Abstract
Deconica is a relatively small genus, with only 90 names recorded in previous research. In this study, four new species of Deconica have been identified based on morphological and phylogenetic evidence from subtropical regions of China. This represents the first discovery of new species of Deconica in China. Morphologically, D. austrosinensis is characterized by medium-sized spores that are elliptical to elongated-ellipsoid in face view, and fusiform to sublageniform and slightly thick-walled pleurocystidia; D. furfuracea is identified by a well-developed and evanescent veil, medium-sized spores that are rhomboid to mitriform in face view, and fusiform to subclavate pleurocystidia that are rare and subacute at apex; D. fuscobrunnea is recognized by dark brown pileus, medium-sized spores that are rhomboid to mitriform in face view, an ixocutis pileipellis, lageniform cheilocystidia with a long neck and lacks pleurocystidia; D. ovispora is distinguished from other Deconica species by medium-sized spores that are ovoid in face view, an ixocutis pileipellis, lageniform cheilocystidia with a long to short neck, and lacks pleurocystidia. Their distinct taxonomic status is confirmed by the positions of the four new species in ITS + LSU phylogenetic trees. Detailed descriptions and morphological photographs of four new species are presented.
Keywords: basidiomycetes, four new taxa, taxonomy, phylogeny
1. Introduction
Deconica (W.G. Sm.) P. Karst. is characterized by usually mycenoid, collybioid, omphaloid, or crepidotoid basidiomata; conical, convex or hemispherical, dry or viscid pileus with or without an umbo; brown to purple-brown, adnate to broadly adnate lamellae with a decurrent tooth in most species; and a slender stipe when present; they often grow on rotten wood, grasses, mosses, trunks, and dung [1,2]. For a long time, all species of Deconica were considered part of Psilocybe (Fr.) P. Kumm until phylogenetic studies were conducted [3]. Morphologically, Deconica differ from Psilocybe by the absence of hallucinogenic compounds, the broadly attached lamellae with a decurrent tooth in most species, and never blackish purple lamellae [1,2].
Deconica is not a very species-rich genus: only 90 names (55 species and 2 varieties), including synonyms and subspecies, were listed in Index Fungorum, since Smith established the subgenus Deconica under Agaricus L. [4,5]. Based on the study of Guzmán, there could be around 133 species of Deconica worldwide [6]. Noordeloos pointed out that there are 24 species in Europe and Ramírez-Cruz treated 47 taxa worldwide [1,2,7]. In previous studies, only six species of Deconica were reported in China [8,9]. As part of the study on Chinese macrofungi species, four new species were discovered, during our investigations in subtropical regions of China, and this also marks the first discovery of new species of this genus in China. In this paper, detailed information on the new taxa is presented.
2. Materials and Methods
2.1. Morphological Studies
Specimens were collected from Fujian, Hubei, Jiangxi, and Zhejiang provinces of China between 2019 and 2024 and were deposited in the Herbarium of Fungi, Jiangxi Agricultural University (HFJAU). Macroscopic descriptions were based on detailed field notes of fresh basidiomata and photos. Colour codes follow the Methuen Handbook of Colour [10]. Microscopic structures were observed and measured from dried specimens mounted in water, 5% KOH. Congo red was used as a stain when necessary [11]. Patent Blue V 0.1% was used to detect chrysocystidia [12]. At least 20 basidiospores, basidia, and cystidia were measured for each collection. The range of spore size is expressed as the form (a) b–c (d), in which “a” and “d” represent the minimum and maximum values, 90% of the spores fall within the range ‘b–c’. The meanings of the other spore characteristics are as follows: “Q” stands for the ratio of length and width [13,14].
2.2. DNA Extraction, PCR Amplification, and Sequencing
DNA was extracted from dried specimens with the NuClean Plant Genomic DNA kit (CWBIO, China). Two regions (ITS, LSU) were selected for the study and were amplified using the primer pairs ITS1/ITS4 [15], LR0R/LR7 [16,17], respectively. PCR was performed using a touchdown program for all regions: initial 95 °C for 5 min, and then 14 cycles of denaturing at 95 °C for 30 s, annealing at 65 °C for 45 s (−1 °C per cycle), extension at 72 °C for 1 min; then 30 cycles of denaturing at 95 °C for 30 s, annealing at 52 °C for 30 s, extension at 72 °C for 1 min; final extension at 72 °C for 10 min [13,18]. The PCR products were sequenced by Qing Ke Biotechnology Co., Ltd. (Wuhan, China).
2.3. Alignment and Phylogenetic Analyses
Sequence reads were assembled and edited using Sequencher v.5.4 and were deposited in GenBank (GB) database. In total, 32 sequences, including ITS and LSU regions, were generated from our collected specimens. Based on the research by Ramírez-Cruz and colleagues [1,17], and the similarity of these new species to the most closely related sequences identified in the BLAST results of ITS, 85 nucleotide DNA sequences in NCBI GenBank were downloaded. The Kuehneromyces brunneoalbescens (Y.S. Chang & A.K. Mills) J.A. Cooper was chosen as an outgroup taxon according to the results of Ramírez-Cruz and colleagues [1]. Details are presented in Table 1.
ITS and LSU sequence datasets were separately aligned on the MAFFT v.7 [19]. Bayesian inference (BI) and maximum likelihood (ML) phylogenetic analyses of the aligned concatenated dataset were respectively carried out in MrBayes v.3.2.7a and IQtree v.2.1.2, respectively [20,21]. The best-fit models of BI were determined by PartitionFinder, complying with the Corrected Akaike information criterion (AICc) [22]. The ML analysis was conducted using the ultrafast bootstrap option with 1000 replicates and allowing partitions to have different seeds (--p). For the BI analysis, four Monte Carlo Markov chains were run for 5 million generations, sampling every 100th generation, with the first 25% of trees discarded as burn-in. The branches of Bayesian posterior probability (BI-PP) ≥ 0.95 and ML bootstrap support (ML-BP) ≥ 75% are considered statistical supports and are shown in the tree (Figure 1). All alignments for phylogenetic analyses and tree were deposited in TreeBASE (ID: TB2:S31681).
Figure 1.
Phylogram of Deconica spp. generated with maximum likelihood (ML) analysis based on ITS and LSU, rooted with Kuehneromyces brunneoalbescens. Bayesian inference (BI-PP) ≥ 0.95 and ML bootstrap proportions (ML-BP) ≥ 75 are indicated as PP/BP. The new taxa are marked in bold.
Table 1.
Details of sequences used in the phylogenetic analyses.
| Species | Voucher | Country | ITS | LSU | References |
|---|---|---|---|---|---|
| D. aequatoriae | F1018172 holotype | Ecuador | MT622203 | [1] | |
| D. angustispora | Ps-7 | USA | MT622205 | [1] | |
| D. argentina | XAL | Italy | KC669307 | [23] | |
| D. austrosinensis | HFJAU2439 | China | PP759373 | PQ282655 | This study |
| D. austrosinensis | HFJAU2562 holotype | China | PP759375 | PQ282656 | This study |
| D. austrosinensis | HFJAU3577 | China | PP759374 | PQ282659 | This study |
| D. austrosinensis | HFJAU3859 | China | PQ282646 | PQ282661 | This study |
| D. austrosinensis | HFJAU4162 | China | PQ282647 | PQ282662 | This study |
| D. austrosinensis | HFJAU4451 | China | PQ282648 | PQ282663 | This study |
| D. austrosinensis | HFJAU1108 | China | MN622718 | This study | |
| D. austrosinensis (as Basidiomycete from a bamboo in GB) | WZ9201 | China | U65602 | [24] | |
| D. austrosinensis (as D. sp. in GB) | Mushroom Observer290081 | Mexico | MH159224 | Unpublished | |
| D. baylisiana | PDD105444 | New Zealand | KM975393 | [23] | |
| D. bullacea | WU-4849 | Austria | MT622207 | [1] | |
| D. castanella | EA0619 | Netherlands | MT622208 | [1] | |
| D. chionophila | CBS 655.87 holotype | France | MH862111 | NG_068969 | [3] |
| D. chionophila | Zu105 | Netherlands | MT622209 | [1] | |
| D. citrispora | PDD87522 | New Zealand | KM975431 | [23] | |
| D. cokeriana | TENN–F–067047 | USA | KC669315 | [23] | |
| D. cokeriana | PRM 922477 | USA | MK965914 | Unpublished | |
| D. coprophila | HFJAU2912 | China | PQ282645 | PQ282657 | This study |
| D. coprophila | V. Ramirez-Cruz 114 | Mexico | KC669308 | KC669336 | [17] |
| D. coprophila | Argentina | JX235960 | [25] | ||
| D. coprophila | MHHNU30335 | China | MK214386 | Unpublished | |
| D. coprophila | ICN-154223 | Brazil | MT622210 | [1] | |
| D. coprophila | TENN–F–061255 | USA | MT622211 | [1] | |
| D. coprophila | MHHNU7935 | China | OP862790 | Unpublished | |
| D. coprophila | MHHNU7937 | China | OP862791 | Unpublished | |
| D. crobula | CBS:193.39 | MH855980 | [26] | ||
| D. crobula | DAMu078 | Netherlands | MT622213 | [1] | |
| D. crobula | MENu020 | Germany | MT622214 | [1] | |
| D. crobula | Vu124 | UK | MT622215 | [1] | |
| D. esperancensis | Ps-493 holotype | Mexico | MT622216 | [1] | |
| D. fuegiana | Ps-275 | Finland | MT622217 | [1] | |
| D. furfuracea | HFJAU3793 | China | PP759377 | PQ282660 | This study |
| D. furfuracea | HFJAU5181 holotype | China | PP759378 | PQ282664 | This study |
| D. furfuracea (as D. phyllogena in GB) | SFC20160714-66 | Korea | MF437002 | [27] | |
| D. furfuracea (as D. phyllogena in GB) | Mushroom Observer # 282800 | USA | MK607529 | Unpublished | |
| D. furfuracea [as D. rhombispor (=D. phyllogena) in GB] | SCM678-H1 | USA | FJ596920 | [28] | |
| D. furfuracea [as D. rhombispor (=D. phyllogena) in GB] | SCM678 | USA | FJ596921 | [28] | |
| D. furfuracea (as D. sp. in GB) | TENN–F–008938 | USA | MT622256 | [1] | |
| D. fuscobrunnea | HFJAU3517 holotype | China | PP759379 | PQ282658 | This study |
| D. fuscobrunnea | HFJAU5706 | China | PQ282652 | This study | |
| D. horizontalis | P.S. Silva 253/10 (ICN) | Brazil | KC669309 | KC669337 | [17] |
| D. horizontalis | ECV1919 | Netherlands | MT622219 | [1] | |
| D. horizontalis | ECV1883 | Netherlands | MT622220 | [1] | |
| D. inquilina | CBS:197.39 | MH855982 | Unpublished | ||
| D. inquilina | EVu194 | Netherlands | MT622221 | [1] | |
| D. inquilina | CUu196 | Netherlands | MT622222 | [1] | |
| D. inquilina | MEN179 | Netherlands | MT622223 | [1] | |
| D. inquilina | EAu188 | Netherlands | MT622224 | [1] | |
| D. inquilina | SVu190 | Netherlands | MT622225 | [1] | |
| D. magica | Vu123 | UK | MT622226 | [1] | |
| D. magica | Vu237 | UK | MT622227 | [1] | |
| D. merdaria | F-1015258 | UK | MT622228 | [1] | |
| D. milvispora | UT-1606 | Australia | KC669314 | NG_081284 | [1] |
| D. montana | VNSu069 | Netherlands | MT622231 | [1] | |
| D. montana | MENu186 | Netherlands | MT622232 | [1] | |
| D. montana var. macrospora | CBS 101983 holotype | Netherlands | MH862774 | MH874369 | [26] |
| D. aff. montana | Ps–370 | Mexico | KC669311 | [1] | |
| D. aff. montana | Ps–96 | USA | MT622229 | [1] | |
| D. neorhombispora | ICN-154462 | Brazil | MT622234 | [1] | |
| D. neorhombispora (as P. subrunneocystidata in GB) | Ps–279 isotype | Brazil | MT622233 | [1] | |
| D. novae-zelandiae | PDD87768 | New Zealand | KM975401 | [1] | |
| D. overeemii | DED 8328 (SFSU) | São Tomé | KX017212 | [29] | |
| D. ovispora | HFJAU1915 | China | PQ282644 | PQ282654 | This study |
| D. ovispora | HFJAU5429 | China | PQ282649 | This study | |
| D. ovispora | HFJAU5476 holotype | China | PQ282650 | This study | |
| D. ovispora (as D. sp. in GB) | TENN–F–060416 | USA | MT622259 | [1] | |
| D. ovispora (as D. sp. in GB) | iNat86992341 | USA | ON774784 | Unpublished | |
| D. ovispora (as D. sp. in GB) | iNat94505915 | USA | OP270521 | Unpublished | |
| D. ovispora (as D. sp. in GB) | iNat179755933 | USA | OR825659 | Unpublished | |
| D. pegleriana | Ps-153 | Brazil | MT622235 | [1] | |
| D. phyllogena | JD72159 | Netherlands | MT622236 | [1] | |
| D. pratensis | MNu189 | Netherlands | MT622238 | [1] | |
| D. protea | BAP 596 (SFSU) | São Tomé | KX017213 | [29] | |
| D. pseudobullacea | Ps-171 | Nepal | MT622239 | [1] | |
| D. semi-inconspicua | Ps-181 isotype | USA | MT622240 | [1] | |
| D. cf. singeriana | Ps-418 | Brazil | MT622241 | [1] | |
| D. subcoprophila | Ps-206 | USA | MT622242 | [1] | |
| D. submaritima | Ps-217 | Italy | MT622243 | [1] | |
| D. subviscida | MENu013 | Netherlands | MT622244 | [1] | |
| D. thailandensis | Ps-430 isotype | Thailand | MT622245 | [1] | |
| D. umbrina | Ps-429 isotype | Malaysia | MT622246 | [1] | |
| D. vorax | PDD87407 | New Zealand | KM975441 | [1] | |
| D. xeroderma | Oswald s.n. (WU) | Austria | KC669312 | KC669340 | [17] |
| D. xeroderma | AHv221 | Austria | MT622248 | [1] | |
| D. xeroderma | MEN200420 | Austria | MT622249 | [1] | |
| D. xeroderma (type of D. alpestris) | WU-784 holotype | Austria | MT622204 | [1] | |
| D. xeroderma (as D. velifera in GB) | SIu210 | Austria | MT622247 | [1] | |
| D. sp.1 | HFJAU1745 | China | PQ282643 | PQ282653 | This study |
| D. sp.1 | HFJAU5705 | China | PQ282651 | This study | |
| D. sp.2 | WU5476 | Austria | MT622261 | [1] | |
| D. sp.3 | Ps–269 | Mexico | MT622265 | [1] | |
| D. sp.4 | TENN–F–062588 | USA | KC669316 | [1] | |
| D. sp.5 | ICN–154712 | Brazil | MT622254 | [1] | |
| D. sp.6 (as D. phyllogena in GB) | ZMU197 | China | MW724279 | Unpublished | |
| D. sp.7 | LXYZF1 | China | MZ452395 | [1] | |
| D. sp.8 | TENN–F–062238 | USA | KC669313 | [1] | |
| Kuehneromyces brunneoalbescens | PDD97054 | New Zealand | KM975426 | KM975380 | [23] |
| K. brunneoalbescens | Ps1608 | Australia | MK965912 | [1] |
3. Results
3.1. Phylogenetic Analysis
A total of 1798 characters from 101 taxa were used in phylogenetic analyses (ITS, 624 bp; LSU, 1174 bp), of which 227 sites were variable and 163 were parsimony informative for ITS, and 56 sites were variable and 34 were parsimony informative for LSU. Due to the different number of models supported by Mrbayes and IQtree, the best models are calculated separately, and the results are as follows: the best models for Bayesian analysis were GTR + I + G for the ITS, HKY + I + G for the LSU; the best models for ML analysis were: TPM2u + F + I + G4 for the ITS, K2P + I + G4 for the LSU. For the Bayes analysis, the average standard deviation of split frequencies was less than 0.01 after 1.57 million generations.
As shown in the ML tree in Figure 1, four new species formed distinct and stable branches, respectively. D. furfuracea groups together with two unknown species (D. sp.8 and D. sp.6), but this grouping has unstable support in Bayesian analysis. Similarly, D. ovispora groups together with D. xeroderma, and this grouping also has unstable support in Bayesian analysis.
3.2. Taxonomy
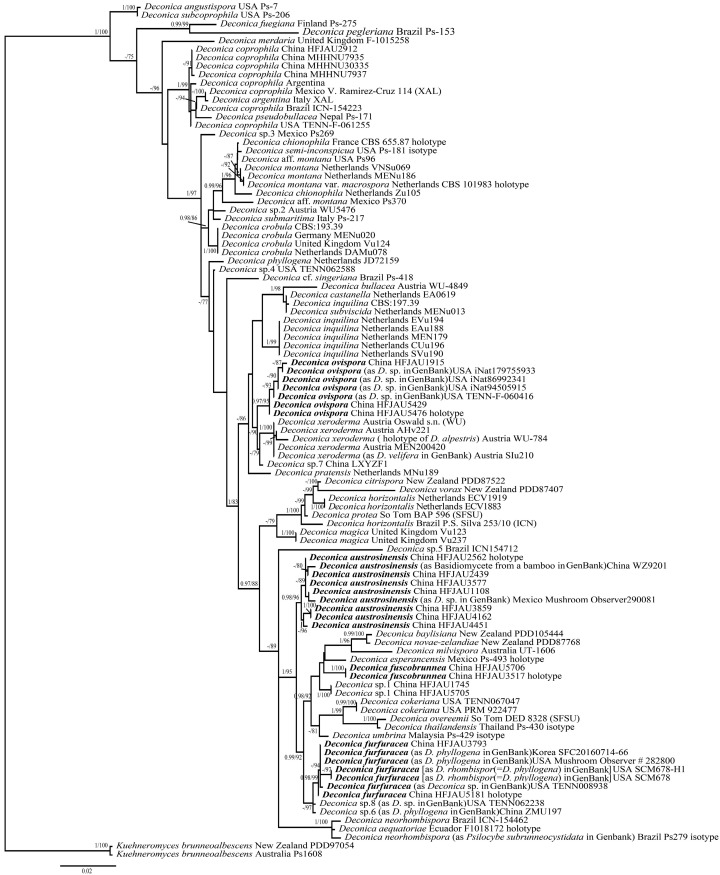
Deconica austrosinensis J.Q. Yan, S.N. Wang, and H. Zeng sp. nov. (Figure 2).
Figure 2.
Morphological structures of Deconica austrosinensis. (A–D) Basidiomata. (E) Pileipellis. (F–J) Pleurocystidia. (K) Spores. (L) Cheilocystidia. Scale bars: (A–D) 10 mm, (E–L) 10 μm. All microstructures were observed in 5%KOH. Structures of (F–I,L) were stained with 1%Congo red.
MycoBank: 855638
Etymology. “austrosinensis” refers to its type specimen originating from the southern regions of China.
Holotype. China, Fujian Province, Wuyishan National Park, 18 June 2021, collected by Jun-Qing Yan, and Sheng-Nan Wang, HFJAU2562.
Diagnosis. Deconica austrosinensis is mainly characterized by very small basidiomata; hygrophanous, reddish brown, striate pileus; ellipsoid to elongated-ellipsoid spores in face view, (5.4) 5.8–7.0 × 3.7–4.5 (4.8) µm; fusiform to sublageniform, slightly thick-walled pleurocystidia; clavate to pyriform, rarely fusiform, thin-walled cheilocystidia. It differs from D. cokeriana by clavate to pyriform, rarely fusiform cheilocystidia, and lack of chrysocystidia.
Description. Basidiomata very small. Pileus 11–20 mm, plano-convex to plane, rarely with unobviously obtuse umbo, hygrophanous, reddish brown (8D5–8E5), striate up to center from the margin, becoming grayish red (8C5) to dull red (8B4–8B5) as drying. Veil dull red (8B4–8B5), scattered or only remains on the edge of the pileus, fibrillose, evanescent. Context thin, 1.0 mm at the center. Lamellae 1.5–2.0 mm, decurrent to short decurrent, distant, unequal, dull red (8B4–8B5), edge even to slightly serrate, white. Stipe 10–30 mm long, 1.5–2.0 mm thick, central, cylindric, equal, dull red (8B4–8B5), gradually darkening toward the base, covered with white and evanescent fibrillose.
Spores (5.4) 5.8–7.0 × 3.7–4.5 (4.8) µm, Q = (1.3) 1.4–1.8, ellipsoid to elongated-ellipsoid in face view, 3.5–4.0 (4.4) µm broad, elongated-ellipsoid in profile, slightly thick-walled, smooth, brownish-yellow, germ pore distinct, 0.8–1.5 µm broad. Basidia 13–20 (23) × (5.0) 5.5–7.2 µm, subcylindric to clavate, pale yellow to hyaline, 4-spored. Pleurocystidia (30) 35–53 (56) × (8.0) 9.0–13 (16) µm, sublageniform with or without amorphous deposits at apex, fusiform with or without mucronate, slightly thick-walled, hyaline. Cheilocystidia 15–30 × (5.0) 7.4–12 µm, clavate to pyriform, rarely fusiform, thin-walled, hyaline. Pileipellis a cutis, hyphae 3.0–7.0 µm broad, pale yellow to brown-yellow. Clamp connections present.
Habitat. Solitary to scattered on rotten wood or humus in broad-leaved forests.
Additional specimens examined: China, Fujian Province, Wuyishan National Park, 18 June 2021, collected by Jun-Qing Yan, and Sheng-Nan Wang, HFJAU2439, HFJAU2562; 7 June 2022, collected by Bin-Rong Ke and Ya-Ping Hu, HFJAU3577; 27 June 2022, collected by Bin-Rong Ke and Cheng-Feng Nie, HFJAU3859, HFJAU3906; 8 July 2022, collected by Zhi-Heng Zeng, Hui Zeng HFJAU4451; Jiangxi Province, Lushan National Nature Reserve, collected by Jun-Qing Yan,10 July 2019, HFJAU1108; Zhejiang Province, Qingtian County, Lishui City, 8 August 2021, collected by Jun-Qing Yan, HFJAU2971.
Notes. This species forms an independent branch in the phylogenetic tree, showing clear genetic differentiation from other known Deconica spp. Morphologically, among the known species of the Deconica, few have a similar combination of characteristics as D. austrosinensis, that is, spores that are elliptical to elongated-ellipsoid in face view, with a length concentrated between 6.0–7.0 μm, and present of pleurocystidia. However, they can be clearly distinguished from D. austrosinensis: The pleurocystidia of D. hartii (Ammirati) Ammirati & Redhead are cylindrical to ventricose, and shorter than 25 μm [30]; D. cokeriana (A.H. Sm. & Hesler) Ram.-Cruz & A. Cortés-Pérez has chrysocystidia that are thin-walled and shorter than 40 μm, and its cheilocystidia are lageniform to widely utriform [23].
In addition, D. austrosinensis forms a clade with two unknown sequences from Mexico (MH159224) and Sichuan Province, China (U65602), and shares over 99% ITS similarity with them. It is highly likely that these two unknown sequences represent the distribution of the species in Mexico and Yunnan Province, China.
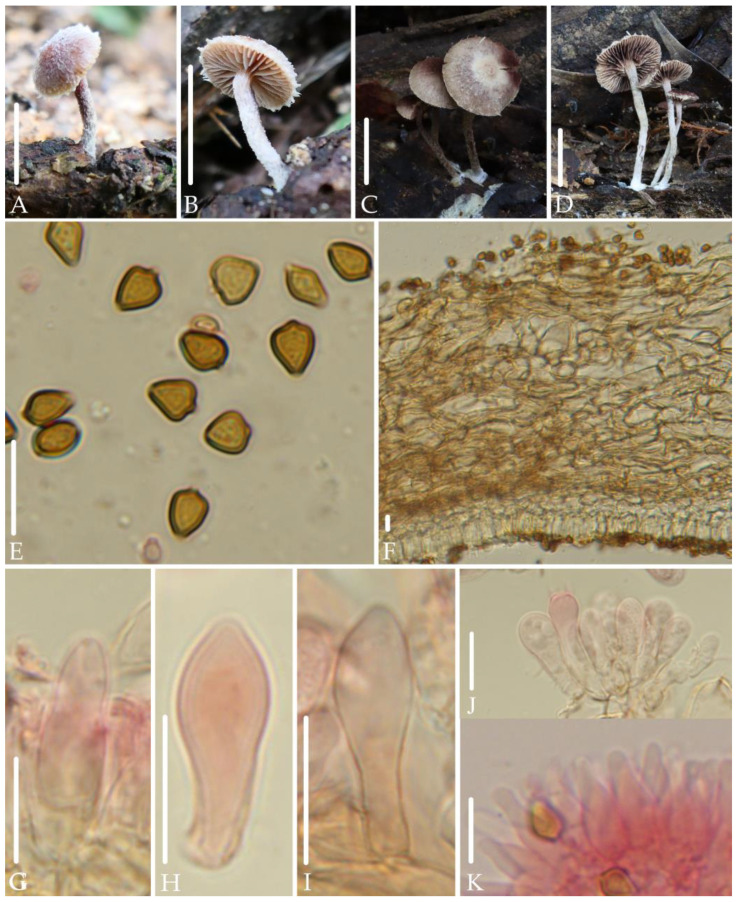
Deconica furfuracea J.Q. Yan, S.N. Wang, and H. Zeng sp. nov. (Figure 3).
Figure 3.
Morphological structures of Deconica furfuracea. (A–D) Basidiomata. (E) Spores. (F) Pileipellis. (G–I) Pleurocystidia. (J) Hymenium. (K) Cheilocystidia. Scale bars: (A–D) 10 mm, (E–K) 10 μm. All microstructures were observed in 5%KOH. Structures of (G–K) were stained with 1%Congo red.
MycoBank: 855639
Etymology. “furfuracea” refer to its well-developed veil in early stage.
Holotype. China, Fujian Province, Wuyishan National Park, 8 June 2022, collected by Cheng-Feng Nie, Sheng-Nan Wang, HFJAU5181.
Diagnosis. Deconica furfuracea is mainly characterized by very small basidiomata; hygrophanous, dull red pileus; well-developed and evanescent veil; rhomboid to mitriform spores in face view, (5.0) 5.5–6.5 (7.5) × 4.5–5.2 (6.0) µm; thin-walled pleurocystidia that are small, rarely, fusiform to subclavate with subacute apex; fusiform to sublageniform and thin-walled cheilocystidia. It differs from D. thailandensis by lack of chrysocystidia.
Description. Basidiomata very small. Pileus 6.5–12 mm, convex to plano-convex, with or without an inconspicuous obtuse umbo in the center, hygrophanous, dull red (8C5–8C6), gradually paler toward the margin, grayish orange to brownish orange (6B6–6C6), becoming brownish orange (6B6–6C6) as drying. Veil well developed in early stage, white, fibrillose, evanescent. Context thin, 1.0 mm at the center. Lamellae 2.0 mm broad, adnate to decurrent, distant, unequal, light orange to grayish orange (6A5–6B5), edge white. Stipe 9.0–20 mm long, 1.0–2.0 mm thick, central, cylindric, equal, dull red (8C5–8C6), covered by abundant white fibrillose.
Spores (5.0) 5.5–6.5 (7.5) × 4.5–5.2 (6.0) µm, Q = (1.1) 1.2–1.5 (1.6), rhomboid to mitriform in face view, ellipsoid to elongated-ellipsoid in profile, 3.5–4.0 (4.5) µm broad, slightly thick-walled, smooth, brownish-yellow, germ pore distinct, 1.0–1.5 µm broad. Basidia (13) 15–21 × 4.4–6.7 µm, subclavate to cylindric, pale yellow to hyaline, 4-spored. Pleurocystidia 15–23 (26) × 5.3–7.7 µm, rarely, fusiform to subclavate, thin-walled, subacute at apex, hyaline. Cheilocystidia (9.0) 15–18 (20) × (4.3) 4.9–6.7 (7.8) µm, fusiform to sublageniform, rarely mucronate at apex, thin-walled, hyaline. Pileipellis a cutis, hyphae (4.0) 5.0–11 (12) µm broad, cylindric, slightly gelatinous, pale yellow to pale yellow-brown. Clamp connections present.
Habitat. Solitary to scattered on rotten wood in mixed coniferous and broad-leaved forests.
Additional specimens examined: China, Jiangxi Province, Jiangxi Agricultural University, 2 June 2019, collected by Jun-Qing Yan, HFJAU1250; Fujian Province, Wuyishan National Park, 26 June 2022, collected by Zhi-Heng Zeng, Hui Zeng, HFJAU3793; 8 June 2022, collected by Cheng-Feng Nie, Sheng-Nan Wang, HFJAU5181.
Notes. This species forms an independent branch in the phylogenetic tree, showing clear genetic differentiation from other known Deconica spp. Morphologically, the pleurocystidia of D. furfuracea in question are similar in size to the basidioles, and due to their rarity, they are easily overlooked and confused with basidioles. However, the apex of basidioles of this species are obtuse to broadly obtuse, while the apex of pleurocystidia are subacute. This phenomenon of confusion between pleurocystidia and basidioles is not unique within this genus. D. thailandensis (E. Horak, Guzmán & Desjardin) Ram.-Cruz & Guzmán also has pleurocystidia that are subclavate, thin-walled, and shorter than 25 µm in length. However, the presence of annulus and chrysocystidia can clearly distinguish D. thailandensis from D. furfuracea [31].
Among the known species of the Deconica, few have a similar combination of characteristics as D. furfuracea, that is, spores that are rhomboid to mitriform in face view, with a length concentrated between 5.5–6.5 μm, and have pleurocystidia. However, they can be clearly distinguished from D. furfuracea: D. esperancensis Ram.-Cruz & Glez.-Adam, D. overeemii (E. Horak & Desjardin) Desjardin & B.A. Perry, D. umbrina (E. Horak, Guzmán & Desjardin) Ram.-Cruz & Guzmán have chrysocystidia [1,17,31]; the pleurocystidia of D. flocculosa (Bas & Noordel.) Noordel. are ventricose-rostrate or lageniform (Bas and Noordeloos, 1996).
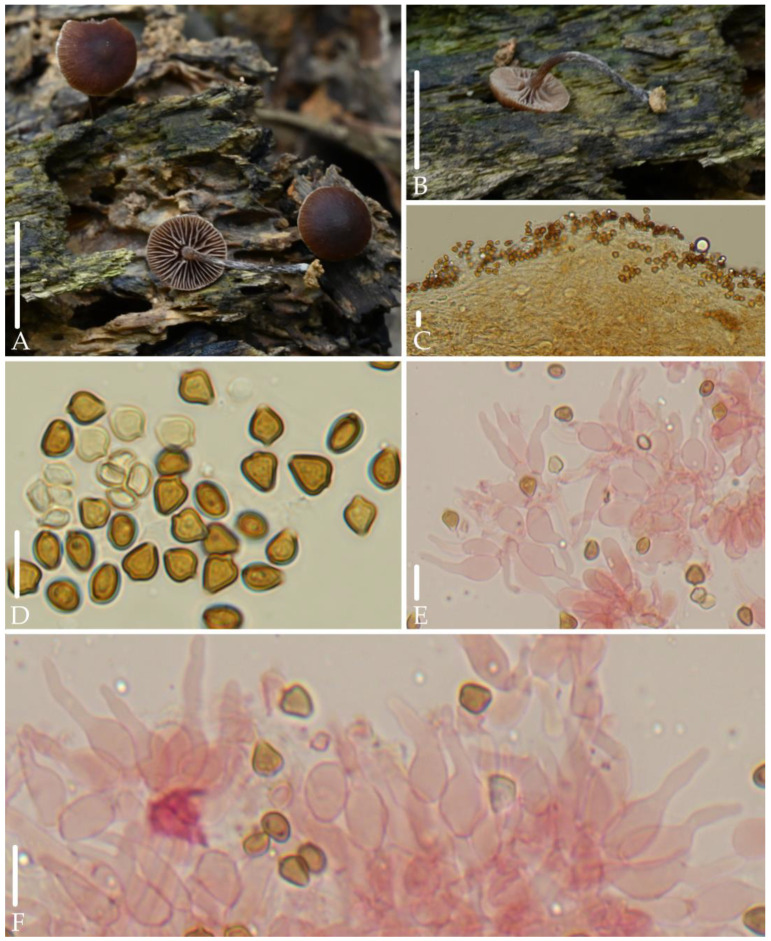
Deconica fuscobrunnea J.Q. Yan, S.N. Wang, and H. Zeng sp. nov. (Figure 4).
Figure 4.
Morphological structures of Deconica fuscobrunnea. (A,B) Basidiomata. (C) Pileipellis. (D) Spores. (E,F) Cheilocystidia. Scale bars: (A,B) 10 mm, (C–F) 10 μm. All microstructures were observed in 5%KOH. Structures of (E,F) were stained with 1%Congo red.
MycoBank: 855640
Etymology. “fuscobrunnea” refer to its dark brown pileus.
Holotype. China, Fujian Province, Wuyishan National Park, 19 May 2022, collected by Jun-Qing Yan, Hu Zeng, Zhi-Heng Zheng, HFJAU3517.
Diagnosis. Deconica fuscobrunnea is mainly characterized by very small basidiomata; hygrophanous, dark brown pileus; decurrent and distant lamellae; rhomboid to mitriform spores in face view, 5.0–6.5 (7.0) × 4.5–5.3 (5.6) µm; lageniform cheilocystidia with long neck; ixocutis of pileipellis. It differs from D. esperancensis by lack of pleurocystidia.
Description. Basidiomata very small. Pileus 5.0–6.0 mm, plano-convex to plane, with unobviously obtuse umbo in the center, hygrophanous, dark brown (8F7–8F8) to reddish brown (8E7–8E8), gradually paler toward the margin, striate up to 1/2 from the margin. Veil only remains on the edge of the pileus, fibrillose, white, evanescent. Context thin, 1.0 mm at the center. Lamellae 1.0–1.5 mm broad, decurrent, distant, unequal, rarely forked, grayish orange (6B4–6C4), edge white. Stipe 9.0–20 mm long, 1.3–2.0 mm thick, central, cylindric, equal, reddish brown (8D5–8E5), gradually darkening toward the base, covered with white and evanescent fibrillose.
Spores 5.0–6.5 (7.0) × 4.5–5.3 (5.6) µm, Q = 1.0–1.3 (1.5), rhomboid to mitriform in face view, 3.6–4.3 (4.6) µm broad, ellipsoid to elongated-ellipsoid in profile, slightly thick-walled, smooth, brownish-yellow, germ pore distinct, 0.6–1.5 µm broad. Basidia 13–22 × 5.0–6.6 µm, subclavate to cylindric, pale yellow to hyaline, 4-spored. Pleurocystidia absent. Cheilocystidia 12–25 (28) × 4.0–8.0 (9.0) µm, lageniform, with long neck, rarely forked, thin-walled, pale yellow to hyaline; Pileipellis an ixocutis, hyphae 3.6–5.3 (6.4) µm broad, cylindric, gelatinous, pale yellow. Clamp connections present.
Habitat. Scattered on rotten wood in broad-leaved forests.
Additional specimens examined: China, Fujian Province, Wuyishan National Park, 19 May 2022, collected by Jun-Qing Yan, Hu Zeng, Zhi-Heng Zheng, HFJAU3517; 19 May 2023, collected by Jun-Qing Yan, Sheng-Nan Wang, HFJAU5706.
Notes. This species forms an independent branch and is grouped with D. baylisiana (E. Horak) J.A. Cooper, D. esperancensis, D. milvispora Ram.-Cruz & Matheny, D. novae-zelandiae (Guzmán & E. Horak) J.A. Coope and D. sp.1 from China. However, spores of D. baylisiana, D. milvispora, and D. novae-zelandiae are longer than 7.5 µm [1,32,33]; D. esperancensis has chrysocystidia [1]; D. sp.1 has chrysocystidia and spores that are ellipsoid to ovoid, rarely rhomboid in face view.
Morphologically, among the known species of the Deconica, only D. deconicoides (E. Horak, Guzmán & Desjardin) Guzmán and D. xeroderma share a similar combination of characteristics as D. fuscobrunnea, that is, spores that are rhomboid to mitriform in face view, with a length concentrated between 5.0–6.5 μm, and lacks pleurocystidia. However, the two former species do not have ixocutis of pileipellis, and their cheilocystidia are fusiform [34,35].
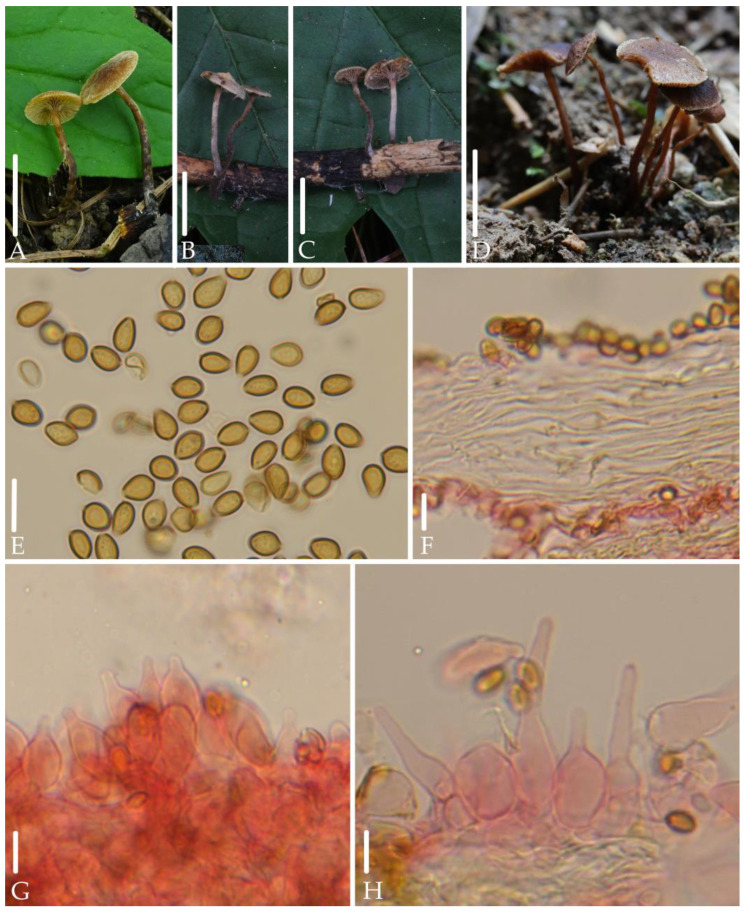
Deconica ovispora J.Q. Yan, S.N. Wang, and H. Zeng sp. nov. (Figure 5).
Figure 5.
Morphological structures of Deconica ovispora. (A–D) Basidiomata. (E) Spores. (F) Pileipellis. (G,H) Cheilocystidia. Scale bars: (A–D) 10 mm, (E–H) 10 μm. All microstructures were observed in 5%KOH. Structures of (F–H) were stained with 1%Congo red.
MycoBank: 855641
Etymology. “ovispora” refer to its ovoid spores in face view.
Holotype. Hubei Province, Xingshan County, Yichang City, 2 July 2024, collected by Jun-Qing Yan, Bin-Rong Ke, HFJAU5476.
Diagnosis. Deconica ovispora is mainly characterized by very small basidiomata; hygrophanous, reddish brown to dark brown pileus; decurrent and distant lamellae; ovoid spores in face view, 6.0–7.0 × 4.0–4.5 µm; lageniform cheilocystidia with long to short neck, ixocutis of pileipellis. It differs from D. xeroderma by ixocutis of pileipellis and lageniform cheilocystidia.
Description. Basidiomata very small. Pileus 5.0–6.0 mm, plano-convex to plane, or umbonate to unobviously bullate in the center, hygrophanous, reddish brown to dark brown (8D7–8E7), gradually paler toward the margin, striate up to 1/2 from the margin, becoming reddish gray to dull red (8B2–8B3) as drying. Veil white, fibrillose, scattered, evanescent. Context thin, 1.0 mm at the center. Lamellae 0.5–1.0 mm broad, decurrent, distant, unequal, brownish (6C5–6D5), edge white. Stipe 10–20 mm long, 1.0–2.0 mm thick, central, cylindric, equal, brown (7E5–7E6), gradually darkening toward the base, covered with white and evanescent fibrillose.
Spores 6.0–7.0 × 4.0–4.5 µm, Q = 1.4–1.6 (1.7), ovoid in face view, ellipsoid to elongated-ellipsoid in profile 3.5–4.0 µm broad, slightly thick-walled, smooth, brownish-yellow, germ pore distinct, 0.8–1.3 µm broad. Basidia 14–24 × 4.5–6.0 µm, subclavate, hyaline, 4-spored. Pleurocystidia absent. Cheilocystidia 21–36 × 5.5–14µm, lageniform, with long to short neck, thin-walled, pale yellow to hyaline; Pileipellis an ixocutis, hyphae 2.5–4.5 µm broad, cylindric, subgelatinous, pale yellow. Clamp connections present.
Habitat. Solitary, scattered or gregarious on rotten wood or humus in broad-leaved forest and mixed coniferous and broad-leaved forests.
Additional specimens examined: China, Zhejiang Province, Qingyuan County, 6 July 2020, collected by Jun-Qing Yan, Sheng-Nan Wang, HFJAU1915; Hubei Province, Xingshan County, Yichang City, 2 July 2024, collected by Jun-Qing Yan, Bin-Rong Ke, HFJAU5429, HFJAU5476.
Notes. Deconica ovispora and the unknown species collected from the USA (ON774784, OP270521, MT622259) form independent branches, and their ITS similarity is over 99%, which also suggests that this species is distributed in the USA. In addition, this species groups together with D. xeroderma and an unknown sequence from China. However, D. xeroderma does not have ixocutis of pileipellis, and its cheilocystidia are fusiform [34].
Morphologically, among the known species of the Deconica, few have a similar combination of characteristics as D. ovispora, that is, spores that are ovoid in face view, with a length concentrated between 6.0–7.0 μm, and lack pleurocystidia. However, they can be clearly distinguished from D. ovispora: D. micropora (Noordel. & Verduin) Noordel. has spores that are up to 6.0 µm broad, with indistinct germ pores, and it is symbiotic with mosses [36]; D. musacearum (Singer) Cortez & P.S. Silva has cutis of pileipellis, and smaller cheilocystidia, measuring 16–17.5 × 4.0–5.5 µm [37]; D. phillipsii (Berk. & Broome) Noordel. has pleurotoid basidiomata and narrowly fusiform cheilocystidia [38].
4. Discussion
Deconica was one of the largest genera with an unsequenced generic type, and due to this limitation, the diversity of species within this genus may be underestimated. In Ramírez-Cruz’s study of 61 samples on a fairly global basis, there are as many as 18 undescribed species [1]. In this article, the publication of two new species, D. ovispora and D. furfuracea, addresses two undescribed species proposed by Ramírez. However, it also reveals the potential for more new species, such as D. sp.1 from China. Due to the quality of the specimens, we did not observe the cheilocystidia, but based on the other observed morphological features, D. sp.1 can be well distinguished from the known species of Deconica.
The confusion in species names in GenBank further complicates the recognition of species within this genus. For example, the branch formed by the new species D. furfuracea in the phylogenetic tree includes sequences from South Korea (MF437002), the United States (MK607529, FJ596920, FJ596921), which were previously named as D. phyllogena or its synonyms, and shares over 99% ITS similarity with them. However, it is well known that D. phyllogena has a viscid and smooth pileus, lacks pleurocystidia, spores that can be as wide as 6.5 μm, and cheilocystidia longer than 25 μm, which can be distinctly differentiated from D. furfuracea [39,40]. The photograph of the pileus with a well-developed veil corresponding to the sequence MK607529 further confirms that these sequences were incorrectly identified. Therefore, the diversity within the species of the Deconica deserves more attention.
The most recent infrageneric classification of Deconica was the work by Noordeloos, who divided Deconica into three sections: Deconica (with two subsections: Deconica and Inquilinae), Melanotus, and Merdariae [2]. However, the infrageneric classification only considered species from Europe and was not supported by molecular phylogenetic research [1].
The results of our phylogenetic analysis were somewhat consistent with previous studies by Ramírez-Cruz et al., showing that some clades were strongly supported but difficult to morphologically characterize [1]. The clade containing D. citrispora (E. Horak) J.A. Cooper and D. neorhombispora (Guzmán) P.S. Silva, Ram.-Cruz & Guzmán includes over 20 species and has a strongly supported (BI = 0.97, ML = 88). This clade, pointed out by Ramírez-Cruz et al., corresponds to Psilocybe sect. Chrysocystidiatae sensu Singer, and is predominantly composed of taxa with chrysocystidia [17]. Nevertheless, more than ten species including the three new species, D. austrosinensis, D. furfuracea, and D. fuscobrunnea, discovered in this study, all lack chrysocystidia. Therefore, the morphological characterization viewpoint of this clade is difficult to sustain. Fortunately, the species diversity of this genus may be much richer than we previously thought. As more species are discovered, we believe that morphologically synapomorphies will be better resolved in the phylogenetic tree.
Acknowledgments
The authors are very grateful for the assistance of Hong Chen and Ling Ding in the field specimen collection and the anonymous reviewers of the manuscript.
Author Contributions
Conceptualization, J.-Q.Y.; data curation, S.-N.W.; formal analysis, B.-R.K.; funding acquisition, S.-N.W., Y.-P.H. and Z.-H.Z.; H.Z. investigation, J.-Q.Y., S.-N.W., C.-F.N., Z.-H.Z. and H.Z.; methodology, J.-Q.Y.; project administration, J.-Q.Y.; software, S.-N.W.; writing—original draft, J.-Q.Y. and S.-N.W.; Writing—review and editing, Z.-H.Z. and H.Z. All authors have read and agreed to the published version of the manuscript. J.-Q.Y. and S.-N.W. contributed equally to this work.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The sequences generated in this study are available in NCBI GenBank under the accession numbers shown in Table 1. The specimens studied in this study were deposited in the Herbarium of Fungi, Jiangxi Agricultural University (HFJAU). All alignments for phylogenetic analyses were deposited in TreeBASE (ID: TB2:S31681); the following links were available: http://purl.org/phylo/treebase/phylows/study/TB2:S31681?x-access-code=a06fbd515d1b8da177f23c6f45f2aaf2&format=html (accessed on 5 September 2024).
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This research was funded by the National Natural Science Foundation of China, grant numbers 32460326 and 31960008; Jiangxi Provincial Natural Science Foundation, grant number 20224BAB205003; Fujian Provincial Natural Science Foundation, grant number 2023J01379; the Project of FAAS, grant number XTCXGC2021007; and Yichang Biodiversity Conservation Initiative, grant number 202309076.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Ramírez-Cruz V., da Silva P.S., Villalobos-Arámbula A.R., Matheny P.B., Noordeloos M., Morgado L., da Silveria R.M.B., Guzmán-Dávalos L. Two new species of Deconica (Agaricales, Basidiomycota) from Australia and Mexico. Mycol. Prog. 2020;19:1317–1328. doi: 10.1007/s11557-020-01629-w. [DOI] [Google Scholar]
- 2.Noordeloos M.E. Strophariaceae s.l. Fungi Europeai No. 13. Canduso Edizione; Saronno, Italy: 2011. [Google Scholar]
- 3.Moncalvo J.-M., Vilgalys R., Redhead S.A., Johnson J.E., James T.Y., Aime M.C., Hofstetter V., Verduin S.J., Larsson E., Baroni T.J. One hundred and seventeen clades of euagarics. Mol. Phylogenet. Evol. 2002;23:357–400. doi: 10.1016/S1055-7903(02)00027-1. [DOI] [PubMed] [Google Scholar]
- 4.Smith W.G. Clavis Agaricinorum: An Analytical Key to the British Agaricini, with Characters of the Genera and Subgenera. L. Reeve & Company; London, UK: 1870. [Google Scholar]
- 5.Karsten P.A. Rysslands, Finlands och den Skandinaviska Halföns Hattsvampar. Finska Litteratur-Sällskapets Tryckeri; Helsinki, Finland: 1879. [Google Scholar]
- 6.Guzmán G. Species diversity in the genus Psilocybe (Basidiomycotina, Agaricales, Strophariaceae) of world mycobiota, with special attention to hallucinogenic properties. Int. J. Med. Mushrooms. 2005;7:305–331. doi: 10.1615/IntJMedMushr.v7.i12.280. [DOI] [Google Scholar]
- 7.Kalichman J., Kirk P.M., Matheny P.B. A compendium of generic names of agarics and Agaricales. Taxon. 2020;69:425–447. doi: 10.1002/tax.12240. [DOI] [Google Scholar]
- 8.Bau T. Flora Fungorum Sinicorum, Vol. 49 Strophariaceae. Beijing Science & Technology Press Co., Ltd.; Beijing, China: 2014. [Google Scholar]
- 9.Ma T. Ph.D. Thesis. Chinese Academy of Forestry; Beijing, China: 2014. Taxonomy of Psilocybe s.l. and Panaeolus in Yunnan, Southwest China, with notes on related genus Protostropharia. [Google Scholar]
- 10.Kornerup A., Wanscher J.H.K. The Methuen Handbook of Colour. 3rd ed. Eyre Methuen Ltd. Reprint.; London, UK: 1978. p. 252. [Google Scholar]
- 11.Horak E. Röhrlinge und Blätterpilze in Europa: Bestimmungsschlüssel für Polyporales (pp), Boletales, Agaricales, Russulales. Spektrum Akad. Verlag; Heidelberg, Germany: 2005. p. 575. [Google Scholar]
- 12.Jahnke K. A simple technique for staining chrysocystidia with patent blue V. Mycologia. 1984;76:940–943. doi: 10.1080/00275514.1984.12023932. [DOI] [Google Scholar]
- 13.Yan J.-Q., Zeng Z.-H., Hu Y.-P., Ke B.-R., Zeng H., Wang S.-N. Taxonomy and multi-gene phylogeny of Micropsalliota (Agaricales, Agaricaceae) with description of six new species from China. Front. Microbiol. 2022;13:1011794. doi: 10.3389/fmicb.2022.1011794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen L.-G., Ding L., Chen H., Zeng H., Zeng Z.-H., Wang S.-N., Yan J.-Q. Seven New Species of Entoloma Subgenus Cubospora (Entolomataceae, Agaricales) from Subtropical Regions of China. J. Fungi. 2024;10:594. doi: 10.3390/jof10080594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.White T.J., Bruns T.D., Lee S.B., Taylor J.W., Innis M.A., Gelfand D.H., Sninsky J.J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. Academic Press; San Diego, CA, USA: 1990. pp. 315–322. [Google Scholar]
- 16.Hopple J.J., Vilgalys R. Phylogenetic relationships in the mushroom genus Coprinus and dark-spored allies based on sequence data from the nuclear gene coding for the large ribosomal subunit RNA: Divergent domains, outgroups, and monophyly. Mol. Phylogenet. Evol. 1999;13:1–19. doi: 10.1006/mpev.1999.0634. [DOI] [PubMed] [Google Scholar]
- 17.Ramírez-Cruz V., Guzmán G., Villalobos-Arámbula A.R., Rodríguez A., Matheny P.B., Sánchez-García M., Guzmán-Dávalos L. Phylogenetic inference and trait evolution of the psychedelic mushroom genus Psilocybe sensu lato (Agaricales) Botany. 2013;91:573–591. doi: 10.1139/cjb-2013-0070. [DOI] [Google Scholar]
- 18.Wang S.N., Fan Y.G., Yan J.Q. Iugisporipsathyra reticulopilea gen. et sp. nov.(Agaricales, Psathyrellaceae) from tropical China produces unique ridge-ornamented spores with an obvious suprahilar plage. MycoKeys. 2022;90:147–162. doi: 10.3897/mycokeys.90.85690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen L.T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2014;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 22.Lanfear R., Frandsen P.B., Wright A.M., Senfeld T., Calcott B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017;34:772–773. doi: 10.1093/molbev/msw260. [DOI] [PubMed] [Google Scholar]
- 23.Ramírez-Cruz V., Cortés-Pérez A., Borovička J., Villalobos-Arámbula A.R., Matheny P.B., Guzmán-Dávalos L. Deconica cokeriana (Agaricales, Strophariaceae), a new combination. Mycoscience. 2019;61:95–100. doi: 10.1016/j.myc.2019.07.001. [DOI] [Google Scholar]
- 24.Zhang W., Wendel J.F., Clark L.G. Bamboozled again! Inadvertent isolation of fungal rDNA sequences from bamboos (Poaceae: Bambusoideae) Mol. Phylogenet. Evol. 1997;8:205–217. doi: 10.1006/mpev.1997.0422. [DOI] [PubMed] [Google Scholar]
- 25.Masiulionis V.E., Weber R.W., Pagnocca F.C. Foraging of Psilocybe basidiocarps by the leaf-cutting ant Acromyrmex lobicornis in Santa Fé, Argentina. SpringerPlus. 2013;2:254. doi: 10.1186/2193-1801-2-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vu D., Groenewald M., De Vries M., Gehrmann T., Stielow B., Eberhardt U., Al-Hatmi A., Groenewald J.Z., Cardinali G., Houbraken J. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 2019;92:135–154. doi: 10.1016/j.simyco.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park M.S., Cho H.J., Kim N.K., Park J.Y., Lee H., Park K.H., Kim M.-J., Kim J.-J., Kim C., Lim Y.W. Ten new recorded species of macrofungi on Ulleung Island, Korea. Mycobiology. 2017;45:286–296. doi: 10.5941/MYCO.2017.45.4.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hughes K.W., Petersen R.H., Lickey E.B. Using heterozygosity to estimate a percentage DNA sequence similarity for environmental species’ delimitation across basidiomycete fungi. New Phytol. 2009;182:795–798. doi: 10.1111/j.1469-8137.2009.02802.x. [DOI] [PubMed] [Google Scholar]
- 29.Desjardin D., Perry B. Dark-spored species of Agaricineae from Republic of São Tomé and Príncipe, West Africa. Mycosphere. 2016;7:359–391. doi: 10.5943/mycosphere/7/3/8. [DOI] [Google Scholar]
- 30.Ammirati J., Traquair J., Martin S., Gillon W., Ginns J. A new Melanotus from gold-mine timbers in Ontario. Mycologia. 1979;71:310–321. doi: 10.1080/00275514.1979.12021013. [DOI] [Google Scholar]
- 31.Horak E., Guzmán G., Desjardin D. Four new species of Psilocybe from Malaysia and Thailand, with a key to the species of sect. Neocaledonicae and discussion on the distribution of the tropical and temperate species. Sydowia. 2009;61:25–37. [Google Scholar]
- 32.Horak E. Contributions to the knowledge of the Agaricales s.l. (Fungi) of New Zealand. N. Z. J. Botan. 1971;9:463–493. doi: 10.1080/0028825X.1971.10430194. [DOI] [Google Scholar]
- 33.Guzmán G., Horak E. New species of Psilocybe from Papua New Guinea, New Caledonia and New Zealand. Sydowia. 1979;31:45–55. [Google Scholar]
- 34.Huijsman H. Sur trois Psilocybe. Persoonia. 1961;2:91–95. [Google Scholar]
- 35.Guzmán G., Ramírez-Guillén F., Hyde K., Karunarathna S. Psilocybe s.s. in Thailand four new species and a review of previously recorded species. Mycotaxon. 2012;119:65–81. doi: 10.5248/119.65. [DOI] [Google Scholar]
- 36.Noordeloos M.E. Notulae ad Floram agaricinam neerlandicam—XXXIV. Further notes on Psilocybe. Persoonia. 1999;17:245–257. [Google Scholar]
- 37.Singer R. New taxa and new combinations of Agaricales (Diagnoses fungorum novorum Agaricalium IV) Fieldiana Bot. New Ser. 1989;21:1–133. [Google Scholar]
- 38.Horak E. The genus Melanotus Pat. Persoonia. 1977;9:305–327. [Google Scholar]
- 39.Noordeloos M.E. Studies in Psilocybe sect. Psilocybe. Österr. Z. Pilzk. 2001;10:115–180. [Google Scholar]
- 40.Knudsen H., Vesterholt J. Agaricoid, Boletoid, Cyphelloid and Gasteroid Genera. Nordsvamp; Copenhagen, Denmark: 2012. Funga Nordica. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequences generated in this study are available in NCBI GenBank under the accession numbers shown in Table 1. The specimens studied in this study were deposited in the Herbarium of Fungi, Jiangxi Agricultural University (HFJAU). All alignments for phylogenetic analyses were deposited in TreeBASE (ID: TB2:S31681); the following links were available: http://purl.org/phylo/treebase/phylows/study/TB2:S31681?x-access-code=a06fbd515d1b8da177f23c6f45f2aaf2&format=html (accessed on 5 September 2024).