Abstract
Recent reports that isolated Treponema pallidum outer membranes contain an ortholog for glycerophosphodiester phosphodiesterase (GlpQ) (D. V. Shevchenko, D. R. Akins, E. J. Robinson, M. Li, O. V. Shevchenko, and J. D. Radolf, Infect. Immun. 65:4179–4189, 1997) and that this protein is a potential opsonic target for T. pallidum (C. E. Stebeck, J. M. Shaffer, T. W. Arroll, S. A. Lukehart, and W. C. Van Voorhis, FEMS Microbiol. Lett. 154:303–310, 1997) prompted a more detailed investigation of its physicochemical properties and cellular location. [14C]palmitate radiolabeling studies of a GlpQ-alkaline phosphatase fusion expressed in Escherichia coli confirmed the prediction from DNA sequencing that the protein is lipid modified. Studies using Triton X-114 phase partitioning revealed that the protein’s amphiphilicity is due to lipid modification and that a substantial portion of the polypeptide is associated with the T. pallidum peptidoglycan sacculus. Three different approaches, i.e., (i) proteinase K treatment of intact treponemes, (ii) indirect immunofluorescence analysis of treponemes encapsulated in agarose beads, and (iii) opsonophagocytosis of treponemes incubated with antiserum against recombinant GlpQ by rabbit peritoneal macrophages, confirmed that GlpQ is entirely subsurface in T. pallidum. Moreover, rabbits hyperimmunized with GlpQ were not protected against intradermal challenge with virulent treponemes. Circular dichroism spectroscopy confirmed that the recombinant form of the polypeptide lacked discernible evidence of denaturation. Finally, GlpQ was not radiolabeled when T. pallidum outer membranes were incubated with 3-(trifluoromethyl)-3-(m-[125I]iodophenyl)-diazarene, a photoactivatable, lipophilic probe which promiscuously labels both proteins and lipids within phospholipid bilayers. Taken as a whole, these studies indicate that the T. pallidum GlpQ ortholog is a periplasmic protein associated predominantly with the spirochete’s peptidoglycan-cytoplasmic membrane complex.
Syphilis, a sexually transmitted disease caused by the spirochetal pathogen Treponema pallidum subsp. pallidum, begins as an ulcer (chancre) at the site of inoculation (usually in the genital area) and, when untreated, may progress through secondary (disseminated), latent, and tertiary (recrudescent) stages (61). Despite the availability of effective antimicrobial therapy since the 1940s, syphilis remains a global public health problem (40, 44). Most recently, genital ulcers caused by syphilis have been shown to facilitate the sexual transmission of human immunodeficiency virus (24, 63). These epidemiological trends underscore the importance of vaccine development as a cornerstone of strategies to curtail syphilis transmission. Unfortunately, the identification of vaccine candidates has been impeded by a number of factors, most notably the inability to cultivate T. pallidum on artificial medium and the syphilis spirochete’s unusual outer membrane ultrastructure (50).
In recent years, the quest for outer membrane (OM) proteins of T. pallidum as potential virulence determinants and vaccine candidates has become a major focus of syphilis research (50). In this regard, we recently reported that OMs isolated from T. pallidum by using a plasmolysis-based procedure contain a 38.5-kDa putative lipoprotein with sequence relatedness to glycerophosphodiester phosphodiesterase (GlpQ) (56), an enzyme which hydrolyzes deacylated phospholipids to alcohol plus glycerol-3-phosphate (37, 39). Although GlpQ is periplasmic in Escherichia coli (37), the Haemophilus influenzae ortholog is surface exposed and capable of inducing bactericidal antibodies (29, 55). Consistent with this, Stebeck and coworkers reported that the treponemal ortholog is a potential opsonic target for motile T. pallidum (60). These findings prompted a detailed investigation of the physicochemical properties and cellular location of this protein. Here we report that, as with GlpQ of H. influenzae, the T. pallidum GlpQ protein is lipid modified but that, unlike its H. influenzae counterpart, the treponemal polypeptide has a subsurface location. Similar to other treponemal lipoprotein immunogens, GlpQ appears to be associated predominantly with the peptidoglycan-cytoplasmic membrane (CM) complex. Moreover, contrary to the recent report by Cameron et al. (16) demonstrating attenuated lesion development when GlpQ-immunized rabbits were intradermally inoculated with virulent T. pallidum, we did not observe any evidence of protective immunity in GlpQ-hyperimmunized rabbits. These results are consistent with the antigen’s lack of surface exposure and suggest that GlpQ will play a limited role in the development of a syphilis vaccine.
MATERIALS AND METHODS
Bacterial strains.
T. pallidum (Nichols) was propagated by intratesticular inoculation of adult New Zealand White rabbits as previously described (52). Spirochetes were separated from testicular tissue debris by low-speed centrifugation (350 × g for 10 min) and, when necessary, purified by Percoll density gradient centrifugation (30). For opsonophagocytosis assays, organisms were extracted from infected testes in medium 199 (M199) (Mediatech, Herndon, Va.) supplemented with 20% heat-inactivated fetal bovine serum (HIFBS) (heated for 30 min at 56°C) (Mediatech) and gassed with 3% O2–5% CO2 overnight at 37°C. Spirochetes were enumerated by dark-field microscopy with a Petroff-Hausser counting chamber (Hausser Scientific Company, Horsham, Pa.). E. coli DH5α was the recipient strain for all recombinant constructs and was grown in Luria-Bertani broth with appropriate antibiotic supplementation.
Production and purification of a recombinant, nonlipidated GlpQ (rGlpQ).
The portion of the glpQ gene encoding the mature (i.e., processed) protein was PCR amplified from T. pallidum genomic DNA by using the forward and reverse primers 5′-GCGGGATCCTGTGCGTCCGAACGTATGATAGTTG-3′ (BamHI site plus nucleotides 61 to 85) and 5′-GCGGAATTCTCAATAGCGGGCGGGTTTGCCC-3′ (complementary to nucleotides 1049 to 1071 plus EcoRI site), respectively (56). The amplified product was cloned into pProEX-HTb (Gibco BRL, Gaithersburg, Md.) to generate a polyhistidine (His)-tagged fusion protein. The IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible fusion protein (designated rGlpQ-His) was purified from E. coli cell supernatant on an Ni-nitrilotriacetic acid agarose matrix (Qiagen, Inc., Santa Clarita, Calif.) according to the manufacturer’s instructions. The His tag was removed by digestion with recombinant tobacco etch virus protease (Gibco BRL) as described by the manufacturer.
Immunologic reagents.
Immune rabbit sera (IRS) were obtained approximately 10 months following intratesticular inoculation of rabbits with motile T. pallidum. Normal rabbit serum (NRS) was obtained from animals that were nonreactive in Venereal Disease Research Laboratory and fluorescent treponemal antibody tests and was filter sterilized. A pool of normal human sera was created by combining equal volumes of sera from five healthy adult volunteers. A pool of human syphilitic sera (HSS) was created by combining sera from five human immunodeficiency virus-seronegative individuals with classical dermatological manifestations of secondary syphilis and reactive nontreponemal and treponemal serological tests.
Rat anti-rGlpQ antiserum was generated by priming 6-week-old Sprague-Dawley rats by intraperitoneal injection with 30 μg of purified, cleaved protein in a 1:1 mixture of phosphate-buffered saline (PBS) (pH 7.4) and complete Freund’s adjuvant; 1 month and 6 weeks later, the animals received 15-μg booster doses in a 1:1 mixture of PBS and incomplete Freund’s adjuvant. Rabbit anti-rGlpQ antisera were generated by immunizing adult New Zealand White rabbits subcutaneously with 100 μg of protein in a 1:1 mixture of PBS and complete Freund’s adjuvant; 1 month and 6 weeks later, the animals were boosted with 50 μg of protein in incomplete Freund’s adjuvant. A murine monoclonal antibody specific for E. coli alkaline phosphatase (PhoA) was obtained from Caltag Laboratories (Burlingame, Calif.). Rabbit antisera directed against T. pallidum endoflagella (TpEf) (34) and native 47-kDa lipoprotein (Tp47) (22) were described previously.
To affinity purify anti-GlpQ antibodies from IRS, 80 μg of rGlpQ was coupled to 100 μl of 1,1′-carbonyldiimidazole-activated 6% cross-linked beaded agarose (Reacti-Gel 6×; Pierce, Rockford, Ill.) according to the manufacturer’s instructions. The Reacti-Gel matrix was equilibrated with 250 mM Tris (pH 7.4) and incubated for 2 h at 4°C with 250 μl of IRS. The adsorbed anti-GlpQ antibodies were eluted from the matrix in 200-μl fractions with 0.5 M acetic acid and neutralized with 100 μl of 1 M Tris base. Both the anti-GlpQ and the resulting IRS depleted of anti-GlpQ antibodies were tested by Western blot analysis for their ability to recognize the native GlpQ (nGlpQ) protein in T. pallidum lysates as described above. To quantitatively remove immunoglobulin G (IgG) antibodies from IRS, IRS was passed over a GammaBind G Sepharose matrix (Pharmacia Biotech, Alameda, Calif.). The adsorbed IgG antibodies were eluted in 500-μl fractions with 0.5 M acetic acid and neutralized with 0.25 ml of 1 M Tris base. The protein concentrations of the affinity-purified anti-GlpQ antibodies and total IgG fractions were determined with the Bio-Rad protein assay (Bio-Rad Laboratories, Hercules, Calif.).
SDS-polyacrylamide gel electrophoresis (SDS-PAGE), immunoblot analysis, and two-dimensional gel electrophoresis.
Samples were boiled for 5 min in final sample buffer and subjected to electrophoresis through sodium dodecyl sulfate (SDS)–12.5% polyacrylamide gels as previously described (56). Gels were stained with either Coomassie brilliant blue or silver or transferred electrophoretically to 0.2-μm-pore-size nitrocellulose (Schleicher and Schuell, Keene, N.H.) for immunoblotting. Immunoblots were incubated with various dilutions of HSS, rabbit or rat anti-rGlpQ antisera, affinity purified anti-nGlpQ antibodies, or IRS depleted of anti-GlpQ antibodies, followed by incubation with 1:1,000 dilutions of horseradish peroxidase-conjugated goat anti-human, -rabbit, or -rat immunoglobulins (Zymed Laboratories, Inc., South San Francisco, Calif.). Blots were developed by using 4-chloro-1-naphthol as a substrate. Two-dimensional gel electrophoresis was performed according to the method of O’Farrell (43).
Enzyme-linked immunosorbent assay.
An Immulon II 96-well U-bottom plate (Dynatech, Chantilly, Va.) was coated with 25 ng of rGlpQ in 0.1 M Na2CO3 buffer (pH 9.4) per well and incubated overnight at 4°C. The wells were washed four times with PBS containing 0.05% Tween 20 and blocked with PBS containing 1% bovine serum albumin (BSA) (Sigma Chemical Company, St. Louis, Mo.) for 2 h at 37°C. To the wells were added HSS, IRS, affinity purified anti-GlpQ antibodies, and IRS depleted of anti-GlpQ antibodies serially diluted in PBS containing 1% BSA plus 0.05% Tween 20. Following incubation for 1 h at 37°C, the wells were washed four times, followed by the addition of a 1:1,000 dilution of goat anti-human, goat anti-rat, or goat anti-rabbit IgG (Zymed)–horseradish peroxidase conjugate. After incubation for 1 h at 37°C, the wells were washed four times and incubated for 20 min at room temperature with TMB peroxidase substrate (Kirkegaard & Perry Laboratories, Gaithersburg, Md.). The reaction was quenched by addition of 2 N H2SO4, and the optical densities in the wells were read immediately at 450 nm.
Glycerophosphodiester phosphodiesterase assay.
The enzymatic activity of rGlpQ was examined at 25°C by measuring glycerol-3-phosphate production in a coupled spectrophotometric assay performed as described by Larson et al. (37) with minor modifications. The 1-ml assay mixture contained 0.9 ml of 1 M hydrazine hydrate in 1.5% glycine buffer (pH 9.0), 0.5 mM NAD, 10 mM CaCl2, 20 U of glycerol-3-phosphate dehydrogenase/ml, and one of the following substrates at 0.5 mM: glycerophosphorylcholine, glycerophosphorylethanolamine, glycerophosphorylserine, or glycerophosphorylinositol (all purchased from Sigma). Glycerophosphorylcholine phosphodiesterase (Sigma catalog no. G1642) and glycerophosphorylcholine were used as a positive control. The rate of NAD reduction was measured by recording the increase in absorbance at 340 nm. One unit of phosphodiesterase activity is defined as the amount of enzyme required to hydrolyze 1 μmol of glycerophosphodiester min−1 under these conditions. A molar absorbance coefficient of 6,300 M−1 cm−1 at 340 nm was used for NADH.
CD spectroscopy.
Circular dichroism (CD) spectra were obtained on an AVIV (Lakewood, N.J.) model 62DS spectropolarimeter with sample temperatures regulated by a Hewlett-Packard model 89100A temperature controller; measurements were taken at 25 ± 0.1°C. CD spectra were obtained with 4 μM samples of rGlpQ or recombinant gonococcal porin P1A (25). CD spectroscopic analysis of rGlpQ was performed with 10 mM phosphate buffer (pH 7.3) with or without 5 mM EDTA and also with 10 mM phosphate buffer containing 0.5% SDS, 0.5% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, or 0.6% octylglucoside. CD spectra were obtained for the recombinant gonococcal porin P1A in 10 mM phosphate buffer (pH 7.3) containing 0.6% octylglucoside. All CD spectra were baseline corrected and smoothed with software provided by AVIV Associates. The self-consistent algorithm of Sreerama and Woody (58, 59) was used to calculate the percentages of various types of secondary structures.
Triton X-114 phase partitioning.
Percoll-purified T. pallidum (5 × 109 organisms) was solubilized overnight in 1 ml of PBS containing 2% Triton X-114. The insoluble material was then removed by centrifugation for 15 min in a microcentrifuge, and the supernatant was phase separated as previously described (12, 15, 52). rGlpQ was subjected to phase partitioning under identical conditions. The proteins in the aqueous and detergent phases were concentrated by acetone precipitation prior to SDS-PAGE.
[14C]palmitate labeling of a GlpQ-PhoA fusion protein.
The primers 5′-TCTAGTCTAGAAGAATGCTTGCGTGTGGCGAAGG-3′ (XbaI site plus nucleotides −399 to −365) and 5′-GCGGGATCCGTGCTCGGGCACATATCCTGCAGC-3′ (complementary to nucleotides 96 to 120 plus BamHI site), were used to generate a 519-bp PCR product which encoded the first 40 amino acids of GlpQ plus upstream flanking DNA (including the putative promoter). The resulting amplicon was cloned upstream from and in frame with the E. coli alkaline phosphatase gene (phoA) lacking the leader peptide by using the previously described vector pSKI/pho (1). Single colonies of E. coli DH5α containing the resulting plasmid (designated pGlpQ-Pho), along with control plasmids expressing PhoA fusions with the N terminus of LacZ (1) and the N terminus of Tp47 (64), were inoculated into 30-ml portions of one-half-strength Luria-Bertani broth containing ampicillin (100 μg/ml) and grown to an optical density at 600 nm of 0.1. Fifteen microcuries of [U-14C]-palmitic acid (500 mCi/mmol; New England Nuclear, Boston, Mass.) was added to each culture. The cultures were then incubated with shaking to an optical density at 600 nm of 0.6, after which they were harvested and washed twice with PBS prior to SDS-PAGE, immunoblot analysis, and autoradiography.
Nucleotide sequence analysis.
Nucleotide sequencing of the various fusion constructs was performed with an Applied Biosystems Inc. (Foster City, Calif.) model 373A automated DNA sequencer and the PRISM ready-reaction DyeDeoxy Terminator cycle sequencing kit according to the manufacturer’s instructions.
Accessibility of nGlpQ to digestion with PK.
The accessibility of nGlpQ to digestion with proteinase K (PK) was determined as described by Barbour et al. (8). Freshly isolated T. pallidum organisms (5 × 109) were centrifuged at 20,000 × g for 15 min and resuspended in 0.1 ml of PBS containing 5 mM MgCl2. PK was added to a final concentration of 0.4 mg/ml, and the spirochetes were incubated at room temperature for 30 min. Proteolytic digestion was stopped by the addition of phenylmethylsulfonyl fluoride to a final concentration of 1 mg/ml. Lysates of PK-treated and untreated T. pallidum were processed for SDS-PAGE and immunoblot analysis as described above.
Localization of nGlpQ by indirect immunofluorescence of T. pallidum encapsulated in gel microdroplets.
T. pallidum cell suspensions were encapsulated in gel microdroplets as previously described (21). Encapsulated organisms were probed with specific antibodies by a three-step indirect immunofluorescence technique. Briefly, rat anti-rGlpQ serum was diluted 1:20 and added directly to small aliquots of beads (0.3 to 0.4 ml) in 1 ml of T. pallidum cultivation medium. In samples incubated with detergent, a 1% (vol/vol) Triton X-100 stock solution in PBS was added to the beads immediately after the addition of the primary antibody to produce a final concentration of 0.05% (vol/vol). Samples were incubated for 2 h with gentle mixing in a 34°C water bath. The beads were washed three times by low-speed centrifugation (100 × g), resuspended in 3 ml of T. pallidum culture medium (23), and then incubated for 1 h at 34°C with mouse anti-rat IgG. Beads were washed as described above, resuspended in 3 ml of T. pallidum culture medium, and incubated for 1 h at 34°C with 3 μg of goat anti-mouse IgG coupled to R-phycoerythrin (Molecular Probes, Eugene, Oreg.). The beads were washed a final time and then viewed on glass slides with a Nikon Optiphot-2 fluorescence microscope equipped with 15× oculars, a dark-field condenser, and a fluorescein filter. Samples were observed with either a 40× or a 100× oil immersion objective. For each condition (i.e., antibody and detergent concentration) three slides were prepared, and then approximately 100 organisms were scored for labeling (fluorescence). Statistical comparisons with the preimmune sera were performed by using a two-tailed Student t test; P values of ≤0.5 were considered significant.
Opsonophagocytosis assay for surface exposure of nGlpQ.
Opsonophagocytosis assays were performed as described by Baker-Zander et al. (4–6). Rabbit peritoneal macrophages were elicited by intraperitoneal injection of 10 ml of 15% sterile Proteose Peptone no. 3 (Difco Laboratories, Detroit, Mich.). Cells were harvested at 3 to 5 days postinjection by peritoneal lavage with FA buffer (Difco) containing 10 U of heparin (Sigma) per ml, centrifuged at 900 × g for 10 min, and resuspended in M199 supplemented with 20% HIFBS (Mediatech), 100 U of penicillin/ml, and 100 μg of streptomycin/ml (M199-complete). They then were counted with a hemacytometer and dispensed into 24-well cluster tissue culture plates containing 12-mm-diameter microscope coverslips (catalog no. 12-545-82; Fisher Scientific, Pittsburgh, Pa.) at a density of 5 × 105 cells/ml. After incubation for 2 h at 37°C, nonadherent cells were removed by washing the adherent monolayers twice with M199-complete. The cells were maintained overnight at 37°C in a reduced-oxygen atmosphere containing 3% O2 and 5% CO2. The following day, the adherent macrophages were washed twice with gassed M199-complete. The cells were then incubated for 4 h at 37°C in a reduced-oxygen atmosphere (3% O2 and 5% CO2) with T. pallidum (10 organisms per macrophage) in the presence of 20% HIFBS plus 10% heat-inactivated (56°C for 30 min) NRS, IRS, rabbit anti-rGlpQ, rabbit anti-TpEf, or rabbit anti-Tp47. In separate reconstitution experiments, the opsonic activities of affinity-purified anti-GlpQ antibodies, IgG antibodies from IRS, and IRS depleted of either anti-GlpQ or total IgG antibodies were evaluated. The affinity-purified antibodies were added to the wells in amounts equivalent to those recovered from 100 μl of IRS. Following incubation with T. pallidum, coverslips were washed twice with serum-free M199 medium and fixed for 15 min in 95% ethanol. Fixed cells on coverslips were rehydrated with PBS containing 1% BSA for 10 min at room temperature or stored overnight at 4°C.
To visualize T. pallidum, the cells were blocked for 30 min at room temperature with CMRL medium containing 10% HIFBS and then incubated for 1 h at room temperature with HSS diluted 1:100 in CMRL plus 10% HIFBS. Coverslips were washed three times with FA buffer and blocked for 30 min at room temperature with CMRL plus 10% HIFBS. After blocking, cells were incubated for 30 min at room temperature with goat anti-human IgG conjugated to fluorescein isothiocyanate (Zymed) diluted 1:1,000 in CMRL plus 10% HIFBS. Following incubation, coverslips were washed three times with FA buffer and once with distilled H2O, allowed to air dry, and inverted on microscope slides with fluorescence mounting medium. Cell monolayers were examined with an Olympus BH2-RFCA fluorescence microscope equipped with 10× oculars, a dark-field condenser, and a fluorescein filter. Samples were observed with a DPLANApo 40× UV objective. Duplicate specimens were prepared for each test serum, and approximately 100 macrophages in each of two randomly chosen fields were counted for each coverslip. Coverslips were scored in a blinded fashion, and data are expressed as the mean percentage of cells with labeled inclusion bodies (internalized T. pallidum). Statistical comparisons between groups were made by using a two-tailed Student t test, with P values of ≤0.05 considered to be statistically significant. In some instances, the association of fluorescently labeled treponemes with macrophages was examined by confocal laser microscopy with an MRC-1024 laser confocal imaging system (Bio-Rad). Specimens were imaged by using 10× oculars with a 63× objective and 4× zoom. A series of 10 to 15 optical sections (0.5 μm) through the macrophages were collected digitally with a resolution of 0.076 μm per pixel.
Intradermal challenge of rabbits immunized with rGlpQ.
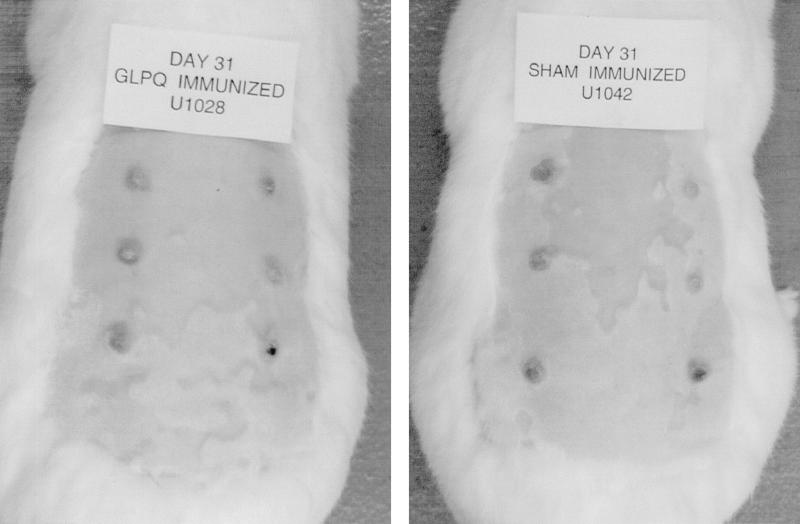
In the first challenge experiment, two rabbits were immunized with rGlpQ as described above. Ten days after the second boost, the rGlpQ-immunized and two serologically nonreactive control rabbits were challenged with 104 virulent T. pallidum organisms by intradermal injection on their shaved backs at each of six sites. In a second challenge experiment, rabbits (four animals/group) were immunized with rGlpQ or sham immunized. Ten days after the second boost, they were challenged with 103 virulent T. pallidum organisms by intradermal injection on their shaved backs at each of six sites. In both experiments animals were examined daily to monitor the development, morphologic appearance, and progression of lesions. Lesions were scored on the basis of erythema, induration, and ulceration. Additionally, photographs were taken of the lesions to document the extent of the reaction to challenge. At 17 days postchallenge, two lesions from each rabbit were injected with 100 μl of PBS, which was then aspirated for dark-field microscopy.
Radiolabeling of isolated T. pallidum outer membranes with ([125I]TID).
Outer membranes isolated from 1010 T. pallidum organisms as previously described (54) were resuspended in cross-linking buffer consisting of 0.01 M HEPES, 0.15 M NaCl, and 5 mM MgCl2 (pH 7.5). After the addition of 5 μCi of 3-(trifluoromethyl)-3-(m-[125I]iodophenyl)-diazarene ([125I]TID) (Amersham, Arlington Heights, Ill.), the sample was photoactivated by exposure for 30 min to long-wave (366-nm) UV light. The sample was then pelleted in an Airfuge (Beckman, Palo Alto, Calif.) and washed twice with cross-linking buffer prior to SDS-PAGE and autoradiography.
RESULTS AND DISCUSSION
The T. pallidum GlpQ ortholog is a lipoprotein.
We originally identified the T. pallidum GlpQ ortholog in OM fractions as a protein of approximately 38 kDa (by SDS-PAGE) whose N terminus was blocked to automated Edman degradation (56). Nucleotide sequence analysis of the cloned glpQ gene provided a potential explanation for this result; the encoded polypeptide possesses a 20-amino-acid signal peptide terminated by a putative lipoprotein processing and modification site (L-V-A-G-C) (32, 56, 60). We began the present study by conducting in vivo radiolabeling experiments with [14C]palmitate to confirm that the protein is indeed lipid modified. Although such studies are ideally performed with T. pallidum, radiolabeling of treponemal lipoproteins is complicated by the spirochete’s extremely slow rate of protein turnover (17). Consequently, here, as in previous studies (1, 2, 45, 48), we exploited the fact that lipid modification and processing signals are highly conserved among eubacteria (47) and used E. coli as a T. pallidum surrogate for labeling experiments. PhoA fusions are particularly advantageous for experiments of this type because they tend to be well expressed in E. coli without toxicity, and they can be distinguished easily from the large majority of native E. coli lipoproteins, which migrate by SDS-PAGE with apparent molecular masses of well below 30 kDa (the PhoA reporter has a molecular mass of 46.6 kDa) (1, 45). As shown in Fig. 1 (lane 3), a 49.5-kDa chimera in which the GlpQ ortholog’s first 40 amino acids were fused to an unexported PhoA reporter was strongly radiolabeled with [14C]palmitate. PhoA fusions with Tp47 (64) and LacZ (1) served as positive and negative controls (Fig. 1, lanes 1 and 2, respectively). Although these results do not prove unequivocally that the native form of the protein is similarly lipidated, we are unaware of any instances in which spirochetal proteins were shown to be lipidated in E. coli but not in T. pallidum.
FIG. 1.
Lipid modification of GlpQ. E. coli transformants harboring plasmids expressing PhoA fusions with the N termini of Tp47 (lane 1), LacZ (lane 2), and GlpQ (lane 3) were metabolically labeled with [14C]palmitate and then analyzed by SDS-PAGE and autoradiography. All three polypeptides also reacted with a monoclonal antibody specific for E. coli PhoA (data not shown). Sizes of molecular mass standards in kilodaltons are shown at the left.
General membrane topology of the native GlpQ ortholog.
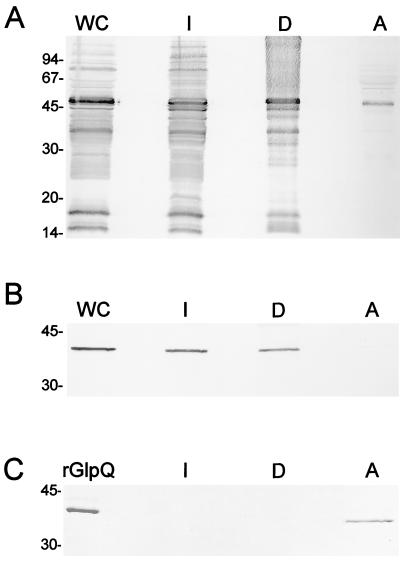
We next used Triton X-114 phase partitioning (12, 15) to assess the contribution of acylation to the amphiphilicity of nGlpQ; this method also served as a convenient starting point for efforts to determine the protein’s cellular location. Percoll-purified T. pallidum, solubilized in 2% Triton X-114 at 4°C, was phase partitioned, and the resulting fractions were immunoblotted against pooled HSS or rat anti-rGlpQ antiserum. As previously reported (52), the major lipoprotein immunogens recognized by HSS are approximately equally distributed between the detergent-enriched phase and the Triton X-114-insoluble material, which contains the peptidoglycan sacculus (53). An identical immunoblot probed with the anti-rGlpQ antiserum revealed that nGlpQ was similarly distributed among the three fractions (Fig. 2B); we interpret these results as indicating that a substantial portion of nGlpQ is intracellular, most likely associated with the peptidoglycan-CM sacculus or its constituents. In contrast to its native counterpart, rGlpQ partitioned exclusively into the aqueous phase (Fig. 2C). Thus, as with other treponemal lipoproteins (50), the amphiphilicity of nGlpQ is not an intrinsic property of the polypeptide but rather is due to lipid modification. This result also is in accord with the prediction by hydropathy analysis that the GlpQ protein is extremely hydrophilic (56).
FIG. 2.
Amphiphilicity of native and recombinant GlpQ. (A and B) T. pallidum was solubilized in 2% Triton X-114 in PBS and phase partitioned as described in Materials and Methods. Whole cells (WC), Triton X-114-insoluble material (I), the detergent-enriched phase (D), and the aqueous phase (A) were subjected to immunoblot analysis with 1:100 and 1:1,000 dilutions of HSS (A) or rat anti-rGlpQ antiserum (B), respectively. (C) SDS-PAGE analysis and Coomassie blue staining of rGlpQ after Triton X-114 phase partitioning. Sizes of molecular mass standards in kilodaltons are shown on the left.
Evidence that nGlpQ is not surface exposed.
Spirochetal lipoproteins consist of hydrophilic polypeptides which are lipid anchored to membranes; because the protein components are extrinsic to the lipid bilayer, they do not form particles when membranes are subjected to freeze-fracture analysis (35, 51). Thus, even if it is OM associated, it is unlikely that nGlpQ would be among the rare proteins visualized in freeze-fractured T. pallidum OMs (49, 50, 62). On the other hand, the studies thus far did not exclude the possibility that some nGlpQ is exposed on the T. pallidum surface in a manner analogous to the outer surface lipoproteins of the Lyme disease spirochete Borrelia burgdorferi (7, 51). In fact, B. burgdorferi is a particularly important precedent for the studies here, because prior work has shown that portions of at least three borrelial outer surface lipoproteins are CM anchored in addition to being exposed on the bacterial surface (20). In addressing this issue experimentally, it should be noted that surface localization studies of T. pallidum are complicated by a number of factors, most notably the lability of the treponemal OM and the lack of antibodies against authentic OM proteins to serve as positive controls (50). In light of these caveats and the fact that biological and microscopy-based assays for surface localization of treponemal antigens have not always correlated (50), we used multiple approaches to determine whether nGlpQ can be detected on the T. pallidum surface.
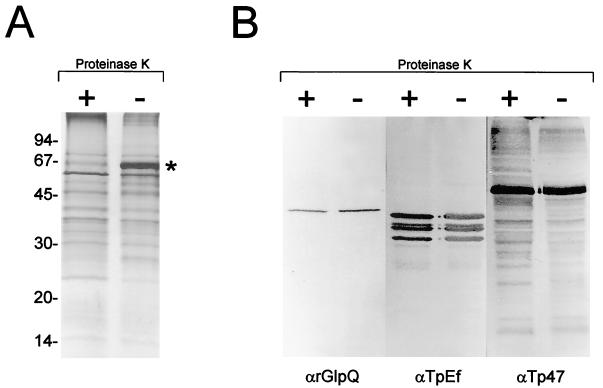
(i) Initially, we examined the accessibility of nGlpQ to digestion with 0.4 mg of PK per ml, a method which has often been used to identify surface-exposed antigens of B. burgdorferi (9, 42, 46). Consistent with the contention that T. pallidum has a paucity of surface-exposed proteins (22, 49, 50), only minor differences, none of which were in the molecular weight range of nGlpQ, in the polypeptide profiles of treponemes incubated with or without PK were observed (Fig. 3A). The most obvious difference, the absence of the albumin band in the PK-treated organisms, confirmed that the spirochetes were exposed to active enzyme. Immunoblot analysis of the same samples did not reveal any obvious difference in the amounts of nGlpQ (Fig. 3B). The surface specificity of the enzyme treatment was confirmed by the observation that PK-treated and untreated organisms contained similar amounts of the periplasmic markers Tp47 and TpEf (Fig. 3B).
FIG. 3.
Inaccessibility of nGlpQ to PK digestion in intact T. pallidum. (A) SDS-PAGE and silver staining of T. pallidum incubated with (+) or without (−) PK (0.4 mg/ml). The asterisk alongside the sample without PK designates albumin. Sizes of molecular mass standards in kilodaltons are shown at the left. (B) Immunoblot analysis of the same samples following incubation with anti-rGlpQ (αrGlpQ), anti-TpEf, or anti-Tp47.
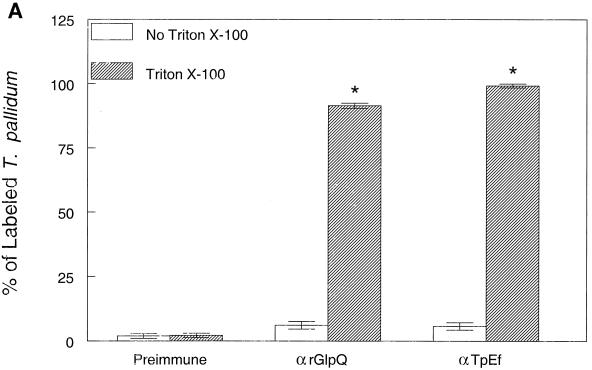
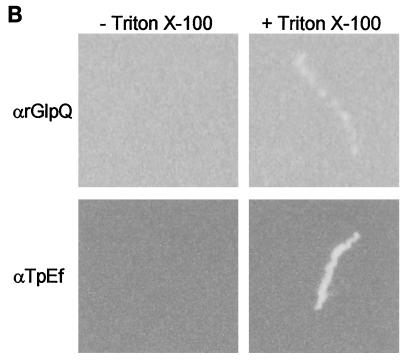
(ii) A second strategy involved immunofluorescence analysis of spirochetes encapsulated in agarose beads (gel microdroplets) (20, 21). This method preserves the integrity of fragile T. pallidum OMs during immunolabeling studies but also enables the detection of intracellular antigens when organisms are incubated with antibodies in the presence of a concentration of Triton X-100 sufficient to permeabilize the OM (>0.03%). The bar graphs in Fig. 4A depict the combined results of three independent experiments. Intact spirochetes did not fluoresce following incubation with an extremely low (1:20) dilution of rat anti-rGlpQ antiserum. In separate immunoblot experiments, this antiserum was capable of detecting nGlpQ in whole-cell lysates at titers of greater than 1:10,000 (data not shown). By contrast, nearly 100% of the organisms were labeled with this same antiserum when the OMs were permeabilized with 0.05% Triton X-100. Virtually identical results were obtained when the encapsulated organisms were incubated with anti-TpEf in the absence or presence of detergent. Qualitative differences in immunolabeling, on the other hand, were observed with the anti-rGlpQ and anti-TpEf antisera. In addition to being consistently less intense, the anti-rGlpQ antibodies produced a beaded labeling pattern as opposed to the more uniform labeling produced by anti-TpEf antibodies (Fig. 4B).
FIG. 4.
Localization of GlpQ in T. pallidum encapsulated in gel microdroplets. (A) Encapsulated treponemes were probed with pooled preimmune sera from rats immunized with rGlpQ, with rat anti-rGlpQ antiserum (αrGlpQ), or with rabbit anti-TpEf antiserum in the absence or presence of 0.05% Triton X-100. Labeling was determined by comparing immunofluorescence with dark-field microscopy. Each bar graph represents the mean ± standard deviation of determinations (involving approximately 100 organisms) in three separate experiments. Asterisks indicate statistically significant results (P ≤ 0.05) compared to those with preimmune sera. (B) Immunofluorescent labeling patterns of treponemes incubated with anti-GlpQ or anti-TpEf antiserum in the absence or presence of 0.05% Triton X-100.
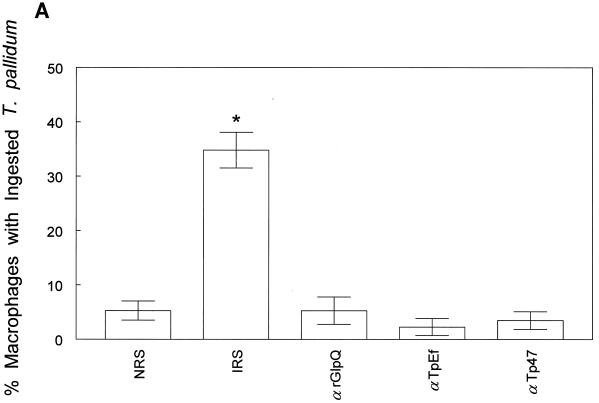
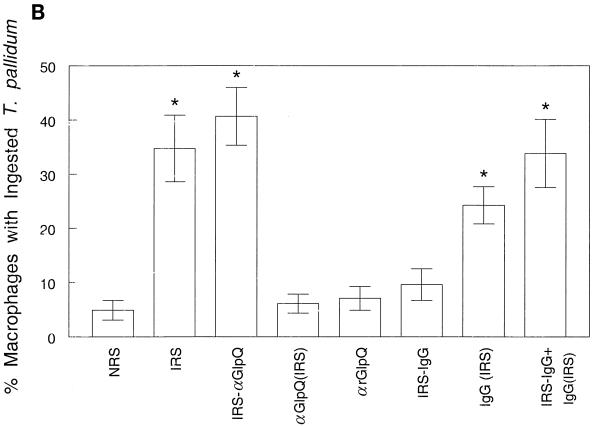
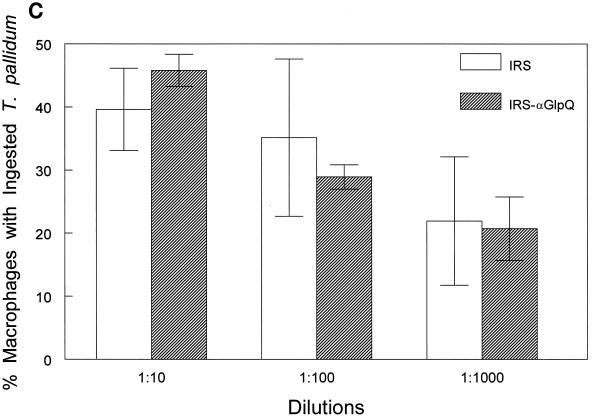
(iii) Stebeck and coworkers (60) identified GlpQ as a potential opsonic target for T. pallidum by differentially screening a genomic expression library with opsonic and nonopsonic IRS. Although they did not directly determine whether anti-GlpQ antibodies promote phagocytosis of motile treponemes in their original communication, more recently this same group reported that antibodies against rGlpQ were not opsonic (16). Here we also undertook a comprehensive examination of the T. pallidum opsonic activity of anti-GlpQ antibodies. In comparison with NRS, anti-TpEf, and anti-Tp47 negative controls, IRS significantly enhanced phagocytosis of motile treponemes, as previously reported (4–6, 38); no enhancement, on the other hand, was observed with anti-rGlpQ antiserum (Fig. 5A). To rule out the possibility that the anti-GlpQ antibodies in IRS and the anti-rGlpQ antisera are qualitatively different with respect to opsonic activity, the assays were repeated with IRS from which anti-GlpQ antibodies had been adsorbed. Preliminary immunoblot and enzyme-linked immunosorbent assay studies confirmed that the adsorbed IRS contained only trace amounts of anti-GlpQ antibodies (data not shown). In three independent experiments, no significant differences in the opsonic activities of IRS and the same IRS depleted of anti-nGlpQ antibodies were observed (Fig. 5B). The affinity-purified anti-GlpQ antibodies by themselves also failed to enhance opsonic activity (Fig. 5B). We also considered the possibility that the trace amounts of residual anti-GlpQ antibodies in the adsorbed IRS were still capable of contributing to its opsonic activity. This possibility was excluded by observing that serial dilutions of IRS and IRS lacking anti-GlpQ antibodies of as high as 1:1,000 possessed essentially identical opsonic activities (Fig. 5C). As an additional control for these adsorption experiments, we also investigated the opsonic activity of IRS depleted of total IgG. In contrast to the results obtained when the IRS was depleted of anti-GlpQ antibodies, IgG adsorption markedly diminished the serum’s opsonic activity, as previously noted (5), and this activity was fully restored when the IgG antibodies were added back to the adsorbed serum (Fig. 5B).
FIG. 5.
Anti-rGlpQ antiserum does not promote phagocytosis of virulent T. pallidum by rabbit peritoneal macrophages. (A) Opsonophagocytosis of virulent treponemes by rabbit peritoneal macrophages 4 h after incubation of organisms in the presence of 10% heat-inactivated antisera. (B) Opsonic activity of anti-GlpQ antibodies and total IgG affinity adsorbed from IRS. IRS-αGlpQ, IRS depleted of anti-GlpQ antibodies; αGlpQ (IRS), anti-GlpQ antibodies affinity purified from IRS; IRS-IgG, IRS depleted of total IgG antibodies; IgG (IRS), affinity-purified total IgG antibodies from IRS; (IRS-IgG)+IgG (IRS), IRS depleted of total IgG antibodies reconstituted with affinity purified IgG. (C) Opsonic activity of serial dilutions of IRS and IRS depleted of anti-GlpQ antibodies. In all panels, asterisks indicate results which were significantly greater than those for NRS controls. Results are means and standard errors.
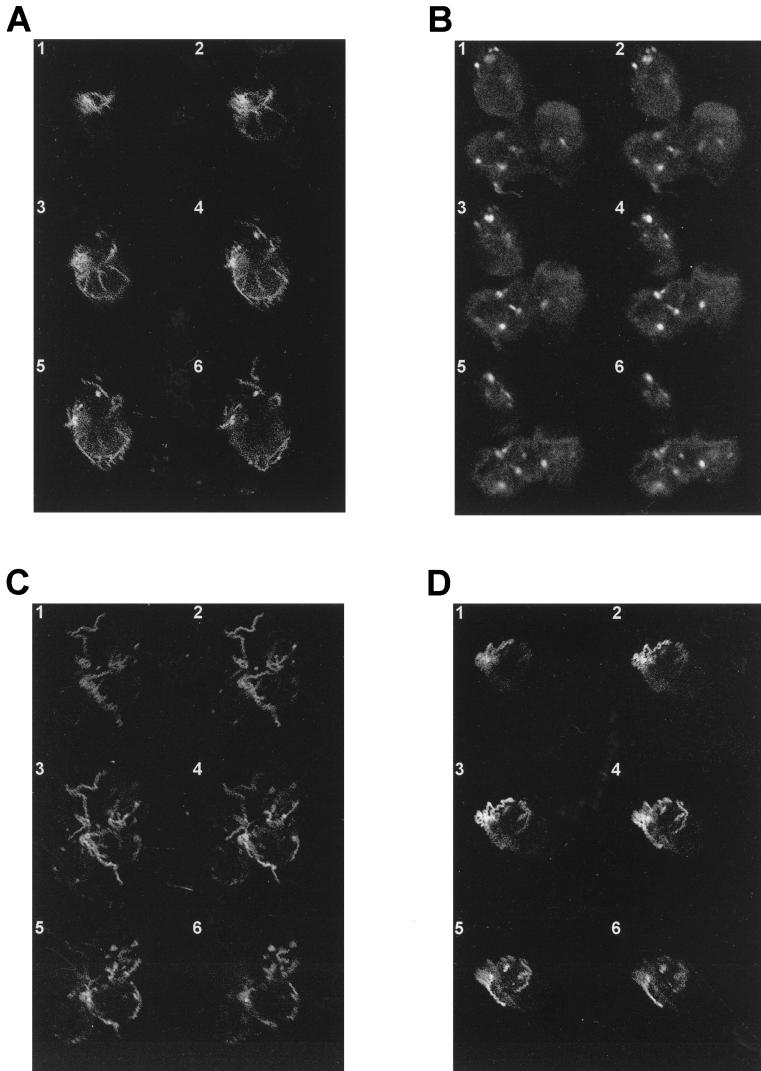
As a sidelight to the present study, we noted an intriguing dichotomy when macrophages incubated with treponemes with or without opsonic antibodies were examined by confocal laser microscopy. Opsonized treponemes were visualized almost exclusively as intracellular inclusions (Fig. 6B), representing degraded organisms within lysosomal vacuoles (38), whereas nonopsonized treponemes were visualized as intact organisms bound to macrophage surfaces (Fig. 6A, C, and D). Even in the absence of opsonizing antibodies, therefore, macrophages appear to express receptors for surface-exposed treponemal ligands. Unlike antibody-Fc receptor interactions, however, these receptor-ligand interactions fail to trigger signaling pathways which promote bacterial uptake. This observation underscores the notion that T. pallidum is an extracellular pathogen with little proclivity for taking up residence within phagocytic cells and emphasizes as well the treponeme’s innate ability to avoid uptake and killing by macrophages.
FIG. 6.
Confocal laser microscopy of rabbit peritoneal macrophages incubated with treponemes in the presence of opsonic or nonopsonic antibodies. Serial optical sections through rabbit peritoneal macrophages incubated with virulent treponemes in the presence or absence of NRS (A), IRS (B), rabbit anti-rGlpQ antiserum (C), or rabbit anti-TpEf antiserum (D) are shown. T. pallidum antigens were visualized by indirect immunofluorescence with HSS.
(iv) Finally, we investigated whether immunization with rGlpQ alters the course of infection in the rabbit model of experimental syphilis. Consistent with the above-described assays demonstrating a lack of surface exposure for nGlpQ, in two separate experiments involving a total of 12 rabbits, no differences in the time course of lesion development and resolution, in gross lesion appearance, or in the presence of treponemes in lesion aspirates from GlpQ-immunized and control animals were observed (Fig. 7). This finding is in contrast to the recent demonstration by Cameron et al. (16) of attenuated lesion development when rGlpQ immunized rabbits were intradermally challenged at each of six sites with 103 treponemes. This modest degree of protective immunity probably reflects differences in immunization protocols. It is also important to note that comparable levels of partial protection have previously been observed following immunization with T. pallidum proteins which are known to be intracellular (13, 18, 65), most likely reflecting a cellular component in the protective immune response.
FIG. 7.
Immunization with rGlpQ does not protect rabbits against experimental syphilis. Sham-immunized rabbits or rabbits immunized with rGlpQ were challenged by intradermal inoculation at each of six sites with 103 organisms. The photographs depict typical animals in one of two separate experiments.
CD spectroscopic analysis of rGlpQ secondary structure.
Although the simplest interpretation of the above results was that nGlpQ is not surface exposed, they also could have reflected a lack of antibodies against conformational, surface-exposed epitopes in the anti-rGlpQ antisera. Arguing against this was the observation from the immunofluorescence studies that the antisera recognized native antigen in organisms whose OMs were disrupted by mild detergent treatment. Nevertheless, it was clearly desirable to learn more about the recombinant protein’s conformational state. Our initial efforts along these lines were unsuccessful in that we were unable to detect significant enzymatic activity in either recombinant E. coli whole-cell lysates or the purified polypeptide by using a variety of substrates. We then turned to CD spectroscopy as an alternative means of examining the recombinant protein’s secondary structure. Consistent with the secondary structures predicted by both the Chou-Fasman (19) and Robson-Garnier (28) algorithms (data not shown), the molar ellipticity values in the far-UV region (195 to 260 nm) indicated that rGlpQ was predominantly α-helical. Equally important was the absence of discernible evidence for denaturation (Table 1); admittedly, we cannot rule out that more subtle structural abnormalities resulted in the loss of enzymatic activity. As a control, we also examined the CD spectrum of recombinant porin P1A from Neisseria gonorrhoeae (3). As previously reported (3), the gonococcal porin contains the relatively high degree of β-sheet structure typical of a gram-negative bacterial outer membrane protein (Table 1) (41).
TABLE 1.
Secondary structure of GlpQ calculated from CD spectroscopy
| Protein | % of secondary structurea
|
|||
|---|---|---|---|---|
| α-Helix | β-Sheet | Turn | Other | |
| GlpQ | 68 | 5 | 13 | 14 |
| Porin P1A | 12 | 45 | 22 | 21 |
Radiolabeling of isolated T. pallidum OMs with ([125I]TID).
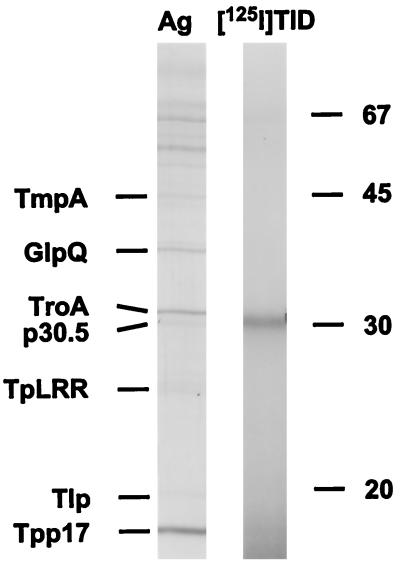
Even with the above-described findings, it was still possible that at least some nGlpQ was anchored via its N-terminal lipids to the inner leaflet of the OM, similar to the case for murein lipoprotein of E. coli (47). In fact, this orientation could explain how the protein could be present in isolated OMs but not surface exposed in intact treponemes. To address this final question, we incubated isolated OMs with the photoactivatable probe [125I]TID, an extremely lipophilic reagent which promiscuously labels proteins and lipids within the hydrophobic interiors of membranes (14, 33, 36). Labeling with this reagent would indicate that the protein was physically integrated into the OM bilayer rather than just peripherally associated with it. As shown in Fig. 8, a single 30.5-kDa polypeptide (designated p30.5 [gene TP0453] [27]) was radiolabeled when isolated OMs were incubated with the photoactivatable cross-linker. Two-dimensional gel electrophoresis confirmed that this was the same protein as was recently identified in T. pallidum whole cells incubated with this probe (3). Two findings argue against the concern that this method may not be sensitive enough to label low-abundance and/or lipid-anchored integral OM proteins: (i) p30.5 is considerably less abundant than nGlpQ (Fig. 8), and (ii) we recently have found that p30.5 also is a lipoprotein (57). Furthermore, Everett and Hatch (26) showed with Chlamydia trachomatis that [125I]TID is capable of labeling bacterial OM lipoproteins. Finally, we also considered the unlikely possibility that nGlpQ detached from OMs during the fractionation process. This was excluded by the finding that p30.5 was the only protein labeled when motile treponemes were incubated with [125I]TID prior to OM isolation (data not shown).
FIG. 8.
nGlpQ associated with T. pallidum OM fractions is not radiolabeled by [125I]TID. Outer membranes from 1010 treponemes were incubated with [125I]TID prior to SDS-PAGE and then silver staining (Ag) or autoradiography ([125I]TID). Labels to the left of the silver-stained lane designate T. pallidum proteins previously identified in isolated OMs. Numbers on the right are molecular masses in kilodaltons.
Final topological assignment and conclusion.
The many difficulties inherent in the molecular characterization of T. pallidum OM proteins (50) have spawned a variety of experimental approaches for accomplishing this important but elusive objective (10, 11, 31, 54, 60). In this regard, it is striking that two dissimilar strategies, OM isolation (54) and differential genomic library screening to identify potential T. pallidum opsonic targets (60), yielded the same candidate OM protein, nGlpQ. Nevertheless, the studies here present a substantial and consistent body of evidence that nGlpQ is a periplasmic protein anchored predominantly, if not exclusively, to the CM, thereby accounting for its inability to induce a strongly protective immune response. Although Cameron et al. (16) disagree with this conclusion, it is consistent with their inability to detect opsonic activity in anti-GlpQ antisera as well as their difficulty in obtaining additional evidence for surface localization by other approaches, such as the gel microdroplet assay. This assignment also agrees with the presumptive physiological role of GlpQ in phospholipid metabolism and the fact that it appears to be transcriptionally, and perhaps physiologically, linked with the gene encoding phosphotidylglycerophosphate synthase, a polytopic CM protein which mediates a key step in phospholipid synthesis (27, 56). With this topology one can envision two potential explanations for the protein’s presence as a contaminant in OM fractions (56) or its removal with OMs during extensive washing of treponemes (16). The first is that one or more domains of the nGlpQ polypeptide stably associate with the OM’s periplasmic leaflet. The second is that a portion of the protein dissociates from the peptidoglycan-CM complex during the isolation procedure and becomes trapped within OM vesicles. While these are not easily distinguished, we believe that the relatively low concentration of Triton X-100 needed for complete immunolabeling of nGlpQ favors the former.
Two other outcomes of this study are noteworthy. The first concerns the parallels between nGlpQ and another lipoprotein immunogen, Tpp17. Both subsurface antigens are antibody accessible at relatively low detergent concentrations, and both produce beaded labeling patterns when lightly detergent-treated treponemes are incubated with specific antibodies (21). The latter suggests that both proteins are nonuniformly distributed along the protoplasmic cylinder. Additionally, both proteins are major constituents in isolated OMs. Based upon its relative abundance in their OM preparations, Blanco and coworkers (11) proposed that Tpp17 is lipid anchored to the OM inner leaflet. As with nGlpQ, the [125I]TID labeling results, coupled with earlier results from gel microdroplet studies (21), argue against this topology. The second noteworthy outcome concerns the observation that the p30.5 lipoprotein was the only protein identified when isolated OMs or intact treponemes were subjected to radiolabeling with [125I]TID. Several years ago, we proposed a model for T. pallidum molecular architecture in which lipoproteins are exclusively CM anchored (22). Although its precise topology remains to be determined, the current data indicate that p30.5 is likely to be an exception to this generalization.
ACKNOWLEDGMENTS
This research was supported in part by U.S. Public Health Service grants AI-26756 and AI-38894 to J.D.R. T.J.S. was supported by NRSA postdoctoral fellowship award AI-09973 and by a research fellowship from the Arthritis Foundation.
REFERENCES
- 1.Akins D R, Porcella S F, Popova T G, Shevchenko D, Baker S I, Li M, Norgard M V, Radolf J D. Evidence for in vivo but not in vitro expression of a Borrelia burgdorferi outer surface protein F (OspF) homolog. Mol Microbiol. 1995;18:507–520. doi: 10.1111/j.1365-2958.1995.mmi_18030507.x. [DOI] [PubMed] [Google Scholar]
- 2.Akins D R, Purcell B K, Mitra M, Norgard M V, Radolf J D. Lipid modification of the 17-kilodalton membrane immunogen of Treponema pallidum determines macrophage activation as well as amphiphilicity. Infect Immun. 1993;61:1202–1210. doi: 10.1128/iai.61.4.1202-1210.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akins D R, Robinson E, Shevchenko D V, Elkins C, Cox D L, Radolf J D. Tromp1, a putative rare outer membrane protein, is anchored by an uncleaved leader peptide to the Treponema pallidum cytoplasmic membrane. J Bacteriol. 1997;179:576–586. doi: 10.1128/jb.179.16.5076-5086.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker-Zander S A, Lukehart S A. Macrophage-mediated killing of opsonized Treponema pallidum. J Infect Dis. 1992;165:69–74. doi: 10.1093/infdis/165.1.69. [DOI] [PubMed] [Google Scholar]
- 5.Baker-Zander S A, Shaffer J M, Lukehart S A. Characterization of the serum requirement for macrophage-mediated killing of Treponema pallidum ssp. pallidum: relationship to the development of opsonizing antibodies. FEMS Immunol Med Microbiol. 1993;6:273–279. doi: 10.1111/j.1574-695X.1993.tb00339.x. [DOI] [PubMed] [Google Scholar]
- 6.Baker-Zander S A, Shaffer J M, Lukehart S A. VDRL antibodies enhance phagocytosis of Treponema pallidum by macrophages. J Infect Dis. 1993;167:1100–1105. doi: 10.1093/infdis/167.5.1100. [DOI] [PubMed] [Google Scholar]
- 7.Barbour A G, Hayes S F. Biology of Borrelia species. Microbiol Rev. 1986;50:381–400. doi: 10.1128/mr.50.4.381-400.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barbour A G, Tessier S L, Hayes S F. Variation in a major surface protein of Lyme disease spirochetes. Infect Immun. 1984;45:94–100. doi: 10.1128/iai.45.1.94-100.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barbour A G, Tessier S L, Stoenner H G. Variable major proteins of Borrelia hermsii. J Exp Med. 1982;156:1312–1324. doi: 10.1084/jem.156.5.1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blanco D R, Giladi M, Champion C I, Haake D A, Chikami G K, Miller J N, Lovett M A. Identification of Treponema pallidum subspecies pallidum genes encoding signal peptides and membrane-spanning sequences using a novel alkaline phosphatase expression vector. Mol Microbiol. 1991;5:2405–2415. doi: 10.1111/j.1365-2958.1991.tb02086.x. [DOI] [PubMed] [Google Scholar]
- 11.Blanco D R, Reimann K, Skare J, Champion C I, Foley D, Exner M M, Hancock R E W, Miller J N, Lovett J N. Isolation of the outer membranes from Treponema pallidum and Treponema vincentii. J Bacteriol. 1994;176:6088–6099. doi: 10.1128/jb.176.19.6088-6099.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- 13.Borenstein L A, Radolf J D, Fehniger T E, Blanco D R, Miller J N, Lovett M A. Immunization with recombinant Treponema pallidum surface antigen 4D alters the course of experimental syphilis. J Immunol. 1988;140:2415–2421. [PubMed] [Google Scholar]
- 14.Brunner J. Photochemical labeling of apolar phase of membranes. Methods Enzymol. 1989;172:628–687. doi: 10.1016/s0076-6879(89)72037-1. [DOI] [PubMed] [Google Scholar]
- 15.Brusca J S, Radolf J D. Isolation of integral membrane proteins by phase partitioning with Triton X-114. Methods Enzymol. 1994;228:182–193. doi: 10.1016/0076-6879(94)28019-3. [DOI] [PubMed] [Google Scholar]
- 16.Cameron C E, Castro C, Lukehart S A, Van Voorhis W C. Function and protective capacity of Treponema pallidum subsp. pallidum glycerophosphodiester phosphodiesterase. Infect Immun. 1998;66:5763–5770. doi: 10.1128/iai.66.12.5763-5770.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chamberlain N R, Brandt M E, Erwin A L, Radolf J D, Norgard M V. Major integral membrane protein immunogens of Treponema pallidum are proteolipids. Infect Immun. 1989;57:2872–2877. doi: 10.1128/iai.57.9.2872-2877.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Champion C I, Miller J N, Borenstein L A, Lovett M A, Blanco D R. Immunization with Treponema pallidum endoflagella alters the course of experimental rabbit syphilis. Infect Immun. 1990;58:3158–3161. doi: 10.1128/iai.58.9.3158-3161.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chou P Y, Fasman G D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- 20.Cox D L, Akins D R, Bourell K W, Lahdenne P, Norgard M V, Radolf J D. Limited surface exposure of Borrelia burgdorferi outer surface lipoproteins. Proc Natl Acad Sci USA. 1996;93:7973–7978. doi: 10.1073/pnas.93.15.7973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cox D L, Akins D R, Porcella S F, Norgard M V, Radolf J D. Treponema pallidum in gel microdroplets: a novel strategy for investigation of treponemal molecular architecture. Mol Microbiol. 1995;15:1151–1164. doi: 10.1111/j.1365-2958.1995.tb02288.x. [DOI] [PubMed] [Google Scholar]
- 22.Cox D L, Chang P, McDowall A, Radolf J D. The outer membrane, not a coat of host proteins, limits the antigenicity of virulent Treponema pallidum. Infect Immun. 1992;60:1076–1083. doi: 10.1128/iai.60.3.1076-1083.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cox D L, Riley B, Chang P, Sayahtaheri S, Tassell S, Hevelone J. Effects of molecular oxygen, oxidation-reduction potential, and antioxidants upon in vitro replication of Treponema pallidum subsp. pallidum. Appl Environ Microbiol. 1990;56:3063–3072. doi: 10.1128/aem.56.10.3063-3072.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dickerson M C, Johnston J, Delea T E, White A, Andrews E. The causal role for genital ulcer disease as a risk factor for transmission of human immunodeficiency virus. An application of the Bradford Hill criteria. Sex Transm Dis. 1997;23:429–440. doi: 10.1097/00007435-199609000-00015. [DOI] [PubMed] [Google Scholar]
- 25.Elkins C, Barkley K B, Carbonetti N H, Coimbre A J, Sparling P F. Immunobiology of purified recombinant outer membrane porin protein I of Neisseria gonorrhoeae. Mol Microbiol. 1994;14:1059–1075. doi: 10.1111/j.1365-2958.1994.tb01339.x. [DOI] [PubMed] [Google Scholar]
- 26.Everett K D, Hatch T P. Architecture of the cell envelope of Chlamydia psittaci 6BC. J Bacteriol. 1995;177:877–882. doi: 10.1128/jb.177.4.877-882.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fraser C M, Norris S J, Weinstock G M, White O, Sutton G C, Dodson R, Gwinn M, Hickey E K, Clayton R, Ketchum K A, Sodergren E, Hardham J M, McLeod M P, Salzberg S, Peterson J, Khalak H, Richardson D, Howell J K, Chidambaram M, Utterback T, McDonald L, Artiach P, Bowman C, Cotton M D, Fujii C, Garland S, Hatch B, Horst K, Roberts K, Sandusky M, Weidman J, Smith H O, Venter J C. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science. 1998;281:375–388. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- 28.Garnier J, Osguthorpe D J, Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978;120:97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- 29.Green B A, Farley J E, Quinn-Dey T, Deich R A, Zlotnick G W. The e (P4) outer membrane protein of Haemophilus influenzae: biologic activity of anti-e serum and cloning and sequencing of the structural gene. Infect Immun. 1991;59:3191–3198. doi: 10.1128/iai.59.9.3191-3198.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanff P A, Norris S J, Lovett M A, Miller J N. Purification of Treponema pallidum, Nichols strain, by Percoll density gradient centrifugation. Sex Transm Dis. 1984;11:275–286. doi: 10.1097/00007435-198410000-00003. [DOI] [PubMed] [Google Scholar]
- 31.Hardham J M, Stamm L V. Identification and characterization of the Treponema pallidum tpn50 gene, an ompA homolog. Infect Immun. 1994;62:1015–1025. doi: 10.1128/iai.62.3.1015-1025.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayashi S, Wu H C. Lipoproteins in bacteria. J Bioenerget Biomembr. 1990;22:451–471. doi: 10.1007/BF00763177. [DOI] [PubMed] [Google Scholar]
- 33.Hoppe J, Brunner J, Jorgensen B B. Structure of the membrane-embedded F0 part of F1F0 ATP synthase from Escherichia coli as inferred from labeling with 3-(trifluoromethyl)-3-(m-[125I]iodophenyl)diazirine. Biochemistry. 1984;23:5610–5616. doi: 10.1021/bi00318a035. [DOI] [PubMed] [Google Scholar]
- 34.Isaacs R D, Hanke J H, Guzman-Verduzco L-M, Newport G, Agabian N, Norgard M V, Lukehart S A, Radolf J D. Molecular cloning and DNA sequence analysis of the 37-kilodalton endoflagellar sheath protein of Treponema pallidum. Infect Immun. 1989;57:3403–3411. doi: 10.1128/iai.57.11.3403-3411.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones J D, Bourell K W, Norgard M V, Radolf J D. Membrane topology of Borrelia burgdorferi and Treponema pallidum lipoproteins. Infect Immun. 1995;63:2424–2434. doi: 10.1128/iai.63.7.2424-2434.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jorgensen P L, Brunner J. Labeling of intramembrane segments of the alpha-subunit and beta-subunit of pure membrane-bound (Na+ + K+)-ATPase with 3-trifluoromethyl-3-(m-[125I]iodophenyl)diazirine. Biochim Biophys Acta. 1983;735:291–296. doi: 10.1016/0005-2736(83)90304-8. [DOI] [PubMed] [Google Scholar]
- 37.Larson T J, Ehrmann M, Boos W. Periplasmic glycerophosphodiester phosphodiesterase of Escherichia coli, a new enzyme of the glp regulon. J Biol Chem. 1983;258:5428–5432. [PubMed] [Google Scholar]
- 38.Lukehart S A, Miller J N. Demonstration of the in vitro phagocytosis of Treponema pallidum by rabbit peritoneal macrophages. J Immunol. 1978;121:2014–2024. [PubMed] [Google Scholar]
- 39.Munson R S, Jr, Sasaki K. Protein D, a putative immunoglobulin D-binding protein produced by Haemophilus influenzae, is glycerophosphodiester phosphodiesterase. J Bacteriol. 1993;175:4569–4571. doi: 10.1128/jb.175.14.4569-4571.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakashima A K, Rolfs R T, Flock M L, Kilmarx P, Greenspan J R. Epidemiology of syphilis in the United States, 1941–1993. Sex Transm Dis. 1996;23:16–23. doi: 10.1097/00007435-199601000-00006. [DOI] [PubMed] [Google Scholar]
- 41.Nikaido H. Porins and specific diffusion channels in bacterial outer membranes. J Biol Chem. 1994;269:3905–3908. [PubMed] [Google Scholar]
- 42.Norris S J, Carter C J, Howell J K, Barbour A G. Low-passage-associated proteins of Borrelia burgdorferi B31: characterization and molecular cloning of OspD, a surface-exposed, plasmid-encoded lipoprotein. Infect Immun. 1992;60:4662–4672. doi: 10.1128/iai.60.11.4662-4672.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Farrell P H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 44.Perine P L. Sexually transmitted diseases in the tropics. Med J Aust. 1994;160:358–363. [PubMed] [Google Scholar]
- 45.Porcella S F, Popova T G, Akins D R, Li M, Radolf J D, Norgard M V. Borrelia burgdorferi supercoiled plasmids encode multiple tandem open reading frames and a lipoprotein gene family. J Bacteriol. 1996;178:3293–3307. doi: 10.1128/jb.178.11.3293-3307.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Probert W S, Allsup K M, LeFebvre R B. Identification and characterization of a surface-exposed 66-kilodalton protein from Borrelia burgdorferi. Infect Immun. 1995;63:1933–1939. doi: 10.1128/iai.63.5.1933-1939.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Purcell B K, Swancutt M A, Radolf J D. Lipid modification of the 15 kilodalton major membrane immunogen of Treponema pallidum. Mol Microbiol. 1990;4:1371–1379. doi: 10.1111/j.1365-2958.1990.tb00716.x. [DOI] [PubMed] [Google Scholar]
- 49.Radolf J D. Role of outer membrane architecture in immune evasion by Treponema pallidum and Borrelia burgdorferi. Trends Microbiol. 1994;2:307–311. doi: 10.1016/0966-842x(94)90446-4. [DOI] [PubMed] [Google Scholar]
- 50.Radolf J D. Treponema pallidum and the quest for outer membrane proteins. Mol Microbiol. 1995;16:1067–1073. doi: 10.1111/j.1365-2958.1995.tb02332.x. [DOI] [PubMed] [Google Scholar]
- 51.Radolf J D, Bourell K W, Akins D R, Brusca J S, Norgard M V. Analysis of Borrelia burgdorferi membrane architecture by freeze-fracture electron microscopy. J Bacteriol. 1994;176:21–31. doi: 10.1128/jb.176.1.21-31.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Radolf J D, Chamberlain N R, Clausell A, Norgard M V. Identification and localization of integral membrane proteins of Treponema pallidum subsp. pallidum by phase partitioning with the nonionic detergent Triton X-114. Infect Immun. 1988;56:490–498. doi: 10.1128/iai.56.2.490-498.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Radolf J D, Moomaw C, Slaughter C A, Norgard M V. Penicillin-binding proteins and peptidoglycan of Treponema pallidum subsp. pallidum. Infect Immun. 1989;57:1248–1254. doi: 10.1128/iai.57.4.1248-1254.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Radolf J D, Robinson E J, Bourell K W, Akins D R, Porcella S F, Weigel L M, Jones J D, Norgard M V. Characterization of outer membranes isolated from Treponema pallidum, the syphilis spirochete. Infect Immun. 1995;63:4244–4252. doi: 10.1128/iai.63.11.4244-4252.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ruan M R, Akkoyunlu M, Grubb A, Forsgren A. Protein D of Haemophilus influenzae. A novel bacterial surface protein with affinity for human IgD. J Immunol. 1990;145:3379–3384. [PubMed] [Google Scholar]
- 56.Shevchenko D V, Akins D R, Robinson E J, Li M, Shevchenko O V, Radolf J D. Identification of homologs for thioredoxin, peptidyl prolyl cis-trans isomerase, and glycerophosphodiester phosphodiesterase in outer membrane fractions from Treponema pallidum, the syphilis spirochete. Infect Immun. 1997;67:4179–4189. doi: 10.1128/iai.65.10.4179-4189.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shevchenko, D. V., D. R. Akins, E. J. Robinson, and J. D. Radolf. 1998. Unpublished observations.
- 58.Sreerama N, Woody R W. A self-consistent method for the analysis of protein secondary structure from circular dichroism. Anal Biochem. 1996;209:32–44. doi: 10.1006/abio.1993.1079. [DOI] [PubMed] [Google Scholar]
- 59.Sreerama N, Woody R W. Poly(Pro)II helices in globular proteins: identification and circular dichroic analysis. Biochemistry. 1994;33:10022–10025. doi: 10.1021/bi00199a028. [DOI] [PubMed] [Google Scholar]
- 60.Stebeck C E, Shaffer J M, Arroll T W, Lukehart S A, Van Voorhis W C. Identification of the Treponema pallidum susp. pallidum glycerophosphodiester phosphodiesterase homologue. FEMS Microbiol Lett. 1997;154:303–310. doi: 10.1111/j.1574-6968.1997.tb12660.x. [DOI] [PubMed] [Google Scholar]
- 61.Tramont E C. Treponema pallidum (syphilis) In: Mandell G L, Bennett J E, Dolin R, editors. Principles and practice of infectious diseases. New York, N.Y: Churchill Livingstone, Inc.; 1994. pp. 2117–2133. [Google Scholar]
- 62.Walker E M, Zampighi G A, Blanco D R, Miller J N, Lovett M A. Demonstration of rare protein in the outer membrane of Treponema pallidum subsp. pallidum by freeze-fracture analysis. J Bacteriol. 1989;171:5005–5011. doi: 10.1128/jb.171.9.5005-5011.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wasserheit J N. Epidemiological synergy. Interrelationships between human immunodeficiency virus infection and other sexually transmitted diseases. Sex Transm Dis. 1992;19:61–77. [PubMed] [Google Scholar]
- 64.Weigel L M, Radolf J D, Norgard M V. The 47-kDa major lipoprotein immunogen of Treponema pallidum is a penicillin-binding protein with carboxypeptidase activity. Proc Natl Acad Sci USA. 1994;91:11611–11615. doi: 10.1073/pnas.91.24.11611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wicher K, Schouls L M, Wicher V, Van Embden J D, Nakeeb S S. Immunization of guinea pigs with recombinant TmpB antigen induces protection against challenge infection with Treponema pallidum Nichols. Infect Immun. 1991;59:4343–4348. doi: 10.1128/iai.59.12.4343-4348.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]