Abstract
Mutations at the Nramp1 locus in vivo cause susceptibility to infection by unrelated intracellular microbes. Nramp1 encodes an integral membrane protein abundantly expressed in the endosomal-lysosomal compartment of macrophages and is recruited to the phagosomal membrane following phagocytosis. The mechanism by which Nramp1 affects the biochemical properties of the phagosome to control microbial replication is unknown. To devise an in vitro assay for Nramp1 function, we introduced a wild-type Nramp1G169 cDNA into RAW 264.7 macrophages (which bear a homozygous mutant Nramp1D169 allele and thus are permissive to replication of specific intracellular parasites). Recombinant Nramp1 was expressed in a membranous compartment in RAW264.7 cells and was recruited to the membrane of Salmonella typhimurium and Yersinia enterocolitica containing phagosomes. Evaluation of the antibacterial activity of RAW264.7 transfectants showed that expression of the recombinant Nramp1 protein abrogated intracellular replication of S. typhimurium. Studies with a replication-defective S. typhimurium mutant suggest that this occurs through an enhanced bacteriostatic activity. The effect of Nramp1 expression was specific, since (i) it was not seen in RAW264.7 transfectants overexpressing the closely related Nramp2 protein, and (ii) control RAW264.7 cells, Nramp1, and Nramp2 transfectants could all efficiently kill a temperature-sensitive, replication-defective mutant of S. typhimurium. Finally, increased antibacterial activity of the Nramp1 RAW264.7 transfectants was linked to increased phagosomal acidification, a distinguishing feature of primary macrophages expressing a wild-type Nramp1 allele. Together, these results indicate that transfection of Nramp1 cDNAs in the RAW264.7 macrophage cell line can be used as a direct assay to study both Nramp1 function and mechanism of action as well as to identify structure-function relationships in this protein.
In inbred mouse strains, susceptibility to infection with Mycobacterium, Salmonella, and Leishmania is controlled by the Bcg/Ity/Lsh locus (20). The genetic advantage of resistant versus susceptible strains is expressed by a differential bacterial growth observed in spleen and liver during the early phase of the infection (20). In vivo experiments with mutant strains of mice, with bone marrow radiation hybrids, and with macrophage poisons suggest that the macrophage is the cell type that phenotypically expresses the genetic difference at Bcg/Ity/Lsh (19). Differential growth rates of Mycobacterium bovis (32), Mycobacterium smegmatis (11), Mycobacterium avium (10, 33), Mycobacterium intracellulare (16), Salmonella typhimurium (26), and Leishmania donovani (8) in vitro in primary macrophages have confirmed that this cell type is affected by Bcg/Ity/Lsh. It has been proposed that Bcg/Ity/Lsh either affects the bactericidal and bacteriostatic activity of the macrophage (26) or affects priming for activation (3, 5).
The positional cloning of Bcg/Ity/Lsh led to the identification of the Nramp1 gene (natural-resistance-associated macrophage protein 1) gene (37). Nramp1 mRNA expression is restricted to spleen and liver and is abundant in macrophage populations purified from these organs (37). Nramp1 expression can be further upregulated by exposure to bacterial lipopolysaccharide and gamma interferon (IFN-γ), as well as by exposure to an inflammatory stimuli (17). Amino acid sequence analysis of the predicted Nramp1 protein sequence reveals features suggestive of an integral membrane protein with transport function, including 12 highly hydrophobic membrane-spanning segments (7, 37), a glycosylated extracellular loop (38), and a consensus “transport signature” previously detected in several prokaryotic and eukaryotic transport proteins (9, 25, 37). In Bcgs inbred strains, susceptibility to infection is associated with a glycine-to-aspartic acid substitution at position 169 (G169D) within predicted transmembrane domain 4 (TM4) (27). The identity of Nramp1 as Bcg/Ity/Lsh has been verified in vivo in transgenic animals bearing either a null (36) or a gain-of-function (18) allele at Nramp1. Recently, polymorphic variants at the human NRAMP1 gene have been associated with susceptibility to tuberculosis (2, 31) and leprosy (1) in populations from areas where these diseases are endemic.
In macrophages, biochemical studies with specific anti-Nramp1 antibodies showed that Nramp1 is a 90- to 110-kDa membrane phosphoglycoprotein expressed in an endomembrane compartment (38). Colocalization studies have shown that Nramp1 is expressed in Lamp-1 positive lysosomal compartments (22). Moreover, studies with phagosomes containing latex beads (22) have shown that, upon phagocytosis, Nramp1 is rapidly recruited to the membrane of the phagosome and remains associated with this organelle throughout phagolysosome biogenesis. Association of Nramp1 with the phagosome suggests that Nramp1 may modify the phagosomal microenvironment to affect microbial replication. Recently, it has been shown that targeting of Nramp1 to the M. bovis-containing phagosomes results in increased acidification of the phagosome compared to phagosomes from identical macrophages where the Nramp1 gene had been disrupted (24). This effect was specific for bacterial phagosomes, was not seen in phagosomes containing either inert latex particles or dead mycobacteria, and was associated with reduced activity or recruitment of the vacuolar H+-ATPase (24). These results have suggested that Nramp1 may directly or indirectly influence the intraphagosomal pH to alter microbial proliferation.
A second Nramp protein, Nramp2 (78% protein identity), exists in mammals and is ubiquitously expressed in several tissues (21). Nramp2 was shown to be the major transferrin-independent iron uptake system of mammals and is mutated in two animal models of microcytic anemia and deficiency in intestinal iron uptake (14). In addition, studies with oocytes have shown that Nramp2 can transport a number of divalent cations, such as Fe2+, Zn2+, Mn2+, and others (23). Nramp also defines a highly conserved family of proteins with members identified in insects (65% identity), plants (52% identity), yeast (40% identity), and even in several bacterial species, including mycobacteria (35% identity) (6, 7). The yeast SMF1 homologue was shown to be a Mn2+ transporter (34), and mammalian Nramp2 can functionally complement an SMF mutant in yeast (29). Together, these results suggest that the Nramp family encodes a family of divalent cation transporters, implicating Nramp1 in this capacity as well.
However, in the absence of a functional assay in vitro, the exact mechanism of action and substrate of Nramp1 in the phagosomal membrane remains difficult to assess. In the current study, we expressed a recombinant Nramp1 protein in the RAW264.7 macrophage cell line, which contains an endogenous, nonfunctional mutant allele in Nramp1. The recombinant protein is properly expressed in RAW264.7 cells and is targeted to the phagosomal membrane. Nramp1 expression in RAW264.7 cells was capable of overcoming innate susceptibility to infection with S. typhimurium in vitro and caused enhanced acidification of the intraphagosomal space. These results suggest that transfection and overexpression of Nramp1 in RAW264.7 macrophages can constitute a convenient in vitro assay for structure-function studies in this important host resistance molecule.
MATERIALS AND METHODS
S. typhimurium infections in mice.
Normal inbred mouse strain 129sv (Nramp1G169/G169) and 129sv mice bearing a null allele at Nramp1 (Nramp1null) (36) were infected with S. typhimurium Keller, originally obtained from Hugh Robson (Royal Victoria Hospital, Montreal, Canada). Mice were inoculated in the caudal tail vein with 0.2 ml of physiological saline containing 0.8 × 103 live Salmonella. Bacterial replication was assessed by determining the number of CFU in liver and spleen homogenates at predetermined time intervals, as previously described (20). The inoculum of S. typhimurium was prepared from a culture during exponential growth phase (2 h at 37°C) in trypticase soy broth, and the exact size of the infectious inoculum was determined by CFU counts of serial 10-fold dilutions plated on trypticase soy agar.
Cell culture and transfection.
The RAW264.7 cell line (ATCC TIB 71) is an immortalized macrophage clone isolated from BALB/c mice (Nramp1D169/D169) transformed with Abelson leukemia virus (30). These cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) high-glucose formulation (supplemented with 10% fetal bovine serum [FBS], 200 mM l-glutamine, and 10 mM HEPES). To obtain a macrophage cell clone expressing the resistant Nramp1G169 allele, RAW264.7 cells were transfected with the pCB6 expression vector encoding the Nramp1G169 protein fused in frame to a c-Myc epitope tag at the carboxyl terminus (22). The pCB6 vector uses cytomegalovirus promoter-enhancer sequences to direct high-level expression of Nramp1; it also contains the neo gene as a selection marker for transfection. For transfections, 40 μg of plasmid DNA was precipitated and resuspended in 50 μl of phosphate-buffered saline (PBS). DNA was added to a 0.4-cm electroporation cuvette containing 20 × 106 RAW264.7 cells resuspended in 0.75 ml of DMEM supplemented with 10% FBS. A Gene Pulser (Bio-Rad) was used to electroporate the sample at 300 V and 960 μF using a capacitance extender. Cells were replated in a 140-mm dish and allowed to recover for 48 h, followed by a 7- to 10-day selection in medium containing 200 μg of geneticin (G418; Gibco-BRL)/ml. Individual colonies growing in G418 were individually picked, expanded in culture, and frozen in 90% FBS and 10% dimethyl sulfoxide. These clones were then screened for expression of the Nramp1-cMyc recombinant protein.
Immunoblot analysis of Nramp1 expression in RAW264.7 cells.
Enriched membrane fractions were prepared from RAW264.7 controls and from the Nramp1 transfected clones in accordance with a published protocol (13). Briefly, cell monolayers were removed from plastic surfaces by gentle scraping, followed by three consecutive washes in PBS. Cells were then homogenized in hypotonic medium by using a Dounce homogenizer, followed by elimination of unbroken cells and nuclei by low-speed centrifugation (2,000 × g, 10 min) and pelleting of the crude membrane fraction by centrifugation of the supernatant (100,000 × g, 30 min). The final membrane pellet was resuspended in TNE (10 mM Tris, NaCl, and 1 mM EDTA), 30% glycerol, and protease inhibitors. Equal amounts (20 μg) of protein were electrophoresed on a sodium dodecyl sulfate (SDS)–7.5% polyacrylamide gel, followed by transfer to nitrocellulose membrane. Equal loading on gel and equal transfer to the immunoblot was verified by light staining of the membrane with Ponceau red. For immunodetection of Nramp1-cMyc proteins, blots were incubated in blocking solution (150 mM NaCl, 10 mM Tris [pH 8.0], 0.1% Tween 20, and 5% nonfat dry milk) for 16 h at 4°C. After blocking, membranes were incubated with a rabbit polyclonal antibody raised against an N-terminal epitope of Nramp1 (used at a 1:100 dilution). Membranes were then washed three times with TBST buffer (NaCl, Tris [pH 8.0], and 0.01% Tween 20) before incubation with a goat anti-rabbit secondary antibody conjugated to a horseradish peroxidase. After 30 min, the membrane was washed four times with TBST, and specific immune complexes were revealed by enhanced chemiluminescence (Amersham).
Immunofluorescence.
S. typhimurium SL14028s expressing green fluorescent protein (GFP) (provided by Olivia Steele-Mortimer) and Yersinia enterocolitica E40 (pYV40) were used for infection of RAW264.7 macrophages in vitro and immunofluorescence. Strain SL14028s-GFP was passaged on Luria-Bertani (LB) agar plates containing 15 μg of tetracycline/ml at 37°C, while strain E40 (pYV40) was passaged on brain heart infusion (BHI) agar (Difco Laboratories, Detroit, Mich.) plates at 30°C. Preparation of bacterial inoculum for infection was as previously described (28). Briefly, S. typhimurium SL14028s-GFP was grown with shaking for 16 h in LB broth containing 15 μg of tetracycline/ml at 37°C. The following morning, strain SL14028s-GFP was subcultured 1:33 into LB broth and grown for 2.5 h with shaking at 37°C to late log phase. Y. enterocolitica E40 (pYV40) was grown for 16 h in BHI broth with constant agitation at 30°C. The following morning, E40 (pYV40) was subcultured 1:25 into BHI broth and grown for 3 h with constant agitation at 30°C. In all cases, the bacteria were harvested by centrifugation (1,000 × g) and resuspended in PBS which was diluted with Earle’s buffered salt solution (pH 7.4). Twenty hours in advance of infection, RAW264.7 macrophages were seeded at 2.5 × 105 cells per 12-mm glass coverslip in DMEM supplemented with 10% fetal calf serum (FCS) in 24-well tissue culture plates. Cells were washed once with PBS, and bacteria were added to the cell samples at a multiplicity of infection (MOI) of 15 S. typhimurium and 2 Y. enterocolitica per macrophage. Following a 15-min infection period, cells were washed twice with PBS, and growth medium containing 12.5 μg of gentamicin/ml was added to further eliminate extracellular bacteria.
Cells were processed for immunofluorescence microscopy as previously described (28). After 2 h, infected cells were washed twice with PBS and fixed with 2.5% paraformaldehyde (wt/vol) for 15 min. The cells were then blocked for 30 min in SS-PBS (PBS containing 0.2% saponin and 25% normal goat serum). The cells were stained with primary antibody in SS-PBS to permeabilize the cells for 60 min at room temperature (RT). Mouse monoclonal anti-Myc antibody 9E10 (Santa Cruz Biotechnology Inc.) was used at a 1:50 dilution to identify the Nramp1-cMyc fusion protein. Rabbit anti-Y. enterocolitica O antiserum (group O:9) (Accurate Chemical, Westbury, N.Y.) was used at a 1:200 dilution. The cells were then washed three times with PBS and incubated for 30 min in SS-PBS followed by the addition of the respective secondary goat anti-mouse antibodies diluted in SS-PBS for 60 min in the dark at 20°C. Anti-mouse Alexa-594 (Molecular Probes Inc.) was used at a dilution of 1:200, and anti-rabbit Alexa-488 (Molecular Probes Inc.) was used at a dilution of 1:400. Cells were then washed three times with PBS and mounted onto slides for epifluorescence microscopy. Cells were photographed with a Zeiss Axioscope microscope under oil immersion (×1,000 magnification).
Replication of S. typhimurium in RAW264.7 macrophages.
Control RAW264.7 macrophages and RAW264.7 Nramp transfectants were grown to 70% confluency in normal DMEM without geneticin (for approximately 48 h). Cells were harvested, seeded at 5 × 105 cells per well in 24-well tissue culture plates, allowed to adhere, and exposed to recombinant IFN-γ (100 U/ml) (Genzyme, Cambridge, Mass.) for 24 h. Cultures of S. typhimurium SL1344 and of the temperature-sensitive mutant (TSΔ27 [26]) were prepared from frozen stocks the day prior to infection. Frozen stocks were diluted in LB broth and grown for 18 h at 37°C (or at RT for TSΔ27). For infection, the bacterial cultures were diluted in DMEM supplemented with 10% heat-inactivated FBS to an optical density at 595 nm of ∼0.13 (approximately 107 bacilli/ml) and incubated on ice for 30 min. Cell samples were washed twice with Hank’s buffered saline solution (HBSS; Gibco-BRL) and then overlaid with 0.4 ml of bacterial suspension containing 5 × 106 bacilli (MOI of 10). Phagocytosis of bacteria occurred for 30 min at 37°C in 5% CO2. At this point, cell cultures were gently washed three times with HBSS to remove nonadherent bacteria. The infection was then continued in the presence of DMEM containing 12.5 μg of gentamicin (Gibco-BRL)/ml to prevent the replication of extracellular bacteria. At predetermined time intervals, cell monolayers were washed twice with HBSS and treated with 0.5 ml of 0.01% bovine serum albumin (BSA) to osmotically lyse the macrophages. After pipetting the cells up and down 10 times, serial dilutions were plated on LB agar plates for CFU counts (minimum of three independent measurements). Results are expressed as the level of infection, which represents CFU counts at each time interval (CFUt) compared to CFU counts after initial phagocytosis (CFU0) and is presented as a percentage.
Measurement of phagosomal pH and microfluorescence imaging.
Measurements of phagosomal pH were obtained through the combined application of video microscopy and fluorescence ratio imaging. Nontransfected and transfected RAW264.7 cells were grown as described in the previous section. M. bovis BCG (substrain Montreal) was obtained from Armand Frappier Institute (Laval, Quebec, Canada) and maintained as described previously (15). Cells were grown overnight on acid-washed glass coverslips to semiconfluency (approximately 70%). BCG was resuspended in DMEM containing 10% FCS and added to cells in six-well plates at an MOI of 10 bacteria per cell and further incubated for 1 h at 37°C. Cells were then washed extensively with DMEM to remove nonadherent bacteria. Cover slips were then transferred to the stage of a Leica inverted microscope in a Leiden chamber controlled at 37°C for measurement of phagosomal pH, as described previously (24).
RESULTS AND DISCUSSION
S. typhimurium infection in 129sv and 129sv.Nramp1nullmutants in vivo.
Although the study of Nramp family members suggests a transport function in the macrophage phagosomal membrane for Nramp1, its antimicrobial mechanism of action remains unknown. Functional understanding of Nramp1 has so far relied on in vivo and in vitro studies that show that Nramp1 mutations cause susceptibility to intracellular infections by impairing the ability of macrophages to restrict microbial replication. Our approach to devising an in vitro functional assay for Nramp1 consisted in (i) transfecting a recombinant Nramp1 protein (Nramp1G169) into immortalized macrophages carrying a mutant allele (Nramp1D169), (ii) assessing whether the protein is properly expressed and targeted in these cells, and (iii) determining if Nramp1G169 expression can correct the permissive phenotype of these cells to infection with intracellular pathogen.
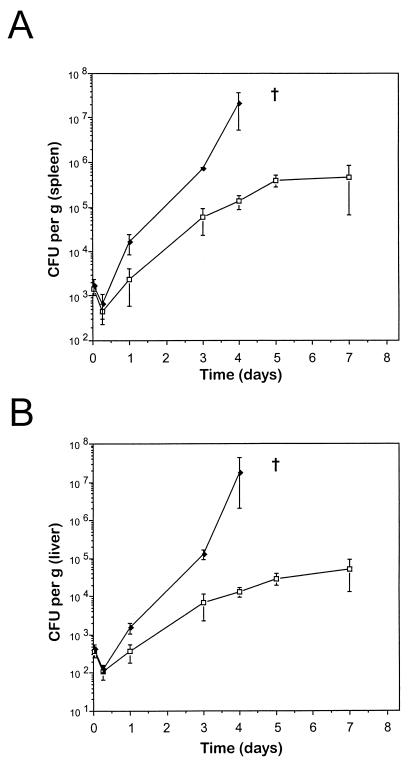
Although the effect of Nramp1 mutations in vivo on the replication of S. typhimurium, M. bovis, and L. donovani has been well documented (18, 36), the latter two organisms are slow replicating and not ideal for short-term in vitro infections using replication-competent macrophage cell lines. Thus, we opted to use S. typhimurium as an infectious agent in these studies. Previous studies with inbred and congenic mouse strains bearing Nramp1r and Nramp1s alleles, designated Ityr and Itys at the time, suggested Nramp1 had an effect on the levels of S. typhimurium as early as 24 h after infection (35). We first verified the effect of loss of Nramp1 function on the rate of early replication of our isolate of S. typhimurium in spleen and liver in vivo. For this, we used a pair of 129sv mouse strains that are genetically identical except for Nramp1, which has been disrupted by homologous recombination (129sv.Nramp1null) (36). These animals were infected intravenously with 0.8 × 103 S. typhimurium, and bacterial replication (CFU counts/gram of tissue homogenate) in the spleen and liver of these mice was measured at 1 and 6 h as well as 1, 3, and 4 days postinfection (Fig. 1). Measurements were made at 5 and 7 days postinfection in 129sv mice but not in 129sv.Nramp1null mice because none of the latter survived longer than 4 days postinfection. The kinetics of infection were quite similar in the spleen and liver. In both organs, an initial Nramp1-independent reduction in CFU counts was noted during the first 6 h. This phase was followed by an active replication of the bacilli in spleen and liver of both strains between 6 h and 3 days; however, S. typhimurium replication was more extensive in the Nramp1null mice, resulting in a 10- to 20-fold difference in CFU counts at 3 days. Between days 3 and 4, continuous and exponential replication was seen in mutant Nramp1null mice which ultimately led to uniform mortality in this group by day 5 (data not shown). In contrast, in 129sv mice, S. typhimurium replication peaked at day 3 (100- and 1,000-fold interstrain difference in CFU counts in spleen and liver, respectively) and remained constant for later time points. In the 129sv group, no mortality was observed during a 15-day observation period, despite continuous bacillar presence in the spleen and liver (data not shown). Bacterial clearance in the latter phase of infection is Nramp1 independent and is controlled by genes of the major histocompatibility complex (4). These experiments verify the key role of Nramp1 in acute S. typhimurium infection in vivo and show the effects of Nramp1 can be detected as rapidly as a few hours postinfection in the spleen (1.5- to 2-fold difference). In addition, these differences become more pronounced with time and are visibly distinct by 24 h (5- to 10-fold difference), a finding which parallels results described by Swanson and O’Brien (35). These results indicate that S. typhimurium would be a suitable infectious agent for in vitro infection studies of immortalized macrophages.
FIG. 1.
Effect of Nramp1 on in vivo replication of S. typhimurium in spleen and liver. Mouse strains 129sv (□) and 129sv.Nramp1null (⧫) were infected intravenously with 0.8 × 103 S. typhimurium Keller CFUs. At 1 h, 6 h, 1 day, 3 days, 4 days, 5 days, and 7 days postinfection, spleens (A) and livers (B) were removed, weighed, and homogenized for CFU counts. The results are expressed as CFU per gram of tissue. All 129sv.Nramp1null mice died from infection prior to the 5-day time point, which is denoted by a cross. A minimum of three to five mice was used for each time point, and the results are shown as means ± standard deviations.
Creation of a transfected cell line RAW264.7 expressing Nramp1G169.
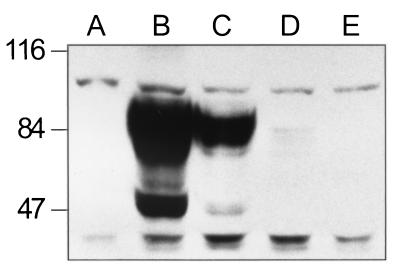
To test the activity of the Nramp1 gene in transfection assays, we used the immortalized macrophage cell line RAW264.7 as a recipient cell. The RAW264.7 cell line was initially derived by Abelson leukemia virus transformation from the BALB/c mouse strain (30) and is homozygous for the susceptible Nramp1D169 allele (27) that encodes a nonfunctional protein rapidly targeted for degradation in macrophages (36). The wild-type allele of Nramp1 (Nramp1G169) was cloned in the mammalian expression vector pCB6 and introduced by transfection in RAW264.7 cells. To facilitate identification of the recombinant Nramp1G169 protein in transfected cells, we modified Nramp1 cDNA by adding a c-Myc epitope tag fused in frame at the C terminus of the protein that can be identified using a commercial monoclonal anti-tag antibody (9E10; Babco Inc.). RAW264.7 cells were transfected by electroporation with the pCB6 Nramp1-cMyc construct followed by selection in G418 for 2 weeks, at which time 16 clones were picked, expanded in culture, and analyzed for expression of the Nramp1-cMyc fusion protein. Enriched membrane fractions were prepared from positive clones and further analyzed by Western blotting using a rabbit anti-Nramp1 polyclonal antiserum (22). Figure 2 shows an immunoblot of three positive clones which express different amounts of the recombinant protein. The Nramp1-cMyc protein is detected both as a 50-kDa species and as a diffuse band of approximately 90 kDa. The apparent molecular mass of the lower band is in agreement with the predicted mass of Nramp1 from the primary amino acid sequence, while the 90-kDa band corresponds to the highly glycosylated mature form of the protein. These characteristics are in agreement with the previously observed mobility of the Nramp1 phosphoglycoprotein expressed in primary peritoneal macrophages (38). Clone 13 (Fig. 2, lane B) expresses very high amounts of the protein, clone 15 (lane C) expresses a lesser amount, and clone 2.2 (lane D) expresses only a small amount of immunoreactive protein. The immunoreactive species were not detected in either nontransfected RAW macrophages or RAW264.7 clones transfected with Nramp2 (lanes A and E, respectively). A parallel analysis by immunoprecipitation produced similar results (data not shown). These results establish that recombinant Nramp1 proteins can be expressed by transfection in RAW264.7 macrophages.
FIG. 2.
Expression of recombinant Nramp1G169-cMyc fusion protein in transfected RAW264.7 clones. Enriched membrane fractions were prepared from RAW264.7 cells (lane A), Nramp1-cMyc-transfected RAW264.7 clone 13 (lane B), clone 15 (lane C), clone 2.2 (lane D), and Nramp2-transfected RAW264.7 cells (lane E). Equal amounts of protein (20 μg per sample) were loaded on an SDS–7.5% polyacrylamide gel, followed by transfer to a nitrocellulose membrane and immunoblotting with an isoform-specific anti-Nramp1 polyclonal antibody (22). After washing, a mouse anti-rabbit secondary antibody conjugated with horseradish peroxidase was used to reveal specific immune complexes.
Localization of Nramp1-cMyc protein to bacterial phagosomes in transfected RAW264.7 macrophages.
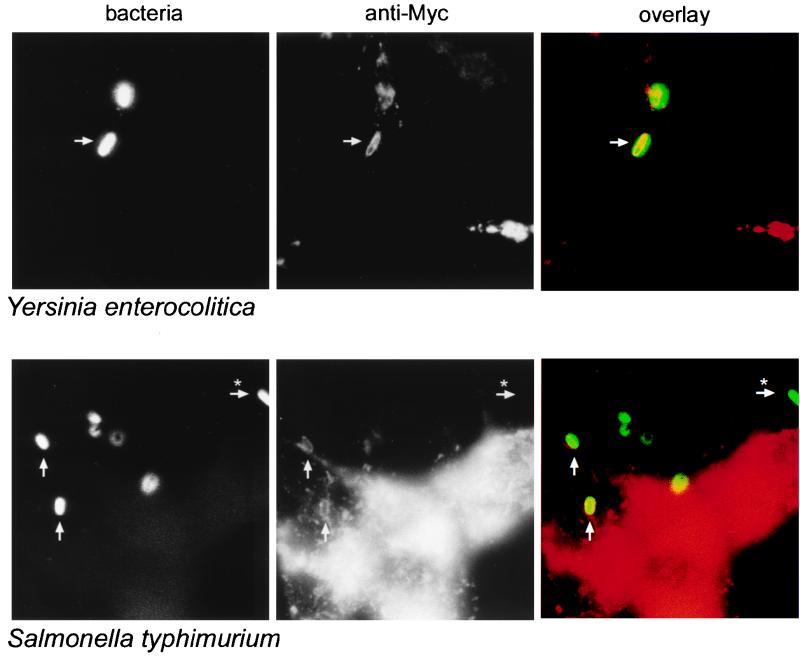
We next determined whether the recombinant Nramp1 protein was properly targeted to the phagosomal membrane in transfected RAW264.7 macrophages. For this, Nramp1-cMyc-transfected RAW264.7 cells were infected in vitro with either S. typhimurium (SL14028s-GFP) or Y. enterocolitica E40 (pYV40) for 2 h at 37°C. Cells were then fixed and stained with the 9E10 anti-cMyc monoclonal antibody or anti-Y. enterocolitica O antiserum. Fluorescence microscopy was used to visualize the bacterial phagosome and the transfected Nramp1 fusion protein. Results in the left panels of Fig. 3 show the immunofluorescence staining of Y. enterocolitica (upper series) and fluorescence emitted by S. typhimurium (SL14028s-GFP) (lower series). The middle panels show the immunofluorescence staining of the infected cells for the recombinant Nramp1-cMyc protein. As previously observed for the wild-type protein in primary cells, Nramp1-cMyc localizes not to the plasma membrane but rather to a subcellular membranous compartment, which appears as an intense punctate staining (compatible with an endosomal-lysosomal staining). In addition, we also observed association of Nramp1-cMyc with larger vesicular structures. Superimposition of the images (right panels) strongly suggests colocalization (yellow) of these large, Nramp1-cMyc-positive vesicular structures (red) with the internalized bacteria (green) detected by anti-Yersinia antibody or emission by Salmonella GFP. Extracellular bacteria emit only a green fluorescence, which indicates that the anti-c-Myc staining is specific to the RAW264.7 macrophages and that there is no bleed-through of fluorescence from the anti-Yersinia secondary antibody (anti-rabbit Alexa-488) or from GFP to the anti-c-Myc secondary antibody (anti-mouse Alexa-594). Together, these results indicate that the recombinant Nramp1-cMyc protein is targeted to the bacterial phagosome in transfected RAW264.7 macrophages in a manner similar to that observed in primary macrophages (22).
FIG. 3.
The recombinant Nramp1-cMyc protein localizes to Y. enterocolitica- and S. typhimurium-containing phagosomes in RAW264.7 macrophages. Nramp1-cMyc-expressing RAW264.7 macrophages were infected with Y. enterocolitica (upper panels) or S. typhimurium (lower panels). Two hours postinfection, cells were fixed with paraformaldehyde and analyzed by immunofluorescence. Y. enterocolitica were identified by staining with an anti-Yersinia antibody plus Alexa-488 secondary antibody (upper left panel). S. typhimurium used in this experiment express GFP and were identifiable in the same channel as the Alexa-488 antibody (lower left panel). An anti-c-Myc monoclonal antibody (9E10) was used to identify the c-Myc tag fused in frame at the C terminus of Nramp1 (middle panels). The respective images are superimposed in the right panels to show colocalization (yellow staining) with red staining representing Nramp1 expression and green staining representing bacteria. Arrows indicate colocalized bacteria and Nramp1, while arrows plus ∗ indicate extracellular bacteria as determined by phase contrast (not shown).
Anti-Salmonella activity of Nramp1-cMyc RAW264.7 transfectants.
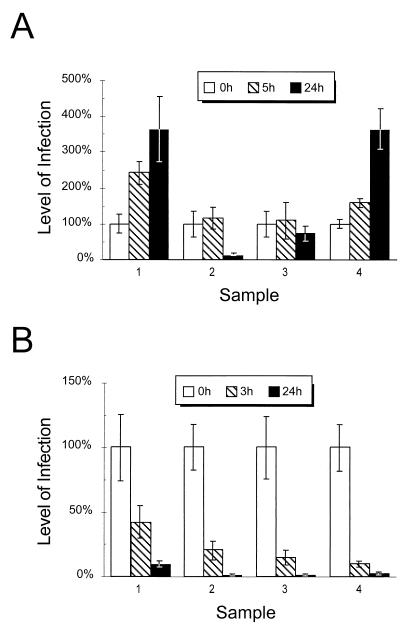
Having observed targeting of the cMyc-Nramp1 recombinant protein to the Salmonella phagosome in RAW264.7 transfectants, we next tested the consequences of Nramp1 expression on intracellular replication of S. typhimurium. For this, we used an in vitro infection assay previously described for primary macrophages by Lissner et al. (26) that we adapted for use with the immortalized RAW264.7 transfectants. RAW264.7 macrophages were infected with S. typhimurium and, at predetermined times, the number of viable CFUs recovered from lysed cells was monitored. For these assays, we used two independent RAW264.7 Nramp1-cMyc transfectants (Fig. 4, sample 2 and sample 3) as well as control nontransfected RAW264.7 cells (Fig. 4, sample 1) and a RAW264.7 transfectant expressing the second member of the Nramp family, Nramp2 (Fig. 4, sample 4). Nramp2 is not known to play a role in resistance to infection. In the assay, we used both a highly virulent strain of S. typhimurium (SL1344) as well as a replication-defective, temperature-sensitive mutant, TSΔ27 (26). Several preliminary experiments to establish optimal experimental conditions for phagocytosis, replication, and bacilli recovery from infected cells were carried out. Experimental conditions to eliminate extracellular bacterial replication during this assay period were also established. In the final experimental protocol (see Materials and Methods), control RAW264.7 cells and RAW264.7 transfectants treated with IFN-γ (24 h) were allowed to phagocytose bacilli for 30 min. Cell cultures were then washed extensively to remove extracellular bacteria, and medium containing gentamicin was added to prevent replication of any remaining extracellular bacilli. Under these conditions, an average of 0.4 bacteria/macrophage was obtained. At 0, 5, and 24 h postinfection, macrophages were lysed with hypotonic medium (0.01% BSA), and cell extracts were plated on LB agar for CFU counts. The results in Fig. 4 show the increase in CFU/well compared to CFU/well at initial phagocytosis and are expressed as percentages. Figure 4 shows a representative experiment. For each macrophage population, the average of five independent wells per time point is shown.
FIG. 4.
Effect of recombinant Nramp1-cMyc protein expression on antibacterial activity of RAW264.7 macrophages. Nontransfected RAW264.7 cells (sample 1), Nramp1-cMyc-transfected RAW264.7 macrophage clone 13 (sample 2), clone 2.2 (sample 3), and a Nramp2-transfected RAW264.7 clone (sample 4) were seeded at 5 × 105 cells per well (5 wells per sample). Cells were infected with either S. typhimurium SL1344 (A) or the temperature-sensitive, replication-defective mutant TSΔ27 (B). After an initial 30-min phagocytosis period (T0), cell cultures were lysed at predetermined time intervals, and CFU counts were determined. The level of infection was determined by dividing the number of CFUt for each well at individual time points by the CFU0 (at T0) and is expressed as a percentage. The standard deviations for each time point are shown. The ranges in individual values for the samples at 24 h in panel A were as follows: sample 1, 4.4- to 3.0-fold increase; sample 2, 10 to 5%; sample 3, 81 to 57%; and sample 4, 4.2- to 3.1-fold increase. For panel B, the ranges for the samples at 24 h were as follows: sample 1, 11.5 to 8.5%; sample 2, 2.7 to 0.7%; sample 3, 1.6 to 0.3%; and sample 4, 4.2 to 2.1%.
Using this protocol, the rate of phagocytosis of the infectious inoculum by nontransfected and transfected RAW264.7 cells was very similar and did not vary by more than twofold in each experiment (data not shown). After 5 h, bacterial replication was already apparent in RAW264.7 controls (2.4-fold increase) and in Nramp2 transfectants (1.6-fold increase) but was absent in Nramp1 transfectants. Twenty-four hours after phagocytosis, there was robust replication of the inoculum in the RAW264.7 controls (3.6-fold increase) and in the Nramp2 transfectants (3.5-fold increase), while either no replication (25% reduction) or active elimination (90% reduction) of the bacterial inoculum was seen in the Nramp1-transfected clones 2.2 and 13, respectively. Comparison of CFU counts recovered at 24 h from RAW264.7 cells and from Nramp1-transfected cell clone 13 (sample 2) revealed a minimum of 50-fold difference. Cell survival over the course of infection in Nramp1-positive versus Nramp1-negative cells did not significantly differ as determined by the amount of protein per well. At 24 h, untransfected and control (Nramp2) samples had averages of 7,840 and 6,960 CFU/μg of protein, respectively, whereas the Nramp1-transfected clones had averages of 180 and 662 CFU/μg of protein. Similar results were obtained in three independent experiments. These results clearly indicate that expression of a wild-type recombinant Nramp1 protein in RAW264.7 macrophages corrects the inability of these cells to control the replication of an infectious inoculum of S. typhimurium. This effect cannot be due to cell death associated with Nramp1 function, as differences in total protein per sample from either cell population were not significant. Interestingly, expression of the Nramp2-cMyc protein in the same cells (sample 4) is without effect and does not correct the susceptible phenotype of RAW264.7 macrophages.
Parallel experiments were conducted with replication-defective, temperature-sensitive mutant TSΔ27 of S. typhimurium (Fig. 4B). These experiments were included to determine whether the active replication of virulent S. typhimurium in control RAW264.7 cells and in Nramp2 transfectants (Fig. 4A) was not due to an inherent defect of these clones in bactericidal mechanisms unrelated to Nramp. When TSΔ27 was used as an infectious agent, control RAW macrophages, Nramp1, and Nramp2 transfectants all rapidly killed this inoculum. A reduction in CFU counts of between 60 and 90% was observed by 3 h, and an 85 to 99% reduction of viable CFUs was seen by 24 h (Fig. 4B). Again, cell survival over the course of infection in Nramp1-positive versus Nramp1-negative cells did not significantly differ as determined by the level of protein per well. At 24 h, untransfected and control (Nramp2) samples had averages of 11 and 6 CFU/μg of protein, respectively, whereas, the Nramp1-transfected clones had averages of 2 and 1 CFU/μg of protein. These results suggest that both nontransfected and transfected RAW264.7 clones are capable of comparable bactericidal activity against this target.
These experiments show that differences in the ability of the virulent S. typhimurium inoculum to survive in the host (Fig. 4A) are caused by functional expression of Nramp1 in these cells. In addition, studies with the S. typhimurium temperature-sensitive mutant suggest that Nramp1 transfectants show increased bacteriostatic activity (Fig. 4B). Previous studies with primary macrophages isolated from inbred Nramp1r and Nramp1s as well as Nramp1 congenic mouse strains suggested that Nramp1 increases the bactericidal activity of these cells towards S. typhimurium (26). Possible explanations for differences in results include the observation that RAW264.7 macrophages used in this study were treated with IFN-γ for 16 h prior to infection, which may have enhanced the bactericidal activity of control and transfected cells against the TSΔ27 mutant compared to primary macrophages. It is also possible that RAW macrophages (controls) die during the infection, and some of the S. typhimurium CFUs from these cells may have escaped our detection. However, we feel that this is unlikely since we did not detect significant protein loss from the wells after 24 h, and similar results were obtained when replication was expressed as CFU/microgram of protein (as opposed to CFU/well). Finally, genetic differences between the mouse strains used by Lissner et al. (26) in addition to Nramp1 alleles may have also modulated the activity of primary macrophages from these mice.
Effect of Nramp1 on the pH of BCG-containing mycobacterial phagosomes.
We have previously reported that the absence of functional Nramp1 in primary macrophages results in impaired acidification of bacterial phagosomes containing M. bovis (BCG) (24). We therefore attempted to determine whether functional expression of the recombinant Nramp1-cMyc protein in RAW264.7 macrophages could also modulate pH of bacterial phagosomes. For these experiments, we used microfluorescence and imaging techniques to monitor the internal pH of individual BCG phagosomes formed in control RAW264.7 macrophages and in RAW264.7 Nramp1-cMyc transfectants. In these experiments, live M. bovis (BCG) cells were covalently labeled with fluorescent, pH-sensitive dyes that emit signals detectable by ratio imaging. Two dyes with different H+ affinity were used in combination, fluorescein (pKa = 6.4) and Oregon green (pKa = 4.7), which together allow a range of pH measurement from 4.0 to 7.5 (24). The procedure used to label the bacteria had no effect on their viability (data not shown). Phagocytosis was allowed to take place for 1 h at 37°C, followed by extensive washing of the inoculum before microfluorescence imaging of individual phagosomes. Three criteria were used to verify that the imaged phagosomes corresponded to internalized mycobacteria as opposed to bacterial cells adhering to the surface of the macrophage. These were (i) abrupt alteration of the extracellular pH, which alters fluorescence of extracellular but not intracellular bacteria; (ii) exposure to the ionophore nigericin and NH4Cl, both of which affect intraphagosomal pH but have no effect on extracellular pH; and (iii) adding bafilomycin, which impairs vacuolar H+-ATPase and blocks phagosome acidification (reference 24 and data not shown).
As summarized in Table 1 the pH of phagosomes containing live M. bovis (BCG) was found to be significantly more acidic in RAW264.7 Nramp1-cMyc transfectants than in control nontransfected RAW264.7 cells. The former showed an intraphagosomal pH of 5.09 ± 0.06 (n = 13 phagosomes tested) as opposed to a pH of 5.8 ± 0.1 (n = 9) for the controls (P < 0.05). Importantly, the difference was specific for live mycobacteria and was not seen for phagosomes containing inert latex particles, which acidified normally in both cell types to approximately 5.1 ± 0.1 (n = 8). This confirms that the phagosomal acidification mechanisms are competent in both cell types and that they differ only by their Nramp1-mediated responsiveness to live mycobacteria (BCG). Together, these results demonstrate that expression of recombinant Nramp1-cMyc protein in RAW264.7 macrophages recapitulates another known functional characteristic of Nramp1, the enhanced acidification of phagosomes containing live mycobacteria.
TABLE 1.
Effect of Nramp1 expression in RAW264.7 macrophages on bacterial phagosome acidification
| Cell type | Phagosome particlea | nb | pH (mean ± SD) |
|---|---|---|---|
| Nontransfected RAW264.7 | Latex bead | 8 | 5.1 ± .1 |
| BCG | 9 | 5.8 ± .1 | |
| Transfected Nramp1 RAW264.7 | Latex bead | 8 | 5.1 ± 1 |
| BCG | 13 | 5.09 ± .06 |
Denotes contents of the phagosome being analyzed.
Denotes number of phagosomes analyzed within each sample.
In conclusion, the transfection and expression of Nramp1-cMyc recombinant protein in RAW264.7 macrophages provides a functional assay for the antimicrobial activity of Nramp1 at a cellular level. Such an assay can now be used to study the functional relevance of predicted structural features of Nramp1, which have been deduced from its primary sequence, through the use of site-directed mutagenesis of the cDNA. Sites of interest include the predicted consensus transport motif that is conserved in many eukaryotic and prokaryotic transporters (including the permeation loop of the shaker K+ channel [39] and one of the subunits of the vacuolar ATPase [12]), the unusual charged residues identified within predicted transmembrane domains, the predicted sites for phosphorylation by casein kinase II and protein kinase C as well as several other sites. In addition, functional expression of Nramp1 in a cell line such as RAW264.7, which can be grown to large numbers, should prove very useful for producing large amounts of protein and for studying its biochemical activity in the phagosomal membrane. In particular, this assay can now be used to further define the parameters of the antimicrobial action of Nramp1 in intact cells but also in isolated phagosome preparations. This can be achieved by using standard biochemical methods to monitor the effect of Nramp1 expression on the level of various antimicrobial molecular species inside the phagosome. Finally, the molecular basis of antimicrobial action of Nramp1 can be studied with this in vitro assay, using a variety of Salmonella mutant strains that are defective in certain biochemical pathways. Mutants with such attenuated virulence are difficult to use in vivo because of their reduced virulence but can provide valuable information on the bacterial biochemical pathways affected by Nramp1 in the type of in vitro assay developed in this study.
ACKNOWLEDGMENTS
This work was supported by a research grants to P.G. from NIAID (AI35237) and to S.G. and B.F. from the Medical Research Council of Canada. P.G., S.G., and B.F. are International Research Scholars of the Howard Hughes Medical Institute. P.G. is supported by a Senior Scientist Award from the Medical Research Council of Canada.
REFERENCES
- 1.Abel L, Sanchez F O, Oberti J, Thuc N V, Hoa L V, Lap V D, Skamene E, Lagrange P H, Schurr E. Susceptibility to leprosy is linked to the human NRAMP1 gene. J Infect Dis. 1998;177:133–145. doi: 10.1086/513830. [DOI] [PubMed] [Google Scholar]
- 2.Bellamy R, Ruwende C, Corrah T, McAdam K P, Whittle H C, Hill A V. Variations in the NRAMP1 gene and susceptibility to tuberculosis in West Africans. N Engl J Med. 1998;338:640–644. doi: 10.1056/NEJM199803053381002. [DOI] [PubMed] [Google Scholar]
- 3.Blackwell J M, Roach T I A, Atkinson S E, Ajioka J W, Barton C H, Shaw M A. Genetic regulation of macrophage priming activation: the Lsh gene story. Immunol Lett. 1991;30:241–248. doi: 10.1016/0165-2478(91)90032-6. [DOI] [PubMed] [Google Scholar]
- 4.Brett S, Orrell J M, Swanson Beck J, Ivanyi J. Influence of H-2 genes on growth of Mycobacterium tuberculosis in the lungs of chronically infected mice. Immunology. 1992;76:129–132. [PMC free article] [PubMed] [Google Scholar]
- 5.Buschman E, Taniyama T, Nakamura R, Skamene E. Functional expression of the Bcg gene in macrophage. Res Immunol. 1989;140:793–797. doi: 10.1016/0923-2494(89)90035-7. [DOI] [PubMed] [Google Scholar]
- 6.Cellier M, Belouchi A, Gros P. Resistance to intracellular infections: comparative genomic analysis of Nramp. Trends Genet. 1996;12:201–204. doi: 10.1016/0168-9525(96)30042-5. [DOI] [PubMed] [Google Scholar]
- 7.Cellier M, Prive G, Belouchi A, Kwan T, Rodrigues V, Chia W, Gros P. Nramp defines a family of membrane proteins. Proc Natl Acad Sci USA. 1995;92:10089–10093. doi: 10.1073/pnas.92.22.10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crocker P R, Blackwell J M, Bradley D J. Expression of the natural resistance gene Lsh in resident liver macrophages. Infect Immun. 1984;43:1033–1040. doi: 10.1128/iai.43.3.1033-1040.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dassa E, Hofnung M. Sequence of gene malG in E. coli K12: homologies between integral membrane components from binding protein-dependent transport systems. EMBO J. 1985;4:2287–2293. doi: 10.1002/j.1460-2075.1985.tb03928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Chastellier C, Frehel C, Offredo C, Skamene E. Implication of phagosome-lysosome fusion in restriction of Mycobacterium avium growth in bone marrow macrophages from genetically resistant mice. Infect Immun. 1993;61:3775–3784. doi: 10.1128/iai.61.9.3775-3784.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denis M, Forget A, Pelletier M, Gervais F, Skamene E. Killing of Mycobacterium smegmatis by macrophages from genetically susceptible and resistant mice. J Leukoc Biol. 1990;47:25–30. doi: 10.1002/jlb.47.1.25. [DOI] [PubMed] [Google Scholar]
- 12.Descoteaux S, Yu Y, Samuelson J. Cloning of Entamoeba genes encoding proteolipids of putative vacuolar proton-translocating ATPases. Infect Immun. 1994;62:3572–3575. doi: 10.1128/iai.62.8.3572-3575.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Devault A, Gros P. Two members of the mouse mdr gene family confer multidrug resistance with overlapping but distinct drug specificities. Mol Cell Biol. 1990;10:1652–1663. doi: 10.1128/mcb.10.4.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleming M D, Trenor C C, 3rd, Su M A, Foernzler D, Beier D R, Dietrich W F, Andrews N C. Microcytic anaemia mice have a mutation in Nramp2, a candidate iron transporter gene. Nat Genet. 1997;16:383–386. doi: 10.1038/ng0897-383. [DOI] [PubMed] [Google Scholar]
- 15.Forget A, Skamene E, Gros P, Miailhe A C, Turcotte R. Differences in response among inbred mouse strains to infection with small doses of Mycobacterium bovis BCG. Infect Immun. 1981;32:42–47. doi: 10.1128/iai.32.1.42-47.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goto Y, Buschman E, Skamene E. Regulation of host resistance to Mycobacterium intracellulare in vivo and in vitro by the Bcg gene. Immunogenetics. 1989;30:218–221. doi: 10.1007/BF02421210. [DOI] [PubMed] [Google Scholar]
- 17.Govoni G, Gauthier S, Billia F, Iscove N N, Gros P. Cell-specific and inducible Nramp1 gene expression in mouse macrophages in vitro and in vivo. J Leukoc Biol. 1997;62:277–286. doi: 10.1002/jlb.62.2.277. [DOI] [PubMed] [Google Scholar]
- 18.Govoni G, Vidal S, Gauthier S, Skamene E, Malo D, Gros P. The Bcg/Ity/Lsh locus: genetic transfer of resistance to infections in C57BL/6J mice transgenic for the Nramp1Gly169 allele. Infect Immun. 1996;64:2923–2929. doi: 10.1128/iai.64.8.2923-2929.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gros P, Skamene E, Forget A. Cellular mechanisms of genetically controlled host resistance to Mycobacterium bovis (BCG) J Immunol. 1983;131:1966–1972. [PubMed] [Google Scholar]
- 20.Gros P, Skamene E, Forget A. Genetic control of natural resistance to Mycobacterium bovis (BCG) in mice. J Immunol. 1981;127:2417–2421. [PubMed] [Google Scholar]
- 21.Gruenheid S, Cellier M, Vidal S, Gros P. Identification and characterization of a second mouse Nramp gene. Genomics. 1995;25:514–525. doi: 10.1016/0888-7543(95)80053-o. [DOI] [PubMed] [Google Scholar]
- 22.Gruenheid S, Pinner E, Desjardins M, Gros P. Natural resistance to infection with intracellular pathogens: the Nramp1 protein is recruited to the membrane of the phagosome. J Exp Med. 1997;185:717–730. doi: 10.1084/jem.185.4.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gunshin H, Mackenzie B, Berger U V, Gunshin Y, Romero M F, Boron W F, Nussberger S, Gollan J L, Hediger M A. Cloning and characterization of a mammalian proton-coupled metal-ion transporter. Nature. 1997;388:482–488. doi: 10.1038/41343. [DOI] [PubMed] [Google Scholar]
- 24.Hackam D J, Rotstein O D, Zhang W, Gruenheid S, Gros P, Grinstein S. Host resistance to intracellular infection: mutation of natural resistance-associated macrophage protein 1 (Nramp1) impairs phagosomal acidification. J Exp Med. 1998;188:351–364. doi: 10.1084/jem.188.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kerppola R E, Ames G F-L. Topology of the hydrophobic membrane-bound components of the histidine periplasmic permease: comparison with other members of the family. J Biol Chem. 1992;267:2329–2336. [PubMed] [Google Scholar]
- 26.Lissner C R, Swanson R N, O’Brien A D. Genetic control of the innate resistance of mice to Salmonella typhimurium: expression of the Ity gene in peritoneal and splenic macrophages isolated in vitro. J Immunol. 1983;131:3006–3013. [PubMed] [Google Scholar]
- 27.Malo D, Vogan K, Vidal S, Hu J, Cellier M, Schurr E, Fuks A, Bumstead N, Morgan K, Gros P. Haplotype mapping and sequence analysis of the mouse Nramp gene predict susceptibility to infection with intracellular parasites. Genomics. 1994;23:51–61. doi: 10.1006/geno.1994.1458. [DOI] [PubMed] [Google Scholar]
- 28.Mills S D, Ruschkowski S R, Stein M A, Finlay B B. Trafficking of porin-deficient Salmonella typhimurium mutants inside HeLa cells: ompR and envZ mutants are defective for the formation of Salmonella-induced filaments. Infect Immun. 1998;66:1806–1811. doi: 10.1128/iai.66.4.1806-1811.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pinner E, Gruenheid S, Raymond M, Gros P. Functional complementation of the yeast divalent cation transporter family SMF by NRAMP2, a member of the mammalian natural resistance-associated macrophage protein family. J Biol Chem. 1997;272:28933–28938. doi: 10.1074/jbc.272.46.28933. [DOI] [PubMed] [Google Scholar]
- 30.Raschke W C, Baird S, Ralph P, Nakoinz I. Functional macrophage cell lines transformed by Abelson leukemia virus. Cell. 1978;15:261–267. doi: 10.1016/0092-8674(78)90101-0. [DOI] [PubMed] [Google Scholar]
- 31.Shaw M A, Collins A, Peacock C S, Miller E N, Black G F, Sibthorpe D, Lins-Lainson Z, Shaw J J, Ramos F, Silveira F, Blackwell J M. Evidence that genetic susceptibility to Mycobacterium tuberculosis in a Brazilian population is under oligogenic control: linkage study of the candidate genes NRAMP1 and TNFA. Tubercle Lung Dis. 1997;78:35–45. doi: 10.1016/s0962-8479(97)90014-9. [DOI] [PubMed] [Google Scholar]
- 32.Stach J L, Gros P, Forget A, Skamene E. Phenotypic expression of genetically-controlled natural resistance to Mycobacterium bovis (BCG) J Immunol. 1984;132:888–892. [PubMed] [Google Scholar]
- 33.Stokes R W, Orme I M, Collins F M. Role of mononuclear phagocytes in expression of resistance and susceptibility to Mycobacterium avium infections in mice. Infect Immun. 1986;54:811–819. doi: 10.1128/iai.54.3.811-819.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Supek F, Supekova L, Nelson H, Nelson N. A yeast manganese transporter related to the macrophage protein involved in conferring resistance to Mycobacteria. Proc Natl Acad Sci USA. 1996;93:5105–5110. doi: 10.1073/pnas.93.10.5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swanson R N, O’Brien A D. Genetic control of the innate resistance of mice to Salmonella typhimurium: Ity gene is expressed in vivo by 24 hours after infection. J Immunol. 1983;131:3014–3020. [PubMed] [Google Scholar]
- 36.Vidal S, Tremblay M L, Govoni G, Gauthier S, Sebastiani G, Malo D, Skamene E, Olivier M, Jothy S, Gros P. The Ity/Lsh/Bcg locus: natural resistance to infection with intracellular parasites is abrogated by disruption of the Nramp1 gene. J Exp Med. 1995;182:655–666. doi: 10.1084/jem.182.3.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vidal S M, Malo D, Vogan K, Skamene E, Gros P. Natural resistance to infection with intracellular parasites: isolation of a candidate for Bcg. Cell. 1993;73:469–485. doi: 10.1016/0092-8674(93)90135-d. [DOI] [PubMed] [Google Scholar]
- 38.Vidal S M, Pinner E, Lepage P, Gauthier S, Gros P. Natural resistance to intracellular infections: Nramp1 encodes a membrane phosphoglycoprotein absent in macrophages from susceptible (Nramp1D169) mouse strains. J Immunol. 1996;157:3559–3568. [PubMed] [Google Scholar]
- 39.Wood M W, VanDongen H M, VanDongen A M. Structural conservation of ion conduction pathways in K+ channels and glutamate receptors. Proc Natl Acad Sci USA. 1995;92:4882–4886. doi: 10.1073/pnas.92.11.4882. [DOI] [PMC free article] [PubMed] [Google Scholar]