Abstract
Asp f 2 is a major Aspergillus fumigatus allergen involved in allergic bronchopulmonary aspergillosis. Knowledge of the B-cell epitopes may contribute to the understanding of immunoregulation and immunodiagnosis. To elucidate the immunoglobulin E (IgE) binding epitopes in the linear sequence of Asp f 2, we synthesized decamer peptides spanning the whole molecule of Asp f 2 on derivatized cellulose membranes and evaluated IgE binding in ABPA patient and control sera. Peptides three to five amino acids long were synthesized based on amino acid sequences within the IgE binding regions and evaluated for the specificity of epitope antibody interactions. Nine IgE binding regions were recognized in this protein of 268 amino acid residues. Of the nine epitopes, seven (ATQRRQI, RKYFG, HWR, YTTRR, DHFAD, ALEAYA, and THEGGQ) are present in the hydrophilic regions of Asp f 2. Immunologic evaluation of the three recombinant fragments, Asp f 2A encompassing the N-terminal epitope region, Asp f 2B without N- and C-terminal regions of the protein, and Asp f 2C representing C-terminal epitopes, revealed that either the N- or C-terminal region of the protein is essential for the correct folding and conformation for IgE antibody binding.
Allergic inhalant diseases such as asthma, allergic rhinitis, and conjunctivitis affect about 20% of the population in the industrialized countries worldwide (16, 31, 40). Fungal spores are universally recovered both indoors and outdoors and are recognized as an important cause of respiratory allergy (8, 19). In the presence of major histocompatibility complex molecules, the allergen protein encountering the host immune system is recognized by its conformational and linear structures by immunoglobulin (B-cell epitopes) or after ingestion and processing by antigen-presenting cells as small peptide fragments by T cells (T-cell epitopes). Identification and characterization of B- and T-cell epitopes of fungal allergens from various sources are essential for the understanding of pathophysiology of the allergic reactions and to develop sensitive and specific diagnosis and treatment of these diseases.
Over the last few years, several proteins with allergenicity have been cloned and expressed by molecular biology techniques (14, 18, 34). These recombinant allergens and polypeptides are now the important source of reliable and standardized antigens for improved diagnosis and immunotherapy. Despite the rapidly increasing number of recombinant allergens, very few immunoglobulin E (IgE)-reactive B cell epitopes have been identified for various allergens from different sources (34). In several studies, enzymatically cleaved antigens or synthetic overlapping peptides were immunologically evaluated for defining IgE epitopes of allergens from pollen, mite, milk, and codfish (2, 14, 15, 17, 30, 36, 38). Recently recombinant DNA techniques have been used also to express a series of overlapping cDNAs for identification of the epitopes, using mouse antibodies and sera from sensitized patients (38, 41).
A. fumigatus, a ubiquitous fungus present in nature, causes a variety of respiratory disorders, including allergic bronchopulmonary aspergillosis (ABPA). A. fumigatus antigens are diverse in their physicochemical and immunological characteristics; the molecular structures and biological functions of most of them are poorly understood (6, 7, 26). By using molecular cloning techniques, some of the major allergens of A. fumigatus have been cloned and sequenced (1, 4, 5, 12, 13, 21, 29). Among the recombinant A. fumigatus allergens, Asp f 1 and Asp f 3 exhibited IgE antibody binding with sera from ABPA patients as well as from A. fumigatus skin prick test-positive allergic asthma patients. On the other hand, intracellular allergens Asp f 4 and Asp f 6 are reported to demonstrate distinct IgE binding exclusively with sera from ABPA patients (12). The distinct IgE binding properties of A. fumigatus allergens indicate that the characteristic features of A. fumigatus-sensitized allergic disorders, to some extent, depend on the proteins involved and their cellular localization, as well as on their structures and conformations. However, information about the B- and T-cell epitopes of these major A. fumigatus allergens is still not available.
Recently we have reported another major allergen of A. fumigatus, Asp f 2, which reacts with IgE antibodies from ABPA patients, especially those with central bronchiectasis. More than 90% of the patients with ABPA used in this study displayed significant IgE reactivity with recombinant Asp f 2 (3). To investigate the role of Asp f 2 in IgE-mediated immune responses of the patients, we have extended our study by analyzing the major IgE binding epitopes and recombinant constructs of Asp f 2 fragments containing various epitopes. The immunological evaluation of Asp f 2 overlapping peptides revealed nine distinct IgE binding epitopes varying in length from 3 to 7 amino acids (aa) throughout the molecule. However, the recombinant fragments representing several epitopes evaluated in this study showed distinct differences in IgE binding properties, indicating that in the recombinant peptides, these epitopes are under conformational constraints and need to be expressed in appropriate conformation to react with IgE antibody in patient.
MATERIALS AND METHODS
Subjects.
Three groups of subjects were included in this study. Serum samples were collected from the patients with ABPA (n = 24) attending the Allergy-Immunology Clinics of Medical College of Wisconsin Affiliated Hospitals and the Allergy-Immunology Clinic of Northwestern University Medical School. These patients fulfilled the criteria for the disease as described by Rosenberg et al. (32). The serum samples from 10 patients with asthma and immediate wheal and flare skin reactivity to A. fumigatus antigens but without clinical features of ABPA and from 10 normal subjects with no history of respiratory disease were also tested. The institutional review committee had approved all human studies.
Solid-phase peptide synthesis.
Peptide synthesis was carried out on derivatized cellulose membranes, 9-fluorenylmethoxy carbonyl-derived amino acids (Fmoc-amino acids) as specified by the manufacturer (SPOTs; Genosys Biotechnologies Inc., The Woodlands, Tex.). The free amino functional group present on the spot was used in the synthesis of the various peptides, and the carboxyl group of the N-terminal amino acid of the peptide was linked with the NH2 group of the Fmoc-amino acid on the particular spot on the membrane (24). Based on published amino acid sequence of Asp f 2, 259 decapeptides were synthesized at an offset of 1 aa to span the complete Asp f 2 molecule (4). Once the IgE binding regions of Asp f 2 were identified, in order to evaluate the specificity of the epitope-IgE interaction, small overlapping peptides of 3 to 5 aa encompassing the identified epitopes RKYFG (epitope 2), HWR (epitope 3), and YTTRR (epitope 4) were synthesized. All of these synthetic peptides were then evaluated with sera from ABPA patients and controls for IgE antibody binding.
Antibody binding of the synthetic peptides.
After synthesis of various Asp f 2 peptides, the membranes were blocked overnight before incubation with pooled sera from patients with ABPA (25). Washings were carried out between incubations with 0.1% Tween 20 in Tris-buffered saline (T-TBS). Biotinylated anti-human IgE (Vector Laboratories, Burlingame, Calif.) diluted 1:1,000 in T-TBS was then added to the membranes and incubated for 1 h at room temperature. The membranes were then treated with 1:27,000-diluted streptavidin-alkaline phosphatase (Sigma, St. Louis, Mo.) for 45 min and washed as before. The color was developed by using as a substrate nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate (Bio-Rad Laboratories, Richmond, Calif.) as described previously (23). The intensities of the color reactions were analyzed by densitometric scanning of the SPOTs membranes with an AMBIS (San Diego, Calif.) image acquisition and analysis instrument. Various identified epitopes were then synthesized as described above and treated with pooled normal sera at a dilution of 1:25 for specific IgE binding. The subsequent steps were the same as described above. The sequence similarity searches were done with the BLAST program of the National Center for Biotechnology Information and with the Genetics Computer Group (Madison, Wis.) package (11). The hydrophilic and hydrophobic regions of Asp f 2 were also predicted with the Seq Vu 1.0 program (Garvan Institute of Medical Research, Sydney, New South Wales, Australia) with a window size of 7 aa.
Competitive ELISA using linear peptides and solid-phase coated Asp f 2.
The IgE antibody binding of Asp f 2 and synthetic peptides were studied by inhibition enzyme-linked immunosorbent assay (ELISA). Wells of a 96-well microtiter plate were coated with recombinant Asp f 2 at 5 μg/ml in phosphate-buffered saline (PBS) and incubated at 4°C overnight. The plate was then washed with PBS, and the wells were incubated with blocking buffer (0.3% gelatin in PBS) for 2 h at room temperature. A second plate was incubated with blocking buffer in the same manner. Serum samples from ABPA patients 1 and 2 were diluted 1:250 and those from patient 3 were diluted 1:25 in blocking buffer and allowed to react with various concentrations of peptides PA (aa 29 to 43; NCALEGWGGHWRGAN), PB (aa 292 to 310; TIDVPSNCHTHEGGQLHCT), and Asp f 2 from 0.05 to 50 μg/ml in the second plate for 2 h at room temperature. The reaction mixtures then were added to the Asp f 2-coated plate and incubated overnight at 4°C. The binding of free antibodies in the preincubated sera to Asp f 2 on the solid phase was detected by a procedure previously described (4). Two individual sera (1:25) from each group of allergic asthma patients and healthy controls were also included in this study. The ELISA IgE absorbance of Asp f 2 with uninhibited sera was considered to represent 100% binding. The percentages of inhibition of IgE antibody binding of the peptides and antigen-incubated sera were calculated in comparison to the binding with uninhibited sera.
Preparation of full-length Asp f 2 (aa 1 to 268) and recombinant constructs Asp f 2A (aa 1 to 203), Asp f 2B (aa 68 to 203), and Asp f 2C (aa 68 to 268).
After identification of sequential IgE-reactive epitopes, we constructed recombinant polypeptides representing different epitopes in order to analyze the involvement of conformational constraints of Asp f 2 in expressing these epitopes.
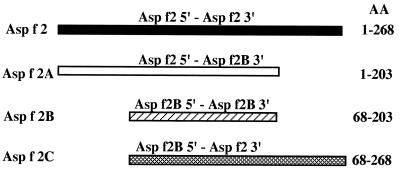
The genes encoding different fragments of Asp f 2 were amplified by PCR. The template DNA used was the clone encoding Asp f 2. Primers used for amplification of the complete Asp f 2 were sense 5′ GACGCTGGCGCGGTGACCTCGT 3′ (Asp f 2 5′) and antisense 5′ AGTGCAATGAAGCTGTCCACCTTC 3′ (Asp f 2 3′). The other primers used were sense 5′ GTCATCGTGAACGGGGACAAG 3′ (Asp f 2B 5′) and antisense 5′ TCCGACACCGGGAGCAGCAATAT 3′ (Asp f 2B 3′). A schematic diagram of the sizes of different constructs is shown in Fig. 1. The amplified fragments were subcloned into pCR 2.1 vector (Invitrogen, San Diego, Calif.).
FIG. 1.
Schematic representation of four cDNA constructs of Asp f 2 encoding various fragments containing IgE binding epitopes. These DNA clones encoding recombinant polypeptides were overexpressed in pET-23b(+) vector (for details, see Materials and Methods).
Expression and purification of recombinant fragments of Asp f 2.
Plasmids encoding the recombinant polypeptides of Asp f 2 were sequenced by primer walking using the dideoxy-chain termination method (33). All sequences were read and recorded manually. The primers used at different stages of sequencing were synthesized by Biosynthesis Inc., Lewisville, Tex. Escherichia coli HMS174 was transformed with pET-23b(+) vectors with or without inserts, and final expression was carried out in E. coli BL21(DE3)(PLYsS) as described before (4).
Antigen-specific ELISA.
The binding of specific IgE antibodies in sera from various subjects to the recombinant constructs of Asp f 2 was analyzed by a solid-phase ELISA as described before (3, 22).
Statistical analysis.
The different groups of patients, healthy subjects, and antigens were compared for reactivity with the antibody by unpaired t test by using Statworks software (Computer Associates International, Islandia, N.Y.).
RESULTS
Identification of linear IgE binding epitopes of Asp f 2.
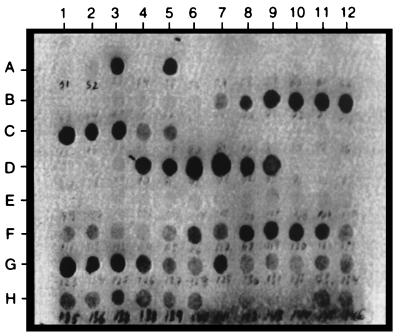
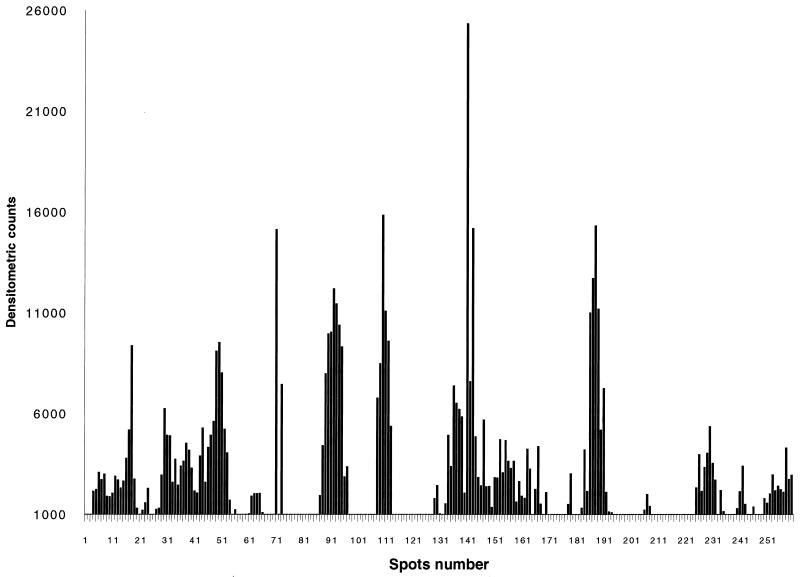
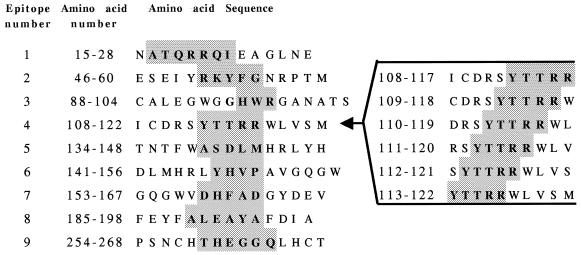
From the IgE antibody binding to the various decapeptides of Asp f 2, we identified several regions of strong reactivity with the pooled sera of patients with ABPA (Fig. 2). The specificity of IgE antibody binding was ascertained from the lack of reaction to peptides with random sequences and the synthetic peptides representing nonepitope regions. Densitometric scanning of the peptide-antibody reactions on the membranes is shown in Fig. 3. Nine IgE binding regions were identified in this allergen, as determined from the densitometric counts of the spots on the cellulose membrane. Based on the primary structure of the protein, amino acid residues of the overlapping decapeptides involved in antibody binding were selected as shown in Fig. 4. The six IgE binding overlapping decapeptides on spots 4 to 9 of row D (Fig. 2) have common amino acid residues YTTRR (aa 113 to 117), indicating that these 5 aa are involved in IgE binding with sera from ABPA patients and hence represent an IgE binding epitope region (epitope 4) of Asp f 2 (Fig. 4, right). Similarly, the amino acid residues of the remaining epitopes were also identified and listed in Fig. 4. The lengths of these IgE binding regions ranged from 3 (HWR; epitope 3) to 7 (ATQRRQI; epitope 1) aa.
FIG. 2.
IgE binding of synthetic peptides of Asp f 2 with sera from ABPA patients. Linear decapeptides were synthesized on derivatized cellulose membranes with an offset of 1 aa. On each spot on the membrane marked for peptide synthesis, a free amino functional group was available for coupling with the carboxyl group of the N-terminal amino acid of the peptide. Ten cycles of amino acid addition were carried out for synthesis of decapeptides. After completion of the peptide synthesis, the protecting groups on the side chains of the amino acids were removed. The IgE binding epitopes were identified at various regions of Asp f 2 based on the antibody peptide reaction demonstrated by enzyme immunostaining (for details, see Materials and Methods).
FIG. 3.
Densitometric counts of the intensity of IgE peptide interaction on the cellulose membrane were determined; based on the counts of the individual spots, the IgE binding regions of Asp f 2 were identified.
FIG. 4.
Amino acid sequences representing nine IgE binding epitopes of Asp f 2. The continuous linear epitopes were identified by synthesizing overlapping peptides spanning the complete protein. The length of the epitopes varied from 3 (HWR; epitope 3) to 7 (ATQRRQI; epitope 1) aa. Various amino acids in epitope 4 are shown at the right as an example of how the epitope was derived by synthesizing various overlapping peptides.
Specificity of IgE binding to the B-cell epitopes of Asp f 2.
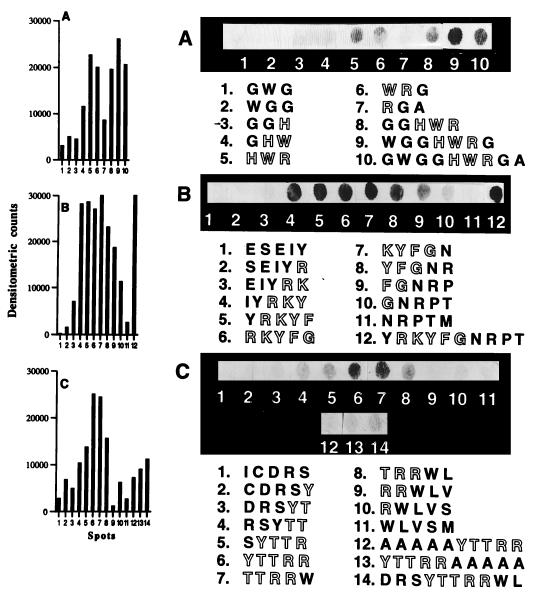
The specificity of IgE binding regions was further evaluated by synthesizing overlapping peptides of various lengths of 3 to 10 aa and tested for IgE binding with sera from ABPA patients. The effects of adjacent amino acids on IgE antibody binding of these epitope regions of Asp f 2 are shown in Fig. 5. Spots 1 to 7 in Fig. 5A represent 3-aa-long peptides of Asp f 2, synthesized at an offset of 1 aa. Amino acid sequence of spot 5 represents the selected epitope HWR (epitope 3). The open-lettered amino acids are the part of the IgE binding region. There is a gradual increase in IgE antibody binding from spots 4 to 6, and the peptides at spots 1, 2, and 3 did not show IgE binding, indicating the specificity of the epitope-antibody interaction. As shown on spots 8, 9, and 10 (Fig. 5A), the additional amino acids at the N- and C-terminal ends of HWR had increased the intensity of the reaction. The WGGHWRG at spot 9 (Fig. 5A) with flanking amino acids WGG and G at the N- and C-terminal ends, respectively, showed the strongest reaction with patient sera. Detailed analysis of IgE antibody binding specificity of 5-aa-long RKYFG (epitope 2) is shown in Fig. 5B. Spot six represents the selected epitope 2 of Asp f 2. Marked differences were seen in the IgE binding when substitution of one amino acid residue at a time was made in the selected epitope. The peptide KYFGN at spot 7 (Fig. 5B) also showed a strong antibody binding reaction, whereas no reactivity was seen on spot 11, representing peptide NRPTM without any amino acid residues from the identified epitope region. The decapeptide on spot 12 (Fig. 5B) with a flanking amino acid Y at the N-terminal end and amino acid residues NRPT at the C-terminal end also exhibited strong IgE antibody binding. Similarly, another 5-aa-long epitope YTTRR (epitope 4) was evaluated for IgE antibody binding (Fig. 5C). The selected epitope on spot six shows distinct IgE binding with patient sera. The addition of five alanine molecules at the N-terminal and C-terminal ends (Fig. 5C, spots 12 and 13, respectively) have shown reduced intensity indicating a weaker IgE antibody binding of this peptides.
FIG. 5.
(A) Specificity of IgE binding of 3-aa-long peptides of Asp f 2, synthesized at an offset of 1 aa encompassing epitope 3 HWR (spot 5). Open-lettered amino acids are the part of the selected epitopes. (B) IgE antibody binding of 5-aa-long peptides synthesized at an offset of 1 aa; spot 6 represents the selected epitope 2 (RKYFG) of Asp f 2. (C) IgE binding of overlapping peptides encompassing epitope 4 (YTTRR) of Asp f 2. All peptides were evaluated for IgE binding by using pooled ABPA sera at a dilution of 1:10. The left panel represents densitometric scanning of the spots as a measure of intensity of IgE-peptide reaction (for details, see Materials and Methods).
Peptide-mediated inhibition of Asp f 2 IgE binding.
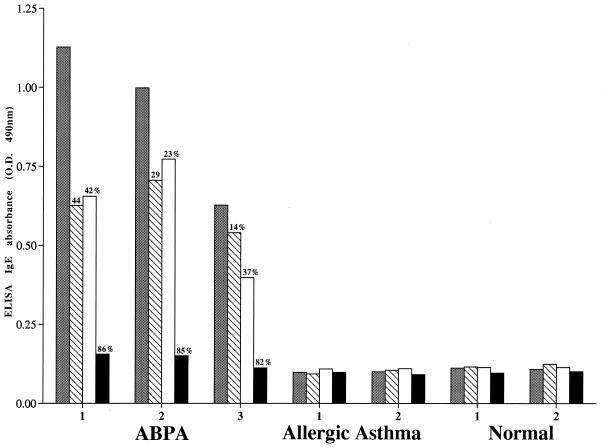
Synthetic peptides representing two of the above-identified epitopes were further evaluated for their structural and conformational similarities with the B-cell epitopes present on the Asp f 2 molecule, using competitive ELISA. As shown in Fig. 6, peptides PA and PB, representing epitopes 3 and 9 of Asp f 2, respectively, demonstrated significant IgE binding with serum from an ABPA patient. This is evident from 86% inhibition in IgE binding with serum 1 when the percentages of inhibition by the two peptides are taken together and is comparable to the inhibition noted with homologous allergen Asp f 2. Similarly, peptide-preincubated sera 2 and 3 resulted in 52 and 51% inhibition in IgE antibody binding, respectively, to solid-phase coated Asp f 2. However, normals and allergic asthmatic sera showed only very low IgE binding, and no significant inhibition was noted.
FIG. 6.
Competitive inhibition ELISA of IgE binding of the peptides with sera from ABPA and allergic asthma patients and from healthy controls. ELISA plates were coated with Asp f 2. The IgE absorbance (O.D., optical density) of uninhibited sera is shown as gray bars; also shown are ELISA absorbances of preincubated sera with peptide with PB-(TIDVPSNCHTHEGGQLHCT) (hatched bars), PA-(NCALEGWGGHWRGAN) (open bars), and with Asp f 2 (closed bars). Peptides and protein were used at a concentration of 50 μg/ml for the inhibition study.
N-terminal/C-terminal region of Asp f 2 is essential for IgE antibody binding.
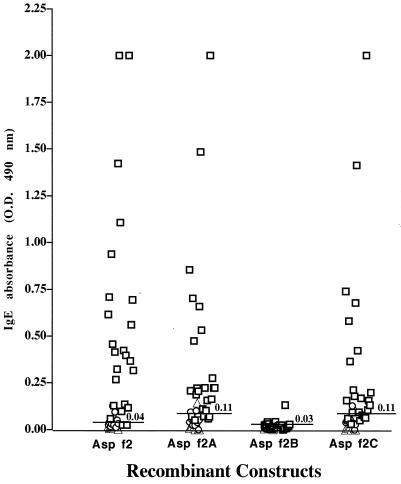
After determining the precise amino acid residues interacting with antibodies, we devised cDNA fragments encoding these epitopes to examine the role of properly folded polypeptides in antigen-antibody binding. Recombinant constructs Asp f 2A (aa 1 to 203), Asp f 2B (aa 68 to 203), and Asp f 2C (aa 68 to 268) as well as the complete Asp f 2 (aa 1 to 268) were cloned and expressed in the pET expression system (Fig. 1). The expressed proteins were purified by Ni2+-agarose affinity chromatography. The complete allergen Asp f 2 expressed a protein of 37 kDa, whereas recombinant polypeptides Asp f 2A, Asp f 2B, and Asp f 2C demonstrated proteins at 24, 17, and 28.6 kDa, respectively (data not shown). In an indirect ELISA, 24 individual sera from ABPA patients and 10 each from Aspergillus skin prick test-positive patients with asthma and healthy controls were tested for IgE binding with the recombinant peptides of Asp f 2 (Fig. 7). The mature allergen Asp f 2 (aa 1 to 268) recognized IgE antibodies in the sera of 22 of 24 ABPA patients (92%). Asp f 2A (aa 1 to 203) expressing the polypeptide without the one C-terminal-end epitope (epitope 9) reacted with 17 of 24 patients (71%), whereas Asp f 2C (aa 68 to 268), without the N-terminal-end epitopes 1 and 2, demonstrated reaction with 14 of 24 ABPA patients (58%). Levels of IgE binding of Asp f 2 and Asp f 2A with ABPA in comparison to healthy controls are statistically significant (P < 0.05). Although six of the nine epitopes are common in all three fragments, Asp f 2B failed to show IgE antibody binding with sera from patients with ABPA. None of these recombinant polypeptides exhibited significant IgE binding with sera from Aspergillus skin prick test-positive patients with asthma and from healthy controls (Fig. 7).
FIG. 7.
IgE antibody binding of sera from patients with ABPA (n = 24; □), allergic asthma (n = 10; ○) and from healthy controls (n = 10; ▵) to purified recombinant Asp f 2, Asp f 2A (aa 1 to 203), Asp f 2B (aa 68 to 203), and Asp f 2C (aa 68 to 268). The cutoff level for positive reaction (mean optical density [O.D.] + 2 standard deviations of the control values) is indicated by the solid line. The statistical significance of IgE antibody binding of the recombinant Asp f 2 and the fragments with sera from ABPA in comparison to healthy controls are as follows: Asp f 2, P < 0.008; Asp f 2A, P < 0.05); Asp f 2B, P < 0.116); and Asp f 2C, P < 0.06.
DISCUSSION
Insight into the allergen-mediated humoral reactions in allergic diseases may facilitate the development of more efficient immunodiagnosis and treatment of these diseases. Thus, the present study was undertaken to identify and characterize the IgE binding epitopes of a major A. fumigatus allergen, Asp f 2, using solid-phase peptide synthesis and recombinant DNA technology. The epitope mapping of linear peptides synthesized on derivatized cellulose membranes demonstrated distinct IgE binding regions throughout the molecule. However, the recombinant fragment Asp f 2B (aa 68 to 203) without the N- and C-terminal regions of Asp f 2 failed to bind IgE antibody from ABPA patients, indicating the possible involvement of various regions of the protein and other structural constraints in maintaining the proper folding and three-dimensional structure of Asp f 2. In contrast to the epitopes from the majority of allergens, which reside in the C-terminal regions of the proteins, both N- and C-terminal regions of Asp f 2 represent IgE binding epitopes.
The identified epitopes are 3 (HWR) to 7 (ATQRRQI) aa long, and seven of these identified epitopes are present in the hydrophilic regions of Asp f 2. The specificities of these IgE binding epitopes were studied further by synthesizing 3- to 10-aa peptides. A representative result as shown in Fig. 5 indicates the specificity of antibody binding of the epitope RKYFG, a 5-aa-long peptide, with a gradual increase in the intensity of the reaction by the incorporation of the amino acids from the identified epitope region (Fig. 5B, spots 5 and 6) and the decrease in the IgE binding with the deletion of the amino acids comprising the epitope region (Fig. 5B, spots 10 and 11). In addition, with similar small peptides without the putative amino acids of the epitopes, no significant IgE binding was detected, as with the tripeptides in spots 1, 2, and 3 in Fig. 5A. The present findings, thus, indicate that IgE binding epitopes of Asp f 2 are specifically directed to a few amino acid residues and involve amino acid sequences arranged in a predefined way as seen in the primary structure of the protein.
A protein data bank search for the conserved amino acid sequences representing these epitopes showed significant sequence homology to three other fungal proteins but not to any other known allergen. These proteins are ASPND1, a cytosolic protein from A. nidulans, and the fibrinogen binding and the pH-regulated proteins from Candida albicans. The amino acid residues of Asp f 2 representing IgE binding epitopes and the homologous sequences of ASPND1 and C. albicans proteins are shown in Table 1. However, these three proteins are not yet known to contribute to IgE-mediated fungal diseases; there are reports of ABPA-like diseases caused by other members of the genus Aspergillus as well as by other fungi such as Candida, Curvularia, Stephylium, and Helminthosporium spp. (26). Recently we have reported significant IgE antibody binding of ASPND1 in sera from A. fumigatus-sensitized patients. Although we have not evaluated the IgE antibody binding of the two proteins from C. albicans, all the three proteins have conserved amino acid sequences as in IgE binding regions of Asp f 2 (5, 9, 27, 35). The cross-reactivity among allergens or antigens from phylogenetically unrelated fungi may be determined by further investigating the roles of these defined epitopes in the immune mechanism of the disease process.
TABLE 1.
Sequence homology of identified IgE epitopes of Asp f 2 with proteins from A. nidulans and C. albicans
| Epitope | Sequence
|
|||
|---|---|---|---|---|
| Asp f 2 (A. fumigatus) | Asp n d1 (A. nidulans) | Fibrinogen binding protein (C. albicans) | pH-regulated protein (C. albicans) | |
| 1 | ATQRRQI | ATEQRQL | ATQYNQL | ATQYNQL |
| 2 | RKYFG | RKYFG | RKYFG | RKYFG |
| 3 | HWR | HWR | YWR | YWR |
| 4 | YTTRR | YTTRR | FVTRR | FVTRR |
| 5 | ASDLM | AGDLL | AGDLL | AGDLL |
| 6 | YHVP | YHMP | WHLK | WHLK |
| 7 | DHFAD | EHYAD | EHYAD | EHYAD |
| 8 | ALEAYA | ALEVYA | ALDVYA | ALDVYA |
| 9 | THEGGQ | THEGGE | THADGE | THADGE |
Immunological evaluation of the decapeptides enables the identification of the epitopes with the minimum number of amino acid residues precisely interacting with antibodies. To evaluate the immunoreactivity of the epitopes in solution, we carried out competitive inhibition ELISA. The extents of inhibition in IgE binding to solid-phase coated Asp f 2 by two peptides representing epitopes 3 (HWR) and 9 (THEGGQ) were studied in sera from patients with ABPA and allergic asthma and from healthy controls. Although the percentages of inhibition differed for the two peptides, together they showed >50% inhibition in IgE binding with all three sera from ABPA patients. The IgE epitopes identified in the present study failed to show significant IgG antibody binding with ABPA (data not shown), indicating the specificity of IgE and IgG binding epitopes in Asp f 2.
IgE epitope analyses of various allergens have revealed that depending on the primary amino acid sequences, the epitopes may be continuous as in Der p 2, Fel d 1, Phl p 1, Lol p VB, β-lactoglobulin from bovine milk, Myr p 1 from ant venom, and Poa p 9, the Kentucky blue grass pollen allergen. On the other hand, for allergens such as Der p 1 and Bet v 1, the IgE epitopes are reported to be discontinuous, whereas both continuous and discontinuous epitopes are present in all vespid allergens (17, 20, 34, 38, 39, 41). For Asp f 2, we have demonstrated the presence of both continuous and discontinuous epitopes. As fragment Asp f 2B with six of nine identified epitopes appeared to be under conformational constraints and may be due to incorrect folding, the epitopes are not available on the surface to interact with IgE antibodies. At the same time, the identified linear epitopes when used as synthetic peptides compete with the epitopes present in Asp f 2 in competitive ELISA, indicating structural and conformational similarities between them.
Not much work has been carried out on defining B- and T-cell epitopes of major A. fumigatus allergens; recently Asp f 1-specific T-cell clones have been established from ABPA patients (10). These HLA-DR 2/5-restricted T-cell clones are mainly directed against two immunodominant epitopes of Asp f 1 in the region from aa 106 to 125. In line with these findings, recently we have reported two immunodominant epitopes of Asp f 1 representing the C-terminal region of the protein (aa 115 to 149) with strong proliferative responses as well as IgE antibody binding with sera from patients with ABPA (24). Hence, the assembly of multimers of short defined epitopes or chimeric construction of B- and T-cell epitopes as a multiple antigen-presenting system may be a useful approach for the design of a polyvalent subunit immunogen for fungal diseases (28, 37).
In conclusion, using solid-phase peptide synthesis and molecular cloning techniques, we have identified the immunodominant B-cell epitopes of a major A. fumigatus allergen, Asp f 2. Although linear IgE binding regions are identified throughout the protein by synthetic peptide analysis of the complete protein, it is evident that these epitopes are presented in a conformational manner. Evaluations of these IgE epitopes and recombinant polyepitopes suggest the usefulness of multimeric epitopes as possible candidates for the development of specific serodiagnosis and probably in the treatment of allergic aspergillosis.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant AI42349, by Department of Veterans Affairs Medical Research funds, and by an Ernest S. Bazley grant to Northwestern Memorial Hospital and Northwestern University.
We thank Laura Castillo and Nancy Elms for technical assistance and Donna Schrubbe for editorial assistance.
REFERENCES
- 1.Arruda L K, Mann B J, Chapman M D. Selective expression of a major allergen and cytotoxin, Asp fI, in Aspergillus fumigatus. Implications for the immunopathogenesis of Aspergillus-related diseases. J Immunol. 1992;149:3354–3359. [PubMed] [Google Scholar]
- 2.Atassi H, Atassi M Z. Antibody recognition of ragweed allergen Ra3: localization of the full profile of the continuous antigenic sites by synthetic overlapping peptides representing the entire protein chain. Eur J Immunol. 1986;16:229–235. doi: 10.1002/eji.1830160304. [DOI] [PubMed] [Google Scholar]
- 3.Banerjee B, Kurup V P, Greenberger P A, Hoffman D R, Nair D S, Fink J N. Purification of a major allergen, Asp f 2 binding to IgE in allergic bronchopulmonary aspergillosis, from culture filtrate of Aspergillus fumigatus. J Allergy Clin Immunol. 1997;99:821–827. doi: 10.1016/s0091-6749(97)80017-6. [DOI] [PubMed] [Google Scholar]
- 4.Banerjee B, Kurup V P, Phadnis S, Greenberger P A, Fink J N. Molecular cloning and expression of a recombinant Aspergillus fumigatus protein Asp f II with significant immunoglobulin E reactivity in allergic bronchopulmonary aspergillosis. J Lab Clin Med. 1996;127:253–262. doi: 10.1016/s0022-2143(96)90093-1. [DOI] [PubMed] [Google Scholar]
- 5.Banerjee B, Greenberger P A, Fink J N, Kurup V P. Immunological characterization of Asp f 2, a major allergen from Aspergillus fumigatus associated with allergic bronchopulmonary aspergillosis. Infect Immun. 1998;66:5175–5182. doi: 10.1128/iai.66.11.5175-5182.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bardana E J., Jr The clinical spectrum of aspergillosis. Part 1. Epidemiology, pathogenicity, infection in animals and immunology of Aspergillus. Crit Rev Clin Lab Sci. 1981;13:21–83. doi: 10.3109/10408368009106444. [DOI] [PubMed] [Google Scholar]
- 7.Bardana E J., Jr The clinical spectrum of aspergillosis. Part 2. Classification and description of saphorophytic, allergic and invasive variants of human disease. Crit Rev Clin Lab Sci. 1981;13:85–159. doi: 10.3109/10408368009106445. [DOI] [PubMed] [Google Scholar]
- 8.Beaumont F, Kauffman H F, Sluiter H J, De Vries K. Sequential sampling of fungal air spores inside and outside the homes of mould-sensitive asthmatic patients: a search for a relationship to obstructive reactions. Ann Allergy. 1985;55:740–746. [PubMed] [Google Scholar]
- 9.Calera J A, Ovejero M C, Lopez-Medrano R, Segurado M, Puente P, Leal F. Characterization of the Aspergillus nidulans aspnd1 gene demonstrates that the ASPND1 antigen, which it encodes, and several Aspergillus fumigatus immunodominant antigens belong to the same family. Infect Immun. 1997;65:1335–1344. doi: 10.1128/iai.65.4.1335-1344.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chauhan B, Knutsen A P, Hutcheson P S, Slavin R G, Bellone C J. T cell subsets, epitope mapping, and HLA-restriction in patients with allergic bronchopulmonary aspergillosis. J Clin Investig. 1997;97:2324–2331. doi: 10.1172/JCI118675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chou P Y, Fasman G D. Prediction of the secondary structure of proteins from their amino acid sequences. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- 12.Crameri R. Recombinant Aspergillus fumigatus allergens: from the nucleotide sequences to clinical applications. Int Arch Allergy Immunol. 1998;115:99–114. doi: 10.1159/000023889. [DOI] [PubMed] [Google Scholar]
- 13.Crameri R, Faith A, Hemmann S, Jaussi R, Ismail C, Menz G, Blaser K. Humoral and cell-mediated autoimmunity in allergy to Aspergillus fumigatus. J Exp Med. 1996;184:265–270. doi: 10.1084/jem.184.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dolecek C, Vrtala S, Laffer S, Steinberger P, Kraft D, Scheiner O, Valenta R. Molecular characterization of Phl p 2, a major timothy grass (phleum pratense) pollen allergen. FEBS Lett. 1993;335:299–304. doi: 10.1016/0014-5793(93)80406-k. [DOI] [PubMed] [Google Scholar]
- 15.Elsayed S, Apold J. Immunochemical analysis of cod fish allergen M: locations of the immunoglobulin binding sites as demonstrated by the native and synthetic peptides. Allergy. 1983;38:449–459. doi: 10.1111/j.1398-9995.1983.tb02353.x. [DOI] [PubMed] [Google Scholar]
- 16.Gergen P J, Turkeltaub P C, Kovar M G. The prevalence of allergic skin test reactivity to eight common aeroallergens in the U.S. population: results from the second National Health and Nutrition Examination Survey. J Allergy Clin Immunol. 1987;80:669–679. doi: 10.1016/0091-6749(87)90286-7. [DOI] [PubMed] [Google Scholar]
- 17.Geysen H M, Rodda S J, Mason T J, Tribbick G, Schoofs P G. Strategies for epitope analysis using peptide synthesis. J Immunol Methods. 1987;102:259–274. doi: 10.1016/0022-1759(87)90085-8. [DOI] [PubMed] [Google Scholar]
- 18.Ghosh B, Perry M P, Marsh D G. Cloning the cDNA encoding the Amb t V allergen from giant ragweed (Ambrosia trifida) Gene. 1991;101:231–238. doi: 10.1016/0378-1119(91)90416-9. [DOI] [PubMed] [Google Scholar]
- 19.Horner W E, Helbling A, Salvaggio J E, Lehrer S B. Fungal allergens. Clin Microbiol Rev. 1995;8:161–179. doi: 10.1128/cmr.8.2.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.King T P, Lu G, Gonzalez M, Qian N, Soldatova L. Yellow jacket venom allergens, hyaluronidase and phospholipase: sequence similarity and antigenic cross-reactivity with their hornet and wasp homologs and possible implications for clinical allergy. J Allergy Clin Immunol. 1996;98:588–600. doi: 10.1016/s0091-6749(96)70093-3. [DOI] [PubMed] [Google Scholar]
- 21.Kumar A, Reddy L V, Sochanik A, Kurup V P. Isolation and characterization of a recombinant heat shock protein of Aspergillus fumigatus. J Allergy Clin Immunol. 1993;91:1024–1030. doi: 10.1016/0091-6749(93)90215-2. [DOI] [PubMed] [Google Scholar]
- 22.Kurup V P. Enzyme-linked immunosorbent assay in the detection of specific antibodies against Aspergillus in patients sera. Zentbl Bakteriol Mikrobiol Hyg. 1986;261:509–516. doi: 10.1016/s0176-6724(86)80084-0. [DOI] [PubMed] [Google Scholar]
- 23.Kurup V P, Alenius H, Kelly K J, Castillo L, Fink J N. A two-dimensional electrophoretic analysis of latex peptides reacting with IgE and IgG antibodies from patients with latex allergy. Int Arch Allergy Clin Immunol. 1996;109:58–67. doi: 10.1159/000237232. [DOI] [PubMed] [Google Scholar]
- 24.Kurup V P, Banerjee B, Murali P S, Greenberger P A, Krishnan M, Hari V, Fink J N. Immunodominant peptide epitopes of allergen Asp f 1 from the fungus Aspergillus fumigatus. Peptides. 1998;19:1469–1477. doi: 10.1016/s0196-9781(98)00113-2. [DOI] [PubMed] [Google Scholar]
- 25.Kurup V P, Greenberger P A, Fink J N. Antibody response to low-molecular-weight antigens of Aspergillus fumigatus in allergic bronchopulmonary aspergillosis. J Clin Microbiol. 1989;27:1312–1316. doi: 10.1128/jcm.27.6.1312-1316.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurup V P, Kumar A. Immunodiagnosis of aspergillosis. Clin Microbiol Rev. 1991;4:439–456. doi: 10.1128/cmr.4.4.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lopez-Ribot J L, Sepulveda P, Cervera A M, Roig P, Gozalbo D, Martinez J P. Cloning of a cDNA fragment encoding part of the protein moiety of the 58-kDa fibrinogen-binding mannoprotein of Candida albicans. FEMS Microbiol Lett. 1997;157:273–278. doi: 10.1111/j.1574-6968.1997.tb12784.x. [DOI] [PubMed] [Google Scholar]
- 28.Lowenadler B, Lycke N, Svanholm C, Svennerholm A M, Krook K, Gidlund M. T and B cell responses to chimeric proteins containing heterologous T helper epitopes inserted at different positions. Mol Immunol. 1992;29:1185–1190. doi: 10.1016/0161-5890(92)90054-2. [DOI] [PubMed] [Google Scholar]
- 29.Moser M, Crameri R, Menz G, Schneider T, Dudler T, Virchow C, Gmachl M, Blaser K, Suter M. Cloning and expression of recombinant Aspergillus fumigatus allergen I/a (rAsp f I/a) with IgE binding and Type I skin test activity. J Immunol. 1992;149:454–460. [PubMed] [Google Scholar]
- 30.Nakajima-Adachi H, Hachimura S, Ise W, Honma K, Nishiwaki S, Hirota M, Shimojo N, Katsuki T, Ametani A, Kohno Y, Kaminogawa S. Determinant analysis of IgE and IgG4 antibodies and T cells specific for bovine alpha (S) 1-casein from the same patients allergic to cow’s milk: existence of alpha (S) 1-casein-specific B cells and T cells characteristic in cow’s milk allergy. J Allergy Clin Immunol. 1998;101:660–671. doi: 10.1016/s0091-6749(98)70175-7. [DOI] [PubMed] [Google Scholar]
- 31.Pope A M, Patterson R, Burge H. Indoor allergens: assessing and controlling adverse health effects. Washington, D.C: National Academy Press; 1993. [PubMed] [Google Scholar]
- 32.Rosenberg M, Patterson R, Mintzer R, Cooper B J, Roberts M, Harris K E. Clinical and immunologic criteria for the diagnosis of allergic bronchopulmonary aspergillosis. Ann Intern Med. 1977;86:405–414. doi: 10.7326/0003-4819-86-4-405. [DOI] [PubMed] [Google Scholar]
- 33.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scheiner O, Kraft D. Basic and practical aspects of recombinant allergens. Allergy. 1995;50:384–391. doi: 10.1111/j.1398-9995.1995.tb01167.x. [DOI] [PubMed] [Google Scholar]
- 35.Sentandreu M, Elorza M V, Sentandreu R, Fonzi W A. Cloning and characterization of PRA1, a gene encoding a novel pH-regulated antigen of Candida albicans. J Bacteriol. 1998;180:282–289. doi: 10.1128/jb.180.2.282-289.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stewart G A, Thompson P J. The biochemistry of common aeroallergens. Clin Exptl Allergy. 1996;26:1020–1044. [PubMed] [Google Scholar]
- 37.Tam J P. Recent advances in multiple antigen peptides. J Immunol Methods. 1996;196:17–32. doi: 10.1016/0022-1759(96)00066-x. [DOI] [PubMed] [Google Scholar]
- 38.Valenta R, Kraft D. Recombinant allergens for diagnosis and therapy of allergic diseases. Curr Opin Immunol. 1995;7:751–756. doi: 10.1016/0952-7915(95)80043-3. [DOI] [PubMed] [Google Scholar]
- 39.Walsh B J, Howden M E. Epitope mapping of allergens for rapid localization of continuous allergenic determinants. Methods Enzymol. 1991;203:301–311. doi: 10.1016/0076-6879(91)03017-b. [DOI] [PubMed] [Google Scholar]
- 40.Wuthrich B. Epidemiology of the allergic diseases: are they really on the increase? Int Arch Allergy Appl Immunol. 1989;90:3–10. doi: 10.1159/000235067. [DOI] [PubMed] [Google Scholar]
- 41.Zhang L, Olsen E, Kisil F T, Hill R D, Sehon A H, Mohapatra S S. Mapping of antibody binding epitopes of a recombinant Poa p IX allergen. Mol Immunol. 1992;29:1383–1389. doi: 10.1016/0161-5890(92)90175-w. [DOI] [PubMed] [Google Scholar]