Abstract
Sesquiterpenes constitute the principal components of the genus Ainsliaea, encompassing guaiane, germacrane, eudesmane, and polymer sesquiterpene lactones types. These secondary metabolites exhibit diverse pharmacological activities, including antitumor, antibacterial, anti-inflammatory, antiviral, antioxidant, hepatoprotective, and neuroprotective effects. Through a comprehensive literature search of the Web of Science, PubMed, SciFinder, and CNKI databases, it was discovered that there are as many as 145 main sesquiterpenoids in the genus Ainsliaea. However, the nuclear magnetic resonance (NMR) data for the sesquiterpenes in this genus have not been systematically compiled and summarized. Therefore, this review aims to highlight the chemical structures, NMR data, and pharmacological activities of sesquiterpenes in Ainsliaea. By meticulously analyzing published scholarly literature, our goal is to provide a solid foundation for further exploration of new sesquiterpenes and extensive utilization of this genus.
Keywords: Ainsliaea, sesquiterpenes, nuclear magnetic resonance (NMR), structure analysis
1. Introduction
Ainsliaea is a perennial herb, with over seventy species mainly distributed in southeast Asia. In China, there are forty-four species and four varieties. The majority of these plants are found around the Yangtze River Basin, with only one species distributed in the northeast. They are typically harvested in summer and autumn, and the entire plants are used for Chinese medicinal purposes [1]. According to the ‘Supplements to the Compendium of Materia Medica’, it is documented that Ainsliaea has a sweet and mild taste, cold properties, and belongs to the lung, spleen, and large-intestine meridians. The ‘Comprehensive Dictionary of Chinese Herbal Medicine’ states that Ainsliaea has the functions of clearing heat, promoting diuresis, cooling blood, and detoxification. Ainsliaea fragrans Champ. Is the primary ingredient in the national protected Chinese medicine ‘Xingxiang Tu’erfeng’ herbal granules and herbal tablets.
Since the 1980s, several studies have been conducted on the chemical composition and pharmacological activities of the genus Ainsliaea, leading to the discovery of over 400 compounds. The chemical constituents of this genus mainly include sesquiterpenoids, triterpenoids, steroids and their derivatives, phenolic acids, flavonoids, anthraquinones, coumarins, lignans, essential oils, and other components. Chemical studies have revealed that sesquiterpenes are the characteristic components of Ainsliaea plants [2,3]. The investigation has shown that sesquiterpenes in Ainsliaea plants mainly consist of guaiane, germacrane, eudesmane, polymer sesquiterpene lactones, and others [4]. However, NMR spectroscopy data for these sesquiterpenes derivatives have not been reported. This paper aims to provide references for the analysis and identification of new structural compounds by summarizing the 1H- and/or 13C-NMR data of 145 sesquiterpenes from the genus Ainsliaea between 1979 and 2022 through consulting the relevant literature.
2. Guaiane-Type Sesquiterpene
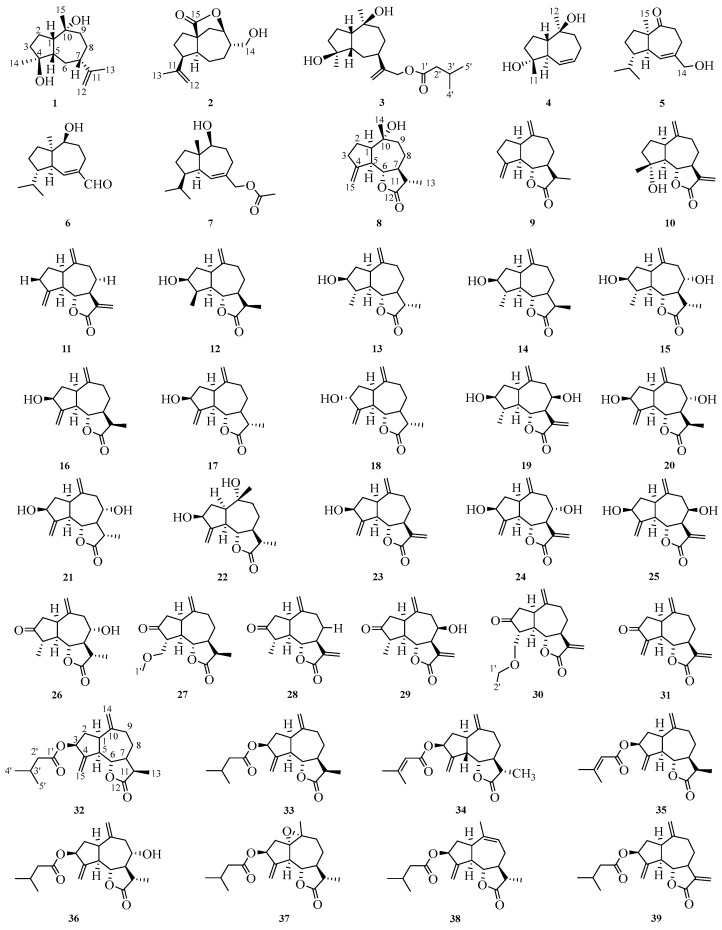
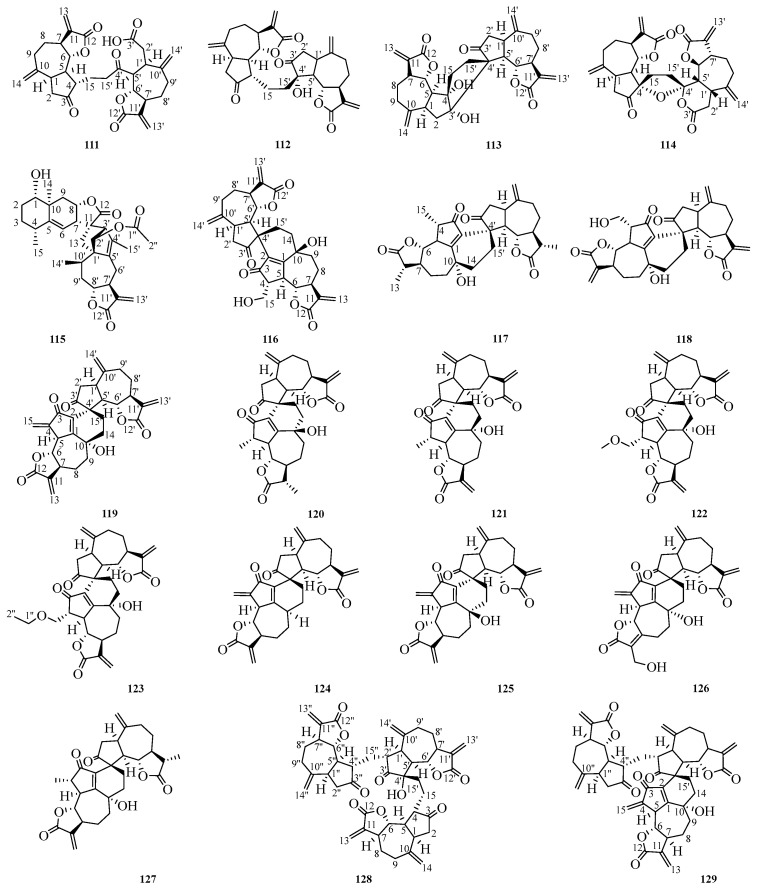
Guaiane sesquiterpenes are a class of compounds with three isoprene units consisting of 5,7 fused rings, which are substituted by 4,10-dimethyl-7-isopropyl moieties as the basic nucleus. These compounds possess antibacterial, anti-inflammatory, antitumor, neuroprotective, and other biological activities [5]. Thus far, a total of 63 guaiane sesquiterpenes have been reported in this genus, mainly 12,6 guaiacan-type sesquiterpene lactones. The structures and detailed information are listed in Figure 1 and Table 1.
Figure 1.
Chemical structures for compounds 1–63.
Table 1.
The compound name, molecular formula, and NMR test reagent of guaiane-type sesquiterpene.
| No. | Compound Name | Molecular Formula | Solvent | Ref. |
|---|---|---|---|---|
| 1 | Epi-guaidiol A | C15H26O2 | CD3OD | [6] |
| 2 | Ainslide A | C15H22O3 | CDCl3 | [7] |
| 3 | Spicatene B | C20H34O4 | CDCl3 | [8] |
| 4 | 4β,10α-Dimethyl-1β,5α-bicycle[3,5,0]dec-6-en-4α,10β-diol | C12H20O2 | CDCl3 | [9] |
| 5 | Aphanamol I | C15H24O2 | CDCl3 | [10] |
| 6 | Aphanamol II | C15H24O2 | CDCl3 | [10] |
| 7 | Yunnanol A | C17H28O3 | CDCl3 | [11] |
| 8 | Ainslide E | C15H22O3 | CDCl3 | [7] |
| 9 | 4(15),10(14)-Guaiadien-12, 6-olide mokkolactone |
C15H20O2 | CDCl3 | [12] |
| 10 | 4-Hydroxy-10(14),11(13)-guaiadien-6,12-olide | C15H20O3 | CDCl3 | [13] |
| 11 | Dehydrocostuslactone | C15H18O2 | CDCl3 | [14] |
| 12 | Ainslide F | C15H22O3 | CDCl3 | [7] |
| 13 | Dihydroestafiatol | C15H22O3 | CDCl3 | [15] |
| 14 | 4β,15,11β,13-Tetrahydrozaluzanin C | C16H22O2 | CDCl3 | [16] |
| 15 | Isolipidiol | C15H22O4 | CD3OD | [17,18] |
| 16 | 11α,13-Dihydrozaluzanin C | C15H20O3 | CDCl3 | [19] |
| 17 | 11β,13-Dihydrozaluzanin C | C15H20O3 | CDCl3 | [15] |
| 18 | 11β,13-Dihydro-3-epizaluzanin C | C15H20O3 | CDCl3 | [20] |
| 19 | 8β-Hydroxy-4β, 15-dihydrozaluzanin C | C15H20O4 | CDCl3 | [21] |
| 20 | 8α-Hydroxy-11α, 13-dihydrozaluzanin C | C15H20O4 | CDCl3 | [22] |
| 21 | 11β,13-Dihydrodesacylynaropicrin | C15H20O4 | CDCl3 | [18,23] |
| 22 | l0α-Hydroxy-10(14),11β(13)-tetrahydroxaluzanin C | C15H22O4 | CDCl3 | [24] |
| 23 | Zaluzanin C | C15H18O3 | CDCl3 | [25] |
| 24 | Desacylcynaropicrin | C15H18O4 | CDCl3 CD3OD/CDCl3 |
[18,26] |
| 25 | 8-Epidesacylcinaropicrin | C15H18O4 | C5D5N | [27] |
| 26 | Isoamberboin | C15H20O4 | CDCl3 | [28] |
| 27 | Ainslide D | C16H22O4 | CDCl3 | [7] |
| 28 | Estafiatone | C15H18O3 | CDCl3 | [29] |
| 29 | 8-Epigrosheimin | C15H18O4 | CDCl3 | [30] |
| 30 | Ainsliaolide B | C17H22O4 | CDCl3 | [31] |
| 31 | Dehydrozaluzanin C | C15H16O3 | CDCl3 | [32] |
| 32 | Diaspanolide A | C20H28O4 | CDCl3 | [33] |
| 33 | Diaspanolide E | C20H28O4 | CDCl3 | [34] |
| 34 | Ainsliaolide A | C24H26O4 | CDCl3 | [35] |
| 35 | Ainsliaolide D | C20H26O4 | CDCl3 | [36] |
| 36 | 8α-Hydroxy-diaspanolide A | C20H28O5 | CDCl3 | [8] |
| 37 | Yunnanolides H | C20H28O5 | CDCl3 | [37] |
| 38 | Yunnanolides I | C20H28O4 | CDCl3 | [37] |
| 39 | Diaspanolide B | C20H26O4 | CDCl3 | [33] |
| 40 | lα-Hydroxy-3-O-isobutyrate | C20H26O5 | CDCl3 | [38] |
| 41 | Ainslide C | C20H24O5 | CDCl3 | [7] |
| 42 | Yunnanolide J | C20H28O6 | CDCl3 | [11] |
| 43 | Spicatene A | C20H26O5 | CDCl3 | [8] |
| 44 | Yunnanolides A | C22H32O6 | CDCl3 | [37] |
| 45 | Yunnanolides C | C22H32O6 | CDCl3 | [37] |
| 46 | Yunnanolides D | C22H32O6 | CDCl3 | [37] |
| 47 | Yunnanolides E | C22H32O5 | CDCl3 | [37] |
| 48 | Yunnanolides F | C22H32O5 | CDCl3 | [37] |
| 49 | Yunnanolides B | C22H32O6 | CDCl3 | [37] |
| 50 | Pertyolide C | C22H30O6 | CDCl3 | [39] |
| 51 | Yunnanolides G | C24H33NO5 | CDCl3 | [37] |
| 52 | 11α, 13-Dihydroglucozaluzanin C | C21H30O8 | CD3OD/C5D5N | [22] |
| 53 | 8α-Hydroxy-11α, 13-dihydroglucozaluzanin C | C21H30O9 | CD3OD | [22] |
| 54 | 13-Ethoxy-4(15),10(14)-dien-guai-6,12-olide-3-O-β-D-glucopyranoside | C23H35O9 | CD3OD | [40] |
| 55 | 11β,13-Dihydro-8α-hydroxyglucozaluzanin C | C21H30O9 | C5D5N | [41] |
| 56 | 4β,15-Dihydrozaluzanin C | C21H30O8 | DMSO | [42] |
| 57 | Glucozaluzanin C | C21H28O8 | CDCl3/C5D5N/ CD3SOCD3 |
[12,22] |
| 58 | Ainsliaside C | C30H34O10 | CD3OD | [43] |
| 59 | Ainsliaside A | C30H34O11 | C5D5N | [44] |
| 60 | 2′-O-E-Caffeoyl-8α-hydroxy-11α,13-dihydro-3-β-O-β-D-glucozaluzanin C | C30H36O12 | CD3OD | [45] |
| 61 | Macrocliniside B | C27H38O13 | DMSO CD3OD |
[25,46] |
| 62 | Macrocliniside I | C33H48O18 | DMSO | [25,46] |
| 63 | ZaluzaninC-3-O-β-glucopyranosyl-(1→3)-β-glucopyranosyl-(1→3)-β-glucopyranosyl-(1→3)-β-glucopyranoside | C39H58O23 | DMSO | [46] |
2.1. NMR Data of Guaiane Sesquiterpenes (1–63)
The 1H and 13C NMR spectroscopy results were summarized in Table 2, Table 3, Table 4, Table 5, Table 6, Table 7, Table 8, Table 9, Table 10 and Table 11. Additionally, this paper provides a summary of the nuclear magnetic data testing instrument used for compounds 1–63. NMR data for compounds 2, 8, 12, 27, and 41 were obtained with Bruker AV-400 HD spectrometers (Bruker, Byersbin, Switzerland). The 1H and 13C data of compounds 3, 7, 36–38, 42–49, and 51 were obtained by a Bruker Ascend-500 spectrometer (Bruker, Nasdaq, New York, NY, USA). For compounds 11, 25, 55, and 59, the NMR data were recorded on a JEOL FX-90Q spectrometer (JEOL, Tokyo, Japan). Compounds 20, 52, 53, and 57 had their 1H and 13C data taken with a Varian Mercury Plus 400 instrument (Varian, Palo Alto, CA, USA). Compounds 23, 61, and 62 were measured by a Varian Inova 400 instrument (Varian, Palo Alto, CA, USA). The 1H and 13C data of compounds 61, 62, and 63 were recorded using a unity Bruker AV500 instrument (Bruker, Switzerland). NMR data of compounds 9 and 57 were obtained using the Bruker AMX 500 (Bruker, Zurich, Switzerland) and Varian Unity Inova 500 instruments (Varian, USA). The 1H and 13C data of compounds 30 and 35 were run on a Bruker Avance 600 spectrometer (Bruker, Germany). The 1H- and 13C-NMR data of compounds 32 and 39 were collected by a Bruker DRX-500 spectrometer (Bruker, Switzerland). Nuclear magnetic data of compounds 1, 4, 14, 15, 16, 18, 28, 31, 33, 54, 56, 58, and 60 were recorded on the following instruments: VNS-600 (Varian, Palo Alto, CA, USA), Bruker ACF-500 NMR (Bruker, Germany), Bruker Avance DRX 500, Bruker Avance II 800 (Bruker, Switzerland), Bruker ARX-300 NMR (Bruker, Switzerland), Bruker Avance 400 (Bruker, Zug, Switzerland), Varian Inova 500 (Varian, Palo Alto, CA, USA), Bruker AC 200 (Bruker, Karlsruhe, Germany), Bruker Advance 500 (Bruker, Germany), Bruker AV-600 (Bruker, Switzerland), Varian 500 MHz (Varian, Palo Alto, CA, USA), and Bruker AV500-III (Bruker, Switzerland), Varian VNS600 (Varian, USA), Bruker Avance 300 (Bruker, Switzerland), and Bruker Avance 500 (Bruker, Switzerland). The 1H and 13C spectrums of compounds 5–6 were tested at 360 and 25 MHz, respectively; 10, 24, and 40 were run at 400MHz; 13 and 17 were recorded at 200 MHz for 1H and 50 MHz for 13C NMR; The 1H-NMR spectra of 19 and 29 were measured on 500.13 MHz; 21 and 26 were tested in the 270 MHz; 22 was taken with 300 MHz; 34 and 50 were collected in the 500 MHz. The carbon spectrum of compound 24 was determined at 100 MHz, compound 19 was recorded at 125.76 MHz, and compound 34 was recorded at 125 MHz. Carbon spectrum data for compounds 10, 21, 22, 26, 40, and 50 have not been reported in the literature.
Table 2.
1H-NMR data of compounds 1–8.
| NO. | 1 [6] | 2 [7] | 3 [8] | 4 [9] | 5 [10] | 6 [10] | 7 [11] | 8 [7] |
|---|---|---|---|---|---|---|---|---|
| CD3OD | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | |
| 1 | 2.54, ddd, 10.8, 8.4, 8.4 | — | 2.74, q, 9.3 | 2.05, m | — | — | — | 2.30, q, 7.6 |
| 2α | 1.81–1.85, m | 1.94–2.02, m | 1.95, m | 1.76, dd, 11.7, 6.4 | 2.08 | — | 1.63, m | 1.78–1.87, m |
| 2β | 1.51–1.54, m | 1.59–1.70, m | 1.49, m | 1.66, dd, 11.7, 7.3 | 1.35 | — | 1.42, m | 1.90–1.98, m |
| 3α | 1.67, br d, 10.4 | 2.47–2.60, m | 1.69, m | 1.70, dd, 7.3, 6.4 | 1.8 | — | 1.43, m | 2.38–2.48, m |
| 3β | 1.66, dd, 10.4, 3.6 | 1.86–1.97, m | 1.35 | |||||
| 4 | — | 2.99–3.08, m | — | — | 1.66 | — | 1.63, m | — |
| 5 | 2.00, ddd, 13.2, 10.8, 3.6 | 2.07, td, 12.1, 2.4 | 2.03, m | 2.24, dd, 11.5, 2.5 | 2.27 | — | 2.03, m | 2.76, t, 8.9 |
| 6α | 1.60, dd, 13.2, 3.6 | 1.76–1.85, m | 1.78, m | 5.74, br d, 11.0 | 5.51 | 6.62, d, 5 | 5.44, d, 4.5 | 4.08, t, 9.9 |
| 6β | 1.35, ddd, 13.2, 13.2, 10.8 | 1.25–1.37, m | 1.64, m | |||||
| 7α | — | 1.86–1.97, m | 2.14, m | 5.80, ddd, 11.0, 5.3, 2.4 | — | — | — | 2.03–2.09, m |
| 7β | 2.12, ddd, 10.8, 10.8, 4.2 | 1.15–1.27, m | — | — | — | — | — | — |
| 8α | 1.72, ddd, 13.8, 7.2, 4.2, 3.6 | — | 1.72, m | 1.98, m | 2.54 | — | 2.17, m | 1.99–2.07, m |
| 8β | 1.49, dddd, 13.8, 13.8 10.8, 4.2 | 2.13–2.22, m | 1.86, m | 2.30, m | 2.29 | — | 1.30–1.39, m | |
| 9α | 1.89, ddd, 13.8, 7.2, 4.2 | 4.77–4.81, m | 1.66, m | 1.61, ddd, 14.0, 9.5, 1.8 | 2.81 | — | 1.76, m | 1.87–1.94, m |
| 9β | 1.61, ddd, 13.8, 13.8, 3.6 | — | 1.78, m | 1.83, ddd, 14.0, 9.5, 2.0 | 2.41 | — | 1.58–1.67, m | |
| 10α | — | 2.47–2.57, m | — | — | — | 3.40, dd, 11, 6 | 3.44, dd, 10.5, 6.0 | — |
| 10β | — | 1.95, d, 13.1 | — | — | — | — | — | — |
| 11 | — | — | — | 1.19, s | 1.57 | — | 1.54, m | 2.17–2.25, m |
| 12α | 4.65, br s | 4.79–4.84, m | 5.03, br s | 1.26, s | 0.9 | 0.92, d, 7 | 0.89, d, 7.0 | — |
| 12β | 4.58, br s | 5.01, br s | — | |||||
| 13 | 1.70, s | 1.68, s | 4.56, s | — | 0.92 | 0.93, d, 7 | 0.89, d, 7.0 | 1.23, d, 6.9 |
| 14α | 1.13, s | 3.62, dd, 10.7, 4.5 | 1.30, s | — | 4.01 | 9.37, s | 4.49, m | 1.13, s |
| 14β | 3.38, dd, 10.7, 7.6 | — | ||||||
| 15α | 1.20, s | — | 1.25, s | — | 1.27 | 1.04, s | 0.97, s | 5.11, s |
| 15β | — | — | 4.95, s | |||||
| 2′ | — | — | 2.22, m | — | — | — | 2.07, s | — |
| 3′ | — | — | 2.11, m | — | — | — | — | — |
| 4′ | — | — | 0.97, d, 6.6 | — | — | — | — | — |
| 5′ | — | — | 0.97, d, 6.6 | — | — | — | — | — |
Table 3.
1H-NMR data of compounds 9–17.
| NO. | 9 [12] | 10 [13] | 11 [14] | 12 [7] | 13 [15] | 14 [16] | 15 [17] | 16 [19] | 17 [15] |
|---|---|---|---|---|---|---|---|---|---|
| CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | C5D5N | CDCl3 | CDCl3 | |
| 1 | 2.89, dt, 8.0, 4.5 | 3.02, br ddd, 12.5, 8, 8 | — | 2.69–2.76, m | — | 2.76, m | 2.79, m | 2.83, m | — |
| 2α | 1.95, m | 1.82, m | — | 1.92–2.06, m | — | 1.75, m | 2.02, m | 2.34, m | — |
| 2β | 1.87, m | — | — | 2.15, m | 2.24, m | — | |||
| 3α | 2.49, m | 1.82, m | — | 4.21–4.28, m | 3.71, m | 3.72, q, 6.4 | 3.91, m | 4.54, t, 6.0 | 4.54, t, 7.4 |
| 3β | 1.92, m | — | — | — | — | — | — | — | |
| 4 | — | — | — | 2.31–2.39, m | — | 1.85, m | 2.14, m | — | — |
| 5 | 2.81, br dd, 9.5, 8.0 | 2.38, dd | — | 2.22–2.31, m | — | 1.75, m | 1.98, m | — | — |
| 6 | 3.93, t, 9.5 | 4.06, dd | 3.98, t, 9 | 4.23, t, 9.7 | 3.93, t, 10 | 3.93, t, 9.7 | 3.93, m | 4.13, t, 9.0 | 4.02, t, 9.5 |
| 7 | 2.12, m | 2.77, ddddd | — | 2.26–2.33, m | — | 1.85, m | 2.18, ddd, 10.4, 10.0, 9.6 | — | — |
| 8α | 1.94, m | 2.26, dddd | — | 1.83–1.91, m | — | 2.10, m | — | 1.41, m | — |
| 8β | 1.32, m | 1.38, dddd | — | 1.33–1.42, m | — | 1.23, m | 3.83, m | — | |
| 9α | 2.22, dd, 12.0, 7.0 | 2.69, ddd | — | 2.62–2.68, m | — | 2.60, dt, 13.0, 4.0 | 2.35, dd, 12.8, 11.8 | — | — |
| 9β | 2.05, dt, 12.0, 5.0 | 1.94, br ddd | — | 1.78–1.88, m | — | 1.85, m | 3.00, dd, 12.8, 4.8 | — | — |
| 11 | 2.49, m | — | — | 2.66–2.71, m | 2.12, m | 2.20, m | 2.77, m | — | 2.14, qd, 11, 6.9 |
| 13α | 1.25, d, 7.0 | 6.24, d | 6.25, d, 3.5 | 1.18, d, 7.8 | 1.21, d, 6.8 | 1.29, d, 7.0 | 1.68, d, 7.2 | 1.16, d, 6.0 | 1.23, d, 6.9 |
| 13β | 5.53, d | 5.51, d, 3.2 | |||||||
| 14α | 4.89, br s | 5.01, br s | 4.91, s | 5.00, s | 4.95, s | 4.92, s | 4.99, br s | 4.95, s | 4.96, s |
| 14β | 4.79, br s | 4.97 br s | 4.84, s | 4.95, s | 5.09, br s | 4.93, s | 4.93, s | ||
| 15α | 5.21, d, 2.0 | 1.32, s | 5.29, br s | 0.97, d, 7.2 | 1.24, d, 6.3 | 1.20, d, 7.3 | 1.44, d, 6.4 | 5.40, s | 5.38, t, 1.9 |
| 15β | 5.06, d, 2.0 | 5.09, br s | 5.31, s | 5.29, t, 1.9 |
Table 4.
1H-NMR data of compounds 18–25.
| NO. | 18 [20] | 19 [21] | 20 [22] | 21 [23] | 22 [24] | 23 [25] | 24 [26] | 25 [27] |
|---|---|---|---|---|---|---|---|---|
| CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CD3OD | C5D5N | |
| 1 | — | 2.83, br ddd, 11.1, 10.6, 6.6 | 2.91, m | 2.92, m | 1.9–2.2, m | — | 2.98, ddd, 10.6, 9.0, 7.1 | — |
| 2α | — | 2.21, ddd, 13.0, 6.6, 6.6 | 1.71, m | 2.25, m | 1.9–2.2, m | — | 1.73, ddd, 12.6, 11.0, 9.0 | — |
| 2β | — | 1.79, ddd, 13.0, 10.6, 8.8 | 2.29, m | 1.77, m | 2.32, ddd, 14, 8, 8 | — | 2.11, ddd, 12.6, 7.2, 7.0 | — |
| 3 | 4.53, t, 7.6 | 3.75, ddd, 8.8, 8.8, 6.6 | 4.52, m | 4.55, br d, 8 | 4.53, br t, 8, 1.9 | 4.53, m | 4.50, br dd, 9.1, 7.5 | — |
| 4 | — | 1.90, m | — | — | — | — | — | — |
| 5 | — | 1.93, m | 2.88, m | 2.85, m | 2.72, br t, 9, 1.9 | — | 2.88, m | — |
| 6 | 4.12, dd, 10.0, 9.2 | 4.27, dd, 9.7, 9.7 | 4.12, dd, 9.9, 9.9 | 4.07, t, 10 | 4.25, t, 9.9 | 4.06, t, 9.2 | 4.17, dd, 10.4, 8.9 | — |
| 7 | — | 2.80, m | 2.35, m | 2.00, q, 10 | 1.9–2.2, m | — | 2.89, m | — |
| 8α | — | 4.34, br m | — | — | 1.4–1.8, m | — | — | — |
| 8β | — | — | 3.78, m | 3.78, t, 9, 4.5 | — | 3.90, ddd, 9.5, 5.0, 4.8 | — | |
| 9α | — | 2.37, dd, 13.5, 3.4 | 2.10, m | 2.72, dd, 14, 5 | 1.4–1.8, m | — | 2.70, dd, 13.6, 5.1 | — |
| 9β | — | 2.69, dd, 13.5, 4.9 | 2.72, dd, 12.5, 4.5 | 2.21, dd, 14, 7 | — | 2.27, dd, 13.6, 4.6 | — | |
| 11 | — | — | 2.87, m | 2.58, m | 2.25, dq, 12, 6.8 | — | — | — |
| 13α | 1.14, d, 8.0 | 6.37, d, 3.6 | 1.29, d, 7.7 | 1.42, d, 3p, 7 | 1.22, d, 6.8 | 6.16, d, 3.2 | 6.15, dd, 3.2, 1.3 | 6.49, d, 3.5 |
| 13β | 5.61, d, 3.2 | 5.46, d, 3.2 | 6.12, dd, 3.5, 1.3 | 5.68, overlapped | ||||
| 14α | 4.94, s | 5.16, br s | 5.08, br s | 5.11, br | 1.17, s | 4.95, br s | 5.08, d, 1.7 | 5.70, s |
| 14β | 4.91, s | 5.03, br s | 5.01, br s | 5.00, br | 4.90, br s | 4.97, d, 2.0 | ||
| 15α | 5.38, s | 1.22, d, 6.3 | 5.42, dd, 1.7, 1.6 | 5.41, t, 1 | 5.24, t, 1.9 | 5.42, br s | 5.37, d, 2.3 | 5.68, overlapped |
| 15β | 5.30, s | 5.32, dd, 1.7, 1.6 | 5.32, t, 1 | 5.20, t, 1.9 | 5.29, br s | 5.30, d, 2.1 | 5.25, br s |
Table 5.
1H-NMR data of compounds 26–33.
| NO. | 26 [28] | 27 [7] | 28 [29] | 29 [30] | 30 [31] | 31 [32] | 32 [33] | 33 [34] |
|---|---|---|---|---|---|---|---|---|
| CDCl3 | CDCl3 | CD3OD | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | |
| 1 | 3.12, m | 3.08, td, 8.5, 3.1 | 3.10, td, 8.5, 2.0 | 3.05, ddd, 8.4, 8.0, 3.1 | 3.10, t, 9.0 | 3.12, ddd | 2.89, m | 2.89–2.94, m |
| 2α | 2.25, c | 2.50–2.55, m | 2.16, t, 8.5 | 2.60, dd, 19.3, 8.4 | 2.48, d, 16.8 | 2.68, dd | 2.46, m | 1.98–2.03, m |
| 2β | 2.25, c | 2.41–2.50, m | 2.54, ddd, 19.3, 3.1, 1.4 | 2.55, dd, 16.8, 9.0 | 2.56, dd | 1.74–1.80, m | ||
| 3 | — | — | — | — | — | — | 5.54, m | 5.56, dd, 6.3, 2.1 |
| 4 | — | 2.31–2.38, m | 2.30, d, 7.5 | 2.33, ddq, 10.3, 6.9, 1.4 | 2.30, m | — | — | — |
| 5 | 2.5, c | 2.78–2.85, m | 2.31, m | 2.28, ddd, 10.3, 9.2, 8.0 | 2.91, q, 9.0 | 3.27, tdd | 2.83, m | 2.81–2.86, m |
| 6 | 3.93, t, 9 | 4.06, t, 9.6 | 4.40, 8.8 | 4.55, dd, 9.2, 9.2 | 3.98, t, 9.0 | 4.01, t | 4.08, t, 9.8 | 4.10, t, 9.7 |
| 7 | 2.05, q, 10 | 2.53–2.61, m | 3.00, dddd, 8.0, 8.0, 3.0, 3.0 | 3.16, dddd, 9.2, 3.5, 3.0, 2.0 | 3.01, m | 3.03, m | 2.67, m | 2.37–2.42, m |
| 8α | — | 1.93–2.02, m | 2.35, m | 4.46, br m | 1.47, m | 3.03, m | — | 1.87–1.94, m |
| 8β | — | 1.38–1.49, m | 1.48, m | 2.33, m | 1.46, m | — | 1.37–1.46, m | |
| 9α | 2.82, dd, 13, 6 | 2.55–2.63, m | 2.63, m | 2.69, dd, 13.9, 3.0 | 2.22, m | 2.20, m | 2.46, m | 2.44–2.49, m |
| 9β | 2.25, c | 2.03–2.12, m | 2.22, m | 2.50, dd, 13.9, 4.2 | 2.61, m | 2.60, m | ||
| 11α | — | 2.71–2.80, m | — | — | — | — | — | 2.66–2.72, m |
| 11β | 2.5, c | — | — | — | — | — | — | — |
| 13α | 1.44, d, 7 | 1.19, d, 7.7 | 6.30, d, 3.5 | 6.45, d, 3.5 | 5.57, d, 3.6 | 6.30, d | 1.15, d, 7.8 | 1.17, d, 7.8 |
| 13β | 5.58, d, 3.5 | 5.68, d, 3.0 | 6.29, d, 3.6 | 5.58, d | ||||
| 14α | 5.06, br | 4.93, s | 5.02, br s | 5.09, br s | 4.66, brs | 4.94, s | 4.89, d, 9.0 | 4.91, s |
| 14β | 4.76, br | 4.63, s | 4.69, br s | 4.84, br s | 4.98, brs | 4.60, s | 4.92, s | |
| 15α | 1.24, d, 7 | 3.84, dd, 8.9, 3.1 | 1.28, d, 6.5 | 1.28, d, 6.9 | 3.71, dd, 9.0, 3.0 | 6.25, dd | 5.25, t, 2.0 | 5.27, t, 2.1 |
| 15β | 3.61, dd, 8.9, 3.1 | 3.98, dd, 9.0, 3.0 | 5.87, dd | 5.39, t, 2.0 | 5.42, t, 2.2 | |||
| 1′ | — | 3.30, s | — | — | 3.48, q, 7.2 | — | — | — |
| 2′ | — | — | — | — | 1.14, t, 7.2 | — | — | 2.24, dd, 7.1, 1.7 |
| 3′ | — | — | — | — | — | — | — | 2.09–2.17, m |
| 4′ | — | — | — | — | — | — | 0.96, d, 6.6 | 0.98, d, 6.6 |
| 5′ | — | — | — | — | — | — | 0.96, d, 6.6 | 0.98, d, 6.6 |
Table 6.
1H-NMR data of compounds 34–42.
| NO. | 34 [35] | 35 [36] | 36 [8] | 37 [37] | 38 [37] | 39 [32] | 40 [38] | 41 [7] | 42 [11] |
|---|---|---|---|---|---|---|---|---|---|
| CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | |
| 1 | 2.89, m | 2.93, m | 2.95, q, 8.4 | — | 2.18, m | 2.94, m | — | 2.44–2.52, m | 2.90, q, 8.0 |
| 2α | 2.46, m | 2.48, m | 2.44, m | 2.23, m | 2.67, m | 1.78, m | 2.43, br dd, 14, 8 | 2.25–2.37, m | 2.49, m |
| 2β | 1.79, m | 1.79, m | 1.77, m | 1.51, m | 2.18, dd, 14, 7 | 1.52–1.62, m | 1.80, m | ||
| 3 | 5.55, dd, 8, 6 | 5.57, ddt, 8, 6, 2 | 5.56, m | 5.72, m | 5.54, m | 5.56, m | 5.75, tt, 8, 7, 1.5 | 5.50, t, 6.7 | 5.56, m |
| 5 | 1.93, m | 2.85, m | 2.82, m | 2.66, d, 11.5 | 2.55, m | 2.85, m | 2.80, br d, 10, 1.5 | 2.89–2.97, m | 2.04, m |
| 6 | 3.99, t, 9 | 4.11, t, 10 | 4.00, t, 9.9 | 3.88, t, 10.5 | 4.07, t, 10.0 | 4.06, dd, 16.9, 7.6 | 3.91, t, 10 | 4.22, t, 9.5 | 4.39, t, 10.0 |
| 7 | 2.81, m | 2.42, m | 2.01, m | 1.69, m | 1.76, m | 2.85, m | 3.05, ddddd, 10, 10, 4, 3.5, 3 | 2.92–3.01, m | 2.76, t, 9.5 |
| 8α | 2.11, m | 1.92, m | — | 1.45, m | 2.40, m | — | 2.30, m, 4, 4 | 2.24–2.33, m | 1.80, m |
| 8β | 1.31, m | 1.42, m | 3.77, m | 1.97, m | — | 1.45, m, 10, 9 | 1.44–1.55, m | ||
| 9α | 2.49, m | 2.52, m | 2.71, dd, 13.1, 5.0 | 1.45, m | 5.54, m | 2.46, m | 2.63, ddd, 12, 9, 4 | 1.88–1.98, m | 2.48, m |
| 9β | 2.03, m | 2.01, m | 2.23, m | — | 2.30, m | 1.78, dt, 14.3, 5.0 | 2.04, m | ||
| 11 | 2.20, m | 2.69, p, 8 | 2.57, m | 2.32, m | 2.29, m | — | — | — | — |
| 13α | 1.22, d, 7 | 1.16, d, 8 | 1.42, d, 7.0 | 1.25, m | 1.23, d, 7.0 | 5.49, d, 3.1 | 6.21, d, 3.5 | 6.27, d, 3.0 | 3.79, d, 12.0 |
| 13β | 6.21, d, 3.5 | 5.49, d, 3 | 5.55, d, 3.0 | 3.65, d, 11.0 | |||||
| 14α | 4.91, s | 4.91, br s | 5.05, s | 1.35, s | 1.80, br s | 4.97, d, 6.4 | 5.20, br s | 2.75, d, 4.4 | 4.93, d, 6.5 |
| 14β | 4.92, br s | 4.99, s | 5.09, br s | 2.54, d, 4.4 | |||||
| 15α | 5.26, brt, 2 | 5.28, br t, 2 | 5.43, t, 2.2 | 5.50, s | 5.44, br s | 5.27, t, 2.1 | 5.53, t, 1.5 | 5.45, s | 5.41, br t, 2.0 |
| 15β | 5.38, brt, 2 | 5.41, br t, 2 | 5.28, t, 2.2 | 5.41, s | 5.33, br s | 5.45, t, 2.0 | 5.36, t, 1.5 | 5.25, s | 5.30, br t, 2.0 |
| 2′α | 5.71, s | 5.72, br s | 2.23, m | 2.17, m | 2.55, m | — | 2.59, qq, 7, 7 | 5.69, s | 2.23, dd, 7.5, 2.0 |
| 2′β | 2.18, m | — | |||||||
| 3′ | — | — | 2.13, m | 2.09, m | 2.09, m | — | — | — | 2.13, m |
| 4′ | 1.89, s | 1.91, br s | 0.97, d, 6.6 | 0.95, d, 6.5 | 0.95, d, 6.5 | 0.96, d, 6.6 | 1.20, d | 1.91, s | 0.97, d, 6.5 |
| 5′ | 2.17, s | 2.19, br s | 0.97, d, 6.6 | 0.95, d, 6.5 | 0.95, d, 6.5 | 0.96, d, 6.6 | 1.19, d | 2.18, s | 0.97, d, 6.5 |
Table 7.
1H-NMR data of compounds 43–51.
| NO. | 43 [8] | 44 [37] | 45 [37] | 46 [37] | 47 [37] | 48 [37] | 49 [37] | 50 [39] | 51 [37] |
|---|---|---|---|---|---|---|---|---|---|
| CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | |
| 1 | 2.90, m | 2.86, q, 8.5 | 2.89, m | 2.90, q, 8.5 | 2.90, q, 8.0 | 2.90, q, 3.0 | 2.89, m | 2.88, q, 8.4 | 2.13, m |
| 2α | 2.21, m | 2.44, m | 2.57, dt, 13.0, 4.0 | 2.46, m | 2.03, m | 2.02, m | 2.57, dt, 13.0, 4.0 | 2.44, m | 2.44, m |
| 2β | 1.84, m | 1.76, m | 1.96, m | 1.78, m | 1.80, m | 1.82, m | 1.99, m | 1.76, m | |
| 3 | 5.61, t, 7.6 | 5.54, m | 5.54, m | 5.55, m | 5.57, m | 5.57, m | 5.56, m | 5.56, m | 5.56, m |
| 5 | 2.62, m | 2.71, t, 9.5 | 2.89, m | 2.73, t, 9.5 | 2.80, t, 9.5 | 2.82, t, 9.2 | 2.89, m | 2.75, t, 9.2 | 2.79, t, 9.5 |
| 6 | 4.81, d, 11.2 | 4.29, t, 9.5 | 4.01, t, 9.5 | 4.36, t, 9.5 | 4.08, t, 10.0 | 4.05, t, 9.5 | 3.91, t, 9.5 | 4.37, t, 9.2 | 4.02, t, 9.5 |
| 7 | — | 1.87, m | 2.35, m | 2.07, m | 2.03, m | 2.13, m | 2.34, m | 1.94, m | 2.45, m |
| 8α | 3.05, m | 2.10, m | 2.10, m | 2.12, m | 2.15, m | 2.13, m | 2.12, m | 1.64, m | 2.38, q, 8.0 |
| 8β | 2.50, m | 1.39, m | 1.30, m | 1.39, m | 1.35, m | 1.35, m | 1.81, m | ||
| 9α | 2.52, m | 2.44, m | 2.49, dt, 14.0, 8.0 | 2.46, m | 2.47, m | 2.49, m | 2.50, dt, 14.0, 8.0 | 2.03, m | 2.03, m |
| 9β | 2.17, m | 2.01, m | 1.75, m | 2.07, m | 1.75, m | 2.50, m | 1.78, dt, 14.0, 6.5 | ||
| 11 | — | — | — | — | 2.46, m | 2.49, m | — | — | 2.89, q, 8.0 |
| 13α | 4.38, dd, 16.6, 13.3 | — | 1.76, m | 1.96, m | 1.80, m | 1.82, m | 1.76, m | 2.62, d, 16.7 | 3.69, dd, 4.5, 4.5 |
| 13β | 1.64, dd, 15.0, 2.5 | 1.68, m | 1.78, m | 1.69, m | 1.67, m | 1.57, dd, 15.0, 2.5 | 2.80, d, 16.7 | 3.57, dd, 5.0, 5.0 | |
| 14α | 5.01, s | 4.91, s | 4.93, s | 4.93, s | 4.93, s | 4.92, s | 4.93, s | 4.93, s | 4.92, s |
| 14β | 4.94, s | 4.88, s | 4.89, s | 4.91, s | 4.90, s | 4.91, s | 4.90, s | ||
| 15α | 5.42, br s | 5.41, br t, 1.5 | 5.39, br t, 2.0 | 5.42, br t, 2.0 | 5.39, br t, 2.0 | 5.40, t, 2.0 | 5.41, br t, 2.0 | 5.43, br s | 5.37, br t, 2.0 |
| 15β | 5.27, br t, 1.5 | 5.29, br t, 2.0 | 5.29, br t, 2.0 | 5.27, br t, 2.0 | 5.26, t, 2.0 | 5.29, br t, 2.0 | 5.30, br s | 5.25, br t, 2.0 | |
| 16 | — | 4.97, m | 4.27, m | 4.16, m | 3.97, m | 4.21, m | 4.31, m | — | — |
| 17 | — | 1.22, d, 6 | 1.23, d, 6.0 | 1.30, d, 8.5 | 1.24, d, 6.0 | 1.24, d, 6.0 | 1.23, d, 6.0 | 2.32, s | — |
| 2′ | 2.23, m | 2.21, dd, 7.5, 1.5 | 2.23, dd, 7.5, 2.0 | 2.22, dd, 7.5, 2.0 | 2.22, dd, 8.0, 2.0 | 2.23, dd, 8.0, 1.5 | 2.22, dd, 8.0, 2.0 | 2.23, dd, 7.1, 1.7 | 2.23, dd, 8.0, 1.5 |
| 3′ | 2.11, m | 2.15, m | 2.10, m | 1.96, m | 2.15, m | 2.13, m | 2.12, m | 2.12, m | 2.13, m |
| 4′ | 0.97, d, 6.6 | 0.95, d, 6.5 | 0.97, d, 6.5 | 0.97, d, 6.5 | 0.97, d, 6.5 | 0.97, d, 6.5 | 0.97, d, 6.5 | 0.97, d, 6.5 | 0.96, d, 7.0 |
| 5′ | 0.97, d, 6.6 | 0.95, d, 6.5 | 0.97, d, 6.5 | 0.97, d, 6.5 | 0.97, d, 6.5 | 0.97, d, 6.5 | 0.97, d, 6.5 | 0.97, d, 6.5 | 0.96, d, 7.0 |
Note: The 1H-NMR data of 2″α, 2″β, 3″, and 4″ for 51 were recorded as 2.30, m; 1.32, m; 2.03, m; and 3.45, m, respectively.
Table 8.
1H-NMR data of compounds 52–57.
| NO. | 52 [22] | 52 [22] | 53 [22] | 54 [40] | 55 [41] | 56 [42] | 57 [12] | 57 [22] |
|---|---|---|---|---|---|---|---|---|
| CD3OD | C5D5N | CD3OD | CD3OD | C5D5N | DMSO | CDCl3 | C5D5N | |
| 1 | 2.96, m | 2.75, m | 3.00, m | 2.97, q, 8.4 | — | 2.76, m | 2.80, br t, 10.0 | 2.77, m |
| 2α | 2.35, m | 2.22, m | 2.30, m | 2.36, m, overlapped | — | 1.69, dd, 12, 10 | 2.39, m | 2.30, m |
| 2β | 1.96, m | 1.81, m | 1.96, m | 1.96, dt, 14.0, 6.9 | — | 2.14, dd, 13.5, 7.0 | 1.98, m | 1.92, m |
| 3 | 4.62, m | 4.41, m | 4.61, m | 4.63, dd, 7.8, 5.9 | 4.84, br t, 7 | 3.56, dd, 15.5, 8.0 | 4.65, br dd, 6.0, 6.0 | 4.81, dd, 7.3, 1.5 |
| 4 | — | — | — | — | — | 1.85, m | — | — |
| 5 | 2.79, dd, 9.5, 9.5 | 2.68, m | 2.83, dd, 9.9, 9.9 | 2.75, dd, 9.9, 8.4 | — | 1.91, m | 3.01, dd, 17.5, 8.5 | 2.74, m |
| 6 | 4.33, dd, 9.9, 9.5 | 4.33, dd, 9.9, 9.5 | 4.38, dd, 10.6, 9.9 | 4.23, t, 9.9 | — | 3.93, t, 10 | 4.28, dd, 9.0, 9.0 | 4.26, m |
| 7 | 2.43, m | 2.33, m | 2.36, m | 2.39, m, overlapped | — | 2.78, m | 2.89, m | 2.70, m |
| 8α | 1.88, m | 1.58, m | 2.16, tt, 10.0, 5.0 | — | 2.25, m | 2.28, m | 1.97, m | |
| 8β | 1.44, m | 1.20, m | 3.72, m | 1.39, m | — | 1.24, m | 1.46, m | 1.16, m |
| 9α | 2.03, m | 2.10, m | 2.15, dd, 12.6, 8.0 | 2.08, ddd, 13.7, 9.3, 5.1 | — | 2.54, m | 2.21, m | 2.10, m |
| 9β | 2.53, m | 2.36, m | 2.71, dd, 12.6, 5.0 | 2.52, m, overlapped | — | 1.97, m | 2.52, ddd, 13.0, 6.5, 6.5 | 2.37, m |
| 11 | 2.67, m | 2.64, m | 2.77, m | 2.54, dt, 11.8, 3.7 | — | — | — | — |
| 13α | 1.12, dd, 7.7, 1.5 | 1.05, d, 7.7 | 1.23, d, 7.7 | 3.71, dd, 9.9, 3.7 | 1.65, d, 7 | 5.59, d, 2 | 6.12, d, 3.0 | 5.53, br d, 1.5 |
| 13β | 3.63, dd, 9.9, 3.7 | 6.00, d, 2.5 | 5.57, d, 3.0 | 5.87, br d, 1.5 | ||||
| 14α | 4.99, s | 5.00, s | 5.09, s | 4.99, s | 5.14, br s | 4.95, d, 5 | 5.01, br s | 5.02, d, 1.1 |
| 14β | 4.91, s | 4.83, s | 4.98, s | 4.91, s | 5.01, br s | 4.99, d, 5 | 4.94, br s | 4.83, d, 1.1 |
| 15α | 5.40, s | 5.82, br s | 5.36, d, 1.3 | 5.42, d, 1.7 | 5.87, br s | 1.15, d, 10 | 5.44, br s | 6.23, d, 3.4 |
| 15β | 5.31, s | 5.50, br s | 5.32, d, 1.3 | 5.30, d, 1.7 | 5.54, br s | 5.35, br d, 1.0 | 5.38, d, 3.4 | |
| 1′ | 4.46, d, 7.7 | 5.04, d, 7.9 | 4.44, d, 7.7 | 4.45, d, 7.8 | 5.06, d, 7 | 4.19, d, 7.5 | 4.47, d, 7.5 | 5.05, d, 7.9 |
| 2′ | 3.24, m | 3.94, m | 3.20, m | 3.23, m | — | 2.95, m | 3.20–3.40, m | 3.96, m |
| 3′ | 3.36, m | 4.24, m | 3.31, m | 3.36, m | — | 3.16, m | 3.87, dd, 10.0, 10.0 | 4.24, m |
| 4′ | 3.28, m | 4.06, m | 3.26, m | 3.28, m | — | 3.04, m | 3.20–3.40, m | 4.08, m |
| 5′ | 3.28, m | 4.22, m | 3.24, m | 3.27, m | — | 3.08, m | 3.67, dd, 12.0, 5.5 | 4.22, m |
| 6′α | 3.66, dd, 11.9, 5.0 | 4.36, m | 3.64, dd, 12.0, 6.0 | 3.88, dd, 12.0, 1.9 | — | 3.42, dd, 11.5, 6.0 | 3.20–3.40, m | 4.40, dd, 11.8, 5.5 |
| 6′β | 3.86, br d, 11.9 | 4.56, dd, 11.9, 2.0 | 3.87, dd, 12.0, 2.0 | 3.67, dd, 12.0, 5.2 | — | 3.65, dd, 9.5, 4.5 | 4.57, dd, 11.8, 2.4 |
Note: The 1H-NMR data of 1″ and 2″ for 54 were recorded as 3.51, q, 7.0 and 1.17, t, 7.0.
Table 9.
1H-NMR data of compounds 58–61.
| NO. | 58 [43] | 59 [44] | 60 [45] | 61 [46] | NO. | 58 [43] | 59 [44] | 60 [45] | 61 [46] |
|---|---|---|---|---|---|---|---|---|---|
| CD3OD | C5D5N | CD3OD | DMSO | CD3OD | C5D5N | CD3OD | DMSO | ||
| 1 | 2.99, dd, 16.8, 8.4 | — | 2.99, t, 9.1 | — | 15α | 5.43, br s | 5.45, br s | 5.32, d, 1.1 | 5.38, br s |
| 2α | 2.33, m | — | 2.26, m | — | 15β | 5.35, br s | 5.27, d, 1.1 | 5.20, br s | |
| 2β | 1.96, m | — | 1.98, m | — | 1′ | 4.45, d, 7.2 | — | 4.66, d, 8.1 | 4.40, d, 7.8 |
| 3 | 4.63, m | — | 4.64, m | 4.50, m | 2′ | 3.87, m | 5.65, br t, 10 | 4.85, m | — |
| 5 | 2.78, dd, 9.6, 9.6 | — | 2.85, br d, 9.4 | — | 3′ | 3.37, t, 8.4 | — | 3.58, t, 8.6 | — |
| 6 | 4.26, dd, 9.6, 9.3 | — | 4.04, t, 10.2 | 4.13, dd, 10.2, 8.9 | 4′ | 3.28, m | — | 3.38, m | — |
| 7 | 2.89, m | — | 2.34, dd, 8.1, 2.1 | — | 5′ | 3.26, m | — | — | — |
| 8α | 2.26, m | — | 3.50, m | — | 6′α | 3.86, dd, 12.0, 2.4 | — | 3.91, dd, 12.0, 2.1 | — |
| 8β | 1.45, m | — | — | 6′β | 3.68, dd, 12.0, 5.4 | — | 3.70, dd, 12.0, 5.8 | — | |
| 9α | 2.18, m | — | 2.06, m | — | 1″ | — | — | — | 4.32, d, 7.8 |
| 9β | 2.50, m | — | 2.58, dd, 11.5, 4.7 | — | 2″ | 7.56, d, 7.8 | 6.9–7.5, m | 7.02, d, 2.0 | — |
| 11 | — | — | 2.72, t, 7.8 | — | 3″ | 7.37, d, 7.8 | — | — | — |
| 13α | 6.09, d, 3.0 | — | 1.09, d, 7.7 | 6.02, d, 3.5 | 5″ | 7.37, d, 7.8 | 6.9–7.5, m | 6.78, d, 8.1 | — |
| 13β | 5.56, d, 3.0 | — | 5.61, d, 3.2 | 6″ | 7.56, d, 7.8 | 6.9–7.5, m | 6.94, dd, 8.2, 1.9 | — | |
| 14α | 5.01, s | 5.13, br s | 5.11, br s | 4.91, br s | 2‴ | 6.48, d, 15.6 | 6.37, d, 15 | 6.24, d, 15.9 | — |
| 14β | 4.91, s | 4.90, br s | 4.88, br s | 3‴ | 7.58, d, 15.6 | 7.82, d, 15 | 7.55, d, 15.9 | — |
Table 10.
1H-NMR data of compounds 61–63.
| NO. | 61 [25] | 62 [46] | 62 [25] | 63 [46] | NO. | 61 [25] | 62 [46] | 62 [25] | 63 [46] |
|---|---|---|---|---|---|---|---|---|---|
| CD3OD | DMSO | DMSO | DMSO | CD3OD | DMSO | DMSO | DMSO | ||
| 1 | — | — | 4.88, d, 7.6 | 2.92, m | 1′ | 4.54, d, 8.0 | 4.40, d, 7.8 | 5.01, d, 8.0 | 4.40, d, 7.8 |
| 2α | — | — | — | 2.25, m | 2′ | — | — | — | 3.43, m |
| 2β | — | — | — | 1.77, m | 3′ | — | — | — | 3.40, m |
| 3 | 4. 64, m | 4.50, t, 7.4 | 4.64, m | 4.51, br s | 4′ | — | — | — | 3.21, m |
| 5 | — | — | — | 2.80, m | 5′ | — | — | — | 3.18, m |
| 6 | 4.27, t, 9.6 | 4.13, t, 9.5 | 4.11, t, 9.6 | 4.14, t, 9.6 | 6′α | — | — | — | 3.36, m |
| 7 | — | — | — | 2.91, m | 6′β | — | — | — | 3.29, m |
| 8α | — | — | — | 2.23, m | 1″ | 4.51, d, 8.0 | 4.47, d, 7.8 | 4.98, d, 7.6 | 4.46, d, 7.7 |
| 8β | — | — | — | 1.35, m | 2″ | — | — | — | 3.28, m |
| 9α | — | — | — | 2.11, m | 3″ | — | — | — | 3.47, m |
| 9β | — | — | — | 2.41, m | 4″ | — | — | — | 3.21, m |
| 11 | — | — | — | — | 5″ | — | — | — | 3.28, m |
| 13α | 6.10, d, 3.6 | 6.02, d, 3.5 | 6.01, d, 3.6 | 6.02, d, 3.5 | 6″α | — | — | — | 3.36, m |
| 13β | 5.57, d, 3.6 | 5.61, d, 3.1 | 5.59, d, 3.6 | 5.61, d, 3.5 | 6″β | — | — | — | 3.29, m |
| 14α | 5.02, br s | 4.91, s | 5.19, br s | 4.91, br s | 1‴ | — | 4.36, d, 7.8 | — | 4.52, d, 7.8 |
| 14β | 4.93, br s | 4.88, s | 5.08, br s | 4.88, br s | 2’—OH | — | — | — | 5.10, d, 4.1 |
| 15α | 5.40, br s | 5.38, br s | 5.36, br s | 5.38, br s | 2″—OH | — | — | — | 5.24, d, 3.2 |
| 15β | 5.35, br s | 5.20, br s | 5.20, br s | 2‴—OH | — | — | — | 5.26, d, 3.4 |
Note: The 1⁗, 2⁗, 3⁗, 4⁗, 5⁗, 6⁗α, and 6⁗β data of compound 63 were 4.36, d, 7.9; 3.05, m; 3.18, m; 3.04, m; 3.24, m; 3.42, m; and 3.29, m.
Table 11.
13C-NMR data of compounds 1–63.
| NO. | 1 [6] | 2 [7] | 3 [8] | 4 [9] | 5 [10] | 6 [10] | 7 [11] | 8 [7] | 9 [12] | 11 [14] | 12 [7] | 13 [15] | 14 [16] | 15 [18] | 15 [17] | 16 [19] | 17 [15] | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CD3OD | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CD3OD | C5D5N | CDCl3 | CDCl3 | ||||
| 1 | 52.7 | 53.1 | 52.1 | 50.6 | 59 | 47.9 | 46.3 | 52.5 | 47.3 | 47.6 | 41.4 | 42.1 | 42.2 | 43.6 | 43 | 43.3 | 43.5 | |||
| 2 | 26.4 | 34.2 | 26.2 | 21.6 | 40 | 39.7 | 39.7 | 26 | 30.5 | 32.5 | 34.8 | 38.3 | 38.3 | 35.5 | 39.6 | 38.6 | 38.7 | |||
| 3 | 40.6 | 30.2 | 39.2 | 40.3 | 27.1 | 27 | 27.3 | 31 | 32.8 | 30.2 | 73.9 | 78.3 | 78.4 | 79.1 | 78.1 | 73.6 | 73.5 | |||
| 4 | 81.9 | 49.4 | 83.5 | 80 | 56.1 | 55 | 55.9 | 152 | 152 | 151.1 | 40.6 | 47 | 47 | 48.2 | 47.9 | 153.3 | 153.2 | |||
| 5 | 53.4 | 58.2 | 49.1 | 51.2 | 51.5 | 53.3 | 51.5 | 51.2 | 52.2 | 52 | 47.8 | 50.6 | 52.9 | 52.3 | 51.5 | 49.7 | 49.5 | |||
| 6 | 32.8 | 24.9 | 29.3 | 130.1 | 141.8 | 159.8 | 135 | 84 | 85.6 | 85.1 | 83.4 | 85.9 | 85.9 | 83.9 | 82.5 | 83.7 | 83.7 | |||
| 7 | 48.6 | 28.9 | 39 | 131.6 | 132.7 | 142 | 133.2 | 48.6 | 42.3 | 45.1 | 46.3 | 52.9 | 50.6 | 60 | 59.2 | 39.3 | 50.8 | |||
| 8 | 31.5 | 45.2 | 32 | 23.5 | 25 | 19.2 | 25.3 | 26.5 | 32.8 | 30.8 | 29.2 | 32.7 | 32.8 | 77.1 | 76.3 | 28.7 | 32.3 | |||
| 9 | 42.6 | 78.5 | 36.6 | 42.7 | 67 | 27.5 | 27.7 | 40.6 | 37.9 | 36.1 | 39.4 | 37 | 37 | 48.9 | 48.9 | 36 | 35.9 | |||
| 10 | 75.5 | 36.4 | 75 | 75 | 213.8 | 75.8 | 76.5 | 74.7 | 150.2 | 149.2 | 148.5 | 149.2 | 149.3 | 146.3 | 146.1 | 149 | 148.8 | |||
| 11 | 153.5 | 146.6 | 149.5 | 22.8 | 33 | 32.8 | 33.1 | 42.8 | 50.1 | 139.7 | 39.7 | 42 | 42.1 | 43.5 | 42.9 | 46.3 | 42 | |||
| 12 | 108.3 | 114.2 | 111.1 | 21.7 | 22.1 | 19.9 | 19.8 | 178.8 | 179 | 169.5 | 180.3 | 178.6 | 178.6 | 181.9 | 179.5 | 179.7 | 179.8 | |||
| 13 | 20.3 | 22.7 | 66.1 | — | 24.7 | 22.1 | 22.1 | 13.2 | 13.5 | 120 | 11.8 | 13 | 13.1 | 16.8 | 19 | 11.4 | 13.1 | |||
| 14 | 23.8 | 65 | 25.4 | — | 34.6 | 193 | 68.7 | 27.1 | 112.1 | 112.5 | 111.8 | 112.5 | 112.5 | 115.1 | 114.2 | 113.4 | 113.5 | |||
| 15 | 26.3 | 180.1 | 32.3 | — | 19.9 | 19.8 | 20 | 107.9 | 109.5 | 109.5 | 8.3 | 14.1 | 18.1 | 18.8 | 17 | 111.4 | 111 | |||
| 1′ | — | — | 173 | — | — | — | 171 | — | — | — | — | — | — | — | — | — | — | |||
| 2′ | — | — | 43.6 | — | — | — | 21 | — | — | — | — | — | — | — | — | — | — | |||
| 3′ | — | — | 25.8 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | |||
| 4′ | — | — | 22.6 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | |||
| 5′ | — | — | 22.6 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | |||
| NO. | 18 [20] | 20 [22] | 21 [18] | 23 [25] | 24 [18] | 24 [18] | 25 [27] | 27 [7] | 28 [29] | 30 [31] | 31 [32] | 32 [33] | 33 [34] | 34 [35] | 35 [36] | 36 [8] | 37 [37] | |||
| CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CD3OD | C5D5N | CDCl3 | CD3OD | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | ||||
| 1 | 43.3 | 43.6 | 44.2 | 43.9 | 45.2 | 46 | 44.7 | 40.1 | 39.8 | 40 | 39.6 | 43.7 | 43.7 | 44.1 | 43.8 | 44.6 | 62.9 | |||
| 2 | 38.6 | 38.6 | 39 | 38.7 | 39.2 | 40 | 39.9 | 45.4 | 44 | 45.7 | 44.6 | 36.2 | 36.2 | 36.6 | 36.3 | 36.7 | 38.1 | |||
| 3 | 73.6 | 73.7 | 73.6 | 73.2 | 73.7 | 73.1 | 73.1 | 217.6 | 219.2 | 217.7 | 204.4 | 74.4 | 74.4 | 73.8 | 73.7 | 74.4 | 74 | |||
| 4 | 153.2 | 152.7 | 153 | 152.7 | 152.4 | 154.3 | 155.2 | 53.3 | 50.6 | 52.9 | 144.4 | 148.9 | 148.9 | 148.8 | 148.8 | 148.3 | 145.1 | |||
| 5 | 49.7 | 49.9 | 50.7 | 45.4 | 51.3 | 51.9 | 50.5 | 45.4 | 47.1 | 44.6 | 48.6 | 50.1 | 50.1 | 50.4 | 50 | 51.2 | 53.1 | |||
| 6 | 83.6 | 78.9 | 79.1 | 83.7 | 79 | 80.9 | 78.8 | 88.5 | 88.8 | 88.8 | 86.8 | 83.7 | 83.7 | 83.9 | 83.8 | 79.1 | 81.8 | |||
| 7 | 39.3 | 53.4 | 56 | 49.6 | 51 | 51.7 | 50.1 | 43.7 | 44 | 44.2 | 44 | 45.7 | 45.7 | 50 | 45.6 | 55.8 | 55.5 | |||
| 8 | 28.7 | 69.9 | 74.9 | 28.8 | 71.9 | 74.1 | 66 | 29.3 | 31.8 | 31.8 | 31.6 | 28.7 | 28.7 | 32.4 | 28.7 | 75.1 | 24.9 | |||
| 9 | 36 | 45 | 44.8 | 32.6 | 41.3 | 42.9 | 44.2 | 38.5 | 38.7 | 38.4 | 38.2 | 36.2 | 36.2 | 36.3 | 36.3 | 45.3 | 37.4 | |||
| 10 | 148.9 | 143.1 | 143.2 | 147.8 | 142.7 | 144.7 | 145.1 | 149.6 | 148.7 | 149.1 | 148.2 | 148.4 | 148.4 | 148.8 | 148.9 | 143.1 | 70 | |||
| 11 | 46.3 | 38.1 | 42 | 139.6 | 138.1 | 140.6 | 137.7 | 39.3 | 138.7 | 139 | 138.6 | 39.2 | 39.2 | 42.1 | 39.2 | 42.1 | 42.4 | |||
| 12 | 179.6 | 179.1 | 178.6 | 170.3 | 169.9 | 172 | 170.1 | 179.8 | 169.9 | 169.9 | not detected | 179.6 | 179.6 | 178.4 | 179.6 | 178.6 | 177.6 | |||
| 13 | 11.4 | 11.2 | 15.9 | 120.2 | 123.2 | 122.9 | 120.9 | 11.6 | 121.3 | 121.2 | 121.4 | 11.4 | 11.4 | 13.2 | 11.3 | 16.1 | 12.4 | |||
| 14 | 113.3 | 115.9 | 116.2 | 115.2 | 117.1 | 117 | 115.9 | 112.6 | 113.1 | 113.1 | 113.6 | 113.4 | 113.4 | 113.5 | 113.2 | 116.4 | 20.7 | |||
| 15 | 111.3 | 112.6 | 112 | 110.9 | 113.2 | 112.1 | 109 | 70.5 | 14.2 | 68.1 | 122.1 | 113.2 | 113.2 | 112.9 | 113.1 | 114.3 | 118.3 | |||
| 1′ | — | — | — | — | — | — | — | 59.3 | — | 66.7 | — | 172.8 | 172.8 | 166.3 | 166.2 | 173 | 172.5 | |||
| 2′ | — | — | — | — | — | — | — | — | — | 14.9 | — | 43.6 | 43.6 | 116 | 115.9 | 43.8 | 43.6 | |||
| 3′ | — | — | — | — | — | — | — | — | — | — | — | 25.7 | 25.7 | 157.3 | 157.2 | 25.9 | 25.7 | |||
| 4′ | — | — | — | — | — | — | — | — | — | — | — | 22.4 | 22.4 | 27.4 | 27.4 | 22.6 | 22.3 | |||
| 5′ | — | — | — | — | — | — | — | — | — | — | — | 22.4 | 22.4 | 20.3 | 20.2 | 22.6 | 22.3 | |||
| NO. | 38 [37] | 39 [33] | 41 [7] | 42 [11] | 43 [8] | 44 [37] | 45 [37] | 46 [37] | 47 [37] | 48 [37] | 49 [37] | 50 [39] | 51 [37] | 52 [22] | 52 [22] | 53 [22] | 54 [40] | |||
| CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CD3OD | C5D5N | CD3OD | CD3OD | ||||
| 1 | 43.9 | 44.6 | 43.1 | 44.2 | 45 | 44.2 | 43.5 | 44.3 | 44.1 | 44.1 | 43.5 | 44.3 | 45.9 | 45.8 | 44 | 46.3 | 45.7 | |||
| 2 | 37.3 | 34.6 | 32.2 | 36.2 | 35.7 | 36.2 | 36 | 36.3 | 36.4 | 36.5 | 36.3 | 36.4 | 36.4 | 38.5 | 37.6 | 38.4 | 38.4 | |||
| 3 | 73.5 | 74.3 | 73.4 | 74.3 | 74.3 | 74.3 | 74.4 | 74.3 | 74.3 | 74.3 | 74.4 | 75.5 | 74.3 | 81.7 | 80.4 | 81.6 | 81.4 | |||
| 4 | 148.4 | 147.8 | 148.7 | 148.1 | 147.1 | 148.5 | 149 | 148.3 | 148.4 | 148.6 | 149.2 | 148.4 | 148.6 | 151.5 | 150.7 | 151 | 151.1 | |||
| 5 | 50.4 | 50.2 | 48.8 | 47.8 | 49.7 | 50.3 | 50 | 50.3 | 50 | 50 | 50.1 | 50.3 | 49.9 | 52 | 50.2 | 52.5 | 51.4 | |||
| 6 | 83.6 | 83.9 | 83.9 | 83.7 | 81.5 | 82.2 | 81.2 | 83.3 | 84.8 | 84.3 | 81 | 83 | 84.2 | 85.7 | 83.6 | 81.2 | 85.1 | |||
| 7 | 49.2 | 45.2 | 44.7 | 50.1 | 165.3 | 52.7 | 55 | 51.9 | 48.9 | 47.9 | 53.4 | 52 | 46.7 | 41.4 | 39.6 | 54.7 | 45.6 | |||
| 8 | 29.9 | 30.6 | 26.8 | 25 | 29.9 | 24.9 | 27.4 | 25.5 | 32.4 | 32.2 | 27.1 | 25.2 | 30.6 | 30 | 28.6 | 70.9 | 33.1 | |||
| 9 | 122.2 | 36.6 | 34 | 34.7 | 30.7 | 34.6 | 36 | 34.4 | 35.9 | 36.1 | 36.1 | 34.6 | 36.4 | 37 | 36 | 46 | 36.1 | |||
| 10 | 137 | 148.2 | 57.3 | 148 | 147.9 | 148.2 | 147.7 | 148.1 | 148.3 | 148.5 | 148 | 148.2 | 148.5 | 151.4 | — | 145.9 | 150.8 | |||
| 11 | 42.2 | 139.5 | 139.3 | 75 | 125.7 | 75.7 | 76.1 | 76.4 | 46.4 | 43.5 | 77.5 | 76.5 | 43.9 | 47.7 | 45.5 | 40.5 | 49.5 | |||
| 12 | 178.3 | 170 | 169.9 | 177.5 | 173.4 | 177.2 | 180.3 | 177.7 | 179.6 | 179 | 177.8 | 176 | 176.5 | 183.1 | 179.6 | 182.5 | 178.9 | |||
| 13 | 12.9 | 120.3 | 121 | 64.3 | 55 | 41.2 | 40.9 | 42.8 | 38.3 | 37.1 | 38 | 44.4 | 40.8 | 12.2 | 11.4 | 11.8 | 67.6 | |||
| 14 | 27.9 | 114.3 | 50.3 | 114 | 114.3 | 113.5 | 113.4 | 113.8 | 113.7 | 113.6 | 113.3 | 114 | 113.5 | 114.2 | 113 | 116.4 | 114 | |||
| 15 | 116.8 | 113.4 | 112.6 | 114.1 | 118.3 | 114 | 114.4 | 114.2 | 113.4 | 113.2 | 113.9 | 114.5 | 113 | 114.2 | 112.2 | 115.7 | 113.2 | |||
| 16 | — | — | — | — | — | 64.8 | 63.5 | 64.9 | 67 | 64.9 | 65.7 | 210.4 | — | — | — | — | — | |||
| 17 | — | — | — | — | — | 24.9 | 24.4 | 24.9 | 24.3 | 23.9 | 24.6 | 32.3 | — | — | — | — | — | |||
| 1′ | 173 | 172.8 | 166.2 | 173 | 173.1 | 173 | 172.9 | 172.9 | 172.8 | 172.8 | 172.8 | 173.1 | 172.8 | 103.3 | 103.8 | 102.5 | 103.2 | |||
| 2′ | 43.6 | 43.6 | 115.9 | 43.6 | 43.8 | 43.6 | 43.6 | 43.6 | 43.6 | 43.6 | 43.6 | 43.8 | 43.6 | 75.7 | 75.3 | 75.8 | 75.3 | |||
| 3′ | 25.8 | 25.8 | 157.9 | 25.8 | 25.9 | 25.7 | 25.7 | 25.8 | 25.8 | 25.8 | 25.7 | 25.9 | 25.8 | 78.7 | 78.6 | 78.8 | 78.2 | |||
| 4′ | 22.4 | 22.4 | 27.6 | 22.4 | 22.6 | 22.4 | 22.4 | 22.4 | 22.4 | 22.4 | 22.4 | 22.5 | 22.4 | 72.2 | 71.7 | 72.4 | 71.8 | |||
| 5′ | 22.4 | 22.4 | 20.5 | 22.4 | 22.6 | 22.4 | 22.4 | 22.4 | 22.4 | 22.4 | 22.4 | 22.6 | 22.4 | 78.4 | 78.4 | 78.4 | 77.9 | |||
| 6′ | — | — | — | — | — | — | — | — | — | — | — | — | — | 63.3 | 62.9 | 63.4 | 62.9 | |||
| NO. | 55 a [41] | 56 b [42] | 57 c [12] | 57 a [22] | 57 d [22] | 58 e [43] | 59 a [44] | 60 e [45] | 61 b [46] | 61 e [25] | 62 b [46] | 62 b [25] | 63 b [46] | NO. | 58 e [43] | 59 a [44] | 60 e [45] | 62 b [46] | 62 b [25] | 63 b [46] |
| 1 | 44.4 | 42.1 | 45.4 | 44.5 | 43.7 | 46.2 | 45.9 | 46.1 | 43.5 | 46.2 | 43.4 | 43.4 | 43.9 | 1″ | 130.9 | 126.7 | 127.8 | 103.9 | 103.9 | 103.9 |
| 2 | 37.9 | 37.3 | 38 | 38 | 37.3 | 38.6 | 37.3 | 37.8 | 36.9 | 38.5 | 36.9 | 36.9 | 37.4 | 2″ | 129 | 114.9 | 115.4 | 72.4 | 72.4 | 73.4 |
| 3 | 80.5 | 86.2 | 80.7 | 80.5 | 79.7 | 81.3 | 79.5 | 80.9 | 83.3 | 81.4 | 83.2 | 83.2 | 79.8 | 3″ | 130 | 148.2 | 149.7 | 73.8 | 73.8 | 86.6 |
| 4 | 150.8 | 44.1 | 150.2 | 150.6 | 150.3 | 150.8 | 150.2 | 150.1 | 150 | 150.6 | 149.9 | 150 | 150.4 | 4″ | 150.1 | 147.3 | 146.8 | 79.4 | 79.4 | 68.9 |
| 5 | 50.8 | 49.5 | 50.7 | 50.1 | 49.1 | 51.8 | 51.6 | 52.4 | 48.8 | 51.7 | 48.8 | 48.8 | 49.2 | 5″ | 130 | 116.3 | 116.5 | 76.1 | 76.1 | 73.2 |
| 6 | 79.4 | 86.2 | 84.7 | 83.6 | 83.6 | 85.2 | 83.1 | 80.8 | 76.9 | 85.2 | 86.6 | 86.6 | 83.7 | 6″ | 129 | 122.1 | 123 | 60.8 | 61.1 | 63.5 |
| 7 | 56.3 | 46.8 | 45.8 | 45.1 | 44.6 | 46.4 | 45.9 | 54.4 | 44.3 | 46.5 | 44.3 | 44.3 | 44.7 | 1‴ | 172.3 | 166.4 | 168.3 | 103.4 | 103.4 | 103.5 |
| 8 | 75.1 | 30.6 | 31 | 30.6 | 30.4 | 31.6 | 30.4 | 70.6 | 30.1 | 31.6 | 30.1 | 30.1 | 30.5 | 2‴ | 121.7 | 115.6 | 115.3 | 72.7 | 72.7 | 73 |
| 9 | 46.6 | 35.7 | 33.9 | 34.2 | 34.1 | 34.4 | 33.8 | 46.4 | 33.8 | 34.4 | 33.8 | 33.7 | 34.2 | 3‴ | 144.6 | 145.9 | 147.3 | 76.9 | 76.9 | 87.2 |
| 10 | 145.1 | 149.5 | 149.3 | 148.9 | 149.1 | 150 | 149 | 144.8 | 148.7 | 150 | 148.7 | 148.8 | 149.2 | 4‴ | — | — | — | 68.5 | 68.5 | 69 |
| 11 | 42.3 | 140.1 | 141.5 | 141 | 140.5 | 142.1 | 140.5 | 39.9 | 140.2 | 142.1 | 140.1 | 140.1 | 140.6 | 5‴ | — | — | — | 76.3 | 76.1 | 76.6 |
| 12 | 179 | 169.6 | 171.6 | 170 | 169.9 | 172.3 | 169.9 | 181.9 | 169.6 | 172.2 | 169.5 | 169.5 | 170 | 6‴ | — | — | — | 61.1 | 61 | 61.3 |
| 13 | 16.5 | 119.5 | 120 | 119.4 | 120.1 | 120.4 | 119.5 | 11.5 | 119.8 | 120.4 | 119.7 | 119.9 | 120.2 | — | — | — | — | — | — | |
| 14 | 115.1 | 112.7 | 114.1 | 113.9 | 113.9 | 114.7 | 114.9 | 116.6 | 113.6 | 114.7 | 113.6 | 113.6 | 114 | NO. | 61 b [46] | 61 e [25] | — | — | — | — |
| 15 | 112.3 | 18.1 | 112.8 | 112.1 | 111 | 113.6 | 114.9 | 116.2 | 110.8 | 113.8 | 110.8 | 110.8 | 111.1 | 1″ | 104.1 | 105.3 | — | — | — | — |
| 1′ | 104 | 104.1 | 102.3 | 104.3 | 102.9 | 103.1 | 98.5 | 99.4 | 101.7 | 102.3 | 101.7 | 101.7 | 102.2 | 2″ | 72.3 | 75.6 | — | — | — | — |
| 2′ | 75.3 | 73.6 | 74.3 | 75.3 | 73.9 | 78.2 | 76.1 | 75.3 | 70.2 | 74.6 | 70.1 | 70.1 | 72.8 | 3″ | 73.9 | 78.3 | — | — | — | — |
| 3′ | 78.2 | 76.8 | 77.5 | 78.6 | 77.2 | 75.3 | 74.9 | 76.4 | 88.3 | 88.3 | 88 | 88 | 88.4 | 4″ | 79.4 | 71.5 | — | — | — | — |
| 4′ | 71.8 | 70.2 | 71.1 | 71.7 | 70.5 | 71.8 | 71.8 | 72.1 | 68.5 | 70.2 | 68.4 | 68.4 | 68.9 | 5″ | 76.1 | 77.8 | — | — | — | — |
| 5′ | 78.5 | 76.8 | 77.2 | 78.5 | 77.2 | 77.9 | 78.2 | 78.1 | 76.3 | 77.5 | 76.3 | 76.3 | 76.8 | 6″ | 61.1 | 62.7 | ||||
| 6′ | 62.9 | 61.2 | 62.1 | 62.8 | 61.5 | 62.8 | 62.6 | 62.8 | 60.9 | 62.6 | 60.8 | 60.9 | 63.5 | |||||||
Note: The 1″, 2″, 3″, and 4″ data of compound 51 were 176.3; 32.4; 18.3; and 48.8, respectively. The 13C-NMR data of 1″ and 2″ of 54 were recorded as 67.8 and 15.3. a: Measured in C5D5N. b: Measured in DMSO. c: Measured in CDCl3. d: Measured in CD3SOCD3. e: Measured in CD3OD.
2.2. Bioactivity of Guaiane Sesquiterpenes
2.2.1. Anti-Inflammatory
Nitric oxide (NO) is a related target of inflammation, and inhibiting the release of NO can treat inflammatory diseases. Dihydroestafiatol (13), zaluzanin C (23), and dehydrozaluzanin C (31) strongly inhibited the production of nitric oxide in RAW264.7 macrophages stimulated with lipopolysaccharide (LPS), with IC50 values of 7.11, 2.50, and 0.82 µM [31]. From the bioassay results, three exocyclic double bonds in guaianolides play a key role in the inhibition of the production of nitric oxide (NO), and a reduction in exocyclic double bonds will lower the inhibitory effect. Under the condition of the presence of three exocyclic double bonds, the hydroxylization of C-1 will enhance the inhibitory activity. Zaluzanin C (23) showed a potent inhibitory effect against NO production in LPS-stimulated RAW264.7 macrophages with an IC50 value of 6.54 ± 0.16 μM [34]. It may be speculated that the α-methylene-γ-lactone moiety of zaluzanin C (23) has a key role in its inhibition of NO release. Moreover, other functional groups, especially hydroxyl, have a great influence on the inhibitory effect of NO production. Zaluzanin C (23) showed remarkable inhibition against NO release in LPS-induced RAW264.7 macrophages, possibly because zaluzanin C (23) had an α-methylene-γ-lactone moiety and the large isovaleroxyl at C-3 hinders the binding of the compound to related proteins [37]. Guailactone can be structurally modified to obtain compounds containing the α-methylene-γ-lactone part. It is also used to introduce hydroxyl groups into guaiacols containing three outer-ring double bonds to enhance the inhibitory ability of these compounds against NO production and achieve anti-inflammatory effects.
Ainslide C (41), ainsliaolide A (34), diaspanolide B (39), zaluzanin C (23), and estafiatone (28) inhibit NLRP3 inflammasome activity by inhibiting the LDH release rate. Meanwhile, a Western blot assay showed that compound 41 inhibited the activity of inflammasome by inhibiting the production of Caspase-1 and IL-1β induced by LPS and Nigericin. Among them, the substituents of compounds 41, 34, 39, 23, and 28 are terminal double bonds, and the α-methylene-γ-butyrolactone structure seems to be the key to inhibiting LDH release activity [7]. Glucozaluzanin C (57) and dihydroestafiatol (13) showed significant anti-inflammatory activity by inhibiting the expression of nuclear factor kappa B (NF-κB) in the 293-NF-κB-luciferase reporter cell line and the production of TNF-α, IL-1β, IL-6, and IL-10 in RAW264.7 macrophages induced by lipopolysaccharide (LPS) [47].
8α-Hydroxy-11α,13-dihydrozaluzanin C (20) showed moderate COX-1-inhibiting activity with an IC50 value of 78.8 μM, comparable to that of the representative anti-inflammatory drug aspirin with an IC50 value of 77.2 μM. 8α-Hydroxy-11α,13-dihydrozaluzanin C (20) and 2′-O-E-Caffeoyl-8α-hydroxy-11α,13-dihydro-3-β-O-β-D-glucozaluzanin C (60) displayed potent COX-2 inhibitory activities with IC50 values ranging from 12.5 to 57.9 μM, in comparison with that of aspirin with an IC50 value of 87.6 μM [45].
2.2.2. Antitumor and Cytotoxic
8-Epidesacylcinaropicrin (25) exhibited moderate activity toward the human tumor cell lines MDA-MB-231 and HepG2, with IC50 values of 18.91, and 11.16 μM, respectively [8]. Mokko lactone (9), zaluzanin C (23), and glucozaluzanin C (57) showed non-specific significant cytotoxicity against the A549 (non-small cell lung adenocarcinoma), SK-OV-3 (ovarian), SK-MEL-2 (skin melanoma), XF498 (CNS), and HCT15 (colon) cell lines with ED50 values ranging from 0.36 to 5.54 μg/mL [12]. Dehydrozaluzanin C (31) is a guaiacol lactone, which has significant cytotoxicity to RAW264.7 macrophages. In the presence of three outer-ring double bonds, the carbonylation of C-1 may result in a high cytotoxicity to RAW264.7 macrophages [31].
2.2.3. Antiobesity
Ainsliaside A (59) isolated from Ainsliaea acerifolia had significant inhibitory activity on pancreatic lipase with a semi-inhibitory concentration of 15.3 ± 0.7 μM. In addition, ainsliaside A (59) also exhibited potent inhibitory effects against 3T3-L1 adipocyte cells and can be used as a potential antiobesity agent [43].
3. Germacrane-Type Sesquiterpenes
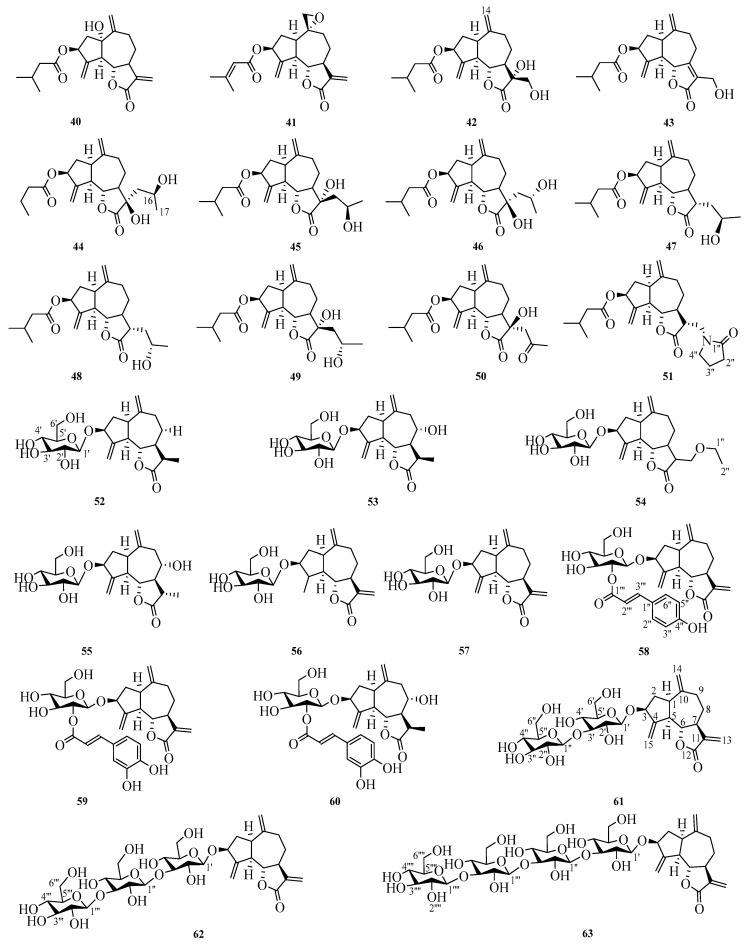
Germacrane sesquiterpenes represent a class of sesquiterpenes that are extensively distributed in Compositae plants, characterized by the formation of a substantial ten-membered ring structure at the 5 and 10 positions. Currently, all twelve germacrane-type sesquiterpenes reported from this genus are lactones, with lactone rings predominantly located at the C-6, C-7 and C-7, C-8 positions. Detailed information is presented in Figure 2 and Table 12.
Figure 2.
Chemical structures for compounds 64–75.
Table 12.
The compound name, molecular formula, and NMR test reagent of germacrane-type sesquiterpenes.
| No. | Compound Name | Molecular Formula | Solvent | Ref. |
|---|---|---|---|---|
| 64 | Isodihydrocostunolide | C15H22O2 | CDCl3 | [48] |
| 65 | Taraxinic acid | C15H18O4 | CDCl3 | [49] |
| 66 | Yunnanolide K | C15H20O4 | CDCl3 | [11] |
| 67 | Germacra-1(10), 4-diene-11α-methyl-12,8α-olide-15-acid | C15H20O4 | DMSO/CD3OD | [34,50] |
| 68 | Germacra-1(10),4,11(13)-triene-12,8α-olide-15-acid | C15H18O4 | CD3OD | [50] |
| 69 | Ainsliaside B | C21H28O9 | C5D5N CD3OD |
[44] |
| 70 | Taraxinsaure-1′-O-β-D-glucopyranoside | C21H28O9 | CD3OD | [51] |
| 71 | Picriside B | C21H30O8 | C5D5N | [27] |
| 72 | Ainsliaolide C | C26H40O12 | DMSO | [31] |
| 73 | Taraxic acid-1′-O-β-D-glucopyranoside | C21H28O9 | CD3OD/C5D5N | [2] |
| 74 | Germacra-1(10),4,11(13)-triene-12,8α-olide-15-oic acid(15-1′)-β-D-glucopyransyl ester | C21H28O9 | CD3OD | [50] |
| 75 | Ainsliaea latifolia A | C21H30O9 | CD3OD | [52] |
3.1. NMR Data of Germacrane-Type Sesquiterpene (64–75)
The NMR spectrum data for both 1H and 13C are presented in Table 13 and Table 14. A summary of the test instruments used to obtain the NMR data for compounds 64–75 is provided. The 1H and 13C data of compounds 67, 68, and 74 were measured with the Bruker Avance III-500 instrument (Bruker, Switzerland). The nuclear magnetic data of compounds 69 and 71 were obtained by the JEOL FX-90Q instrument (JEOL, Duzhao, Japan). The NMR spectra of compounds 65, 66, 67, 72, 73, and 75 were recorded on various instruments including the Brukerspeckospin AC-600P (Bruker, Germany), Bruker Ascend-500 spectrometer (Bruker, Germany), Bruker Advance 500 (Bruker, Germany), Bruker Avance 600 (Bruker, Biel, Switzerland), JNM-FX-100 (JEOL, Japan), and Bruker Avance-500 (Bruker, Karlsruhe, Germany), respectively. The 1H-NMR data of compound 64 were measured at a frequency of 300 MHz; however, no literature reports exist regarding the proton data for compound 70. The carbon spectra for compounds 64 and 70 were acquired at 75 MHz and 25.2 MHz, respectively.
Table 13.
1H-NMR data of compounds 64–75.
| NO. | 64 [48] | 65 [49] | 66 [11] | 67 [34] | 67 [50] | 68 [50] | 69 [44] | 71 [27] | 72 [31] | 73 [2] | 74 [50] | 75 [52] |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CDCl3 | CDCl3 | CDCl3 | DMSO | CD3OD | CD3OD | C5D5N | C5D5N | DMSO | C5D5N | CD3OD | CD3OD | |
| 1 | 1.68–1.40, m | 5.68, dd, 13.0, 3.7 | 5.09, dd, 12.0, 5.0 | 5.06, dd, 11.7, 4.5 | 5.06, dd, 11.7, 4.5 | 5.06, dd, 11.7, 4.5 | 6.72, br t, 7 | — | 4.81, m | 5.64, dd, 11.0, 4.0 | 5.13, m | 5.16, dd, 11.8, 4.7 |
| 2α | 1.68–1.40, m | 3.38, m | 2.31, m | 2.16, t, 12.3, 4.9 | 2.13, m | 2.13, m | — | — | 2.17, m | 2.14, m | — | 2.38–2.42, m |
| 2β | 2.18, m | 2.02–2.06, m | 2.02, m | 2.02, m | — | — | 2.26, m | 3.54, m | — | 2.14–2.20, m | ||
| 3α | 2.10, d, 2.7 | 2.25, m | 2.93, m | 2.66–2.72, overlapped | 2.69, m | 2.69, m | — | — | 1.83, m | 2.3–2.0, m | — | 2.92, dd, 12.5, 3.9 |
| 3β | 1.9–1.7, m | 2.35, m | 1.88, m | 1.78, t, 12.3, 5.2 | 1.78, m | 1.78, m | — | — | 2.61, m | — | 1.90–1.98, m | |
| 5 | 5.15, d, 6 | 4.91, d, 10.0 | 5.51, dd, 11.0, 2.5 | 5.38, dd, 11.1, 2.1 | 5.38, dd, 11.1, 2.1 | 5.38, dd, 11.1, 2.1 | 4.81, d, 10 | — | 4.75, br s | 4.94, dd, 10.0, 1.2 | 5.74, m | 5.65, dd, 11.3, 2.6 |
| 6α | — | — | 3.13, m | 3.01, t, 16.1, 10.8 | 3.01, m | 3.01, m | 4.58, dd, 10, 9 | — | — | — | — | 3.19–3.24, m |
| 6β | 4.8, m | 4.58, dd, 10.0, 9.0 | 2.49, d, 16.5 | 2.41–2.55, overlapped | 2.44, m | 2.44, m | — | — | 4.77, d, 9.6 | 4.72, dd, 10.0, 10.0 | — | 2.55–2.65, m |
| 7 | 3.6, m | 2.56, m | 2.31, m | 1.82–1.90, m | 1.86, m | 1.86, m | — | — | 1.63, m | 2.54, m | — | 1.92–2.01, m |
| 8α | 1.68–1.40, m | 2.19, m | 4.39, t, 11.5 | — | — | — | — | — | 1.72, m | 2.3–2.0, m | — | — |
| 8β | — | 4.19–4.25, m | 4.22, m | 4.22, m | — | — | 2.06, m | 4.27, m | 4.27–4.37, m | |||
| 9α | 1.68–1.40, m | 2.90, m | 2.93, m | 2.66–2.72, overlapped | 2.66, m | 2.66, m | — | — | 2.24, m | 2.86, m | — | 2.83, d, 12.3 |
| 9β | 2.15, m | 2.28, m | 2.27, t, 11.8 | 2.27, m | 2.27, m | — | — | 1.96, m | 2.3–2.0, m | — | 2.33–2.38, m | |
| 10 | 2.5, m | — | — | — | — | — | — | — | — | — | — | — |
| 11 | — | — | 2.78, m | 2.41–2.55, overlapped | 2.51, m | — | — | — | 2.33, m | — | — | 2.54–2.58, m |
| 13α | 6.2, d, 2 | 6.24, d, 3.4 | 1.26, d, 7.5 | 1.11, d, 7.0 | 1.11, d, 7.0 | 6.08, m | — | 5.51, d, 3.2 | 1.09, d, 6.6 | 6.23, d, 3.5 | 6.23, dd, 18.8, 3.0 | 1.28, d, 7.0 |
| 13β | 5.65, d, 2 | 5.51, d, 2.9 | 5.79, m | — | 6.35, d, 3.6 | 5.74, m | ||||||
| 14 | 1.1, d, 8 | — | 1.35, s | 1.27, s | 1.27, s | 1.31, s | — | 1.37, br s | 1.30, s | — | 1.41, s | 1.41, s |
| 15α | 1.2, s | 1.60, s | — | — | — | — | 1.71, br s | — | 3.78, br s | 1.72, d, 1.2 | — | — |
| 15β | — | — | — | — | — | 4.40, br s | — | — | ||||
| 1′ | — | — | — | — | — | — | 6.26, d, 8 | 4.96, d, 7.5 | 4.17, d, 7.8 | 6.18, d, 7.6 | 5.52, d, 7.9 | 5.56, d, 7.8 |
| 2′ | — | — | — | — | — | — | — | — | 2.97, m | 4.44–3.80, m | 3.43, overlapped | |
| 3′ | — | — | — | — | — | — | — | — | 3.13, m | 4.44–3.80, m | 3.43, overlapped | |
| 4′ | — | — | — | — | — | — | — | — | 2.99, m | 4.44–3.80, m | 3.43, overlapped | |
| 5′ | — | — | — | — | — | — | — | — | 3.23, m | 4.44–3.80, m | 3.43, overlapped | |
| 6′α | — | — | — | — | — | — | — | — | 3.44, d, 11.2, 7.2 | 4.44–3.80, m | 3.81, m | 3.84, dd, 12.0, 2.0 |
| 6′β | — | — | — | — | — | — | — | — | 3.81, d, 11.2, 1.8 | 4.44–3.80, m | 3.69, m | 3.75, dd, 12.0, 4.4 |
Note: The 1H-NMR data of 1″, 2″, 4″α, and 4″β and 5″ of 72 were recorded as 4.88, br s; 3.74, m; 3.57, d, 9.6; 3.83, d, 9.6; and 3.33, s, respectively.
Table 14.
13C-NMR data of compounds 64–75.
| NO. | 64 [48] | 65 [49] | 66 [11] | 67 [34] | 67 [50] | 68 [50] | 69 [44] | 70 [51] | 71 [27] | 72 [31] | 73 [2] | 74 [50] | 75 [52] |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CDCl3 | CDCl3 | CDCl3 | DMSO | CD3OD | CD3OD | CD3OD | CD3OD | C5D5N | DMSO | CD3OD | CD3OD | CD3OD | |
| 1 | 16.7 | 149.9 | 128.8 | 128.9 | 128.9 | 130.4 | 143 | 149.7 | 126.9 | 125.9 | 149.6 | 130 | 129.8 |
| 2 | 32.6 | 26.7 | 26.8 | 27.1 | 27.1 | 27.6 | 26.2 | 27.6 | 27.7 | 25.9 | 27.6 | 27.7 | 27.9 |
| 3 | 42.6 | 39.2 | 34.8 | 34.9 | 34.9 | 35.7 | 38 | 39.9 | 36 | 34.9 | 40 | 35.5 | 35.7 |
| 4 | 148.9 | 139.8 | 124.1 | 125.1 | 125.1 | 126.6 | 141.3 | 141.8 | 141 | 138.8 | 141.8 | 126.6 | 125.5 |
| 5 | 118.5 | 126 | 150.7 | 148.6 | 148.6 | 149.2 | 127.1 | 127.2 | 130.1 | 130.1 | 127.1 | 151.6 | 152 |
| 6 | 76.4 | 82.1 | 28.6 | 31.1 | 31.1 | 32.8 | 82.8 | 83.8 | 80.3 | 78.7 | 83.8 | 32.8 | 32.4 |
| 7 | 39.5 | 50.3 | 48.3 | 52.6 | 52.6 | 50.1 | 46.7 | 51.1 | 50.8 | 53.7 | 51.1 | 50.1 | 54.4 |
| 8 | 41.7 | 30.1 | 82.4 | 82.6 | 82.6 | 85.1 | 24.2 | 31.3 | 27.1 | 27 | 31.3 | 85 | 84.5 |
| 9 | 32.7 | 36.7 | 46.2 | 45.5 | 45.5 | 47.2 | 26.4 | 37.2 | 41.1 | 40.7 | 37.2 | 47.2 | 46.8 |
| 10 | 37.5 | 130.3 | 134.2 | 134 | 134 | 134.3 | 135.1 | 131.9 | 137.6 | 137 | 131.9 | 135.1 | 135.6 |
| 11 | 139.9 | 143.2 | 40.7 | 41.4 | 41.4 | 141.2 | 139 | 144.5 | 141.2 | 41 | 144.4 | 141.1 | 43 |
| 12 | 170.4 | 170.6 | 178.7 | 178 | 178 | 171.7 | 172.4 | 172.7 | 170.3 | 178.1 | 172.6 | 171.7 | 180.5 |
| 13 | 121.5 | 120.1 | 11.7 | 13.1 | 13.1 | 121.2 | 119.4 | 120.4 | 119.1 | 12.8 | 120.4 | 121.2 | 13.2 |
| 14 | 22.5 | 173.3 | 16.8 | 16.8 | 16.8 | 17.1 | 167.6 | 169.8 | 16.3 | 15.8 | 167.8 | 17.4 | 17.3 |
| 15 | 28.6 | 17 | 171.9 | 169 | 169 | 167.5 | 17.2 | 17.3 | 67.8 | 65.8 | 17.3 | 167.5 | 167.7 |
| 1′ | — | — | — | — | — | — | 95.7 | 95.3 | 105.3 | 103.1 | 95.3 | 95.7 | 95.7 |
| 2′ | — | — | — | — | — | — | 73.9 | 73.9 | 75.2 | 73.3 | 73.9 | 74 | 74 |
| 3′ | — | — | — | — | — | — | 78.6 | 78.3 | 78.6 | 76.6 | 78.3 | 78.7 | 78.4 |
| 4′ | — | — | — | — | — | — | 71 | 71 | 71.8 | 70.2 | 71 | 71 | 71 |
| 5′ | — | — | — | — | — | — | 77.9 | 78.7 | 78.6 | 75.8 | 78.7 | 78.4 | 78.7 |
| 6′ | — | — | — | — | — | — | 62.4 | 62.3 | 62.9 | 67.5 | 62.4 | 62.2 | 62.2 |
Note: The 13C NMR of 1″, 2″, 3″, 4″, and 5″ of 72 were 109.3; 75.6; 78.7; 73.2; and 63.3, respectively.
3.2. Bioactivity of Germacrane-Type Sesquiterpene
Isodihydrocostunolide (64) showed moderate cytotoxicity against the human cancer cell lines MDA-MB-231 (IC50 = 18.2 μM) and HepG2 (IC50 = 12.2 μM), respectively [11]. Ainsliaea latifolia A (75), isolated from Ainsliaea latifolia, exhibited moderate activity against the HCT116 and SMMC-7721 human tumor cell lines when adriamycin was used as the positive control, with IC50 values of 14.72 and 10.53 μM [52].
4. Eudesmane Sesquiterpenes
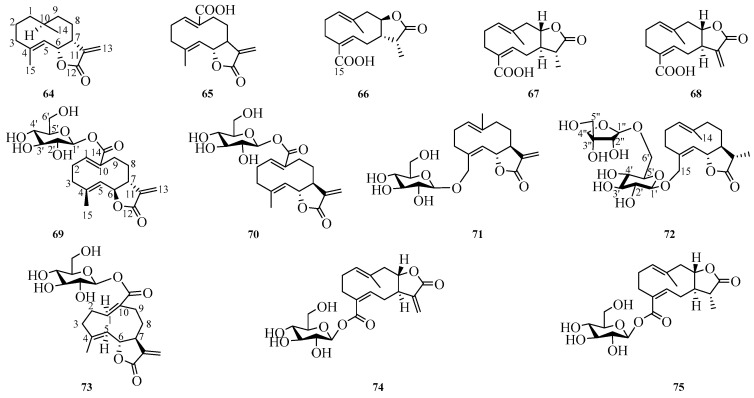
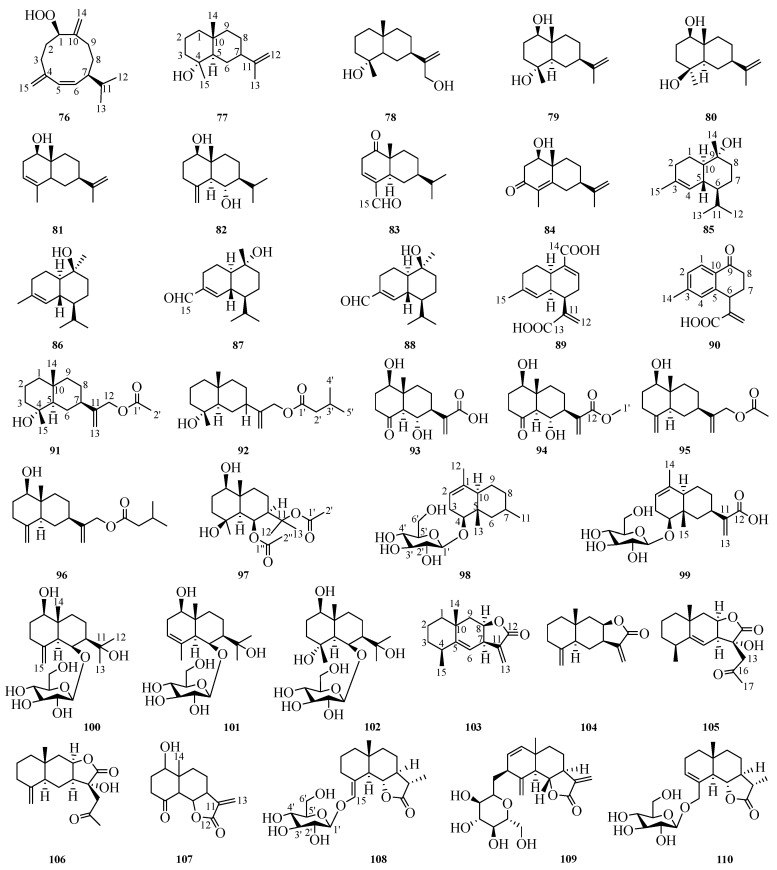
The basic skeleton of eudesmane-type sesquiterpenes consists of two six-membered rings comprising a total of 15 carbon atoms. These compounds exhibit a diverse range of biological activities, including anti-inflammatory, cytotoxic, antibacterial, antimalarial, insecticidal, and neuroprotective activities [53]. Up to now, 35 eudesmane-type sesquiterpenes have been reported within this genus. Their structures and detailed information are shown in Figure 3 and Table 15.
Figure 3.
Chemical structures for compounds 76–110.
Table 15.
The compound name, molecular formula, and NMR test reagent of eudesmane sesquiterpenes.
| No. | Compound Name | Molecular Formula | Solvent | Ref. |
|---|---|---|---|---|
| 76 | 1β-Hydroperoxygermacra-4(15),5,10(14)-triene | C15H24O2 | CDCl3 | [12] |
| 77 | Selin-11-en-4α-ol | C15H26O | CDCl3 | [54] |
| 78 | 4α-Hydroxy-4β-methyldihydrocostol | C15H26O2 | CDCl3 | [55] |
| 79 | Cyperusol C | C15H26O2 | CDCl3 | [56] |
| 80 | 1β,4β-Dihydroxyeudesman-11-ene | C15H26O2 | CDCl3 | [57] |
| 81 | α-Dictyopterol | C15H24O | CDCl3 | [58] |
| 82 | 1β,6α-Dihydroxy-4(15)-eudesmane | C15H26O3 | CDCl3 | [59] |
| 83 | 1-oxo-5α,7αH-eudesma-3-en-15-al | C15H22O2 | CDCl3 | [60] |
| 84 | 1β-Hydroxy-α-cyperone | C15H22O2 | CDCl3 | [61] |
| 85 | (-)-α-Cadinol | C15H26O | CDCl3 | [62,63] |
| 86 | T-Cadinol | C15H26O | CDCl3 | [63,64] |
| 87 | 10-Hydroxyl-15-oxo-α-cadinol | C15H24O2 | CDCl3 | [65,66] |
| 88 | 15-oxo-T-cadinol | C15H24O2 | CDCl3 | [66] |
| 89 | Ainsliaea acid B | C15H18O4 | CD3OD | [67] |
| 90 | 4-Acrylic-6-methyl-α-tetralone | C14H14O3 | CDCl3 | [67] |
| 91 | 4α-Hydroxy-12-acetoxy-eudesm-11(13)-en | C17H28O3 | — | [68] |
| 92 | 4α-Hydroxy-eudesm-11-en-12-isovaleroxyl | C20H34O3 | CDCl3 | [34] |
| 93 | Ainsliatone A acid | C14H20O5 | CD3OD | [42] |
| 94 | Ainsliatone B | C15H22O5 | CDCl3 | [31] |
| 95 | Ainslide B | C17H26O3 | CDCl3 | [7] |
| 96 | Spicatene C | C20H32O3 | CDCl3 | [8] |
| 97 | 6,11-Diacetoxy-1,4-dihydroxyeudesmane | C19H32O6 | CD3OD | [40] |
| 98 | Alatoside N | C20H34O6 | CD3OD | [42] |
| 99 | Alatoside M | C21H32O8 | CD3OD | [42] |
| 100 | Ainsliaside C | C21H36O8 | C5D5N | [69] |
| 101 | Ainsliaside D | C21H36O8 | C5D5N | [69] |
| 102 | Ainsliaside E | C21H38O9 | C5D5N | [69] |
| 103 | Alantolactone | C15H20O2 | CDCl3 | [70] |
| 104 | Isoalantolactone | C15H20O2 | CDCl3 | [70] |
| 105 | Pertyolides B | C17H24O4 | CDCl3 | [39] |
| 106 | Pertyolides A | C17H24O4 | CDCl3 | [39] |
| 107 | Ainsliatone A | C14H18O4 | CDCl3 | [71] |
| 108 | 4(15)-En-eudesma-6,12-olide-15-O-β-D-glucopyranoside | C21H32O8 | CD3OD | [40] |
| 109 | Ixerin W | C22H30O7 | C5D5N | [27] |
| 110 | 3(4)-En-eudesma-6,12-olide-15-O-β-D-glucopyranoside-O-β-D-glucopyranoside | C21H32O8 | CD3OD | [40] |
4.1. NMR Data of Eudesmane-Type Sesquiterpene (76–110)
The 1H and 13C spectrum data are shown in Table 16, Table 17, Table 18, Table 19 and Table 20. An overview of the testing instruments used for the NMR data of compounds 76–110 is provided. The NMR data for compounds 82 and 87 were measured with the Bruker DRX-500 spectrometer (Bruker, Germany). The 1H and 13C spectra of compounds 85 and 86 were obtained by the JEOL MN 100 instrument (JEOL, Japan). For compounds 87 and 88, NMR data were recorded on the Bruker AM 400 spectrometer (Bruker, Switzerland). The NMR data for compounds 93, 98, and 99 were taken with the Varian 500 (Varian, Palo Alto, CA, USA) and Bruker AV500-III instruments (Bruker, Switzerland). Compounds 97, 108, and 110 had their NMR data obtained on the Bruker AV-600 spectrometer (Bruker, Switzerland). The 1H and 13C data of compounds 100, 101, 102, and 109 were measured using the JEOL FX-90Q instrument (JEOL, Japan). The NMR data for compounds 100, 101, and 102 were recorded by the GSX-270 (JEOL, Japan) and GSX-500 NMR instruments (JEOL, Japan). The 1H and 13C spectra of compounds 103 and 104 were analyzed using the Bruker Avance DRX 500 spectrometer (Bruker, Germany). NMR data for compounds 76, 77, 79, 80, 83, 85, 92, 94, 95, and 96 were obtained from various instruments including Bruker AMX 500 (Bruker, Zurich, Switzerland) and Varian Unity Inova 500 (Varian, Palo Alto, CA, USA), Bruker DRX-300 (Bruker, Karlsruhe, Germany), JEOL JNM LA-500 (JEOL, Japan), Varian Mercury-300 BB (Varian, San Jose, USA), Varian Mercury Plus 400 (Varian, USA), NT-200 (University of California, Davis, CA, USA), Bruker Advance 500 (Bruker, Germany), Bruker Avance 600 (Bruker, Biel, Switzerland), Bruker AV-400 HD (Bruker, Byersbin, Switzerland), Bruker Ascend 500 (Bruker, Zurich, Switzerland), and other instruments. Compound 78 was measured at 200 MHz for 1H NMR. Compounds 84 and 107 were tested at 400 MHz. Compounds 89 and 90 were recorded at 600 MHz. Compound 91 was run at 60 MHz. Compounds 105 and 106 were operated at 500 MHz. No test instrumentation has been reported in the literature regarding NMR spectrum data for compound 81. Furthermore, no hydrogen spectrum data of compound 86 have been reported in the existing literature. The carbon spectrum data for compound 84 were measured at 50.32 MHz; the 13C data collection for compound 107 occurred at 100 MHz; compound 86 was recorded at 75 MHz for 13C NMR; while the carbon spectrum data of compounds 89 and 90 were measured at 150 MHz. Compounds 78, 91, 105, and 106 have not been described within any available literature concerning their 13C NMR spectra.
Table 16.
1H-NMR data of compounds 76–84.
| NO. | 76 [12] | 77 [54] | 78 [55] | 79 [56] | 80 [57] | 81 [58] | 82 [59] | 83 [60] | 84 [61] |
|---|---|---|---|---|---|---|---|---|---|
| CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | |
| 1 | 4.15, dd, 12.0, 3.5 | — | — | 3.32, dd, 11.2, 4.4 | 3.27, dd, 11.4, 4.8 | 3.49–3.56, m | 3.43, dd, 11.7, 4.6 | — | 3.83, dd, 13, 5.5 |
| 2α | 2.05, m | — | — | 1.62, m | 1.96, m | 1.45–1.55, m | 1.86, m | 2.54, m | 2.64, dd, 16.5, 5.5 |
| 2β | — | — | 1.72, m | 1.91, m | 1.55, m | 2.56, dd, 16.5, 13 | |||
| 3α | 2.27, ddd, 2.5, 5.5, 13.0 | — | — | 1.52, ddd, 13.5, 12.0, 3.5 | 1.16, m | 5.26, s | 2.07, m | 6.64, br d, 5.2 | — |
| 3β | 2.46, td, 13.0, 5.0 | — | — | 1.79, ddd, 12.0, 3.5, 3.0 | 1.09, m | — | 2.33, m | — | — |
| 5 | 6.04, d, 16.0 | 1.81, dddd | — | 1.28, m | 1.24, dd, 9.6, 3.6 | 1.10–1.38, m | 1.75, m | 2.21, dd, 10.8, 4.8 | — |
| 6α | 5.46, dd,16.0, 10.5 | — | — | 1.26, m | 1.27, m | 1.10–1.38, m | 1.44, m | 2.19, ddd | |
| 6β | — | — | 1.84, m | 1.22, m | 3.72, dd, 9.8, 9.8 | 1.87, m | 2.08, ddd, 14, 12, 1.5 | ||
| 7 | 1.5–1.86, 2.62, m | 1.96, dddd, br | — | 1.94, m | 1.58, m | 1.84–1.92, m | 1.28, m | 1.81, m | 2.02, br dddd, 13.5, 12, 3.5, 2.5 |
| 8α | 1.5–1.86, 2.62, m | — | — | 1.61, m | 1.78, m | 1.84–1.92, m | 1.53, m | 1.58, m | 1.75, m |
| 8β | — | — | 1.38, dddd, 17.0, 13.5, 13.0, 3.5 | 1.71, m | 1.21, m | 1.59, dddd, 13.5, 13.5, 13.5, 3.5 | |||
| 9α | 1.5–1.86, 2.62, m | — | — | 1.13, ddd, 13.5, 13.0, 4.0 | 1.88, m | 1.84–1.92, m | 1.17, m | 2.43, ddd, 14.4, 10.8, 3.6 | 1.35, ddd, 13.5, 13.5, 4 |
| 9β | — | — | 1.90, ddd, 13.5, 3.5, 3.5 | 1.86, m | 1.92, m | 2.86, ddd, 14.4, 5.4, 5.4 | 2.16, ddd, 13.5, 3.5, 3 | ||
| 11 | 1.5–1.86, 2.62, m | — | — | — | — | — | 2.24, m | 1.67, m | — |
| 12α | 0.83, d, 6.5 | — | 4.12, s | 4.72, m | 4.74, br s | 4.71, s | 0.95, d, 6.9 | 0.94, d, 6.8 | 4.78, m |
| 12β | — | 4.71, br s | |||||||
| 13α | 0.92, d, 6.5 | 1.75, s | 5.00, d, 1.4 | 0.89, s | 1.76, s | 1.73, s | 0.87, d, 7.1 | 0.94, d, 6.8 | 1.78, dd |
| 13β | 4.90, d, 1.0 | ||||||||
| 14α | 5.21, br s | 1.12, s | 0.88, s | 1.75, s | 1.05, s | 1.58, s | 0.71, s | 1.33, s | 1.18, s |
| 14β | 5.34, br s | ||||||||
| 15α | 4.89, br s | 0.89, s | 1.09, s | 1.11, s | 1.16, s | 0.76, s | 5.02, s | 9.35, s | 1.73, d, 1 |
| 15β | 4.97, br s | 4.74, s |
Table 17.
1H-NMR data of compounds 85–92.
| NO. | 85 [63] | 86 [63] | 87 [66] | 88 [66] | 89 [67] | 90 [67] | 91 [68] | 92 [34] |
|---|---|---|---|---|---|---|---|---|
| CDCl3 | CDCl3 | CDCl3 | CDCl3 | CD3OD | CDCl3 | CDCl3 | ||
| 1α | — | — | 2.16, m | 2.24, m | 2.34, m | 7.97, d, 8.0 | — | 1.36–1.46, overlapped |
| 1β | — | — | 1.21, m | 1.21, m | 1.69, m | — | 1.12, m | |
| 2α | — | — | 2.06, m | 2.06, m | 1.65, m | 7.17, d, 8.0 | — | 1.53–1.62, overlapped |
| 2β | — | — | 2.46, m | 2.45, m | 1.80, m | — | ||
| 3α | — | — | — | — | — | — | — | 1.77–1.81, m |
| 3β | — | — | — | — | — | — | — | 1.36–1.46, overlapped |
| 4 | 5.29, br s | 5.42, br s | 6.87, s | 6.94, s | 4.99, br s | 6.96, br s | — | — |
| 5 | — | — | 2.01, m | 2.04, m | 2.75, br s | — | — | 1.21–1.29, overlapped |
| 6α | — | — | 1.20, m | 1.20, m | 2.88, d, 12.0 | 4.25, br s | — | 1.89–1.94, m |
| 6β | — | — | — | — | — | — | — | 1.21–1.29, overlapped |
| 7α | — | — | 1.72, m | 1.55, m | 2.26, m | 2.33, m | — | 2.02–2.06, m |
| 7β | — | — | 1.23, m | 1.27, m | 1.99, m | 2.29, m | — | — |
| 8α | — | — | 1.45, m | 1.47, m | 6.90, m | 2.56, m | — | 1.53–1.62, overlapped |
| 8β | — | — | 1.84, m | 1.78, m | — | 2.64, m | — | 1.36–1.46, overlapped |
| 9α | — | — | — | — | — | — | — | 1.21–1.29, overlapped |
| 9β | — | — | — | — | — | — | — | |
| 10 | — | — | 1.36, m | 1.35, m | 2.96, br s | — | — | — |
| 11 | 1.66, br s | 1.61, br s | 2.21, m | 2.35, m | — | — | — | — |
| 12α | — | — | 0.85, d, 6.8 | 0.87, d, 6.9 | 6.34, br s | 6.51, br s | — | 4.55–4.63, m |
| 12β | — | — | 5.54, br s | 5.23, br s | — | |||
| 13α | 0.91, d, 7.2 | 0.90, d, 6.9 | 0.98, d, 6.8 | 0.97, d, 6.9 | — | — | 5.03 | 5.03, d, 1.4 |
| 13β | — | — | 4.6 | 4.98, s | ||||
| 14 | 0.78, d, 7.2 | 0.78, d, 6.9 | 1.14, s | 1.24, s | — | 2.36, s | 0.90, s | 0.89, s |
| 15 | 1.05, s | 1.15, s | 9.43, s | 9.43, s | 1.60, s | — | 1.09, s | 1.10, s |
| 1′ | — | — | — | — | — | — | — | — |
| 2′ | — | — | — | — | — | — | 2.06, s | 2.23, d, 7.2 |
| 3′ | — | — | — | — | — | — | — | 2.10–2.17, m |
| 4′ | — | — | — | — | — | — | — | 0.96, d, 6.6 |
| 5′ | — | — | — | — | — | — | — | 0.96, d, 6.6 |
Table 18.
1H-NMR data of compounds 93–100.
| NO. | 93 [42] | 94 [31] | 95 [7] | 96 [8] | 97 [40] | 98 [42] | 99 [42] | 100 [69] |
|---|---|---|---|---|---|---|---|---|
| CD3OD | CDCl3 | CDCl3 | CDCl3 | CD3OD | CD3OD | CD3OD | C5D5N | |
| 1α | 3.80, dd, 10.0, 5.0 | 3.86, dd, 11.7, 5.1 | 3.43, dd, 11.6, 4.6 | 3.43, dd, 11.6, 4.6 | 3.12, dd, 11.0, 3.4 | — | — | 3.58, dd, 11, 4 |
| 1β | — | — | — | — | — | — | — | — |
| 2α | 2.08, m | 1.89, m | 1.78–1.87, m | 1.57, m | 1.46, m | 5.30, br s | 5.31, br s | — |
| 2β | 1.84, m | 2.14, m | 1.55–1.60, m | 1.82, m | 1.93, m | — | — | — |
| 3α | 2.21, m | 2.42, m | 2.32, ddd, 13.6, 4.9, 2.2 | 2.10, m | 1.49, m | 2.00, m | 2.03, m | — |
| 3β | 2.53, m | 2.06–2.17, m | 2.32, m | 1.58, m | 2.40, m | 2.41, m | — | |
| 4 | — | — | — | — | — | 3.67, dd, 10.0, 6.5 | 3.72, dd, 10.0, 6.5 | — |
| 5 | 1.64, m | 2.24, d, 9.9 | 1.75–1.82, m | 1.77, m | 0.96, br s | — | — | 2.84, d, 6 |
| 6α | — | — | 1.67–1.74, m | 1.25, m | 5.78, s | 1.25, m | 1.26, m | 4.92, br t, 5.5 |
| 6β | 4.14, t, 10.0 | 4.22, dd, 10.2, 9.9 | 1.39–1.46, m | 1.67, m | — | 2.37, m | 2.40, m | — |
| 7 | 2.41, m | 2.54, m | 1.98–2.08, m | 2.00, m | 2.11, d, 13.6 | 1.95, m | 2.02, m | — |
| 8α | 1.64, m | 1.67, m | 1.64–1.71, m | 1.35, m | 1.56, m | 1.57, m | 1.50, m | — |
| 8β | 1.74, m | 1.33–1.40, m | 1.69, m | 1.87, m | 1.72, m | 1.67, m | — | |
| 9α | 1.36, m | 1.40, m | 1.97, t, 3.3 | 1.21, m | 1.09, m | 1.25, m | 1.26, m | — |
| 9β | 1.81, m | 1.87, m | 1.17–1.27, m | 1.98, m | 1.96, m | 1.72, m | 1.84, m | — |
| 10 | — | — | — | — | — | 2.02, m | 2.04, m | — |
| 11 | — | — | — | — | — | 1.13, d, 7.0 | — | — |
| 12 | — | — | 4.59, s | 4.59, s | 1.41, s | 1.58, s | — | 1.57, s |
| 13α | 6.23, s | 6.27, br s | 5.07, d, 0.9 | 5.10, br s | 1.45, s | 0.81, s | 5.58, s | 1.58, s |
| 13β | 5.67, s | 5.69, br s | 5.02, s | 5.00, br s | 6.14, s | |||
| 14 | 0.76, s | 0.88, s | 0.71, s | 0.71, s | 1.31, s | — | 1.58, s | 1.08, s |
| 15α | — | — | 4.77, d, 1.5 | 4.50, br s | 1.28, s | — | 0.85, s | 5.03, br s |
| 15β | — | — | 4.50, d, 1.5 | 4.77, br s | — | 5.17, br s | ||
| 1′ | — | 3.76, s | — | — | — | 4.30, d, 7.5 | 4.30, d, 8.0 | 5.09, d, 8 |
| 2′ | — | — | 2.10, s | 2.23, m | 1.98, s | 3.14, m | 3.14, m | 3.96, t, 8.5 |
| 3′ | — | — | — | 2.13, m | — | 3.31, m | 3.30, m | 4.20, t, 9 |
| 4′ | — | — | — | 0.97, d, 6.6 | — | 3.27, m | 3.27, m | 4.24, t, 9 |
| 5′ | — | — | — | 0.97, d, 6.6 | — | 3.22, m | 3.25, m | 3.77, m |
| 6′α | — | — | — | — | — | 3.67, dd, 12.0, 6.5 | 3.65, dd, 12.0, 6.5 | 4.34, dd, 12, 4 |
| 6′β | — | — | — | — | — | 3.85, dd, 11.5, 2.0 | 3.85, dd, 11.5, 2.0 | 4.37, dd, 12, 2 |
Note: The 1H-NMR data of 2″ of compound 97 were 1.98, s.
Table 19.
1H-NMR data of compounds 101–110.
| NO. | 101 [69] | 102 [69] | 103 [70] | 104 [70] | 105 [39] | 106 [39] | 107 [71] | 108 [40] | 109 [27] | 110 [40] |
|---|---|---|---|---|---|---|---|---|---|---|
| C5D5N | C5D5N | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CD3OD | C5D5N | CD3OD | |
| 1α | 3.78, dd, 8, 7 | 3.63, dd, 10, 5 | 1.40–1.82, m | 1.50–2.20, m | 1.15, m | 1.23, m | 3.96, dd, 11.0, 4.8 | 1.38, m | 5.46, dd, 10, 2 | 1.52, m |
| 1β | — | — | 1.62, m | 1.53, m | — | 1.45, m | — | 1.44, dd, 13.1, 7.0 | ||
| 2α | — | — | 1.40–1.82, m | 1.50–2.20, m | 1.40, m | 1.26, m | 1.91, m | 1.55, m | 5.86, dd, 10, 3 | 2.15, m |
| 2β | — | — | 1.80, m | 1.60, m | 2.21, m | 2.20, m | ||||
| 3α | 5.43, br s | — | 1.40–1.82, m | 1.50–2.20, m | 1.56, m | 2.00, m | 2.40, m | 1.53, m | 6.36, br s | 5.81, s |
| 3β | — | — | 2.34, m | 2.51, m | 3.00, d, 11.7 | 6.36, br s | 5.81, s | |||
| 4 | — | — | 2.43, m | — | 2.46, m | — | — | — | — | — |
| 5 | 3.25, br s | 2.66, d, 3 | — | 2.34, m | — | 1.81, d, 12.4 | 2.62, d, 11.0 | 2.08, d, 11.0 | 2.55, br d, 11 | 2.33, d, 10.5 |
| 6α | 4.91, t, 3 | 5.09, t, 3 | 5.13, d, 8 | 1.71, m | 4.94, d, 3.5 | 1.06, q, 12.4 | — | 4.06, t, 11.0 | 4.22, overlapped | 4.25, t, 10.5 |
| 6β | — | — | 1.24, m | 1.42, m | 4.11, t, 11.0 | |||||
| 7 | — | — | 3.56, m | 2.97, m | 3.01, dd, 5.4, 3.5 | 2.38, m | 2.45, m | 1.65, overlapped | 2.50, m | 1.61, m |
| 8α | — | — | 4.80, m | 4.48, m | 5.13, dt, 5.4, 3.0 | 5.04, m | 1.59, m | 1.79, m | — | 1.81, d, 11.7 |
| 8β | — | — | 2.10, m | 1.58, m | — | 1.63, m | ||||
| 9α | — | — | 2.09, dd, 6, 6 | 1.99, m | 2.14, dd, 14.9, 3.0 | 1.46, dd, 15.6, 4.5 | 1.50, m | 1.55, m | — | 1.36, d, 11.2 |
| 9β | — | — | 1.53, m | 1.39, m | 1.51, dd, 14.9, 3.0 | 2.20, dd, 15.6, 2.6 | 2.06, m | 1.35, m | — | 1.58, m |
| 11 | — | — | — | — | — | — | — | 2.43, dq, 10.6, 6.9 | — | 2.37, m |
| 12 | 1.49, s | 1.52, s | — | — | — | — | — | — | — | — |
| 13α | 1.61, s | 1.63, s | 6.17, d, 4 | 6.11, d, 2 | 2.65, d, 17.5 | 2.64, d, 17.5 | 5.41, d, 3.0 | 1.15, d, 6.9 | 5.36, d, 3.1 | 1.16, d, 6.8 |
| 13β | 5.60, d, 4 | 5.57, d, 2 | 2.95, d, 17.5 | 3.02, d, 17.5 | 6.09, d, 3.0 | 6.15, d, 3.2 | ||||
| 14 | 1.20, s | 1.33, s | 1.17, s | 0.82, s | 1.22, s | 0.78, s | 0.86, s | 0.88, s | 0.92, s | 0.97, s |
| 15α | 2.15, br s | 1.47, s | 1.07, s | 4.76, d, 3 | 1.13, d, 7.7 | 4.79, s | — | 6.16, br s | 5.18, br s | 4.38, d, 11.7 |
| 15β | 4.43, d, 3 | 4.43, s | — | 5.13, br s | 4.15, d, 11.7 | |||||
| 17 | — | — | — | — | 2.33, s | 2.34, s | — | — | — | — |
| 1′ | 5.15, d, 8 | 5.30, d, 8 | — | — | — | — | — | 4.48, d, 7.7 | 5.06, d, 7 | 4.39, d, 7.7 |
| 2′ | 4.01, t, 8.5 | 4.01, t, 8.5 | — | — | — | — | — | 3.28, m | — | 3.17, m |
| 3′ | 4.25, t, 8.5 | 4.19, t, 9 | — | — | — | — | — | 3.35, m | — | 3.33, m |
| 4′ | 4.20, t, 9 | 3.94, t, 9 | — | — | — | — | — | 3.35, m | — | 3.28, m |
| 5′ | 3.92, m | 4.06, dt, 8, 2 | — | — | — | — | — | 3.28, m | — | 3.25, m |
| 6′α | 4.31, dd, 12, 5 | 4.16, dd, 11, 8.5 | — | — | — | — | — | 3.84, dd, 12.1, 2.3 | — | 3.86, dd, 11.9, 2.0 |
| 6′β | 4.42, dd, 12, 2 | 4.67, dd, 11, 1.5 | — | — | — | — | — | 3.69, dd, 12.1, 5.0 | — | 3.67, dd, 11.9, 5.1 |
Table 20.
13C-NMR data of compounds 76–110.
| NO. | 76 [12] | 77 [54] | 78 [55] | 79 [56] | 80 [57] | 82 [59] | 83 [60] | 84 [61] | 85 [62] | 86 [64] | 87 [65] | 88 [66] | 89 [67] | 90 [67] | 92 [34] | 93 [42] | 94 [31] |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CD3OD | CDCl3 | CDCl3 | CD3OD | CDCl3 | |
| 1 | 89.9 | 43.3 | 42.2 | 79.3 | 79.7 | 79.1 | 212.2 | 74.4 | 21.9 | 22.6 | 21.3 | 21.2 | 26.5 | 127.5 | 41 | 77.3 | 77.2 |
| 2 | 29.3 | 20.1 | 20.1 | 28.5 | 25.6 | 31.9 | 38.9 | 42.4 | 30.9 | 30.9 | 22.2 | 22 | 27.4 | 128.6 | 20.1 | 31.6 | 30 |
| 3 | 30.7 | 44.6 | 43.5 | 40.8 | 39.5 | 35.1 | 158.6 | 197.5 | 135 | 134.3 | 142.4 | 141.1 | 138.2 | 145.1 | 43.4 | 40.6 | 39.2 |
| 4 | 146.4 | 72.2 | 72.2 | 71.6 | 71.3 | 146.1 | 143.7 | 129.5 | 122.3 | 122.6 | 151.6 | 152.8 | 119.6 | 129.6 | 72.1 | 212.6 | 212.1 |
| 5 | 129.6 | 54.9 | 55 | 52.9 | 50.4 | 55.9 | 53.1 | 161.9 | 39.8 | 37.7 | 41.4 | 39.3 | 38 | 142 | 54.9 | 62.2 | 61.3 |
| 6 | 138.1 | 26 | 27.3 | 25.7 | 26.4 | 67 | 25 | 32.8 | 46.7 | 46.6 | 45.6 | 45.6 | 38.9 | 40.4 | 26.2 | 67.6 | 67.5 |
| 7 | 52.7 | 46.3 | 41.1 | 45.7 | 46.1 | 49.3 | 55.8 | 45.1 | 22.7 | 19.8 | 22.1 | 26.4 | 26.6 | 27.5 | 42.6 | 48.4 | 46 |
| 8 | 35.6 | 26.8 | 26.6 | 26.4 | 26.8 | 18.1 | 26.8 | 26.5 | 42.2 | 40.3 | 41.8 | 40 | 140.8 | 35 | 27.1 | 28.1 | 26.6 |
| 9 | 36.5 | 41 | 44.7 | 40.5 | 39.3 | 36.3 | 35.1 | 37.7 | 72.4 | 70.7 | 72.1 | 70.6 | 134 | 198.1 | 44.6 | 37.5 | 36 |
| 10 | 148 | 34.6 | 34.7 | 38.9 | 38.9 | 41.7 | 59.6 | 41.3 | 50 | 47.9 | 49.6 | 47.6 | 36.1 | 130.5 | 34.6 | 45.7 | 43.8 |
| 11 | 31.9 | 150.7 | 154.1 | 150.3 | 150.6 | 26 | 32.3 | 148.9 | 25.9 | 26.1 | 26.2 | 28.6 | 143.8 | 144.7 | 148.8 | 125.9 | 141.9 |
| 12 | 20.5 | 108.1 | 65.3 | 108.3 | 108.6 | 21.1 | 19.4 | 109.4 | 21.1 | 21.4 | 21.4 | 21.2 | 124.9 | 130.7 | 65.8 | 170.5 | 167.6 |
| 13 | 20.7 | 22.7 | 107.9 | 21 | 20.7 | 16.2 | 21.9 | 20.6 | 15.1 | 15.2 | 15.2 | 15.2 | 170.5 | 171.3 | 110.7 | 144.1 | 125 |
| 14 | 114.6 | 18.6 | 18.7 | 13 | 12.5 | 11.6 | 19.6 | 16.3 | 20.8 | 28.4 | 20.5 | 19.9 | 171.5 | 21.9 | 18.7 | 12.2 | 11.9 |
| 15 | 113.2 | 21 | 22.7 | 22.7 | 29.7 | 107.8 | 192.7 | 11 | 23.8 | 23.7 | 194.5 | 194.6 | 23.9 | — | 22.7 | — | — |
| 1′ | — | — | — | — | — | — | — | — | — | — | — | — | — | — | 172.8 | — | 51.9 |
| 2′ | — | — | — | — | — | — | — | — | — | — | — | — | — | — | 43.5 | — | — |
| 3′ | — | — | — | — | — | — | — | — | — | — | — | — | — | — | 25.7 | — | — |
| 4′ | — | — | — | — | — | — | — | — | — | — | — | — | — | — | 22.4 | — | — |
| 5′ | — | — | — | — | — | — | — | — | — | — | — | — | — | — | 22.4 | — | — |
| NO. | 95 [7] | 96 [8] | 97 [40] | 98 [42] | 99 [42] | 100 [69] | 101 [69] | 102 [69] | 103 [70] | 104 [70] | 105 [39] | 106 [39] | 107 [71] | 108 [40] | 109 [27] | 110 [40] | |
| CDCl3 | CDCl3 | CD3OD | CD3OD | CD3OD | C5D5N | C5D5N | C5D5N | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CD3OD | C5D5N | CD3OD | ||
| 1 | 79.4 | 79.4 | 81.2 | 136.3 | 136.2 | 79.4 | 76.6 | 79 | 41.7 | 42.5 | 42.3 | 42.2 | 76.4 | 43.2 | 127.1 | 38.7 | |
| 2 | 31.6 | 31.6 | 27.6 | 120.6 | 120.6 | 33.3 | 34.3 | 29.9 | 16.7 | 22.7 | 16.9 | 22.8 | 30.5 | 22.7 | 138.3 | 23.9 | |
| 3 | 34.3 | 34.3 | 42.3 | 29 | 29.9 | 33.8 | 122 | 42.5 | 32.7 | 36.8 | 32.9 | 36.9 | 38.6 | 26.7 | 76.2 | 128.2 | |
| 4 | 148.6 | 148.8 | 72.1 | 82.2 | 82.2 | 148.5 | 136.3 | 72.3 | 37.5 | 148.9 | 38.7 | 149.4 | 206.3 | 118.1 | 141.1 | 134.4 | |
| 5 | 47.7 | 47.7 | 54.6 | 37.9 | 37.9 | 53.2 | 53.6 | 59.4 | 149 | 46.2 | 152.8 | 46.7 | 58.4 | 53 | 49 | 49.9 | |
| 6 | 27 | 27 | 70 | 36.3 | 36.5 | 78 | 79.9 | 80.9 | 118.8 | 27.5 | 113.7 | 21.2 | 76.6 | 81.1 | 77.3 | 82.8 | |
| 7 | 41.5 | 41.6 | 51.1 | 42.5 | 41.3 | 45.4 | 44.8 | 45.5 | 39.4 | 40.5 | 47.1 | 47 | 48.9 | 54.1 | 51.7 | 55.1 | |
| 8 | 29.3 | 29.3 | 20.1 | 25.3 | 28.1 | 19.8 | 17.8 | 18.1 | 76.4 | 76.6 | 77.2 | 77.7 | 20.9 | 24.1 | 20.4 | 23.9 | |
| 9 | 37 | 37.1 | 40.7 | 29.8 | 30.2 | 36.4 | 30.8 | 34.6 | 42.6 | 41.3 | 42.7 | 41.4 | 36.1 | 41 | 39.5 | 40.7 | |
| 10 | 40.4 | 40.4 | 41 | 48.3 | 37.9 | 41.2 | 38.8 | 40.4 | 32.6 | 34.3 | 33.1 | 34.7 | 45.5 | 39.4 | 35.7 | 36.9 | |
| 11 | 148.7 | 148.6 | 85.9 | 14.7 | 147.9 | 72.9 | 72.4 | 72.1 | 139.8 | 142.2 | 79.2 | 79.7 | 138.3 | 42.2 | 139.1 | 41.9 | |
| 12 | 66.4 | 66.1 | 24.6 | 21 | 170.9 | 29.6 | 30.2 | 30.4 | 170.3 | 170.5 | 175.5 | 175.5 | 170.2 | 182.1 | 169.4 | 182.4 | |
| 13 | 111.3 | 111.2 | 25.6 | 10.7 | 123 | 31.3 | 30.7 | 30.7 | 121.5 | 119.9 | 43.6 | 42.2 | 117.2 | 12.6 | 115.1 | 12.6 | |
| 14 | 10.4 | 10.4 | 14.3 | — | 21 | 15.7 | 12.6 | 16.5 | 22.5 | 17.6 | 28.8 | 18 | 12.3 | 18.3 | 19.6 | 17.7 | |
| 15 | 107.2 | 107.2 | 29.8 | — | 10.9 | 107.2 | 23.1 | 23.8 | 28.5 | 106.6 | 23.2 | 106.5 | — | 139.1 | 107.5 | 73.4 | |
| 16 | — | — | — | — | — | — | — | — | — | — | 210.4 | 210.5 | — | — | — | — | |
| 17 | — | — | — | — | — | — | — | — | — | — | 32 | 32 | — | — | — | — | |
| 1′ | 171 | 173 | 172.5 | 101.5 | 101.5 | 104.4 | 105.6 | 105.6 | — | — | — | — | — | 104.5 | 104.2 | 103.8 | |
| 2′ | 21.2 | 43.7 | 22.2 | 75.2 | 75.2 | 75.2 | 74.1 | 75.2 | — | — | — | — | — | 74.9 | 74.4 | 75.3 | |
| 3′ | — | 25.9 | — | 78.2 | 78.2 | 78.8 | 78.7 | 78.8 | — | — | — | — | — | 78.2 | 77.5 | 78.3 | |
| 4′ | — | 22.6 | — | 71.9 | 71.9 | 71.5 | 71.8 | 72.4 | — | — | — | — | — | 71.2 | 70.8 | 71.7 | |
| 5′ | — | 22.6 | — | 77.8 | 77.8 | 78.3 | 78.3 | 78.5 | — | — | — | — | — | 78 | 77.5 | 77.8 | |
| 6′ | — | — | — | 63 | 63 | 62.6 | 62.8 | 63 | — | — | — | — | — | 62.4 | 61.8 | 62.8 | |
Note: The 1″ and 2″ data of compound 97 were 172.7 and 22.5.
4.2. Bioactivity of Eudesman-Type Sesquiterpene
Eucalyptane sesquiterpenes exert anti-inflammatory effects by inhibiting the activity of the NLRP3 inflammasome and inhibiting the production of NO in RAW264.7 macrophages. Cyperusol C (79) derived from Ainsliaea pertyoides can inhibit NLRP3 inflammasome activity by inhibiting the LDH release rate [7]. Ainsliatone B (94) strongly inhibited the production of nitric oxide in RAW264.7 macrophages stimulated with lipopolysaccharide (LPS), with IC50 values of 8.78 µM [31].
Isoalantolactone (104) remarkably inhibited the proliferation of MGC803 cell lines with IC50 values of 2.2 ± 0.2 μM. Double-bond moieties may be necessary for its cytotoxicity [34]. Alantolactone (103) exhibited significant inhibition against the human tumor cell lines A549, HCT116, MGC803, and CCRF-CEM with IC50 values of 3.56, 2.23, 2.89, and 14.67 µM, respectively [39].
5. Polymer Sesquiterpene Lactones
In Ainsliaea plants, polymer sesquiterpene lactones are typically formed through the polymerization of two or three sesquiterpene units; most monomeric precursors belong to guaiane sesquiterpenes. The structures of the 25 reported polymer sesquiterpene lactones can be found in Figure 4 and Table 21.
Figure 4.
Chemical structures for compounds 111–135.
Table 21.
The compound name, molecular formula, and NMR test reagent of polymer sesquiterpene lactones.
| No. | Compound Name | Molecular Formula | Solvent | Ref. |
|---|---|---|---|---|
| 111 | Macrocephadiolide B | C30H34O8 | CD3OD | [72] |
| 112 | Ainsliadimer J | C30H34O7 | CDCl3 | [72] |
| 113 | Ainsliadimer A | C30H34O7 | CDCl3 | [73] |
| 114 | Macrocephadiolide A | C30H32O8 | CDCl3 | [72] |
| 115 | Japonicone A | C32H40O7 | CDCl3 | [74] |
| 116 | Ainsliadimer B | C30H32O8 | CDCl3 | [75] |
| 117 | Ainsliadimer C | C30H36O7 | CDCl3 | [31] |
| 118 | Ainsliadimer D | C30H36O8 | DMSO | [31] |
| 119 | Gochnatiolide A | C30H30O7 | CDCl3 | [76,77] |
| 120 | Ainsliadimer F | C31H36O6 | CDCl3 | [47] |
| 121 | Ainsliadimer I | C31H34O6 | CDCl3 | [47] |
| 122 | Ainsliadimer G | C32H36O7 | CDCl3 | [47] |
| 123 | Ainsliadimer H | C33H38O7 | CDCl3 | [47] |
| 124 | Gochnatiolide C | C30H30O6 | CDCl3 | [76] |
| 125 | Gochnatiolide B | C30H30O7 | CDCl3 | [76] |
| 126 | Gochnatiolide E | C30H30O8 | CDCl3 | [78] |
| 127 | Gochnatiolide F | C30H34O7 | CDCl3 | [78] |
| 128 | Macrocephatriolide B | C45H50O10 | CDCl3 | [79] |
| 129 | Macrocephatriolide A | C45H46O10 | CDCl3 | [79] |
| 130 | Ainsliatriolides A | C45H48O10 | CDCl3 | [80] |
| 131 | Ainsliatriolide C | C45H48O11 | CDCl3 | [81] |
| 132 | Ainsfragolide | C45H46O10 | CDCl3 | [78] |
| 133 | Ainsliatrimer A | C45H44O10 | CDCl3 | [75] |
| 134 | Ainsliatrimer B | C45H44O10 | CDCl3 | [75] |
| 135 | Ainsliatriolides B | C46H50O11 | CDCl3 | [80] |
5.1. NMR Data of Polymer Sesquiterpene Lactones (111–135)
The 1H and 13C NMR spectroscopy are summarized in Table 22, Table 23, Table 24, Table 25 and Table 26. This paper also provides an overview of the nuclear magnetic resonance testing instruments used for compounds 111–135. The 1H and 13C spectra of compounds 111, 112, 114, 130, and 131 were measured using the Bruker Avance III-500 instrument. The NMR data for compounds 113, 117, 118, 120, 121, 122, and 123 were recorded on the Bruker Avance 600 spectrometer. For compounds 115 and 116, the respective 1H and 13C spectra were obtained by the Bruker Avance 400 instrument. Compound 119’s NMR data were acquired on the Bruker AC 200 spectrometer. NMR data for compounds 119, 124, and 125 were acquired using the Bruker AV500 instrument. The 1H and 13C data of compounds 126, 127, and 132 were taken with the Bruker Avance AV500 spectrometer. Data for compounds 128, 129, and 135 were performed on the Bruker Avance III-600 instrument. NMR data collection for compounds 133 and 134 was conducted using the Bruker Avance 400 instrument.
Table 22.
1H-NMR data of compounds 111–118.
| NO. | 111 [72] | 112 [72] | 113 [73] | 114 [72] | 115 [74] | 116 [75] | 117 [31] | 118 [31] |
|---|---|---|---|---|---|---|---|---|
| CD3OD | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | DMSO | |
| 1 | 3.24, dd, 8.0, 2.3 | 3.09, ddt, 20.6, 7.7, 4.2 | 3.15, dd, 12.0, 11.4 | 3.11, dd, 17.0, 7.0 | 3.28, dd, 11.7, 3.8 | — | — | — |
| 2α | 2.63, m | 2.64–2.54, m | 2.38, dd, 13.8, 12.0 | 2.68, m | 1.82, m | — | — | — |
| 2β | 2.43, m | 2.29, tdt, 10.8, 5.0, 3.0 | 1.88–1.92 | 1.62, m | — | — | — | |
| 3α | — | — | — | — | 1.65, m | — | — | — |
| 3β | — | — | — | — | 1.57, m | — | — | — |
| 4 | 2.40, m | 2.29, tdt, 10.8, 5.0, 3.0 | — | — | 2.45, m | 2.68, m | 2.50, m | 2.37, m |
| 5 | 2.54, q, 9.4 | 2.43–2.35, m | 2.55, t, 11.4 | 2.53, t, 10.0 | — | 3.33, dd, 9.8, 4.3 | 2.78, dd, 11.4, 4.2 | 3.37, dd, 11.4, 4.8 |
| 6 | 4.08, t, 9.2 | 3.95, t, 9.2 | 4.07, dd, 11.4, 9.0 | 4.09, dd, 10.0, 8.8 | 5.37, d, 3.0 | 4.35, t, 9.8 | 4.29, t, 10.8 | 4.39, t, 10.8 |
| 7 | 3.16, m | 2.92–2.81, m | 2.72, m | 2.95, tt, 8.8, 3.0 | 2.79, dd, 5.5, 3.2 | 2.80, m | 1.82, overlap | 2.84, m |
| 8α | 2.38, m | 2.12–1.91, m | 2.22–2.24, m | 2.34, m | — | 2.05, m | 2.00, overlap | 2.04, m |
| 8β | 1.49, m | 1.55–1.41, m | 1.37, dq, 12.6, 6.0 | 1.49, m | 4.89, dd, 5.3, 2.7 | 2.10, m | 1.82, overlap | 1.90, m |
| 9α | 2.63, m | 2.64–2.54, m | 2.67, m | 2.71, m | 2.59, dd, 14.8, 3.3 | 1.88, m | 2.00, overlap | 1.68, m |
| 9β | 2.29, td, 12.5, 5.7 | 1.76, ddd, 14.0, 11.6, 5.0 | 1.88–1.92 | 2.16, td, 12.8, 5.0 | 1.53, dd, 15.1, 2.0 | 2.05, m | 1.72, m | 1.81, overlap |
| 11 | — | — | — | — | — | — | 2.36, m | — |
| 13α | 6.20, d, 3.2 | 6.26, dd, 5,1, 3.4 | 6.19, d, 3.0 | 6.26, d, 3.2 | 2.00, m | 5.53, d, 3.0 | 1.27, d, 7.2 | 6.01, d, 3.0 |
| 13β | 5.68, d, 3.2 | 5.55, dd, 5.3, 3.1 | 5.46, d, 3.0 | 5.57, d, 3.2 | 1.88, m | 6.21, d, 3.0 | 5.62, d, 3.0 | |
| 14α | 5.00, s | 5.05, d, 1.1 | 5.15, s | 5.20, s | 1.19, s | 1.95, m | 1.82, overlap | 1.81, overlap |
| 14β | 4.69, s | 4.69, s | 5.03, s | 4.84, s | 1.70, m | 1.69, m | 1.54, m | |
| 15α | 2.09, m | 2.12–1.91, m | 2.25–2.30 | 2.16, m | 1.11, d, 7.6 | 3.94, dd, 11.0, 4.0 | 1.28, d, 7.2 | 3.88, m |
| 15β | 1.90, m | 1.65–1.56, m | 2.16–2.21 | 3.62, m | ||||
| 1′ | 3.22, m | 2.96, ddq, 12.0, 9.3, 3.2 | 3.04–3.06 | 2.97, q, 9.6 | — | 3.22, m | 3.18, t, 9.0 | 3.27, t, 9.0 |
| 2′α | 2.61, dd, 16.2, 5.5 | 2.50–2.46, m | 2.91, dd, 16.8, 9.6 | 3.25, dd, 15.6, 10.2 | 4.60, s | 3.23, m | 3.22, m | 3.27, overlap |
| 2′β | 2.46, dd, 16.2, 9.4 | 2.29, tdt, 10.8, 5.0, 3.0 | 2.45, d, 16.8 | 2.68, m | — | 2.66, m | 2.62, d, 18.6 | 2.49, overlap |
| 3′ | — | — | — | — | 2.86, d, 1.4 | — | — | — |
| 5′ | 3.26, m | 3.09, ddt, 20.6, 7.7, 4.2 | 3.04–3.06 | 2.53, t, 10.1 | — | 3.20, m | 3.12, t, 9.6 | 2.89, t, 9.6 |
| 6′α | — | — | — | — | 3.03, d, 15.5 | — | — | — |
| 6′β | 4.39, t, 9.5 | 4.27, dd, 11.0, 8.8 | 4.12, dd, 10.2, 8.4 | 4.82, dd, 10.1, 9.0 | 2.08, m | 4.12, t, 10.5 | 4.14, t, 9.6 | 4.25, t, 9.6 |
| 7′ | 3.07, m | 2.92–2.81, m | 3.04–3.06 | 2.89, ddd, 11.9, 9.0, 3.1 | 2.82, s | 3.01, m | 2.07, m | 3.18, m |
| 8′α | 2.30, m | 2.12–1.91, m | 2.25–2.30 | 1.56, m | — | 1.49, m | 2.19, overlap | 2.27, m |
| 8′β | 1.53, m | 1.55–1.41, m | 1.50, dq, 12.6, 3.6 | 2.28, m | 4.21, ddd, 12.4, 8.4, 3.2 | 2.30, m | 1.40, m | 1.45, m |
| 9′α | 2.55, m | 2.50–2.46, m | 2.16–2.21 | 2.06, m | 2.36, dt, 13.1, 4.1 | 2.21, m | 2.58, m | 2.49, br s |
| 9′β | 2.32, m | 2.19, td, 12.5, 5.7 | 2.57, m | 2.68, m | 2.00, m | 2.62, m | 2.12, overlap | 2.16, m |
| 10′ | — | — | — | — | 2.15, m | — | — | — |
| 11′ | — | — | — | — | — | — | 2.19, overlap | — |
| 13′α | 6.18, d, 3.2 | 6.26, dd, 5,1, 3.4 | 6.31, d, 3.0 | 6.33, d, 3.2 | 6.22, d, 3.3 | 5.57, d, 3.0 | 1.24, d, 6.6 | 6.04, d, 3.0 |
| 13′β | 5.67, d, 3.2 | 5.55, dd, 5.3, 3.1 | 5.61, d, 3.0 | 5.57, d, 3.2 | 5.54, d, 3.1 | 6.26, d, 3.0 | 5.68, d, 3.0 | |
| 14′α | 5.05, s | 5,14, s | 4.96, s | 5.19, s | 1.05, d, 7.3 | 5.09, s | 5.06, s | 5.83, s |
| 14′β | 4.99, s | 4.99, s | 4.59, s | 5.10, s | 4.73, s | 4.70, s | 5.03, s | |
| 15′α | 2.97, ddd, 18.3, 9.0, 5.3 | 2.72, ddd, 13.2, 4.8, 3.0 | 2.05, dd, 14.4, 5.4 | 2.80, m | 1.67, d, 1.5 | 2.15, m | 2.12, overlap | 2.20, m |
| 15′β | 2.74, ddd,18.3, 9.0, 6.2 | 2.64–2.54, m | 2.01, dd, 14.4, 7.8 | 2.25, m | 2.06, m | 2.05, m | 1.85, m |
Note: The 1H-NMR data of 2″ of 115 were 2.08, s.
Table 23.
1H-NMR data of compounds 119–127.
| NO. | 119 [76] | 120 [47] | 121 [47] | 122 [47] | 123 [47] | 124 [76] | 125 [76] | 126 [78] | 127 [78] |
|---|---|---|---|---|---|---|---|---|---|
| CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | |
| 4 | — | 2.52, dd, 7.2, 4.2 | 2.56, dd, 7.1, 4.2 | 2.63, m | 2.62, m | — | — | — | 2.46, m |
| 5 | 3.92, d, 10.4 | 2.80, dd, 11.1, 4.1 | 2.91, dd, 11.2, 4.2 | 3.52, dd, 12.3, 5.3 | 3.52, dd, 11.3, 4.1 | 3.55, d, 10.4 | 3.89, d, 10.8 | 3.62, d, 10.0 | 3.03, dd, 1.5, 10.2 |
| 6 | 3.76, t, 9.9 | 4.32, dd, 11.0, 9.6 | 4.33, dd, 11.1, 9.7 | 4.38, dd, 11.2, 9.8 | 4.36, dd, 11.3, 9.7 | 3.92, d, 9.9 | 4.36, t, 10.7 | 4.59, d, 10.0 | 3.75 |
| 7 | 3.88, m | 1.84, m | 2.75, mH | 2.87, m | 2.86, m | 3.23–3.29, m | 2.86, m | — | 3.75 |
| 8α | 2.34–2.44, m | 2.05, m | 2.10, m | 2.16, m | 2.11, m | 2.32–2.44, m | 2.04–2.11, m | 3.41 | 2.39, m |
| 8β | 1.60–1.67, m | 1.85, m | 2.02, m | 2.08, m | 2.03, m | 1.68–1.75, m | 2.04–2.11, m | 2.66 | 2 |
| 9α | 2.06–2.16, m | 1.97, m | 1.71, m | 2.00, m | 1.95, m | 2.00–2.07, m | 2.04–2.11, m | 2.22 | 2.07, m |
| 9β | 2.06–2.16, m | 1.70, m | 1.68, m | 1.73, m | 1.73, m | 1.61–1.68, m | 1.84–2.01, m | 2.09 | 2.03 |
| 10 | — | — | — | — | — | 2.74, m | — | — | — |
| 11 | — | 2.35, m | — | — | — | — | — | — | — |
| 13α | 6.21, d, 3.7 | 1.27, d, 6.9 | 6.20, d, 3.2 | 6.23, d, 3.3 | 6.21, d, 3.3 | 6.28, d, 3.4 | 6.23, d, 3.4 | 4.43, dd, 13.3, 19.5 | 6.18, d, 3.2 |
| 13β | 5.49, d, 3.3 | 5.52, d, 3.1 | 5.56, d, 3.0 | 5.54, d, 3.1 | 5.57, d, 3.1 | 5.56, d, 3.1 | 5.45, d, 3.2 | ||
| 14α | 1.87–2.00, m | 1.86, m | 1.86, m | 2.12, m | 2.09, m | 2.00–2.07, m | 1.84–2.01, m | 1.95 | 1.87 |
| 14β | 1.87–2.00, m | 1.69, m | 1.69, m | 1.94, m | 1.93, m | 1.84–1.97, m | 1.74, m | ||
| 15α | 6.22, br s | 1.27, d, 7.1 | 1.30, d, 7.2 | 3.94, dd, 9.5, 2.2 | 3.96, dd, 9.6, 2.3 | 6.18, br s | 6.24, br s | 6.26, s | 1.23, d, 7.6 |
| 15β | 6.03, br s | 3.74, dt, 9.5, 3.2 | 3.76, dd, 9.7, 3.1 | 6.00, br s | 6.16, br s | 6.05, s | |||
| 1′ | 3.25, t, 9.6 | 3.22, m | 3.22, t, 9.0 | 3.22, m | 3.24, t, 9.3 | 3.23–3.29, m | 3.27, m | 3.27 | 3.17, t, 8.9 |
| 2′α | 3.37, t, 10.0 | 3.22, m | 3.23, m | 3.22, m | 2.68, m | 3.33–3.40, m | 3.38, m | 3.32 | 3.23, m |
| 2′β | 2.63, m | 2.63, m | 2.64, m | 2.63, m | 2.62, m | 2.62–2.66, m | 2.70, m | 2.68 | 2.61 |
| 5′ | 3.34, t, 10.5 | 3.20, d, 4.9 | 3.19, t, 8.7 | 3.25, t, 10.2 | 3.17, t, 9.3 | 3.33–3.40, m | 3.28, m | 3.39 | 3.11, t, 9.4 |
| 6′ | 4.24, t, 9.2 | 4.16, t, 9.4 | 4.17, t, 9.2 | 4.19, t, 9.2 | 4.18, t, 9.8 | 4.21, t, 9.5 | 4.21, m | 4.26, t, 9.8 | 4.20, t, 9.4 |
| 7′ | 3.08, m | 3.00, m | 3.01, m | 3.05, m | 3.03, m | 3.06, m | 3.03, m | 3.08, m | 2.15 |
| 8′α | 2.34–2.44, m | 2.31, m | 2.35, m | 2.35, m | 2.33, m | 2.32–2.44, m | 2.34, m | 2.35, m | 2.20, m |
| 8′β | 1.49–1.57, m | 1.47, m | 1.48, m | 1.51, m | 1.51, m | 1.51, m | 1.50, m | 1.54, m | 1.42, m |
| 9′α | 2.66, br s | 2.64, m | 2.65, m | 2.65, m | 2.64, m | 2.62–2.66, m | 2.66, s | 2.66 | 2.57 |
| 9′β | 2.34–2.44, m | 2.20, m | 2.22, m | 2.24, m | 2.23, m | 2.26, m | 2.25, m | 2.26 | 2.13 |
| 11′ | — | — | — | — | — | — | — | — | 2.25, m |
| 13′α | 6.30, d, 3.4 | 6.26, d, 3.4 | 6.27, d, 3.4 | 6.29, d, 3.4 | 6.27, d, 3.4 | 6.29, d, 3.4 | 6.28, d, 3.4 | 6.29, d, 3.0 | 1.26, d, 7.0 |
| 13′β | 5.64, d, 3.1 | 5.58, d, 3.0 | 5.58, d, 3.0 | 5.60, d, 3.0 | 5.59, d, 3.0 | 5.60, d, 3.0 | 5.60, d, 3.1 | 5.65, d, 3.0 | |
| 14′α | 5.10, s | 5.09, br s | 5.09, br s | 5.12, br s | 5.11, br s | 5.08, s | 5.12, s | 5.11, s | 5.05, s |
| 14′β | 4.71, s | 4.72, br s | 4.72, br s | 4.75, br s | 4.74, br s | 4.73, s | 4.75, s | 4.75, s | 4.67, s |
| 15′α | 2.06–2.16, m | 2.13, m | 2.11, m | 2.12, m | 2.12, m | 2.00–2.07, m | 2.04–2.11, m | 2.15 | 2.1 |
| 15′β | 1.87–2.00, m | 2.02, m | 1.97, m | 2.07, m | 2.07, m | 1.84–1.97, m | 2.04–2.11, m | 1.92 | 1.85 |
| 1″ | — | — | — | 3.29, s | 3.42, dd, 7.0, 3.5 | — | — | — | — |
| 2″ | — | — | — | — | 1.06, t, 7.0 | — | — | — | — |
Table 24.
1H-NMR data of compounds 128–135.
| NO. | 128 [79] | 129 [79] | 130 [80] | 131 [81] | 132 [78] | 133 [75] | 134 [75] | 135 [80] | NO. | 128 [79] | 129 [79] | 130 [80] | 131 [81] |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | ||
| 1 | 3.06, m | — | — | — | — | — | — | — | 1′ | 2.72, m | 3.07, dd, 10.6, 5.9 | 3.28, m | 3.28, m |
| 2 | 2.51–2.58, m | — | — | — | — | — | — | — | 2′α | 3.43, q, 7.3 | 3.36, dt, 8.5, 5.9 | 3.27, m | 3.26, m |
| 4 | 2.22, m | — | 2.66, m | — | — | — | — | — | 2′β | 2.62, m | 2.63, m | ||
| 5 | 2.45, m | 3.91, d, 10.3 | 3.23, dd, 11.0, 3.7 | 3.40, m | 3.94, d, 9.0 | 3.43, d, 10.3 | 3.87, d, 10.3 | 3.07, m | 5′ | 2.47, t, 11.1 | 3.43, m | 3.34, m | 3.26, m |
| 6 | 3.92, t, 9.2 | 3.76, dd, 10.3, 10.3 | 4.21, dd, 11.0, 9.8 | 4.47, t, 10.0 | 3.80, d, 9.0 | 3.78, t, 10.3 | 3.64, t, 10.3 | 4.48, t, 10.5 | 6′ | 4.32, dd, 11.1, 8.7 | 4.25, dd, 11.0, 8.6 | 4.12, t, 9.4 | 4.12, t, 10.0 |
| 7 | 2.97, m | 3.85, m | 3.00, m | 3.02, m | 3.89, m | 3.21, m | 3.78, m | 2.66, m | 7′ | 2.79, m | 2.99, m | 3.12, m | 3.12, m |
| 8α | 2.20, m | 2.36, m | 2.06, m | 2.07, m | 2.4 | 1.82, m | 1.80, m | 1.91, m | 8′α | 2.24, m | 2.30, m | 2.32, m | 2.28, m |
| 8β | 1.44, m | 1.56, m | 2.02, m | 1.98, m | 1.61 | 2.35, m | 2.34, m | 1.95, m | 8′β | 1.50, m | 1.46, m | 1.48, m | 1.48, m |
| 9α | 2.56, m | 2.05, m | 2.05, m | 2.03, m | 2.08, m | 2.00, m | 2.05, m | 1.67, m | 9′α | 2.64, m | 2.69, m | 2.61, m | 2.61, m |
| 9β | 2.18, m | 1.92, m | 2.01, m | 1.75, m | 1.95, m | 2.38, m | 2.49, m | 1.92, m | 9′β | 1.97, td, 12.8, 4.5 | 2.18, m | 2.27, m | 2.21, m |
| 10 | — | — | — | — | — | 2.77, m | — | — | 13′α | 6.23, d, 3.3 | 6.22, d, 3.2 | 6.15, d, 3.2 | 6.17, d, 3.4 |
| 13α | 6.15, d, 3.2 | 6.16, d, 3.4 | 6.15, d, 3.2 | 6.11, d, 3.2 | 6.21, d, 3.2 | 5.57, d, 3.0 | 5.49, d, 3.4 | 5.43, d, 3.3 | 13′β | 5.52, d, 3.3 | 5.58, d, 3.2 | 5.50, d, 3.2 | 5.52, d, 3.4 |
| 13β | 5.49, d, 3.2 | 5.45, d, 3.4 | 5.50, d, 3.2 | 5.47, d, 3.2 | 5.48, d, 3.2 | 6.25, d, 3.0 | 6.19, d, 3.4 | 6.10, d, 3.3 | 14′α | 5.37, br s | 5.18, br s | 5.08, br s | 5.08, s |
| 14α | 4.96, br s | 1.98, m | 1.84, m | 1.84, m | 2 | 2.55, m | 2.65, m | 1.78, td, 13.9, 3.7 | 14′β | 5.13, br s | 5.10, br s | 4.71, br s | 4.74, s |
| 14β | 4.66, br s | 1.88, m | 1.70, m | 1.73, m | 1.93, m | 1.64, m | 1.93, m | 1.56, m | 15′α | 2.04, m | 2.28, m | 2.18, m | 2.13, m |
| 15α | 1.98, m | 6.17, br s | 1.91, m | 2.15, m | 6.22, br s | 5.97, s | 5.99, s | 2.51, m | 15′β | 1.85, td, 12.8, 4.7 | 1.65, td, 14.3, 3.7 | 2.05, m | 2.01, m |
| 15β | 1.43, m | 5.97, br s | 1.84, m | 1.86, m | 6.02, br s | 6.26, s | 6.30, s | 2.13, m | |||||
| NO. | 132 [78] | 133 [75] | 134 [75] | 135 [80] | NO. | 128 [79] | 129 [79] | 130 [80] | 131 [81] | 132 [78] | 133 [75] | 134 [75] | 135 [80] |
| CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | ||
| 1′ | 3.10, m | — | — | 3.06, m | 1″ | 3.10, m | 3.11, td, 8.7, 4.5 | 3.06, m | 3.09, m | 3.14, m | 3.14, m | 3.13, m | — |
| 2′α | — | — | — | 3.07, m | 2″α | 2.45–2.55, m | 2.56, m | 2.48, m | 2.47, m | 2.6 | 3.11, m | 3.07, m | — |
| 2′β | 3.39, m | — | — | 2.71, m | 2″β | 2.43, m | 2.46, m | 2.29, m | 2.48 | 2.58, m | 2.60, m | — | |
| 5′ | 3.46, t, 10.7 | 3.19, d, 10.0 | 3.17, d, 9.6 | 3.24, t, 9.3 | 4″ | 2.62, m | 2.52, m | 2.29, m | 2.29, m | 2.55 | — | — | — |
| 6′ | 4.29, dd, 10.7, 8.7 | 4.36, t, 10.0 | 4.35, t, 9.6 | 4,23, t, 9.3 | 5″ | 2.55, m | 2.40, m | 2.51, m | 2.52, m | 2.44 | 2.95, m | 2.94, t, 9.6 | 2.37, m |
| 7′ | 3.04 | 2.65, m | 2.65, m | 2.89, m | 6″ | 3.96, t, 9.1 | 3.98, dd, 9.3, 9.3 | 3.94, t, 9.1 | 3.89, t, 10.0 | 4.02, t, 9.2 | 4.22, t, 9.5 | 4.22, t, 9.6 | 4.19, dd, 11.3, 8.8 |
| 8′α | 2.32 | 1.99, m | 1.99, m | 1.50, m | 7″ | 3.04 | 2.96, m | 2.96, m | 3.03, m | 2.99 | 2.99, m | 3.00, m | 2.77, m |
| 8′β | 1.5 | 2.15, m | 2.08, m | 2.31, m | 8″α | 2.19, m | 2.29, m | 2.27, m | 2.16, m | 2.33 | 1.50, m | 1.52, m | 1.56, m |
| 9′α | 2.73, m | 1.80, m | 1.78, m | 2.16, m | 8″β | 1.47, m | 1.46, m | 1.41, m | 1.39, m | 1.5 | 2.34, m | 2.31, m | 2.18, m |
| 9′β | 2.22 | 1.94, m | 2.10, m | 2.66, m | 9″α | 2.56, m | 2.55, m | 2.56, m | 2.57, m | 2.59 | 2.20, m | 2.31, m | 2.27, m |
| 13′α | 6.27, d, 3.2 | 5.48, d, 3.0 | 5.48, d, 3.2 | 5.59, d, 3.2 | 9″β | 2.18, m | 2.20, m | 2.18, m | 2.19, m | 2.22 | 2.61, m | 2.63, m | 2.35, m |
| 13′β | 5.62, d, 3.2 | 6.10, d, 3.0 | 6.11, d, 3.2 | 6.28, d, 3.2 | 13″α | 6.12, d, 3.2 | 6.23, d, 3.2 | 6.19, d, 3.2 | 6.17, d, 3.2 | 6.28, d, 3.0 | 6.27, d, 3.0 | 6.28, d, 3.2 | 6.16, d, 3.4 |
| 14′α | 5.22, s | 2.10, m | 2.09, m | 5.13, br s | 13″β | 5.47, d, 3.2 | 5.54, d, 3.2 | 5.50, d, 3.2 | 5.52, d, 3.2 | 5.57, d, 3.0 | 5.59, d, 3.0 | 5.59, d, 3.2 | 5.45, d, 3.4 |
| 14′β | 5.14, s | 1.80, m | 1.79, m | 4.79, br s | 14″α | 4.99, br s | 4.98, br s | 4.96, br s | 4.94, s | 5.02, s | 5.09, s | 5.10, s | 4.93, br s |
| 15′α | 2.32 | 2.17, m | 2.17, m | 1.90, m | 14″β | 4.65, br s | 4.73, br s | 4.63, br s | 4.59, s | 4.77, s | 4.76, s | 4.77, s | 4.89, br s |
| 15′β | 1.67 | 1.85, m | 1.99, m | 2.08, m | 15″α | 2.29, m | 2.29, m | 1.76, m | 1.68, m | 2.32 | 2.30, m | 2.30, m | 2.18, m |
| 15″β | 1.67, ddd, 13.9, 7.5, 5.8 | 1.96, m | 1.59, m | 1.48, m | 2.03 | 2.06, m | 2.30, m | 1.99, m |
Table 25.
13C-NMR data of compounds 111–127.
| NO. | 111 [72] | 112 [72] | 113 [73] | 114 [72] | 115 [74] | 116 [75] | 117 [31] | 118 [31] | 119 [77] | 120 [47] | 121 [47] | 122 [47] | 123 [47] | 124 [76] | 125 [76] | 126 [78] | 127 [78] |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CD3OD | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | DMSO | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | |
| 1 | 41 | 38.3 | 41.8 | 36.6 | 80.7 | 173.6 | 171.8 | 172.7 | 170.6 | 172 | 171.6 | 173.2 | 173.1 | 171 | 169 | 169.6 | 174 |
| 2 | 45.3 | 45.9 | 38.1 | 39.8 | 26 | 140.7 | 140.3 | 140.5 | 143.2 | 140.2 | 140.4 | 140.6 | 140.8 | 142.1 | 142.7 | 142.9 | 140.6 |
| 3 | 221 | 218.5 | 90.5 | 213.9 | 29.7 | 207.3 | 208.5 | 206.1 | 194.1 | 208.5 | 208.4 | 206.3 | 206.5 | 193.8 | 193.6 | 193.8 | 210 |
| 4 | 51.6 | 52.1 | 89.1 | 90.8 | 38.2 | 53.6 | 46.9 | 53.6 | 142 | 47 | 47 | 52 | 51.8 | 141.8 | 141.1 | 142.1 | 45 |
| 5 | 49.1 | 48.8 | 53.1 | 49.1 | 149.5 | 49 | 54.9 | 46.9 | 47.3 | 54.8 | 55.1 | 48.3 | 48.5 | 51 | 49.7 | 46.5 | 52.6 |
| 6 | 90.1 | 87.9 | 82.9 | 81.7 | 118.1 | 82.3 | 82.6 | 82.2 | 83.6 | 82.6 | 82.6 | 82.6 | 82.6 | 82.7 | 81.2 | 84 | 85 |
| 7 | 45.1 | 44.4 | 49 | 44.4 | 42.2 | 51.6 | 54.7 | 50.8 | 43.4 | 54.9 | 51.6 | 51.6 | 51.7 | 43.7 | 52.4 | 166 | 43.3 |
| 8 | 32.7 | 31.7 | 31.3 | 31.9 | 75.3 | 21 | 22.5 | 20.7 | 35.3 | 22.5 | 21 | 21 | 21 | 25.3 | 20.8 | 21.7 | 23 |
| 9 | 39 | 37.9 | 37.8 | 39.2 | 39.8 | 36 | 36.3 | 36.7 | 38.4 | 36.2 | 36.1 | 36.2 | 36.2 | 27.7 | 35.9 | 35.2 | 35 |
| 10 | 150.9 | 148.6 | 147.5 | 148 | 38.5 | 68.3 | 68.2 | 67.3 | 71.1 | 68.1 | 68.3 | 68.4 | 68.4 | 33.3 | 68.2 | 69.3 | 71.2 |
| 11 | 141.1 | 139 | 139.5 | 138.4 | 56.8 | 139.3 | 41.9 | 140 | 140.3 | 41.9 | 139.4 | 139.5 | 139.6 | 139.3 | 138.8 | 126 | 140.6 |
| 12 | 171.9 | 169.7 | 170 | 169.1 | 178.8 | 169.7 | 178.5 | 169.6 | 169.9 | 178.6 | 170 | 170.1 | 170.1 | 168.4 | 169.8 | 172.8 | 170 |
| 13 | 121.6 | 121.4 | 119.7 | 121.8 | 36.4 | 119.3 | 12.6 | 118.4 | 119.6 | 12.6 | 119.1 | 119.1 | 119.1 | 120.7 | 119.2 | 54.6 | 119.5 |
| 14 | 113.4 | 113.8 | 113.8 | 114.4 | 21.6 | 36.2 | 36.3 | 36 | 28.3 | 36.3 | 36.3 | 36 | 36 | 31.2 | 36.3 | 37.3 | 38.6 |
| 15 | 23.6 | 22.8 | 33.1 | 28.6 | 23 | 60.6 | 14.3 | 57.5 | 122.5 | 14.3 | 14.2 | 69.2 | 67.2 | 120.9 | 122.4 | 122.3 | 16.5 |
| 1′ | 41.1 | 39.6 | 50.1 | 36.1 | 62.3 | 40 | 40 | 38.7 | 39.9 | 40 | 40 | 40 | 40 | 40 | 40.1 | 39.8 | 39.7 |
| 2′ | 36.5 | 44.4 | 46.9 | 34.9 | 81.9 | 44.7 | 44.8 | 39.3 | 44.9 | 44.7 | 44.7 | 44.8 | 44.8 | 45.1 | 44.7 | 44.7 | 44.7 |
| 3′ | 175.1 | 215.3 | 224.9 | 172.4 | 56.2 | 222 | 222.1 | 220.4 | 219.7 | 222.3 | 222.1 | 222.5 | 222.4 | 220.4 | 222.1 | 219.4 | 219.8 |
| 4′ | 212.5 | 80.1 | 62.2 | 113.2 | 134.2 | 51 | 50.9 | 50.1 | 51 | 50.9 | 50.9 | 50.9 | 50.9 | 51 | 51.1 | 51.1 | 50.7 |
| 5′ | 57.4 | 51.4 | 38.9 | 47.3 | 136.4 | 49.6 | 50 | 50.8 | 49.1 | 49.6 | 49.5 | 49.6 | 49.7 | 49.4 | 49.6 | 48.8 | 49.7 |
| 6′ | 86 | 82.2 | 84 | 81.6 | 26 | 83.8 | 83.6 | 83.6 | 84.4 | 83.9 | 83.9 | 83.9 | 83.8 | 84.1 | 83.9 | 84.3 | 84 |
| 7′ | 44.4 | 40.1 | 43.7 | 45.3 | 45.3 | 43.5 | 47.8 | 42.5 | 43.5 | 43.5 | 43.5 | 43.4 | 43.5 | 43.5 | 43.5 | 43.6 | 47.8 |
| 8′ | 31.1 | 31.5 | 32.8 | 31.9 | 82.5 | 32 | 33 | 31.1 | 31.9 | 31.9 | 31.9 | 32 | 32 | 32 | 32 | 31.9 | 32.9 |
| 9′ | 37.9 | 36.5 | 38.9 | 40.1 | 36.1 | 39.5 | 39.8 | 39.2 | 39.4 | 39.5 | 39.5 | 39.5 | 39.5 | 39.5 | 39.4 | 39.5 | 39.8 |
| 10′ | 148.2 | 147.2 | 150 | 145.8 | 29.8 | 150.1 | 150.6 | 151.2 | 150.1 | 150.1 | 150.1 | 150.2 | 150.2 | 150.6 | 150.1 | 150.1 | 150.4 |
| 11′ | 140.8 | 138.7 | 138.6 | 138.8 | 139.4 | 138.5 | 41.8 | 139.3 | 138.1 | 138.5 | 138.4 | 138.6 | 138.5 | 138.6 | 138.4 | 138.1 | 41.8 |
| 12′ | 171.3 | 169.6 | 169 | 169.8 | 170 | 169.3 | 177.5 | 169.2 | 170.4 | 169.3 | 169.3 | 169.4 | 169.2 | 169.8 | 169.5 | 170.7 | 179 |
| 13′ | 121.4 | 121.4 | 122 | 120.8 | 119.5 | 121.7 | 13.4 | 120.6 | 121.8 | 121.6 | 121.7 | 121.5 | 121.5 | 121.6 | 121.8 | 122.7 | 13.3 |
| 14′ | 116 | 114.3 | 113.7 | 116.2 | 17 | 114.2 | 113.6 | 113.1 | 114.1 | 114.1 | 114.1 | 114.1 | 114.1 | 114 | 114.2 | 114.2 | 113.6 |
| 15′ | 42.9 | 31.2 | 26.3 | 37.1 | 14.3 | 25.8 | 26 | 25.7 | 23 | 25.8 | 25.8 | 25.9 | 25.9 | 26.2 | 25.8 | 27.6 | 28.4 |
| 1″ | — | — | — | — | 170 | — | — | — | — | — | — | 59.2 | 66.8 | — | — | — | — |
| 2″ | — | — | — | — | 21.2 | — | — | — | — | — | — | — | 15 | — | — | — | — |
Table 26.
13C-NMR data of compounds 128–135.
| NO. | 128 [79] | 129 [79] | 130 [80] | 131 [81] | 132 [78] | 133 [75] | 134 [75] | 135 [80] | NO. | 128 [79] | 129 [79] | 130 [80] | 131 [81] | 132 [78] | 133 [75] | 134 [75] | 135 [80] |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | ||
| 1 | 40.1 | 170.4 | 173.4 | 172.9 | 170.1 | 173.8 | 172.4 | 149.6 | 1′ | 44.9 | 46.5 | 40.2 | 39.9 | 46.4 | 171.9 | 172.3 | 40 |
| 2 | 44.4 | 142.8 | 141.6 | 139.4 | 142.7 | 138.2 | 139.1 | 134.8 | 2′ | 42.7 | 48.6 | 44.6 | 44.6 | 48.6 | 140.3 | 140 | 44 |
| 3 | 218.7 | 193.6 | 207.8 | 205.2 | 193.6 | 193.9 | 194.2 | 115.4 | 3′ | 215 | 218.3 | 222.2 | 223 | 218.2 | 207.6 | 207.3 | 222 |
| 4 | 51.2 | 142.3 | 49.2 | 79 | 142.1 | 141.4 | 141.6 | 79.4 | 4′ | 80.9 | 52.8 | 51.2 | 51 | 52.7 | 52.3 | 51.9 | 52.7 |
| 5 | 49.1 | 47.6 | 51.2 | 53.6 | 47.5 | 51.2 | 46.9 | 56.9 | 5′ | 50.4 | 45.9 | 49.5 | 50.2 | 45.8 | 58.1 | 58.3 | 48.2 |
| 6 | 88.1 | 84 | 82.9 | 80.3 | 83.9 | 83.2 | 84 | 80.3 | 6′ | 81.9 | 83.7 | 84.2 | 84.2 | 83.6 | 80.7 | 80.6 | 83.9 |
| 7 | 44.2 | 43.7 | 51.5 | 51.4 | 43.6 | 44 | 43.5 | 50.5 | 7′ | 46.7 | 45.2 | 43.3 | 43.4 | 45 | 52.2 | 52.2 | 44.2 |
| 8 | 31.8 | 23.1 | 21 | 21.9 | 23 | 24.9 | 22.9 | 21.5 | 8′ | 31.3 | 32.1 | 31.9 | 31.8 | 32 | 21.2 | 21.1 | 32 |
| 9 | 38.1 | 35.2 | 35.8 | 36.3 | 35.1 | 28.9 | 34.2 | 36.9 | 9′ | 39.4 | 39.8 | 39.6 | 39.7 | 39.7 | 36.6 | 36.6 | 39.9 |
| 10 | 148.8 | 71.2 | 68.7 | 68 | 71.2 | 34.6 | 72.6 | 67.9 | 10′ | 146.2 | 148.9 | 150.4 | 150.1 | 148.8 | 68.3 | 68.3 | 149.6 |
| 11 | 139.4 | 140.6 | 140.1 | 139.3 | 140.4 | 139.3 | 140.2 | 139.9 | 11′ | 138.7 | 138.4 | 139.1 | 138.8 | 138.2 | 138.8 | 138.6 | 138.5 |
| 12 | 169.8 | 170.1 | 170.4 | 170.3 | 170 | 169.4 | 169.8 | 170.7 | 12′ | 169.8 | 170.9 | 169.5 | 169.6 | 170.8 | 169.4 | 169.3 | 169.9 |
| 13 | 120.8 | 119.6 | 118.7 | 118.5 | 119.5 | 120.5 | 119.6 | 118.4 | 13′ | 121.1 | 122.4 | 121 | 121.3 | 122.3 | 119.3 | 119.5 | 122.1 |
| 14 | 113.6 | 38.6 | 36.5 | 36.7 | 38.5 | 27.1 | 38.2 | 37.1 | 14′ | 114.8 | 114.6 | 114.2 | 114.5 | 114.5 | 36.7 | 36.6 | 114.9 |
| 15 | 23.6 | 121.6 | 25.2 | 31.7 | 121.7 | 122 | 123 | 27.8 | 15′ | 32.2 | 27 | 26.2 | 25.9 | 26.9 | 28.6 | 31.2 | 26.1 |
| NO. | 128 [79] | 129 [79] | 130 [80] | 131 [81] | 132 [78] | 133 [75] | 134 [75] | 135 [80] | |||||||||
| CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | ||||||||||
| 1″ | 40.0 | 40.4 | 39.8 | 39.6 | 40.3 | 39.9 | 39.9 | 46.5 | |||||||||
| 2″ | 44.1 | 43.9 | 44.9 | 44.9 | 43.8 | 44.6 | 44.5 | 45.2 | |||||||||
| 3″ | 218.4 | 218.2 | 218.3 | 218.5 | 218.3 | 218.8 | 218.8 | 109.4 | |||||||||
| 4″ | 49.3 | 49.0 | 51.6 | 51.1 | 48.9 | 50.5 | 50.5 | 95.2 | |||||||||
| 5″ | 48.6 | 49.9 | 46.6 | 46.2 | 49.8 | 50.3 | 50.4 | 54.5 | |||||||||
| 6″ | 88.2 | 87.0 | 89.0 | 89.0 | 86.9 | 84.1 | 84.0 | 79.4 | |||||||||
| 7″ | 44.0 | 44.8 | 44.1 | 43.9 | 44.7 | 43.7 | 43.7 | 43.7 | |||||||||
| 8″ | 31.4 | 31.3 | 31.6 | 31.1 | 31.2 | 31.9 | 31.9 | 27.6 | |||||||||
| 9″ | 38.1 | 36.8 | 38.7 | 38.8 | 36.7 | 39.6 | 39.6 | 27.1 | |||||||||
| 10″ | 148.7 | 148.2 | 148.7 | 148.8 | 148.1 | 150.0 | 149.9 | 146.1 | |||||||||
| 11″ | 139.3 | 139.2 | 139.1 | 138.7 | 139.1 | 138.6 | 138.5 | 139.1 | |||||||||
| 12″ | 169.8 | 169.6 | 169.9 | 169.8 | 169.5 | 169.4 | 169.4 | 169.5 | |||||||||
| 13″ | 120.6 | 121.2 | 120.9 | 121.1 | 121.2 | 121.6 | 121.7 | 120.5 | |||||||||
| 14″ | 113.6 | 114.3 | 113.3 | 113.1 | 114.2 | 114.4 | 114.5 | 116.3 | |||||||||
| 15″ | 29.6 | 32.8 | 22.5 | 20.9 | 32.8 | 25.8 | 25.8 | 22.7 |
5.2. Bioactivity of Polymer Sesquiterpene Lactones
5.2.1. Antitumor and Cytotoxic
Polymer sesquiterpene lactones inhibit the activity of tumor cell lines or cancer cell lines. The dimeric sesquiterpene lactone japonicone A (115) showed significant inhibitory activity against the three tested human tumor cell lines 95D, MDA-MB-231, and HepG2, with IC50 values of 9.10, 3.82, and 1.43 μM, respectively [8]. Ainsliatriolide C (131), ainsliadimer B (116), and ainsliatrimer B (134) were isolated from Ainsliaea yunnanensis and showed very significant selective cytotoxic activities on MDA-MB-468, PANC-1, HEPG2, and A549 cells, and IC50 values from 5.1 μM to 34.4 μM [81]. With DOX (doxorubicin) as the positive control, the antitumor activity of the isolated compounds against A549, LOVO, CEM, and MDA-MB-M-435 (MDA) was detected by an MTT assay. Both Ainsliatrimer A (133) and ainsliatrimer B (134) showed potent cytotoxicites against the LOVO and CEM cell lines [75]. Ainsliatriolide A (130) and ainsliatriolide B (135) exhibited stronger cytotoxicity on A-549, HT-29, BEL-7402, and HL-60 cancer cell lines, especially ainsliatriolide A (130) which displayed potent cytotoxicity with an averaged IC50 value of 1.17 μM against four cancer cell lines [80].
With cisplatin as a positive control, the cytotoxicity of the compounds isolated from Ainsliaea fragrans was tested in the five cancer cell lines of C6 rat glioma cells, Huh1, HCC-LM3 human hepatocellular carcinoma cells, PANC-1 human pancreatic cells, and Hela human cervical cancer cells. The cytotoxicity results showed that ainsfragolide (132) is an unusual guaianolide sesquiterpene trimer, which is generated by a new C2″–C15″ bond and has a significant inhibitory effect on five cancer cells with a half-inhibitory concentration value in the range of 0.4–8.3 μM. Three trimers ainsfragolide (132), ainsliatrimer A (133), and ainsliatrimer B (134) showed more potent cytotoxic effect against the five test cancer cells than the dimers. When compared with ainsliatrimer B (134), the decreased activity of ainsliatrimer A (133) indicated that the introduction of an extra hydroxy group at the C-10 position is important for the resultant cytotoxicity. Similarly, the dimers gochnatiolide A (119) and gochnatiolide B (125) with a C-10 OH group were more cytotoxic than gochnatiolide C (124), respectively. Furthermore, gochnatiolide A (119) with a β-configuration of OH-10 was about 2–10-fold more active than gochnatiolide B (125) with α-OH at C-10. In contrast, ainsliadimer B (116) with 10β–OH and OH-15 groups showed reduced activity [78]. Therefore, we can try to obtain a trimer with a β-configuration hydroxyl at the C-10 position and no hydroxyl at the C-15 position by synthesis or structural modification, so as to improve the cytotoxic activity of polymer sesquiterpene lactones against cancer cells.
5.2.2. Anti-Inflammatory
Macrocephadiolide A (114) and macrocephadiolide B (111) showed a potent inhibitory effect on nitric oxide (NO) production, with IC50 values of 0.99 and 6.13 µM, respectively, on lipopolysaccharide (LPS)-stimulated RAW264.7 macrophages. Macrocephadiolide A (114) dose-dependently suppressed the expression of inducible NO oxidase (iNOS) through inhibiting nuclear factor kappa B (NF-κB) activation [72]. Ainsliadimer A (113) represents an unusual carbon skeleton with a cyclopentane system connecting the two monomeric sesquiterpene lactone units. This unique molecule exerted potent inhibitory activity against the production of nitric oxide in RAW264.7 cells stimulated by LPS, with an IC50 value of 2.41 µg/mL [73]. Gochnatiolide A (119) showed significant anti-inflammatory activity by inhibiting the expression of nuclear factor kappa B (NF-κB) in the 293-NF-κB-luciferase reporter cell line and the production of TNF-α, IL-1β, IL-6, and IL-10 in RAW264.7 macrophages induced by lipopolysaccharide (LPS) [47]. The anti-inflammatory effects of polymer sesquiterpene lactones in Ainsliaea are achieved by inhibiting the production of nitric oxide (NO) in RAW264.7 macrophages, inhibiting the activity of nuclear factor kappa B (NF-κB) in the luciferase 293-NF-κB-luciferase reporter cell line, and inhibiting the expression of TNF-α, IL-1β, IL-6, and IL-10 in RAW264.7 macrophages.
5.2.3. Other Biological Activities
The polymer sesquiterpene lactones also have antibacterial and blood-sugar regulation effects. Ainsliatrimer B (134) also showed a medium inhibiting effect on Bacillus subtilis with a MIC value of 32 μg/mL [81]. Macrocephatriolide B (128) showed potent inhibition against protein tyrosine phosphatase 1B (PTP1B) with an IC50 value of 26.26 ± 0.88 μM. In insulin-stimulated C2C12 myotubes, macrocephatriolide B (128) dose-dependently enhanced glucose uptake by activating the insulin signaling pathway and might represent a new scaffold of insulin sensitizers [79].
6. Other Sesquiterpenoids
Beyond those mentioned above, there exist other categories of sesquiterpenoids and their derivatives, such as myrrhanes, lananes, and carabanes. And further details can be referenced in Figure 5 and Table 27.
Figure 5.
Chemical structures for compounds 136–145.
Table 27.
The compound name, molecular formula, and test reagent of other sesquiterpenoids.
| No. | Compound Name | Molecular Formula | Solvent | Ref. |
|---|---|---|---|---|
| 136 | 1-Oxo-bisabola-2-ene-12-ol | C15H26O2 | CDCl3 | [82] |
| 137 | Pubescone | C14H22O2 | CDCl3 | [83] |
| 138 | Ainsliaea acid A | C16H22O3 | CD3OD | [67] |
| 139 | Curzerenone | C15H18O2 | CDCl3 | [84] |
| 140 | 1-O-Acetyl-6-O-isobutyrylbritannilactone | C20H30O6 | CDCl3 | [85] |
| 141 | 6α-(3-Methylvaleryloxy)-1-hydroxy-4αH-1,10-secoeudesma-5(10),11(13)-dien-12,8β-olide | C21H32O5 | CDCl3 | [86] |
| 142 | Kobusone | C14H21O2 | CDCl3 | [87] |
| 143 | 10-Hydroxy-6,10-epoxy-7(14)-isodaucane | C15H24O2 | CDCl3 | [88] |
| 144 | Clovane-2β,9α-diol | C15H26O2 | CDCl3 | [89] |
| 145 | Caryolane-1,9β-diol | C15H26O2 | CDCl3 | [89] |
6.1. NMR Data of Other Sesquiterpenoids 136–145
The NMR spectra of 1H and 13C are summarized in Table 28 and Table 29. This paper also compiles data regarding the nuclear magnetic resonance testing instruments used for the compounds numbered from 136 to 145. The 1H and 13C data of compound 136 were recorded using a Bruker instrument operating at 300 MHz. For compound 137, NMR data collection was performed with the Bruker-Avance-III-400 instrument (Bruker, Switzerland). For compound 140, the NMR data were conducted on a Bruker AMX 400 instrument (Bruker, Zurich, Switzerland). The NMR data for compound 141 were taken with a Bruker Avance-500 instrument (Bruker, Switzerland). Both the 1H and 13C data of compound 143 were measured using JEOL Lamda 400 (JEOL, Japan) and Lamda 600 instruments (JEOL, Japan). Finally, compounds 144 and 145 had their respective 1H and 13C data collected via the JEOL JNM-GX400 instrument (JEOL, Japan). The 1H data for compounds 138, 139, and 142 were measured at frequencies of 600 MHz, 300 MHz, and 400 MHz, respectively. The 13C NMR of compounds 138, 139, and 142 were tested under 150 MHZ, 75 MHZ, and 100 MHZ, respectively.
Table 28.
1H-NMR data of compounds 136–145.
| NO. | 136 [82] | 137 [83] | 138 [67] | 139 [84] | 140 [85] | 141 [86] | 142 [87] | 143 [88] | 144 [89] | 145 [89] |
|---|---|---|---|---|---|---|---|---|---|---|
| CDCl3 | CDCl3 | CD3OD | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | |
| 1α | — | — | 5.90, dd, 17.8, 11.2 | 5.78, dd, 17.6, 10.6 | 3.85–4.00, m | 3.52, m | — | — | — | — |
| 1β | — | 1.66–1.63, m | — | — | 3.44, m | 1.93, ddd, 10.4, 8.8, 1.2 | — | — | — | |
| 2α | 5.86, m | 1.76–1.63, m | 5.34, dd, 17.8, 1.1 | 4.74–4.99, m | 1.34–1.45, m | 1.32, m | 1.67, m | 1.82 | 3.79, dd, 10.5, 5.5 | 2.22, ddd, 12, 10, 8.5 |
| 2β | 5.25, dd, 18.0, 1.2 | 1.21–1.31, m | 1.13, m | 1.54, m | 1.37 | |||||
| 3α | — | 2.53–2.48, m | 4.90, m | 4.74–4.99, m | 1.21–1.31, m | 1.32, m | 2.15, dt, 13.2, 3.6 | 1.87 | 1.51, dd, 11.5, 10 | 1.49, dd, 10, 9 |
| 3β | — | 0.97–1.08, m | 1.00, m | 0.94, td, 13.2, 4.4 | 1.27 | 1.71, dd, 11.5, 5.5 | 1.54, t, 10 | |||
| 4 | 2.32, m | — | — | — | 2.60–2.76, m | 2.70, m | — | 1.58 | — | — |
| 5α | 1.92, m | 1.05–1.03, m | 2.79, br s | 3.00, s | — | — | — | 1.56 | — | 1.89, ddd, 12, 9, 6 |
| 5β | 1.78, m | — | — | 2.69, dd, 10.0, 5.2 | — | 1.42, m | — | |||
| 6α | 2.15, m | 0.72–0.67, m | — | — | 5.19, d, 1.7 | 5.23, d, 1.7 | 2.40, m | 3.98 | 1.32, m | 1.39, m |
| 6β | — | — | 4.64, br s | — | — | — | 1.44, m | — | 1.35, m | 1.53, m |
| 7α | 2.33, m | — | 2.02, m | — | 3.41–3.49, m | 3.50, m | 2.55, dd, 6.4, 2.0 | — | 1.11, m | 1.15, m |
| 7β | — | — | — | — | — | — | 2.53, d, 6.4 | — | 1.50, m | 1.42, m |
| 8α | 1.26, m | 2.05, dd, 8.8, 3.2 | 1.96, m | — | 4.90–4.98, m | 5.01, m | — | 2.49 | — | — |
| 8β | 1.43, m | — | — | — | — | 2.35 | — | — | ||
| 9α | 1.34, m | 2.14–2.11, m | 1.86, m | 2.83, AB system, 17.6 | 2.60–2.76, m | 2.75, dd, 16.2, 2.1 | 3.05, td, 8.8, 8.8 | 2.02 | — | 3.44, t, 3 |
| 9β | 2.02–1.99, m | 2.45–2.56, m | 2.51, dd, 16.2, 2.3 | — | 1.76 | 3.32, br s | — | |||
| 10α | 1.39, m | 1.88–1.84, m | — | — | — | — | 2.06, dd, 10.4, 8.8 | — | 1.64, m | 1.77, ddt, 15, 5, 3 |
| 10β | 1.12, m | — | — | — | — | — | 1.66, d, 10.4 | — | 1.99, m | 2.04, dddd, 15, 12.5, 5.5, 3 |
| 11α | 1.62, m | 1.80–1.77, m | — | — | — | — | — | 1.51 | 1.07, m | 1.51, m |
| 11β | — | — | 2.36, m | — | — | — | — | — | 1.66, m | 1.64, td, 12.5, 5 |
| 12α | 3.50, dd, 10.6, 5.8 | 0.92, d, 6.8 | — | 7.08, br s | — | — | 1.30, s | 0.89, d, 6.6 | 0.91, br d, 12.5 | 1.42, d |
| 12β | 3.42, dd, 10.6, 5.8 | — | — | — | 1.56, d, 12.5 | 1.47, d | ||||
| 13α | 0.91, d, 6.7 | 0.92, d, 6.8 | 1.24, d, 7.0 | 2.16, br s | 6.37, d, 2.9 | 6.36, d, 2.6 | 1.02, s | 0.92, d, 6.6 | 0.86, s | 1.00, s |
| 13β | 5.94, d, 2.3 | 6.02, d, 2.3 | ||||||||
| 14 | 0.79, d, 6.8 | 2.18, s | — | 1.17, s | 1.80, s | 1.82, s | 1.02, s | 1.15, s | 1.04, s | 1.02, s |
| 15α | 1.93, br s | — | 1.67, s | 1.83, br s | 0.86, d, 6.9 | 0.90, d, 7.0 | — | 4.76, br s | 0.96, s | 0.93, s |
| 15β | — | — | 4.70, br s | |||||||
| 2′α | — | — | — | — | 2.45–2.56, m | 2.32, m | — | — | — | — |
| 2′β | — | — | — | — | 2.11, m | — | — | — | — | |
| 3′ | — | — | — | — | 1.15, d, 6.9 | 1.87, m | — | — | — | — |
| 4′α | — | — | — | — | 1.15, d, 6.9 | 1.37, m | — | — | — | — |
| 4′β | — | — | — | — | — | 1.26, m | — | — | — | — |
| 5′ | — | — | — | — | — | 0.92, t, 7.5 | — | — | — | — |
| 6′ | — | — | — | — | — | 0.94, d, 7.0 | — | — | — | — |
Note: The 1H-NMR data of 2″ of compound 140 were 2.04, s.
Table 29.
13C-NMR data of compounds 136–145.
| NO. | 136 [82] | 137 [83] | 138 [67] | 139 [84] | 140 [85] | 141 [86] | 142 [87] | 143 [88] | 144 [89] | 145 [89] | NO. | 140 [85] |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CDCl3 | CDCl3 | CD3OD | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | ||
| 1 | 201.4 | 34.4 | 135.8 | 145.5 | 64.2 | 61.7 | 51.1 | 54.5 | 44.6 | 70.7 | 1′ | 176.9 |
| 2 | 127.2 | 24.8 | 117.9 | 115.7 | 26.5 | 30.7 | 26.2 | 26.4 | 80.8 | 38 | 2′ | 34.6 |
| 3 | 161.4 | 41.7 | 115.5 | 113 | 31 | 30.3 | 38.8 | 30.6 | 47.5 | 34 | 3′ | 18.7 |
| 4 | 30.6 | 208.7 | 144.4 | 141.1 | 33 | 32.9 | 58.6 | 57.6 | 37.1 | 35 | 4′ | 18.8 |
| 5 | 22.6 | 29.9 | 61.5 | 64.1 | 132.1 | 131.9 | 61.3 | 58.9 | 50.6 | 43.8 | 1″ | 171.2 |
| 6 | 50.1 | 46.4 | 82.2 | 194 | 69 | 68.8 | 24.5 | 86 | 20.7 | 20.3 | 2″ | 21 |
| 7 | 30.9 | 214.2 | 43.3 | 120.2 | 42.9 | 42.7 | 37.4 | 144.8 | 33.2 | 35.3 | ||
| 8 | 34.9 | 32.8 | 25 | 165.6 | 74.9 | 75.6 | 213.8 | 33.2 | 34.7 | 39.3 | NO. | 141 [86] |
| 9 | 24.9 | 22.9 | 34.7 | 33.6 | 34.1 | 34.1 | 52.3 | 35.9 | 75.1 | 72.1 | CDCl3 | |
| 10 | 33.3 | 27.9 | 52.3 | 42.9 | 133.6 | 133.3 | 35 | 105.1 | 26 | 28.1 | 1′ | 173.4 |
| 11 | 35.8 | 30.9 | 43.8 | 119.3 | 136.3 | 136 | 34.2 | 34.1 | 26.4 | 33.3 | 2′ | 41.3 |
| 12 | 68.4 | 19.5 | 179.2 | 139.6 | 169.5 | 170.5 | 16 | 21.5 | 35.6 | 42.4 | 3′ | 31.6 |
| 13 | 16.7 | 19.2 | 15.4 | 9.1 | 124.9 | 125.1 | 29.1 | 20.4 | 25.4 | 20.8 | 4′ | 28.8 |
| 14 | 15.8 | 29.9 | 180 | 24.9 | 20.5 | 19.9 | 22 | 21.6 | 31.4 | 30.5 | 5′ | 10.6 |
| 15 | 24.2 | — | 21.9 | 25 | 18.6 | 18.1 | — | 108.4 | 28.4 | 26.7 | 6′ | 18.7 |
6.2. Bioactivity of Other Sesquiterpenoids
At the concentration of 10 μMol/L, ainsliaea acid A (138) significantly inhibited nuclear factor kappa B (NF-κB) in lipopolysaccharides-induced 293-NF-κB-luciflucidase reporter cell lines with a inhibitory rate of 17.5%. Further experiments showed that ainsliaea acid A (138) exerted anti-inflammatory effects by inhibiting the production of tumor necrosis factor-α (TNF-α), interleukin-1β (IL-1β), IL-6, and IL-10 in RAW264.7 macrophages induced by LPS [67].
7. Conclusions
Sesquiterpenoids derived from plants of the genus Ainsliaea exhibit a wealth of pharmacological activities, demonstrating significant antitumor, anti-inflammatory, antibacterial, and antiobesity effects. Currently, the existing literature primarily focuses on the chemical composition and pharmacological effects of these plants; however, comprehensive NMR data summarizing the related components remain scarce. This paper consolidates the 1H- and/or 13C-NMR data for sesquiterpenes extracted from Ainsliaea species, and the pharmacological activities of sesquiterpenes are summarized, thereby providing a valuable reference for discovering novel sesquiterpenes and differentiating between various types. It also offers essential data support for structural analysis and compound identification. Additionally, exploring new sesquiterpene constituents is crucial for investigating their pharmacodynamic material basis, as well as their structure–activity relationships and mechanisms of action, and enriching the diversity of natural products.
The sesquiterpenoids found in the plants of Ainsliaea are mostly guaiane and eudesmane, and the active ingredients found are mostly guaiane and polymer sesquiterpene lactones. Guaiacanolactone containing a α-methylene-γ-lactone moiety and guaiac lactones containing hydroxyl groups with three outer-ring double bonds have a significant inhibitory effect on NO production. Trimeric sesquiterpene lactones with a β-configuration hydroxyl at the C-10 position and without a hydroxyl group at C-15 have high cytotoxic activity. Therefore, it is of great significance to find compounds containing these components in natural products or to obtain these compounds through synthesis and structural modification to improve the anti-inflammatory activity of guaiane sesquiterpenes and the cytotoxic activity of polymer sesquiterpene lactones. The limited amount of germacrane and other sesquiterpenes reported within this genus renders the data support for structural analysis and identification of these compounds insufficiently convincing. It is essential to conduct further investigations into their chemical constituents, identify additional sesquiterpenes, and enhance the diversity of sesquiterpene types within the genus. Moreover, a comprehensive summary of the NMR spectral data pertaining to triterpenoids, steroids and their derivatives, phenolic acids, flavonoids, anthraquinones, coumarins, lignans, and other components remains lacking and needs to be further summarized.
Author Contributions
Writing—original draft, H.Z. and R.-R.S.; Investigation, Y.-F.L. and X.G.; Methodology, C.-L.L.; Writing—review and editing, Z.-D.N. and Z.-B.J.; Project administration, Z.-D.N. and Z.-B.J. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
All authors have no conflicts of interest to declare.
Funding Statement
This research was funded by the National Natural Science Foundation of China (No. 82160672), the General Project of Ningxia Natural Science Foundation (No. 2024AAC03172), and the Scientific Research Start-up project for Recruitment Talents of North Minzu University in 2021 (No. 2021KYQD35).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Editorial Committee of Chinese Botany, Chinese Academy of Sciences . Chinese Flora. Volume 79. Science Press; Beijing, China: 1996. pp. 68–69. [Google Scholar]
- 2.Hisanori J. Studies on the constituents of Ainsliaea acerifolia SCH.-BIP. Var. Subapoda NAKAI. Yakugaku Zasshi. 1982;102:911–922. doi: 10.1248/yakushi1947.102.10_911. [DOI] [Google Scholar]
- 3.Bohlmann F., Chen Z.L. Guaianolides from Ainsliaea fragrans. Phytochemistry. 1982;21:2120–2122. doi: 10.1016/0031-9422(82)83061-6. [DOI] [Google Scholar]
- 4.Gao Y., Lu Y.H., Xu X., Xin J., Yang M., Lin T., Chen J., Qi X., Feng Y., Zu X. A Review of Chemical Constituents in Ainsliaea Plants and Their Pharmacological Activities. Chin. Arch. Tradit. Chin. Med. 2024;42:142–163. doi: 10.13193/j.issn.1673-7717.2024.02.030. [DOI] [Google Scholar]
- 5.Guo J., Wang J.-P., Peng B., Liu X.-Q., Yang C.-X., Yan L.-H., Wang Z.-M. Research progress on natural guaiane-type sesquiterpenoids and their biological activities. China J. Chin. Mater. Medica. 2023;48:5727–5749. doi: 10.19540/j.cnki.cjcmm.20230731.301. [DOI] [PubMed] [Google Scholar]
- 6.Xu Y., Zhang H., Wan X., Zou Z. Complete assignments of 1H and 13C NMR data for two new sesquiterpenes from Cyperus rotundus L. Magn. Reson. Chem. 2009;47:527–531. doi: 10.1002/mrc.2416. [DOI] [PubMed] [Google Scholar]
- 7.Zhang J., Xu Y., Li R., Zhang Y., Yue C., Bi D., Cheng B., Wu X., Zhang R., Zhang X., et al. Ainslides A-F, six sesquiterpe-noids isolated from ainsliaea pertyoides and their NLRP3-inflammasome inhibitory activity. Chem. Biodivers. 2022;19:e202200135. doi: 10.1002/cbdv.202200135. [DOI] [PubMed] [Google Scholar]
- 8.Shi Z.-R., Zhang X.-Y., Zeng R.-T., Zhuo Z.-G., Feng F., Shen Y.-H., Zhang W.-D. Sesquiterpenoids from Ainsliaea spicata and their cytotoxic and NO production inhibitory Activities. Phytochem. Lett. 2016;18:87–94. doi: 10.1016/j.phytol.2016.09.005. [DOI] [Google Scholar]
- 9.Feng F., Chen M.-H., Xing C.-X., Liu W.-Y., Xie N. Two novel sesquiterpenoids from Ainsliaea fragrans Champ. J. Asian Nat. Prod. Res. 2009;11:856–860. doi: 10.1080/10286020903014944. [DOI] [PubMed] [Google Scholar]
- 10.Nishizawa M., Inoue A., Hayashi Y., Sastrapradja S., Kosela S., Iwashita T. Structure of aphanamols I and II. J. Org. Chem. 1984;49:3660–3662. doi: 10.1021/jo00193a046. [DOI] [Google Scholar]
- 11.Fang X., Zeng R.-T., Zhuo Z.-G., Shen Y.-H., Zhang W.-D. Sesquiterpenoids from Ainsliaea yunnanensis and their cytotoxic activities. Phytochem. Lett. 2018;26:25–29. doi: 10.1016/j.phytol.2018.05.013. [DOI] [Google Scholar]
- 12.Choi S.-Z., Yang M.-C., Choi S.-U., Lee K.-R. Cytotoxic terpenes and lignans from the roots of Ainsliaea acerifolia. Arch. Pharmacal Res. 2006;29:203–208. doi: 10.1007/BF02969394. [DOI] [PubMed] [Google Scholar]
- 13.Tsichritzis F., Jakupovic J., Bohlmann F. Sesquiterpene lactones and farnesol derivatives from Arctotis and Arctotheca species. Phytochemistry. 1990;29:195–203. doi: 10.1016/0031-9422(90)89036-9. [DOI] [Google Scholar]
- 14.Adegawa S., Miyase T., Ueno A. Sesquiterpene lactones from Diaspananthus uniflorus (SCH. BIP.) KITAM. Chem. Pharm. Bull. 1987;35:1479–1485. doi: 10.1248/cpb.35.1479. [DOI] [Google Scholar]
- 15.Kumari G., Masilamani S., Ganesh M., Aravind S. Microbial transformation of zaluzanin D. Phytochemistry. 2003;62:1101–1104. doi: 10.1016/S0031-9422(02)00667-2. [DOI] [PubMed] [Google Scholar]
- 16.Boglarka C.L., Istvan Z., Judit M., Peter F., Judit H. Bioactivity-guided isolation of antiproliferative compounds from the roots of Onopordum acanthium. Nat. Prod. Commun. 2014;9:337–340. doi: 10.1177/1934578X1400900313. [DOI] [PubMed] [Google Scholar]
- 17.Lee J.Y., Cha M.-R., Kim M.R., Lee K., Choi S.-U., Ryu S.Y. Two Novel Guaiane Sesquiterpenes from the Whole Plant of Youngia. Planta Medica Lett. 2015;2:e31–e34. doi: 10.1055/s-0035-1557794. [DOI] [Google Scholar]
- 18.Bruno M., Bancheva S., Rosselli S., Maggio A. Sesquiterpenoids in subtribe centaureinae (Cass.) dumort (tribe cardueae asteraceae): Distribution, 13C NMR spectral data and biological properties. Phytochemistry. 2013;95:19–93. doi: 10.1016/j.phytochem.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 19.Hou P.Y., Huang J., Sun B.-H., Wu L.-J., Gao H.-Y. Isolation and identification of chemical constituents from whole plant of Saussurea involucrata Kar. et kir. J. Shenyang Pharm. Univ. 2011;2:120–123. doi: 10.14066/j.cnki.cn21-1349/r.2011.02.002. [DOI] [Google Scholar]
- 20.Shen T., Weng C.W., Xie W.D., Row K.H. A new guaiane sesquiterpene from Paraixeris pinnatipartite. J. Chem. Res. 2009;10:623–624. doi: 10.3184/030823409X12526892025865. [DOI] [Google Scholar]
- 21.Kisiel W., Michalska K. Sesquiterpenoids and phenolics from Crepis conyzifolia. Z. Für Naturforschung C. 2001;56:961–964. doi: 10.1515/znc-2001-11-1208. [DOI] [PubMed] [Google Scholar]
- 22.Li X., Liu J., Cai J., Cai P. Complete 1H and 13C data assignments of two new guaianolides isolated from Ainsliaea fragrans. Magn. Reson. Chem. 2008;46:1070–1073. doi: 10.1002/mrc.2294. [DOI] [PubMed] [Google Scholar]
- 23.Singhal A.K., Chowdhury P.K., Sharma R.P., Baruah J.N., Herz W. Guaianolides from Tricholepis glaberrima. Phytochemistry. 1982;21:462–463. doi: 10.1016/S0031-9422(00)95292-0. [DOI] [Google Scholar]
- 24.Kisiel W. Guaianolides from Picris altissima. Phytochemistry. 1992;31:328–329. doi: 10.1016/0031-9422(91)83066-T. [DOI] [Google Scholar]
- 25.Liu B., Xie Y.-Z., Li X.-X., Sun Y.-Y., Xu F., Ji T.-F. Chemical constituents of Myripnois dioica. Chin. J. Tradit. Chin. Med. 2016;41:3260–3264. doi: 10.4268/cjcmm20161723. [DOI] [PubMed] [Google Scholar]
- 26.Zong Y., Yu M., Huang L., Chang Y., Wang Y., Che C.-T. Studies of Tibetan medicinal plants, I.I. Antitumour activity of Saussurea eopygmaea. Int. J. Pharmacogn. 1994;32:284–293. doi: 10.3109/13880209409083006. [DOI] [Google Scholar]
- 27.Seto M., Miyase T., Fukushima S. Sesquiterpene Lactones from Ixeris dentata NAKAI. Chem. Pharm. Bull. 1986;34:4170–4176. doi: 10.1248/cpb.34.4170. [DOI] [Google Scholar]
- 28.Das S., Baruah R.N., Sharma R.P., Baruah J.N., Kulanthaivel P., Herz W. Guaianolides from Saussurea affinis. Phytochemistry. 1983;22:1989–1991. doi: 10.1016/0031-9422(83)80030-2. [DOI] [Google Scholar]
- 29.Cho J.-Y., Jeong S.-J., La Lee H., Park K.-H., Hwang D.Y., Park S.-Y., Lee Y.G., Moon J.-H., Ham K.-S. Sesquiterpene lactones and scopoletins from Artemisia scoparia Waldst. & Kit. and their angiotensin I-Converting Enzyme Inhibitory Activities. Food Sci. Biotechnol. 2016;25:1701–1708. doi: 10.1007/s10068-016-0261-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kisiel W., Zielińska K., Joshi S.P. Sesquiterpenoids and phenolics from Crepis mollis. Phytochemistry. 2000;54:763–766. doi: 10.1016/S0031-9422(00)00167-9. [DOI] [PubMed] [Google Scholar]
- 31.Wu Z.-J., Xu X.-K., Zeng H.-W., Shen Y.-H., Tian J.-M., Su J., Li H.-L., Shan L., Liu R.-H., Zhang W.-D. New sesquiterpenoids from Ainsliaea macrocephala and their nitric oxide inhibitory activity. Planta Medica. 2011;77:1545–1550. doi: 10.1055/s-0030-1270930. [DOI] [PubMed] [Google Scholar]
- 32.Fournet A., Muñoz V., Roblot F., Hocquemiller R., Cavé A., Gantier J. Antiprotozoal activity of dehydrozaluzanin C, a sesquiterpene lactone isolated from Munnozia maronii (asteraceae) Phytother. Res. 1993;7:111–115. doi: 10.1002/ptr.2650070203. [DOI] [Google Scholar]
- 33.Dong X.Y., Wen B., Shen Y.H. Chemical constituents of Ainsliaea latifolia. Chin. Tradit. Herbal Drugs. 2014;45:2148–2152. doi: 10.7501/j.issn.0253-2670.2014.15.006. [DOI] [Google Scholar]
- 34.Zeng R., Dong X., Fang X., Yang N., Shi Z., Zhuo Z., Shen Y., Zhang W. Cytotoxic and anti-inflammatory sesquiterpenes from Ainsliaea henryi. Chem. Biodivers. 2017;14:e1600210. doi: 10.1002/cbdv.201600210. [DOI] [PubMed] [Google Scholar]
- 35.Pu J.-X., Zhao J.-F., Yang X.-D., Mei S.-X., Zhang H.-B., Li L. A New Sesquiterpene lactone from Ainsliaea bonatii. Chin. Chem. Lett. 2004;15:1454–1456. [Google Scholar]
- 36.Li X., Wang W., Jaeger F., Kreyenschmidt J. Ainsliaolide D: A new sesquiterpene lactone from Ainsliaea pertyoides. Nat. Prod. Res. 2014;28:115–118. doi: 10.1080/14786419.2013.850688. [DOI] [PubMed] [Google Scholar]
- 37.Fang X., Xu X.-K., Wang G.-W., Zeng R.-T., Tian X.-H., Shi Z.-R., Zhuo Z.-G., Shen Y.-H., Zhang W.-D. Guaianolide sesquiterpenoids from Ainsliaea yunnanensis. Phytochemistry. 2017;139:47–55. doi: 10.1016/j.phytochem.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 38.Zdero C., Bohlmann F., King R., Robinson H. Sesquiterpene lactones and other constituents from Australian helipterum species. Phytochemistry. 1989;28:517–526. doi: 10.1016/0031-9422(89)80045-7. [DOI] [Google Scholar]
- 39.Shi Z.-R., Shen Y.-H., Zhang X.-Y., Fang X., Zeng R.-T., Liu Q.-X., Zhuo Z.-G., Feng F., Zhang W.-D. Structurally novel C17-sesquiterpene lactones from Ainsliaea pertyoides. RSC Adv. 2015;5:91640–91644. doi: 10.1039/C5RA16551B. [DOI] [Google Scholar]
- 40.Zhang C., Zhou W., Lei X., Zhao S. Nitric oxide inhibitory terpenes and its glycosides from Ainsliaea bonatii. Fitoterapia. 2022;156:105028. doi: 10.1016/j.fitote.2021.105028. [DOI] [PubMed] [Google Scholar]
- 41.Miyasw T., Yamada M., Fukushima S. Studies on sesquiterpene glycosides from Prenanthes acerifolia BENTH. Chem. Pharm. Bull. 1987;35:1969–1974. doi: 10.1248/cpb.35.1969. [DOI] [Google Scholar]
- 42.Li J., Li X., Wang X., Zhong X., Ji L., Guo Z., Liu Y., Shang X. Sesquiterpenoids and their antiinflammatory activity: Evaluation of Ainsliaea yunnanensis. Molecules. 2019;24:1701. doi: 10.3390/molecules24091701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim T., Jo C., Kim H.-S., Park Y.-M., Wu Y.-X., Cho J.-H., Kim T.H. Chemical constituents from Ainsliaea acerifolia as potential anti-obesity agents. Phytochem. Lett. 2016;16:146–151. doi: 10.1016/j.phytol.2016.04.005. [DOI] [Google Scholar]
- 44.Miyase T., Fukushima S. Sesquiterpene Lactones from Ainsliaea acerifolia SCH: BIP. A. Dissecta FRANCH. Et SAV. Chem. Pharm. Bull. 1984;32:3043–3046. doi: 10.1248/cpb.32.3043. [DOI] [PubMed] [Google Scholar]
- 45.Wang H., Wu T., Yan M., Liu G., Li P., Zhang X.-Q., Ye W.-C., Zhang L.-Y. Sesquiterpenes from Ainsliae fragrans and their inhibitory activities against cyclooxygenases-1 and 2. Chem. Pharm. Bull. 2009;57:597–599. doi: 10.1248/cpb.57.597. [DOI] [PubMed] [Google Scholar]
- 46.Ding N., Li S.-Y., Li C.-H., Chen L.-H., Zhu Y.-J., Wang J.-Y., Yang Y.-S., Hu L.-H., Wang X.-C. A new guaiane sesquiterpene glycoside from Ainsliaea fragrans. Chin. Tradit. Herb. Drugs. 2020;51:5669–5674. doi: 10.7501/j.issn.0253-2670.2020.22.001. [DOI] [Google Scholar]
- 47.Wu X.-L., Xiong X.-J., Lu W.-Q., Huang H., Shen Y.-H., Wu Z.-J., Chen W.-S. New sesquiterpenenoids from Ainsliaea yunnanensis. Molecules. 2016;21:1031. doi: 10.3390/molecules21081031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson A., Kumar T.V., Sreedhar E., Naidu V., Krishna S.R., Babu K.S., Srinivas P., Rao J.M. A new sesquiterpene lactone from the roots of Saussurea lappa: Structure-anticancer activity study. Bioorg. Med. Chem. Lett. 2008;18:4015–4017. doi: 10.1016/j.bmcl.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 49.Chen Y.-P., Wang S.-L., Shen Y.-H., Wu Z.-J. Chemical constituents from Ainsliaea glabra. Guihaia. 2014;34:402–407. doi: 10.3969/j.issn.1000G3142.2014.03.022. [DOI] [Google Scholar]
- 50.Dong X.Y. Ph.D. Thesis. Fujian University of Traditional Chinese Medicine; Fuzhou, China: 2015. Studies on Chemical Constituents and Bioactivities of the Ainsliaea latifolia (D. Don) Sch. Bip. [Google Scholar]
- 51.Hänsel R., Kartarahardja M., Huang J.-T., Bohlmann F. Sesquiterpenlacton-β-D-glucopyranoside sowie ein neues eudesmanolid aus Taraxacum officinale. Phytochemistry. 1980;19:857–861. doi: 10.1016/0031-9422(80)85126-0. [DOI] [Google Scholar]
- 52.Dong X.-Y., Wang G.-W., Zhuo Z.-G., Lv C., Fang X., Shi Z.-R., Zeng R.-T., Shen Y.-H., Zhang W.-D. Terpenoids from Ainsliaea latifolia their cytotoxic activities. J. Asian Nat. Prod. Res. 2016;18:232–238. doi: 10.1080/10286020.2015.1082550. [DOI] [PubMed] [Google Scholar]
- 53.Wu G.-X., Zhao H.-Y., Peng C., Liu F., Xiong L. Eudesmane-type sesquiterpenoids: Structural diversity and biological activity. Heliyon. 2024;10:e35270. doi: 10.1016/j.heliyon.2024.e35270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chanotiya C., Sammal S., Mathela C. Composition of a new chemotype of Tanacetum nubigenum. Indian J. Chem. 2005;44:1922–1926. doi: 10.1002/chin.200603194. [DOI] [Google Scholar]
- 55.González A.G., Barrera J.B., Méndez J.T., Sánchez M.L., Martínez J.L.E. Sesquiterpene alcohols from Gonospermum fructicosum. Phytochemistry. 1992;31:1816–1817. doi: 10.1016/0031-9422(92)83155-R. [DOI] [Google Scholar]
- 56.Xu F., Morikawa T., Matsuda H., Ninomiya K., Yoshikawa M. Structures of new sesquiterpenes hepatoprotective constituents from the egyptian herbal medicine Cyperus longus. J. Nat. Prod. 2004;67:569–576. doi: 10.1021/np030368k. [DOI] [PubMed] [Google Scholar]
- 57.Li X., Yang M., Han Y.F., Gao K. New sesquiterpenes from Erigeron annus. Planta Medica. 2005;71:268–272. doi: 10.1055/s-2005-837827. [DOI] [PubMed] [Google Scholar]
- 58.Taber D.F., Bhamidipati R.S., Yet L. Phenyldimethylsilyl as an alcohol surrogate in intramolecular Diels-Alder cycloaddition: Synthesis of a-dictyopterol. J. Org. Chem. 1995;60:5537–5539. doi: 10.1021/jo00122a038. [DOI] [Google Scholar]
- 59.Brown G.D., Liang G.Y., Sy L.K. Terpenoids from the seeds of Artemisia annua. Phytochemistry. 2003;64:303–323. doi: 10.1016/S0031-9422(03)00294-2. [DOI] [PubMed] [Google Scholar]
- 60.Gao X., Deng X. A new eudesmane sesquiterpene from Senecio cannabifolius. J. Chem. Res. 2009;2009:457–458. doi: 10.3184/030823409X465303. [DOI] [Google Scholar]
- 61.Sanz J.F., Marco J.A. Sesquiterpene lactones from Artemisia caerulescens Subsp. Gargantae. Phytochemistry. 1990;29:2913–2917. doi: 10.1016/0031-9422(90)87104-3. [DOI] [Google Scholar]
- 62.Bottini A.T., Garfagnoli D.J., Delgado L.S., Dev V., Duong S.T., Kelley C.G., Keyer R., Raffel R., Joshi P., Mathela C.S. Sesquiterpene Alcohols from Hedychium spicatum var. Acuminatum. J. Nat. Prod. 1987;50:732–734. doi: 10.1021/np50052a027. [DOI] [PubMed] [Google Scholar]
- 63.Borg-Karlson A.-K., Norin T., Talvitie A. Configurations and conformations of torreyol (δ-cadinol), α-cadinol, T-muurolol and T-cadinol. Tetrahedron. 1981;37:425–430. doi: 10.1016/S0040-4020(01)92031-9. [DOI] [Google Scholar]
- 64.Appendino G., Jakupovic J., Jakupovic S. Sesquiterpenoids from Pallenis spinosa. Phytochemistry. 1997;46:1039–1043. doi: 10.1016/S0031-9422(97)00386-5. [DOI] [Google Scholar]
- 65.Zhang H.-J., Tan G.T., Santarsiero B.D., Mesecar A.D., Van Hung N., Cuong N.M., Soejarto D.D., Pezzuto J.M., Fong H.H.S. New sesquiterpenes from Litsea verticillate. J. Nat. Prod. 2003;66:609–615. doi: 10.1021/np020508a. [DOI] [PubMed] [Google Scholar]
- 66.Xu J.J., Tan N.H. Sesquiterpenes and cyclopeptides from the aerial parts of Jatropha curcas Linn. Chin. J. New Drugs. 2013;22:713–718. [Google Scholar]
- 67.Chen Y.-P., Tong C., Lu W.-Q., Shen Y.-H., Wu Z.-J., Chen W.-S. Three new sesquiterpenes from Ainsliaea glabra. Nat. Prod. Res. 2019;33:274–279. doi: 10.1080/14786419.2018.1446134. [DOI] [PubMed] [Google Scholar]
- 68.Guerreiro E., Kavka J., Giordano O.S. Sesquiterpenoids and Flavonoids from Flourensia oolepis. Phytochemistry. 1979;18:1235–1237. doi: 10.1016/0031-9422(79)80148-X. [DOI] [Google Scholar]
- 69.Miyase T., Ozaki H., Ueno A. Sesquiterpene glycosides from Ainsliaea cordifolia Franch. et Sav. Chem. Pharm. Bull. 1991;39:937–938. doi: 10.1248/cpb.39.937. [DOI] [Google Scholar]
- 70.Yan H., Haiming S., Cheng G., Xiaobo L. Chemical constituents of the roots of Inula helenium. Chem. Nat. Compd. 2012;48:522–524. doi: 10.1007/s10600-012-0298-x. [DOI] [Google Scholar]
- 71.Wang Y., Xu M.L., Jin H.Z., Fu J.J., Hu X.J., Qin J.J., Yan S.K., Shen Y.H., Zhang W.D. A new nor-sesquiterpene lactone from Ainsliaea fulvioides. Chin. Chem. Lett. 2009;20:586–588. doi: 10.1016/j.cclet.2009.01.016. [DOI] [Google Scholar]
- 72.Ren Y.-M., Zhou S.-Z., Zhang T., Qian M., Zhang R., Yao S., Zhu H., Tang C., Lin L., Ye Y. Targeted isolation of two disesquiterpenoid macrocephadiolides A and B from Ainsliaea macrocephala using a molecular networking-based dereplication strategy. Org. Chem. Front. 2020;7:1481–1489. doi: 10.1039/D0QO00030B. [DOI] [Google Scholar]
- 73.Wu Z.-J., Xu X.-K., Shen Y.-H., Su J., Tian J.-M., Liang S., Li H.-L., Liu R.-H., Zhang W.-D. Ainsliadimer A, a new sesquiterpene lactone dimer with an unusual carbon skeleton from Ainsliaea macrocephala. Org. Lett. 2008;10:2397–2400. doi: 10.1021/ol800656q. [DOI] [PubMed] [Google Scholar]
- 74.Qin J.J., Jin H.Z., Fu J.J., Hu X.J., Wang Y., Yan S.K., Zhang W.D. Japonicones A-D, bioactive dimeric sesquiterpenes from Inula japonica Thunb. Bioorg. Med. Chem. Lett. 2009;19:710–713. doi: 10.1016/j.bmcl.2008.12.043. [DOI] [PubMed] [Google Scholar]
- 75.Wang Y., Shen Y.-H., Jin H.-Z., Fu J.-J., Hu X.-J., Qin J.-J., Liu J.-H., Chen M., Yan S.-K., Zhang W.-D. Ainsliatrimers A and B, the first two guaianolide trimers from Ainsliaea fulvioides. Org. Lett. 2008;10:5517–5520. doi: 10.1021/ol802249z. [DOI] [PubMed] [Google Scholar]
- 76.Ding N. Ph.D. Thesis. Nanjing University of Chinese Medicine; Nanjing, China: 2022. Study on the Chemical Constituents and Biological Activities of Two Medicinal Plants from Ainsliaea Genus. [DOI] [Google Scholar]
- 77.Strapasson R.L.B., Cervi A.C., Carvalho J.E., Ruiz A.L.T.G., Salvador M.J., Stefanello M.A. Bioactivity-guided Isolation of Cytotoxic Sesquiterpene Lactones of Gochnatia polymorpha ssp. floccosa. Phytother. Res. 2012;26:1053–1056. doi: 10.1002/ptr.3693. [DOI] [PubMed] [Google Scholar]
- 78.Ding N., Wang J., Liu J., Zhu Y., Hou S., Zhao H., Yang Y., Chen X., Hu L., Wang X. Cytotoxic guaianolide sesquiterpe-noids from Ainsliaea fragrans. J. Nat. Prod. 2021;84:2568–2574. doi: 10.1021/acs.jnatprod.1c00587. [DOI] [PubMed] [Google Scholar]
- 79.Ren Y.-M., Zhang R., Feng Z., Ke C.-Q., Yao S., Tang C., Lin L., Ye Y. Macrocephatriolides A and B: Two guaianolide trimers from Ainsliaea macrocephala as PTP1B inhibitors insulin sensitizers. J. Org. Chem. 2021;86:17782–17789. doi: 10.1021/acs.joc.1c01996. [DOI] [PubMed] [Google Scholar]
- 80.Zhang R., Tang C., Liu H.-C., Ren Y., Xu C.-H., Ke C.-Q., Yao S., Huang X., Ye Y. Ainsliatriolides A and B: Two guaianolide trimers from Ainsliaea fragrans their cytotoxic activities. J. Org. Chem. 2018;83:14175–14180. doi: 10.1021/acs.joc.8b02346. [DOI] [PubMed] [Google Scholar]
- 81.Zhou N., Li J.-J., Wu Y., Wang Z.-Y., Yin Z.-P., Wang X.-P., Liu Y.-R., Shang X.-Y. New polymerized sesquiterpene lactones from Ainsliaea yunnanensis and their activity evaluation. Nat. Prod. Res. 2021;36:4862–4868. doi: 10.1080/14786419.2021.1904924. [DOI] [PubMed] [Google Scholar]
- 82.Granica S., Lohwasser U., Jöhrer K., Zidorn C. Qualitative and quantitative analyses of secondary metabolites in aerial and subaerial of Scorzonera hispanica L. (black salsify) Food Chem. 2015;173:321–331. doi: 10.1016/j.foodchem.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 83.Wang R., Liu L.L., Shi Y.P. Pubescone, a novel 11 (7→6) abeo-14-norcarabrane sesquiterpenoid from Siegesbeckia pubescens. Helv. Chim. Acta. 2010;93:2081–2085. doi: 10.1002/hlca.201000034. [DOI] [Google Scholar]
- 84.Dekebo A., Dagne E., Sterner O. Furanosesquiterpenes from Commiphora sphaerocarpa and related adulterants of true myrrh. Fitoterapia. 2002;73:48–55. doi: 10.1016/S0367-326X(01)00367-7. [DOI] [PubMed] [Google Scholar]
- 85.Dong S., Tang J.-J., Zhang C.-C., Tian J.-M., Guo J.-T., Zhang Q., Li H., Gao J.-M. Semisynthesis in vitro cytotoxic evaluation of new analogues of 1-O-acetylbritannilactone a sesquiterpene from Inula britannica. Eur. J. Med. Chem. 2014;80:71–82. doi: 10.1016/j.ejmech.2014.04.028. [DOI] [PubMed] [Google Scholar]
- 86.Qin J.-J., Jin H.-Z., Zhu J.-X., Fu J.-J., Zeng Q., Cheng X.-R., Zhu Y., Shan L., Zhang S.-D., Pan Y.-X., et al. New sesquiterpenes from Inula japonica Thunb with their inhibitory activities against LPS-induced NO production in RAW264.7 macrophages. Tetrahedron. 2010;66:9379–9388. doi: 10.1016/j.tet.2010.09.091. [DOI] [Google Scholar]
- 87.Chuang L.-F., Fan T.-Y., Li J.-J., Sung P.-J. Kobusone: Occurrence of a norsesquiterpenoid in the gorgonian coral Rumphella antipathies (Gorgoniidae) Biochem. Syst. Ecol. 2007;35:470–471. doi: 10.1016/j.bse.2007.01.007. [DOI] [Google Scholar]
- 88.Yukawa C., Iwabuchi H., Komemushi S., Sawabe A. Mono- and sesquiterpenoids of the volatile oil of Bursera graveolens. Flavour Fragr. J. 2005;20:653–658. doi: 10.1002/ffj.1523. [DOI] [Google Scholar]
- 89.Heymann H., Tezuka Y., Kikuchi T., Supriyatna S. CConstituents of Sindora sumatrana MIQ: I. isolation and NMR spectral analysis of sesquiterpenes from the dried pods. Chem. Pharm. Bull. 1994;42:138–146. doi: 10.1248/cpb.42.138. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.