Abstract
We isolated the genetic determinant of AF/R1 pilus production in attaching/effacing Escherichia coli RDEC-1 and identified seven genes required for pilus expression and function. DNA sequence analysis of the structural subunit gene afrA corrected an error in the published sequence and extended homology with the F18 pilus subunit of pig edema E. coli strains. AfrB and AfrC, encoded downstream from AfrA, were required for pilus expression. AfrB was related to the usher protein PefC of Salmonella typhimurium plasmid-encoded fimbriae, and AfrC was related to PefD, a chaperone protein. AfrD and AfrE, encoded downstream from AfrC, were not necessary for the expression of AF/R1 pili but were required for ileal adherence as assayed by ileal brush border aggregation. Thus, the adhesive subunit of the AF/R1 pilus is distinct from the structural subunit, as is the case for Pap pili and type 1 pili. AfrD was related to FedE of the F18 fimbrial operon of the E. coli strain that causes edema disease in pigs. AfrE was a novel protein. AfrR and AfrS are encoded upstream from AfrA, in the opposite orientation. AfrR is related to the AraC family of transcriptional regulators, and AfrR and AfrS interact to function in a novel mode of transcriptional activation of afrA. AF/R1 pili mediate the adherence to Peyer’s patch M cells, ileal mucosa, and colonic mucosa in a rabbit model of diarrhea caused by enteropathogenic E. coli. Our observations will facilitate the further study of the phenomena of M-cell adherence.
Escherichia coli RDEC-1 produces diarrhea in rabbits. It attaches to absorptive epithelial cells of the distal ileum, cecum, and colon and to M cells of Peyer’s patches and produces an attaching/effacing (A/E) lesion at the site of colonization (5, 6, 26, 57). The AF/R1 pilus that strain RDEC-1 expresses mediates an early stage of A/E adherence and is necessary for full virulence (7, 25, 61). The pilus confers the ability to adhere to partially purified brush borders and to M cells (7, 25). RDEC-1 also possesses sequences closely related to the genes of the locus of enterocyte effacement (LEE) pathogenicity island of enteropathogenic E. coli (37), the products of which are required for the production of the A/E lesion (16). RDEC-1 infection of rabbits is therefore a relevant, natural model of enteropathogenic E. coli infection of humans.
The production of AF/R1 is encoded on a 132-kb conjugative plasmid of RDEC-1 (11). The gene encoding the structural subunit of AF/R1 pili, afrA, has been isolated, and its nucleotide sequence has been reported (62). We examined the sequence and functions of additional DNA necessary for the function and expression of afrA. We identified two regulatory genes required for afrA transcription. One of these genes has homology to the araC family of transcriptional regulators. We also determined that the adherence function is encoded by one or both of two genes (afrD and afrE), downstream from the structural subunit gene afrA, and genes afrB (usher protein) and afrC (chaperone protein).
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
RDEC-1 is an E. coli O15:K−:H− strain that expresses AF/R1 pili. Shigella D15 and D12 were used as positive and negative strains for pilus expression, respectively, as strain D15 is an exconjugate containing the 86-MDa plasmid encoding AF/R1 pilus expression (11). Strain M129, an RDEC-1 strain cured of the 132-kb plasmid (reference 62 and unpublished observations), and RDEC-1 42-2-37-8, a strain resulting from a temperature-sensitive mutation that does not express AF/R1 pili (27), both served as negative controls for pilus expression. DH5α [F− φ80d lacZDM15 endA1 recA1 hsdR17 supE44 thi-1 l− gyrA96 relA1 D(lacZYA-argF)U169] (Gibco BRL, Gaithersburg, Md.) was used as a host for recombinant plasmid constructions and transposon mutagenesis. Strain JM101 (59) was used as a host for bacteriophage M13 derivatives in nucleotide sequence analysis (59). Multicopy plasmids pUC18 and pUC19 were used as cloning vectors. pKI100, a pACYC derivative (29) compatible with pUC vectors, was used as a vector for complementation experiments with pUC constructs. A KpnI fragment internal to afrA was cloned into pBluescript II KS(+) phagemid (Stratagene Inc., La Jolla, Calif.) and used to prepare the probe for the Northern blots.
Organisms were grown in Luria broth or on Luria agar unless otherwise noted. Penassay broth was used to induce AF/R1 pilus expression in the wild-type RDEC-1 strain (11). Ticarcillin-clavulanic acid (100 mg/ml) was used to select for the presence of pUC derivatives, and kanamycin (25 mg/ml) was used for the selection of pKI100 derivatives.
Brush border preparation and bacterial adherence assay.
Rabbit ileal brush borders were purified according to the method of Cheney et al. (10). Bacterial strains were grown overnight and examined for their ability to adhere to the purified brush borders (10). Bacteria adhering to 50 brush borders were counted, and results were calculated as the average number of bacteria/brush border. It was difficult to count more than 10 bacteria/brush border with any accuracy. For that reason, these brush borders were considered to have 10 bacteria when the average number of adherent bacteria was calculated. Adherence assays were performed on four separate occasions with different brush border preparations each time. Adherence was considered to be present if the results were significantly different from negative controls.
Purification of AF/R1 pili.
AF/R1 pili were purified by a modification of the technique of Isaacson (28). Pili were sheared from bacteria cultured overnight in static Penassay broth by using an Omnimixer (Omni International, Inc., Waterbury, Conn.). The pili were precipitated in an ammonium sulfate solution, resuspended in phosphate-buffered saline, pH 7.5, dialyzed against water, and eluted from a DEAE column in a discontinuous salt gradient. Pilus preparations were confirmed as pilus structures by electron microscopy of negatively stained samples of pili. Pili were electrophoresed in a sodium dodecyl sulfate–13% polyacrylamide gel, and stained with silver (44) for the assessment of purity.
Production of MAb.
Monoclonal antibody (MAb) AFR20 was prepared according to a previously published technique (53), except that RPMI 1640 culture medium was substituted for Dulbecco’s modified Eagle’s medium. Immunoblotting and Western blot analysis with purified pili confirmed MAb activity against AF/R1 pili.
Immunoassays for pili.
Pilus expression in bacterial strains was detected by blotting unfixed bacterial colonies to nitrocellulose (colony immunoblot) or by dot blotting 2 μl of bacterial suspensions or purified pili at a concentration of 1 mg/ml to an NC nitrocellulose, 0.45-μm-pore-size filter (Schleicher and Schuell, Keene, N.H.). The nitrocellulose was air dried and incubated at room temperature in 5% skim milk for 1 h and MAb containing ascites fluid to a dilution of 1:1,000 (dot blot) or 1:10,000 (colony immunoblot) added to the skim milk. The blots were incubated for 6 h on a shaker at room temperature and washed three times in phosphate-buffered saline, pH 7.4. The skim milk solution was made at concentrations up to 1:10,000 in peroxidase-labeled goat anti-mouse immunoglobulin G, and the blots were immersed in the solution on a shaker, overnight at room temperature. The blots were washed three times in phosphate-buffered saline and developed in 0.01 M Tris (pH 7.4), 0.005% O-dianisidine, and 15 μl of hydrogen peroxide per 100 ml. The blots were washed in tap water and air dried.
Oligonucleotide DNA probe for the AF/R1 structural subunit.
A 68-base oligonucleotide with the sequence 5′-CAGGGTGATGTGCAGTTCTTCGGTACCGTGACCGCGAAAACCTGCGATCTGGTGGTGGAATGCGAAGG-3′ based on the N-terminal amino acid sequence of the AF/R1 pilus (9a) was used as a probe for the AF/R1 structural subunit.
Insertion mutagenesis.
The transposon Tn5 was introduced into E. coli DH5α containing RDEC-1 AF/R1 plasmid constructs by infection with bacteriophage lambda b221 rex::Tn5 c1857 Oam8 Pam29 (4). Transductants growing on a medium containing both ampicillin and kanamycin were harvested, and total plasmid DNA was isolated by the method of Birnboim and Doly (3). The resulting pool of plasmid DNA was used to transform strain DH5α. Transformants were selected on a medium supplemented with ampicillin and kanamycin. Transformants were screened for AF/R1 pilus expression with AF/R1 MAb. Plasmid DNA was isolated from individual pilus-negative transformants, and restriction analysis identified the site of transposon insertion.
DNA transformation.
Plasmids of interest were transformed into E. coli HB101 by the method of Hanahan (22).
DNA manipulation and analysis.
Plasmid DNA was isolated by the method of Birnboim and Doly (3) or by cesium chloride-ethidium bromide density gradient centrifugation (45). For screening purposes plasmids were isolated by the method of Kado and Liu (30).
Preparation of subclone expressing afrR.
No restriction site was available to generate a subclone encoding afrR alone. The technique of oligonucleotide-directed in vitro mutagenesis was used to generate a site-specific mutation between DNA encoding afrS and afrR. Specifically, an SphI site was placed at bases 519 to 525 by changing a C to G at position 519 and deleting a C at position 524, in the carboxy-terminal end of afrS. The primer 5′-ACATCTTTCTGCATGCTGA-3′ (SphI site underlined) was used to generate the mutation by using the USB T-7 GEN in vitro mutagenesis system (58) (United States Biochemical Corporation). The mutated subclone was then cloned into pKI100 to produce p598 (Fig. 1), which was shown to produce AfrR in maxicell preparations. The mutation was confirmed by DNA sequencing.
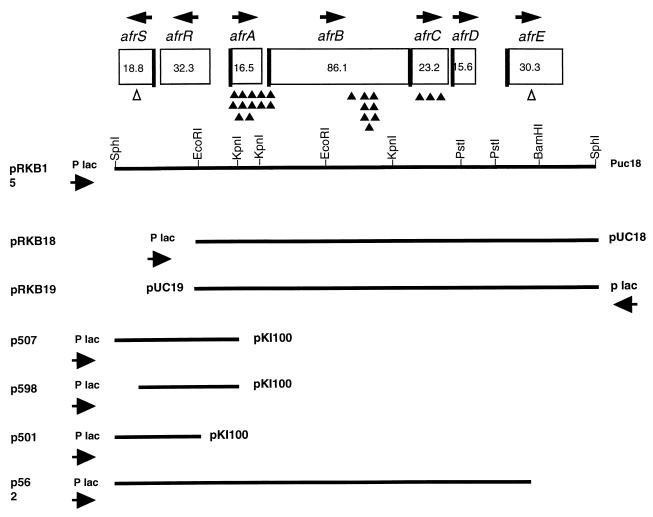
FIG. 1.
Genetic organization of AF/R1 determinants and constructs utilized in complementation and adherence experiments. Boxes indicate peptides encoded by pRKB15. The thick lines on the sides of the boxes indicate the presence and positions of signal peptides. Arrows above the boxes indicate directions of transcription. The numbers in the boxes are the predicted molecular masses (in kilodaltons) of the mature polypeptides. Sites of transposon insertions in pRKB18 are indicated as follows: Tn5 insertions not affecting the expression of AF/R1 pili (white triangles) and Tn5 insertions which abolished the expression of AF/R1 pili (black triangles). All transposon mutagenesis was performed on pRKB18. p598 is described in Materials and Methods. Its construction required mutagenesis to insert an SphI site in the carboxy-terminal end of afrS. The new site allowed the subcloning of an SphI-KpnI fragment that expressed AfrR.
Colony and Southern blot hybridizations.
The synthesized oligonucleotide probe was labeled by phosphorylation with bacteriophage T4 polynucleotide kinase (47) with [γ-32P]ATP (DuPont, NEN Research Products, Boston, Mass.). In the case of colony blots, bacteria were streaked onto Luria agar plates, incubated overnight at 37°C, and blotted onto Whatman 541 filters (Whatman, Bedford, Mass.). Colony and Southern blots were prepared, and hybridizations were conducted under high-stringency conditions (39, 41, 56, 60). Hybridization and wash conditions were unusual due to the length of the oligonucleotide probe and included incubation at 42°C in 20% formamide with 0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) followed by two washes at 40°C in 0.2× SSC. Results were visualized by autoradiography.
Northern hybridizations.
Bacterial RNA was prepared as previously described (20). The electrophoresis of RNA and transfer to nylon membranes were performed as previously described (2). The afrA hybridization probe was prepared as follows. p365 was linearized with BamHI, and a 32P-labeled antisense RNA probe was generated with [α-32P]UTP, T3 RNA polymerase, and reagents provided in kit form (Maxiscript transcription kit; Ambion, Austin, Tex.). Hybridization conditions were as previously described (2). Following hybridization, the nylon membranes were washed in 300 mM sodium chloride or 30 mM sodium citrate at 75°C for 4 h and then incubated at room temperature in the same buffer with 25 μg of RNase A per ml and 10 U of RNase T1 per ml. The membranes were then exposed to X-ray film.
Nucleotide sequence analysis.
Bacteriophages M13mp18 and M13mp19 (43) were used to generate single-stranded DNA templates for the dideoxy-chain termination method of Sanger et al. (49). Modified T7 DNA polymerase (Sequenase, version 2.0; United States Biomedical, Cleveland, Ohio) was used to generate the nucleotide sequence information. Sequences were also obtained by thermal cycle sequencing with Taq DNA polymerase and fluorescent dye-labeled terminators by using the ABI 377 automated sequencer of the Biotechnology Resource Laboratory of the Medical University of South Carolina. Double-stranded plasmid DNA was used as a template.
Expression of plasmid-encoded proteins in maxicells.
Maxicell analysis was performed by a modification of the method described by Sancar et al. (48). Overnight cultures of organisms harboring plasmids of interest grown in K broth (46) at 37°C were diluted 1:50 in the same medium, grown to the exponential phase, and then exposed to UV radiation from a germicidal lamp. Cultures were incubated for 1 h at 37°C following irradiation. Cycloserine was added to a final concentration of 100 mg/ml, and the incubation was continued for 12 to 16 h at 37°C. The bacteria were harvested, washed three times with Hershey salts (48), and incubated for 60 min at 37°C in a Hershey medium (48). [35S]methionine (Dupont, NEN Research Products) was then added to a final concentration of 30 mCi/ml, and the incubation was continued for an additional hour. Bacteria were washed once with Hershey salts and lysed in polyacrylamide gel electrophoresis sample buffer. The radioactive polypeptides were separated on a 13% polyacrylamide gel (35) and visualized, following autoradiography.
Computer analysis.
The Genetics Computer Group sequence analysis software package (14) and DNASTAR DNA analysis software for the Macintosh (DNASTAR; DNASTAR Inc., Madison, Wis.) were used for DNA analysis. Homology searches were performed with the BLAST programs (1) at the National Center for Biotechnology Information, Bethesda, Md. Multiple sequence alignments were performed with the MACAW program (52), which was kindly provided by G. D. Schuler, National Center for Biotechnology Information. Signal peptides were predicted by the method of Nielsen et al. (40).
RESULTS
Isolation of the afr gene cluster encoding the production of AF/R1 pili.
DNA isolated from E. coli RDEC-1 was partially digested with Sau3AI and size fractionated by agarose gel electrophoresis. Fragments of 15 to 20 kb were ligated into BamHI-digested pUC18, and the resulting plasmids were transformed into DH5α. Transformants were examined by colony hybridization for sequences homologous to a 68-base oligonucleotide probe, the sequence of which was derived from a partial amino acid sequence of purified AF/R1 pili. Three of 600 colonies examined hybridized with the probe, and each of these bound the antibody specific for AF/R1 (data not shown). One of these was selected for further study and designated DH5α(pJRC10).
Deletion and subcloning experiments revealed that an 8,314-kb SphI fragment of pJRC10 was sufficient for the expression and function of AF/R1. A pUC18 derivative containing this fragment was designated pRKB15.
Sequence analysis of the afr gene cluster.
The complete nucleotide sequence of pRKB15 was determined, and open reading frames preceded by sequences encoding potential ribosome binding sites (54) were identified corresponding to each of these genes (GenBank accession no. AF050217). Genes were designated afrSRABCDE (Fig. 1). Each gene encoded a predicted product of a size consistent with maxicell expression data, with the exception of afrD, which we were unable to visualize in maxicell gels (data not shown). Genes afrSR were of opposite polarity to afrABCDE. The afrS gene encoded a predicted signal sequence for transmembrane secretion, and a signal peptidase cleavage site is predicted between amino acids 27 and 28. AfrS was predicted to be a mature protein of 18,804 Da. This gene is related to ais, an aluminum-inducible gene of E. coli of unknown function (GenBank accession no. X83874), and prgG, a gene of unknown function of Salmonella typhimurium regulated by the PmrA-PmrB two-component regulatory system associated with resistance to polymyxin (21). The product of afrR, which was the only gene product of the operon that did not possess a predicted signal sequence, was predicted to be a mature protein of 32,359 Da and revealed significant relatedness to a number of transcriptional regulators of the araC family (18). The most closely related proteins were Caf1R (32), a regulatory protein associated with the expression of the F1 envelope protein of Yersinia pestis, and RobA, an E. coli protein which binds to the right border of oriC (12). The N-terminal region of AfrR also had homology with the C terminus of the AraC family of transcriptional regulators (18, 50). The alignment of AfrR with Caf1R, AraC, and other related sequences by the MACAW multiple sequence alignment program (52) is discussed below (see Fig. 3).
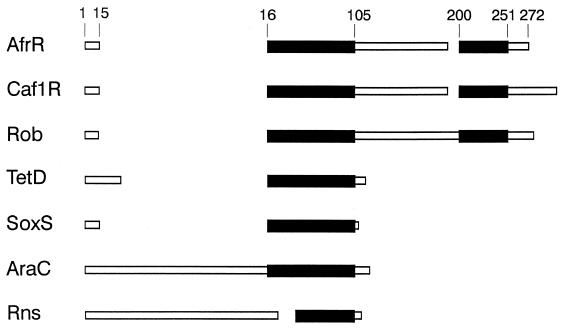
FIG. 3.
Alignment of AfrR and related proteins by the MACAW multiple sequence alignment program (52). Amino acid coordinates are shown for AfrA. Black boxes represent regions of significant homology. White boxes represent regions of the peptides not having significant homology.
The afrA gene encodes a predicted signal sequence for transmembrane secretion, and a signal peptidase cleavage site is predicted between amino acids 21 and 22, in agreement with the N-terminal amino acid sequence determined from purified AF/R1 pili (7a). AfrA is the structural subunit of AF/R1 pili, and the size of the mature protein is predicted to be 16,525 Da. AfrA had significant relatedness to a number of pilus structural proteins. The most closely related protein was the F18 fimbrial subunit of E. coli associated with edema disease in swine (23). afrB was predicted to encode an amino-terminal signal sequence, and a signal peptidase cleavage site is predicted between amino acids 24 and 25. The mature protein is predicted to be 86,126 Da. AfrB is related to a number of usher proteins associated with bacterial pilus biogenesis. The most closely related protein was PefC, the usher protein of plasmid-encoded fimbriae of S. typhimurium (17). afrC encoded a predicted signal sequence, and a signal peptidase cleavage site is predicted between amino acids 23 and 24. The mature protein is predicted to be 23,196 Da and has significant relatedness to the PefD chaperone of Salmonella plasmid-encoded fimbriae (17). afrD encoded a predicted signal sequence with a signal peptidase cleavage site predicted between amino acids 21 and 22. The mature protein was predicted to be 15,652 Da and was related to fedE of the F18 fimbrial operon. FedE is thought to be involved in determining fimbrial length and binding specificity (24). The last open reading frame was afrE, which encoded a predicted signal sequence with a signal peptidase cleavage site predicted between amino acids 32 and 33. The mature protein was predicted to be 30,259 Da. No significant homology exists with proteins entered in sequence databases as determined by BLASTP comparisons (1).
Insertion mutagenesis analysis.
Tn5 insertion mutagenesis was used to assess gene function in DH5α(pRKB18). Transductants were screened for the loss of AF/R1 pilus expression by using AF/R1 MAb. The results are shown in Fig. 1. Insertions in afrABC resulted in a loss of pilus expression. Insertions in afrS and afrE did not affect pilus expression.
AfrS and AfrR encode positive transcriptional regulators of the afr gene cluster.
The sequence relatedness of the predicted product of afrR with the AraC family of transcriptional activators suggested that AfrR may serve to regulate the expression of the afr gene cluster at the transcriptional level. We tested this hypothesis by complementation experiments on the expression of pili and Northern blot analysis to ascertain the effects of complementation. We examined the expression of afrA in the presence and absence of afrR. We also examined the effect of afrS, the open reading frame immediately downstream of afrR, on afrA transcription. Subclones of pRKB15 were constructed which lacked the region encoding afrRS. pRKB18 (Fig. 1), which placed the Plac promoter upstream of afrA, mediated the production of AF/R1 pili as determined by a colony blot assay. In contrast, pRKB19 (Fig. 1), which placed Plac downstream of afrE, did not mediate the production of AF/R1 pili, suggesting that the production of pili by pRKB18 was due to the positioning of the vector promoter upstream of afrA. A plasmid was constructed using a vector compatible with pUC derivatives, pKI100 (29), which included afrRS. This plasmid was designated p507. When p507 was introduced into DH5a(pRKB19), AF/R1 production was restored. A second plasmid, p501, was constructed that encoded only afrS. Maxicell analysis of p501 confirmed the expression of afrS (data not shown). p501 did not complement AF/R1 pilus production. A plasmid encoding only afrR was constructed by using the pKI100 vector. This was designated p598, and afrR expression was confirmed by maxicell analysis (data not shown). p598 did not complement pRKB19 for AF/R1 expression. These results are summarized in Table 1.
TABLE 1.
Pilus expression with various plasmids
| Plasmida | Detection of AF/R1 pilus expression with:
|
|
|---|---|---|
| MAb | mRNA | |
| pRKB18 | + | NDb |
| pRKB19 | − | − |
| pRKB19 + p507 | + | + |
| pRKB19 + p598 | − | − |
| pRKB19 + p501 | − | − |
See Fig. 1 for details of plasmid constructs.
ND, not done.
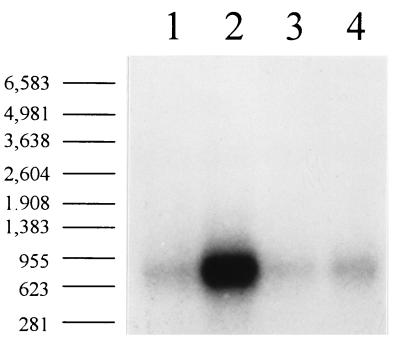
Northern blot analysis confirmed that the results of expression analysis noted above involved transcriptional activation. When pRKB19 was complemented with p507 (Fig. 1), which encodes both afrS and afrR, an eightfold increase in the amount of afrA mRNA was observed (Fig. 2, lane 2) relative to the amount of afrA mRNA produced by E. coli harboring only pRKB19 (Fig. 2, lane 1), as determined by densitometric analysis. When pRKB19 was complemented with either p598 expressing afrR or p501 expressing afrS, no increase in the amount of afrA message was observed. These results are consistent with the hypothesis that AfrR and AfrS interact to activate the transcription of afrA.
FIG. 2.
Northern blot analysis of afrA expression in the presence and absence of afrR and afrS. Total RNA was separated by formaldehyde agarose gel electrophoresis and transferred to a nylon membrane. The blotted RNA was hybridized to a single-stranded RNA probe corresponding to the antisense sequence of a KpnI fragment internal to afrA. Size standards were visualized by UV illumination of the ethidium bromide-stained gel. The mobilities of the size standards, expressed in kilobases, are shown on the left. Lane 1, DH5α(pRKB19); lane 2, DH5α(pRKB19,p507); lane 3, DH5α(pRKB19,p598); lane 4, DH5α(pRKB19,p501).
AF/R1 pilus adherence is encoded in the afrDE region.
A subclone of pRKB15, p562, that deleted afrE (Fig. 1) was prepared. DH5α(pRKB15) and DH5α(p562) were examined for AF/R1 surface expression with MAb and by rabbit intestinal brush border agglutination. AF/R1 antigen was expressed in both cases. However, only DH5α(pRKB15) agglutinated rabbit brush borders (data not shown). This indicates that AfrA does not confer adhesiveness to AF/R1 fimbriae in the absence of AfrE. The role of afrD in AF/R1 pilus expression and function remains unknown. AfrD is highly homologous to FedE, one of two proteins reported to be essential for fimbrial adherence and fimbrial length of the F18 (24).
DISCUSSION
Sequence analysis, maxicell assays, complementation experiments, and Northern blots have defined seven genes associated with the expression and function of AF/R1 pili by the rabbit pathogen E. coli RDEC-1. Our sequence extends the homology displayed between AfrA and FedA, the major structural subunit of F107 (now called F18 [24] fimbriae) (23), through the C termini of the two peptides and is consistent with molecular size data. Comparison of the sequence of AfrA and the predicted sequence of mature FedA suggests that the two peptides have evolved from a common ancestor relatively recently, with an overall identity of 47% and a similarity of 62%.
We report the sequence of a gene downstream of afrA which we have designated afrB. Analysis of the predicted amino acid sequence of the putative gene product revealed significant homology with a number of usher proteins associated with pilus biogenesis. The most closely related was PefC, the usher protein of S. typhimurium plasmid-encoded fimbriae (17). PefC has homology with PapC and FaeD, outer membrane proteins required for the biosynthesis of P and K88 fimbriae of E. coli, respectively (38, 42). These proteins are required for the extracellular localization of fimbrial proteins, and proteins of similar sizes (80 to 100 kDa) are encoded by all E. coli fimbrial determinants studied (15). We hypothesize that afrB encodes an outer membrane protein involved in the assembly of AF/R1 fimbriae. Transposon insertions in afrB prevent the expression of AfrA antigen on the bacterial cell surface, consistent with this hypothesis (Fig. 1).
afrC encodes a protein with homology to periplasmic chaperones required for fimbrial synthesis in a number of systems, the best characterized of which is the pap operon, for which PapD serves as the chaperonin protein. These proteins act to protect fimbrial subunits from proteolytic degradation in the periplasm prior to assembly at the outer membrane (15). Transposon insertions in afrC also prevent the expression of AfrA antigen on the bacterial cell surface (Fig. 1), consistent with this hypothesis. Transposon insertion mutagenesis revealed that only afrABC were necessary for the expression of the AF/R1 pilus.
The functions of the region downstream of afrC, afrDE, which encodes two proteins, appear to include the adhesive properties of AF/R1 fimbriae. When afrE is deleted, AF/R1 fimbriae are produced which are antigenic but which do not mediate the adherence of E. coli to rabbit intestinal brush borders. This indicates that the adhesive subunit is distinct from the major structural subunit of AF/R1 and is likely encoded by afrE. Several fimbrial adhesins are characterized by adhesive subunits distinct from the major fimbrial subunits, including P pili (36), type I pili (34), and others. The characterization of the adhesive subunit of AF/R1 pili will be an important contribution to the understanding of the M cell-specific adherence mediated by these pili.
The regulation of several fimbrial adhesin operons of E. coli involves members of the AraC family of transcriptional activators. Examples include Rns, CfaD, CsvR, and FapR, which are regulatory proteins for the fimbrial adhesins CS1, CS2, CS5, CFA/I, CS5, and 987P, respectively (8, 9, 13, 33). Our observations indicate that the expression of AF/R1 pili requires a protein, AfrR, which displays homology with the AraC family of transcriptional activators (Fig. 3). The homology of AfrR with Rns is rather limited, in contrast with the other AraC-related activators of E. coli fimbrial adhesins, which form a highly homologous group. Rns, CfaD, CsvR, and FapR have extensive regions of similarity and are all related to AraC at the C terminus. AfrR does not have any homology with Rns outside the AraC-related region. Moreover, the region of homology with the C terminus of AraC is at the N terminus of AfrR. AfrR is more closely related to a group of proteins including Caf1R (31) and Rob (55). Caf1R is a positive regulator of the F1 envelope protein of Y. pestis, while Rob is a protein of E. coli which binds to the chromosomal origin of replication. AfrR, Caf1R, and Rob each have homology in their N termini with the C terminus of AraC. These three proteins also have homology with each other in their C-terminal regions. Other AraC-related proteins displaying significant homology with AfrR include SoxS, a regulatory protein involved in the superoxide response to E. coli (63) and the predicted product of an open reading frame downstream of tetA in Tn10 (51).
The close relationship of AfrR with the Y. pestis regulatory protein Caf1R is of interest. The caf1 operon of Y. pestis encodes the production of a proteinaceous capsule, the major component of which is the F1 antigen (19). Although this protein does not appear to form a fimbria-like structure, the operon encodes at least two proteins required for capsule assembly which have homology with fimbrial assembly and periplasmic chaperone proteins (19, 32). Capsule production requires the presence of Caf1R, and the truncated derivative of Caf1R consisting of the N-terminal 81 amino acids of the protein is capable of positively regulating the expression of the capsule (31). This is consistent with the observed homology of the N terminus of Caf1R with the DNA binding domain of AraC. The occurrence of these closely related regulatory proteins in two otherwise unrelated fimbria-like operons suggests that AfrR-mediated expression may represent a third general pattern of regulation of E. coli fimbriae associated with pathogenesis.
AfrS was related to two proteins of unknown function. Thus, the sequence information does not provide an indication of the function. Our data indicate that AfrS is required for the transcription of the afr operon. A transposon insertion in afrS did not affect pilus expression (Fig. 1). However, in this construct, afrA transcription was likely mediated by a vector promoter, relieving the requirement for afrS. Possible modes of action of AfrS include a direct interaction with AfrR, an indirect involvement through interaction with another unknown regulatory protein, or signal transduction, as suggested by the predicted presence of a signal sequence for secretion. Details of the role of AfrS in afr transcriptional regulation await further studies on the localization of the protein and identification of potential interactions with other proteins or with the afr promoter.
ACKNOWLEDGMENTS
The Research Service of the Department of Veterans Affairs supported this work.
We acknowledge C. V. Sciortino for his guidance in developing the MAb that was so useful in our studies and for his careful review of the manuscript. We also thank M. Bratoeva for her review of the manuscript and help with preparing the references. We thank the MUSC Biotechnology Laboratory for providing DNA sequence analysis and synthesizing oligonucleotides.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Bilge S S, Apostol J, Jr, Fullner K J, Moseley S L. Transcriptional organization of the F1845 fimbrial adhesin determinant of Escherichia coli. Mol Microbiol. 1993;7:993–1006. doi: 10.1111/j.1365-2958.1993.tb01191.x. [DOI] [PubMed] [Google Scholar]
- 3.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruijn F J, Lupski J R. The use of transposon Tn5 mutagenesis in the rapid generation of correlated physical and genetic maps of DNA segments cloned into multicopy plasmids—a review. Gene. 1984;27:131–149. doi: 10.1016/0378-1119(84)90135-5. [DOI] [PubMed] [Google Scholar]
- 5.Cantey J R, Blake R K. Diarrhea due to Escherichia coli in the rabbit: a novel mechanism. J Infect Dis. 1977;135:454–462. doi: 10.1093/infdis/135.3.454. [DOI] [PubMed] [Google Scholar]
- 6.Cantey J R, Inman L R. Diarrhea due to Escherichia coli strain RDEC-1 in the rabbit: the Peyer’s patch as the initial site of attachment and colonization. J Infect Dis. 1981;143:440–446. doi: 10.1093/infdis/143.3.440. [DOI] [PubMed] [Google Scholar]
- 7.Cantey J R, Inman L R, Blake R K. Production of diarrhea in the rabbit by mutant of Escherichia coli (RDEC-1) that does not express adherence (AF/R1) pili. J Infect Dis. 1989;160:136–141. doi: 10.1093/infdis/160.1.136. [DOI] [PubMed] [Google Scholar]
- 7a.Cantey, J. R. Unpublished observations.
- 8.Caron J, Coffield L M, Scott J R. A plasmid-encoded regulatory gene, rns, required for expression of the CS1 and CS2 adhesins of enterotoxigenic Escherichia coli. Proc Natl Acad Sci USA. 1989;86:963–967. doi: 10.1073/pnas.86.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caron J, Scott J R. A rns-like regulatory gene for colonization factor antigen I (CFA/I) that controls expression of CFA/I pilin. Infect Immun. 1990;58:874–878. doi: 10.1128/iai.58.4.874-878.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Cassels, F. J., and E. C. Boedeker. Personal communication.
- 10.Cheney C P, Boedeker E C, Formal S B. Quantitation of the adherence of an enteropathogenic Escherichia coli to isolated rabbit intestinal brush borders. Infect Immun. 1979;26:736–743. doi: 10.1128/iai.26.2.736-743.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheney C P, Formal S B, Schad P A, Boedeker E C. Genetic transfer of a mucosal adherence factor (R1) from an enteropathogenic Escherichia coli strain into a Shigella flexneri strain and the phenotypic suppression of this adherence factor. J Infect Dis. 1983;147:711–723. doi: 10.1093/infdis/147.4.711. [DOI] [PubMed] [Google Scholar]
- 12.Crooke E, Hwang D S, Skarstad K, Thöny B, Kornberg A. Escherichia coli minichromosome replication: regulation of initiation at oriC. Res Microbiol. 1991;140:127–130. doi: 10.1016/0923-2508(91)90019-7. [DOI] [PubMed] [Google Scholar]
- 13.de Haan L A, Willshaw G A, van der Zeijst B A, Gaastra W. The nucleotide sequence of a regulatory gene present on a plasmid in an enterotoxigenic Escherichia coli strain of serotype O157:H7. FEMS Microbiol Lett. 1991;67:341–346. doi: 10.1111/j.1574-6968.1991.tb04487.x. [DOI] [PubMed] [Google Scholar]
- 14.Devereaux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dodson K W, Jacob-Dubuisson F, Striker R T, Hultgren S J. Outer-membrane PapC molecular usher discriminately recognizes periplasmic chaperone-pilus subunit complexes. Proc Natl Acad Sci USA. 1993;90:3670–3674. doi: 10.1073/pnas.90.8.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donnenberg M S, Donohue-Rolfe A, Keusch G T. Epithelial cell invasion: an overlooked property of enteropathogenic Escherichia coli (EPEC) associated with the EPEC adherence factor. J Infect Dis. 1989;160:452–459. doi: 10.1093/infdis/160.3.452. [DOI] [PubMed] [Google Scholar]
- 17.Friedrich M J, Kinsey N E, Vila J, Kadner R J. Nucleotide sequence of a 13.9 kb segment of the 90 kb virulence plasmid of Salmonella typhimurium: the presence of fimbria biosynthetic genes. Mol Microbiol. 1993;8:543–558. doi: 10.1111/j.1365-2958.1993.tb01599.x. [DOI] [PubMed] [Google Scholar]
- 18.Gallegos M-T, Schleif R, Bairoch A, Hofmann K, Ramos J L. AraC/XylS family of transcriptional regulators. Microbiol Mol Biol Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galyov E E, Smirnov O Y, Karlishev A V, Volkovoy K I, Denesyuk A I, Nazimov I V, Rubtsov K S, Abramov V M, Dalvadyanz S M, Zav’yalov V P. Nucleotide sequence of the Yersinia pestis gene encoding F1 antigen and the primary structure of the protein. Putative T and B cell epitopes. FEBS Lett. 1990;277:230–232. doi: 10.1016/0014-5793(90)80852-a. [DOI] [PubMed] [Google Scholar]
- 20.Goluszko P, Moseley S L, Truong L D, Kaul A, Williford J R, Selvarangan R, Nowicki S, Nowicki B. Development of an experimental model of chronic pyelonephritis with Escherichia coli O75:K5:H-bearing Dr fimbriae. J Clin Investig. 1997;99:1662–1672. doi: 10.1172/JCI119329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gunn J S, Lim K B, Kreuger J, Kim K, Guo L, Hackett M, Miller S I. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol Microbiol. 1998;27:1171–1182. doi: 10.1046/j.1365-2958.1998.00757.x. [DOI] [PubMed] [Google Scholar]
- 22.Hanahan D. Studies on the transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 23.Imberechts H, De Greve H, Schlicker C, Bouchet H, Pohl P, Charlier G, Bertschinger H, Wild P, Vandekerckhove J, Van Damme J, Van Montagu M, Lintermans P. Characterization of F107 fimbriae of Escherichia coli 107/86, which causes edema disease in pigs, and nucleotide sequence of the F107 major fimbrial subunit gene, fedA. Infect Immun. 1992;60:1963–1971. doi: 10.1128/iai.60.5.1963-1971.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imberechts H, Wild P, Charlier G, De Greve H, Lintermans P, Pohl P. Characterization of F18 fimbrial genes fedE and fedF involved in adhesion and length of enterotoxemic Escherichia coli strain 107/86. Microb Pathog. 1996;21:183–192. doi: 10.1006/mpat.1996.0053. [DOI] [PubMed] [Google Scholar]
- 25.Inman L R, Cantey J R. Peyer’s patch lymphoid follicle epithelial adherence of a rabbit enteropathogenic Escherichia coli (strain RDEC-1). Role of plasmid-mediated pili in initial adherence. J Clin Investig. 1984;74:90–95. doi: 10.1172/JCI111423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inman L R, Cantey J R. Specific adherence of Escherichia coli (strain RDEC-1) to membranous (M) cells of the Peyer’s patch in Escherichia coli diarrhea in the rabbit. J Clin Investig. 1983;71:1–8. doi: 10.1172/JCI110737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inman L R, Cantey J R, Formal S B. Colonization, virulence, and mucosal interaction of an enteropathogenic Escherichia coli (strain RDEC-1) expressing Shigella somatic antigen in the rabbit intestine. J Infect Dis. 1986;154:742–751. doi: 10.1093/infdis/154.5.742. [DOI] [PubMed] [Google Scholar]
- 28.Isaacson R E. K99 surface antigen on Escherichia coli: purification and partial characterization. Infect Immun. 1977;11:272–279. doi: 10.1128/iai.15.1.272-279.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishimoto K S, Lory S. Formation of pilin in Pseudomonas aeruginosa requires the alternative ς factor (RpoN) of RNA polymerase. Proc Natl Acad Sci USA. 1989;86:1954–1957. doi: 10.1073/pnas.86.6.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kado C I, Liu S-T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karlyshev A V, Galyov E E, Abramov V M, Zav’yalov V P. Caf1R gene and its role in the regulation of capsule formation of Y. pestis. FEBS Lett. 1992;305:37–40. doi: 10.1016/0014-5793(92)80650-6. [DOI] [PubMed] [Google Scholar]
- 32.Karlyshev A V, Galyov E E, Smirnov O Y, Guzayev A P, Abramov V M, Zav’yalov V P. A new gene of the f1 operon of Y. pestis involved in the capsule biogenesis. FEBS Lett. 1992;297:77–80. doi: 10.1016/0014-5793(92)80331-a. [DOI] [PubMed] [Google Scholar]
- 33.Klaasen P, de Graaf F K. Characterization of FapR, a positive regulator of expression of the 987P operon in enterotoxigenic Escherichia coli. Mol Microbiol. 1990;4:1779–1783. doi: 10.1111/j.1365-2958.1990.tb00556.x. [DOI] [PubMed] [Google Scholar]
- 34.Krogfelt K A, Bergmans H, Klemm P. Direct evidence that the FimH protein is the mannose-specific adhesin of Escherichia coli type 1 fimbriae. Infect Immun. 1990;58:1995–1998. doi: 10.1128/iai.58.6.1995-1998.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 36.Lund B, Lindberg F, Marklund B I, Normark S. The PapG protein is the alpha-d-galactopyranosyl-(1→4)-beta-d-galactopyranose-binding adhesin of uropathogenic Escherichia coli. Proc Natl Acad Sci USA. 1987;84:5898–5902. doi: 10.1073/pnas.84.16.5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McDaniel T K, Jarvis K G, Donnenberg M S, Kaper J B. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci USA. 1995;92:1664–1668. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mooi F R, Claassen I, Bakker D, Kuipers H, de Graaf F K. Regulation and structure of an Escherichia coli gene coding for an outer membrane protein involved in export of K88ab fimbrial subunits. Nucleic Acids Res. 1986;14:2443–2457. doi: 10.1093/nar/14.6.2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moseley S L, Huq I, Alim A R, So M, Samaspour-Motalebi M, Falkow S. Detection of enterotoxigenic Escherichia coli by DNA colony hybridization. J Infect Dis. 1980;142:892–898. doi: 10.1093/infdis/142.6.892. [DOI] [PubMed] [Google Scholar]
- 40.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 41.Nishibuchi M, Hill W E, Zon G, Payne W L, Kaper J B. Synthetic oligodeoxyribonucleotide probes to detect Kanagawa phenomenon-positive Vibrio parahaemolyticus. J Clin Microbiol. 1986;23:1091–1095. doi: 10.1128/jcm.23.6.1091-1095.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Norgren M, Baga M, Tennent J M, Normark S. Nucleotide sequence, regulation and functional analysis of the papC gene required for cell surface localization of Pap pili of uropathogenic Escherichia coli. Mol Microbiol. 1987;1:169–178. doi: 10.1111/j.1365-2958.1987.tb00509.x. [DOI] [PubMed] [Google Scholar]
- 43.Norrander J, Kempe T, Messing J. Construction of improved M13 vectors using oligonucleotide-directed mutagenesis. Gene. 1982;26:101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- 44.Oakley B R, Kirsch D R, Morris N R. A simplified ultrasensitive silver stain for detecting proteins in polyacrylamide gels. Anal Biochem. 1980;105:361–363. doi: 10.1016/0003-2697(80)90470-4. [DOI] [PubMed] [Google Scholar]
- 45.Portnoy D A, Moseley S L, Falkow S. Characterization of plasmids and plasmid-associated determinants of Yersinia enterocolitica pathogenesis. Infect Immun. 1981;31:775–782. doi: 10.1128/iai.31.2.775-782.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rupp W, Wilde C, Reno D. Exchanges between DNA strands in ultraviolet-irradiated Escherichia coli. J Mol Biol. 1971;61:25–44. doi: 10.1016/0022-2836(71)90204-x. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 48.Sancar A, Hack A M, Rupp W D. A simple method for identification of plasmid-encoded proteins. J Bacteriol. 1979;137:692–693. doi: 10.1128/jb.137.1.692-693.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schleif R. The l-arabinose operon. In: Neidhart F, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1987. pp. 1473–1481. [Google Scholar]
- 51.Schollmeier K, Hillen W. Transposon Tn10 contains two structural genes with opposite polarity between tetA and IS10R. J Bacteriol. 1984;160:499–503. doi: 10.1128/jb.160.2.499-503.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schuler G D, Altschul S F, Lipman D J. A workbench for multiple alignment construction and analysis. Proteins Struct Funct Genet. 1991;9:180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
- 53.Sciortino C V, Yang Z S, Finkelstein R A. Monoclonal antibodies to outer membrane antigens of Vibrio cholerae. Infect Immun. 1985;49:122–131. doi: 10.1128/iai.49.1.122-131.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shine J, Dalgarno L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Skarstad K, Thöny B, Hwang D S, Kornberg A. A novel binding protein of the Escherichia coli chromosome. J Biol Chem. 1992;268:5365–5370. [PubMed] [Google Scholar]
- 56.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 57.Takeuchi A, Inman L R, O’Hanley P D, Cantey J R, Lushbaugh W B. Scanning and transmission electron microscopic study of Escherichia coli O15 (RDEC-1) enteric infection in rabbits. Infect Immun. 1978;19:686–694. doi: 10.1128/iai.19.2.686-694.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vandeyar M A, Weiner M P, Hutton C J, Batt C A. A simple and rapid method for the selection of oligodeoxynucleotide-directed mutants. Gene. 1988;65:129–133. doi: 10.1016/0378-1119(88)90425-8. [DOI] [PubMed] [Google Scholar]
- 59.Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;20:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 60.Wallace R B, Shaffer J, Murphy R F, Bonner J, Hirose T, Itakura K. Hybridization of synthetic oligodeoxyribonucleotides to phi chi 174 DNA: the effect of single base pair mismatch. Nucleic Acids Res. 1979;6:3543–3557. doi: 10.1093/nar/6.11.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wolf M. Characterization of a plasmid from Escherichia coli RDEC-1 that mediates expression of adhesin AF/R1 and evidence that AF/R1 pili promote but are not essential for enteropathogenic disease. Infect Immun. 1988;56:1846–1857. doi: 10.1128/iai.56.8.1846-1857.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wolf M K, Boedeker E C. Cloning of the genes for AF/R1 pili from rabbit enteroadherent Escherichia coli RDEC-1 and DNA sequence of the major structural subunit. Infect Immun. 1990;58:1124–1128. doi: 10.1128/iai.58.4.1124-1128.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu J, Weiss B. Two divergently transcribed genes, soxR and soxS, control a superoxide response regulon of Escherichia coli. J Bacteriol. 1991;173:2864–2871. doi: 10.1128/jb.173.9.2864-2871.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]