by alveolar macrophages. J Lab Clin Med. 1980;95:429–439. [PubMed] [Google Scholar]
- 30.Jones S M, Winter A J. Survival of virulent and attenuated strains of Brucella abortus in normal and gamma interferon-activated murine peritoneal macrophages. Infect Immun. 1992;60:3011–3014. doi: 10.1128/iai.60.7.3011-3014.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kreft B, Marre R, Schramm U, Wirth R. Aggregation substance of Enterococcus faecalis mediates adhesion to cultured renal tubular cells. Infect Immun. 1992;60:25–30. doi: 10.1128/iai.60.1.25-30.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuhn M, Kathariou S, Goebel W. Hemolysin supports survival but not entry of the intracellular bacterium Listeria monocytogenes. Infect Immun. 1988;56:79–82. doi: 10.1128/iai.56.1.79-82.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis C M, Zervos M J. Clinical manifestations of enterococcal infection. Eur J Clin Microbiol Infect Dis. 1990;9:111–117. doi: 10.1007/BF01963635. [DOI] [PubMed] [Google Scholar]
- 34.Lissner C R, Swanson R N, O’Brien A D. Genetic control of the innate resistance of mice to Salmonella typhimurium: expression of the ity gene in peritoneal and splenic macrophages isolated in vitro. J Immunol. 1983;131:3006–3013. [PubMed] [Google Scholar]
- 35.Mackaness G B. Cellular resistance to infection. J Exp Med. 1962;116:381–406. [PubMed] [Google Scholar]
- 36.Maurin M, Raoult D. Optimum treatment of intracellular infection. Drugs. 1996;52:45–59. doi: 10.2165/00003495-199652010-00004. [DOI] [PubMed] [Google Scholar]
- 37.Megran D W. Enterococcal endocarditis. Clin Infect Dis. 1992;15:63–71. doi: 10.1093/clinids/15.1.63. [DOI] [PubMed] [Google Scholar]
- 38.Moellering R C., Jr Emergence of enterococcus as a significant pathogen. Clin Infect Dis. 1992;14:1173–1178. doi: 10.1093/clinids/14.6.1173. [DOI] [PubMed] [Google Scholar]
- 39.Murray B E. The life and times of the enterococcus. Clin Microbiol Rev. 1990;3:46–65. doi: 10.1128/cmr.3.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noskin G A, Peterson L A, Warren J R. Enterococcus faecium and Enterococcus faecalis bacteremia: acquisition and outcome. Clin Infect Dis. 1995;20:296–301. doi: 10.1093/clinids/20.2.296. [DOI] [PubMed] [Google Scholar]
- 41.Olmsted S B, Dunny G M, Erlandsen S L, Wells C L. A plasmid-encoded surface protein of Enterococcus faecalis augments its internalization by cultured intestinal cells. J Infect Dis. 1994;170:1549–1556. doi: 10.1093/infdis/170.6.1549. [DOI] [PubMed] [Google Scholar]
- 42.Poole L B, Claiborne A. Interactions of pyridine nucleotides with redox forms of the flavin-containing NADH peroxidase from Streptococcus faecalis. J Biol Chem. 1986;26:14525–14533. [PubMed] [Google Scholar]
- 43.Portnoy D A, Jacks P S, Hinrichs D J. Role of hemolysin for the intracellular growth of Listeria monocytogenes. J Exp Med. 1988;167:1459–1471. doi: 10.1084/jem.167.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poyart C, Berche P, Trieu-Cuot P. Characterization of superoxide dismutase genes from Gram positive bacteria by polymerase chain reaction using degenerate primers. FEMS Microbiol Lett. 1995;131:41–45. doi: 10.1016/0378-1097(95)00232-t. [DOI] [PubMed] [Google Scholar]
- 45.Ralph P, Prichard J, Cohn M. Reticulum cell sarcoma: an effector cell in antibody-dependent cell-mediated immunity. J Immunol. 1975;114:898–905. [PubMed] [Google Scholar]
- 46.Sansonetti P J, Ryter A, Clerc P, Maurelli A T, Mounier J. Multiplication of Shigella flexneri within HeLa cells: lysis of the phagocytic vacuole and plasmid-mediated contact hemolysis. Infect Immun. 1986;51:461–469. doi: 10.1128/iai.51.2.461-469.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schaberg D R, Culver D H, Gaynes R P. Major trends in the microbial etiology of nosocomial infection. Am J Med. 1991;91(Suppl 3B):72S–75S. doi: 10.1016/0002-9343(91)90346-y. [DOI] [PubMed] [Google Scholar]
- 48.Schwan W R, Demuth A, Kuhn M, Goebel W. Phosphatidylinositol-specific phospholipase C from Listeria monocytogenes contributes to intracellular survival and growth of Listeria innocua. Infect Immun. 1994;62:4795–4803. doi: 10.1128/iai.62.11.4795-4803.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson J. N5-(l-1-Carboxyethyl)-l-ornithine:NADP+ oxidoreductase from Streptococcus lactis. Purification and partial characterization. J Biol Chem. 1989;264:9592–9601. [PubMed] [Google Scholar]
- 50.Van den Broek P J. Antimicrobial drugs, microorganisms, and phagocytes. Rev Infect Dis. 1989;11:213–245. doi: 10.1093/clinids/11.2.213. [DOI] [PubMed] [Google Scholar]
- 51.Wells C L, Feltis B A, Hanson D F, Jechorek R P, Erlandsen S L. Oral infectivity and bacterial interactions with mononuclear phagocytes. J Med Microbiol. 1993;38:345–353. doi: 10.1099/00222615-38-5-345. [DOI] [PubMed] [Google Scholar]
- 52.Wells C L, Jechorek R P, Erlandsen S L. Evidence for the translocation of Enterococcus faecalis across the mouse intestinal tract. J Infect Dis. 1990;162:82–90. doi: 10.1093/infdis/162.1.82. [DOI] [PubMed] [Google Scholar]
- 53.Wells C L, Maddaus M A, Simmons R L. Proposed mechanisms for the translocation of intestinal bacteria. Rev Infect Dis. 1988;10:958–979. doi: 10.1093/clinids/10.5.958. [DOI] [PubMed] [Google Scholar]