Abstract
Streptococcus pyogenes (Group A Streptococcus, GAS) is a human-restricted pathogen that causes a wide range of diseases from pharyngitis and scarlet fever to more severe, invasive infections such as necrotising fasciitis and streptococcal toxic shock syndrome. There has been a global increase in both scarlet fever and invasive infections during the COVID-19 post-pandemic period. The aim of this study was the molecular characterisation of 17 invasive and non-invasive clinical non-emm1 GAS isolates from an Australian tertiary hospital collected between 2021 and 2022. Whole genome sequencing revealed a total of nine different GAS emm types with the most prevalent being emm22, emm12 and emm3 (each 3/17, 18%). Most isolates (14/17, 82%) carried at least one superantigen gene associated with contemporary scarlet fever outbreaks, and the carriage of these toxin genes was non-emm type specific. Several mutations within key regulatory genes were identified across the different GAS isolates, which may be linked to an increased expression of several virulence factors. This study from a single Australian centre provides a snapshot of non-emm1 GAS clinical isolates that are multiclonal and linked with distinct epidemiological markers commonly observed in high-income settings. These findings highlight the need for continual surveillance to monitor genetic markers that may drive future outbreaks.
Keywords: Streptococcus pyogenes, scarlet fever, invasive infection, emm types, superantigen, streptolysin O
1. Introduction
Streptococcus pyogenes, also known as Group A Streptococcus (GAS), is a Gram positive, β-haemolytic, facultatively anaerobic bacterial pathogen. GAS is exquisitely adapted to the human host, causing a wide range of clinical manifestations, ranging from mild infections such as pharyngitis, scarlet fever, and impetigo to more severe invasive diseases including bacteraemia, necrotising fasciitis and streptococcal toxic shock syndrome [1]. Additionally, GAS infection can trigger post-infection autoimmune sequelae, such as acute rheumatic fever and poststreptococcal glomerulonephritis [2]. It is estimated that GAS causes approximately 500,000 deaths worldwide annually [3], predominantly from rheumatic heart disease where there is an inequitable burden impacting lower middle-income countries and Indigenous populations of affluent nations [4,5]. Quantifying the disease burden of GAS infections remains difficult due to under-reporting of both invasive and non-invasive cases, a lack of comprehensive disease registries and reliance on passive surveillance [6]. There is no commercial vaccine currently available against GAS, and the development of a safe and effective vaccine remains a high priority. GAS infections can be treated with antibiotics [7] with GAS remaining remarkably susceptible to β-lactams [8].
A global re-emergence of scarlet fever has been reported since 2011 [9,10,11]. In some cases, GAS pharyngeal infection may result in acute rheumatic fever and rheumatic heart disease with long-term damage to the heart valves [12,13]. In 2022, the World Health Organization (WHO) reported spikes in both scarlet fever and invasive GAS (iGAS) infections in several European countries, including the United Kingdom (UK), France, Ireland, Sweden, the Netherlands and Australia [4,14]. iGAS disease is defined as the isolation of strains from normally sterile body sites, such as blood, pleural and joint fluid or deep tissue [15,16]. Although progression from non-invasive to iGAS infection is rare, invasive infections are associated with mortality rates of up to 25%.
GAS isolates are classified into emm types based on the hypervariable 5′ sequence of the M protein, which is a surface-anchored protein and key virulence determinant [17]. Each GAS isolate harbours an emm gene variant with over 275 emm types documented currently [18]. Typing GAS isolates using emm sequencing remains important in the epidemiological surveillance of GAS infections [11]. Prevalent emm types vary across different geographical locations [19,20,21,22,23,24].
The concerning post-pandemic spike in both scarlet fever and iGAS cases, possibly due to a combination of events including the emergence of the M1UK lineage, viral co-infections, and a potential decrease in the immune status of the general population following the relaxation of SARS-CoV-2 restrictions, has highlighted the requirement for the adequate detection and surveillance of GAS infections [25,26,27,28,29,30]. The presence of the M1UK lineage has already been reported recently in Australia, and its dominance in the GAS population has been reported globally [27,31]. M1UK isolates exhibit an increased production of superantigen SpeA, which may contribute to its fitness [27,31]. Contemporary scarlet fever isolates are also associated with a specific toxin repertoire, including superantigens SpeC and SSA, alongside deoxyribonuclease (DNase) Spd1 [1,20,23,32,33]. Therefore, we aimed to understand the prevalence of toxin repertoires in the population of non-emm1 clinical GAS isolates. Here, we performed emm typing and conducted a molecular analysis of 17 non-emm1 GAS clinical isolates from a tertiary hospital in southeast Queensland, Australia.
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
A total of 17 non-emm1 clinical GAS isolates were collected from the Gold Coast University Hospital between 2021 and 2022 (Table 1). No patients succumbed to infection. Isolates were cultured on horse blood agar (HBA) or in Todd–Hewitt broth supplemented with 1% yeast extract (THY) and incubated at 37 °C.
Table 1.
Group A Streptococcus isolates from patients at Gold Coast University Hospital, 2021–2022.
| GAS Isolate Designation | Age | Sex | Sample Source | Clinical Diagnosis | Non-Invasive/Invasive | emm Type |
|---|---|---|---|---|---|---|
| SP1492 | 13 months | M | Blood | STSS | Invasive | 12 |
| SP1493 | 11 months | M | Fluid lower leg | Bacteraemia | Invasive | 12 |
| SP1494 | 48 years | M | Thigh tissue | Bacteraemia | Invasive | 4 a |
| SP1495 | 17 months | M | Pleural fluid | Bacteriemia | Invasive | 3.93 |
| SP1496 | 15 months | F | Throat | Tonsilitis | Non-invasive | 3.93 |
| SP1497 | 39 years | F | Bursa fluid | Prepatellar bursitis | Invasive | 41.2 |
| SP1498 | 51 years | M | Blood | Bacteraemia | Invasive | 41.2 |
| SP1499 | 3 years | F | Swab throat | Scarlet fever | Non-invasive | 3.93 |
| SP1500 | 37 years | M | Blood | Bacteraemia | Invasive | 53 |
| SP1501 | 39 years | M | Tibia tissue | NF | Invasive | 22 a |
| SP1502 | 8 years | F | Throat | Scarlet fever | Non-invasive | 22 a |
| SP1503 | 6 years | M | Throat | Scarlet fever | Non-invasive | 89 a |
| SP1504 | 4 months | F | Blood | Bacteraemia | Invasive | 12 |
| SP1505 | 5 years | M | Throat | Scarlet fever | Non-invasive | 22 a |
| SP1506 | 56 years | M | Skin | NF | Invasive | 77 |
| SP1507 | 2 years | F | Blood | Bacteraemia | Invasive | 75 |
| SP1508 | 6 years | F | Blood | Bacteraemia | Invasive | 4 a |
F, female; M, male; NF, necrotising fasciitis; STSS, streptococcal toxic shock syndrome. a Acapsular emm types.
2.2. Genome Sequencing, Assembly and Analysis
Genomic DNA (gDNA) was extracted from all isolates and used to create paired-end multiplex libraries, which were sequenced using the Illumina HiSeq 2500 platform at a length read of 150 (bp) (Australian Genome Research Facility, Brisbane, Australia). Read mapping was performed using Shovill V1.1.0 (https://github.com/tseemann/shovill, accessed on 14 May 2024) with an underlying SPAdes assembler [34]. Genome contigs were aligned in FASTA format with gene predictions and annotations generated using PROKKA [35] and streptococcal RefSeq-specific databases. Screening for virulence genes and regulators of interest was performed using screen_assembly v1.2.7 [36]. Methods were followed according to Davies et al. [36].
2.3. Phylogenetic Analysis
Mashtree [37] was used to place the 17 GAS isolates genomes in this study with a previously analysed 2083 global GAS genome sequences from Davies et al. [36].
2.4. Polymerase Chain Reaction (PCR) Screening
GAS toxin genes slo (streptolysin O), speA (streptococcal pyrogenic exotoxin A), ssa (streptococcal superantigen), speC (streptococcal pyrogenic exotoxin C), spd1 (streptococcus pyogenes deoxyribonuclease 1), speB (streptococcal pyrogenic exotoxin B) and hasA (hyaluronan synthase), and macrolide-resistance gene ermB (erythromycin methylase), were each amplified by PCR using the KAPA HiFi PCR kit (Roche, cat no. 07958846001) and gDNA as a template. Primers and reagents parameters are listed in Supplementary Table S1. gDNA from SP1380 [31] served as a positive control for all toxins tested, and gDNA from HKU16 [23] served as a positive control for ermB. Distilled H2O (dH2O) was used as a negative control for all testing. PCR products were resolved on a 2% TAE agarose gel.
2.5. Sodium Dodecyl Sulfate–Polyacrylamide Electrophoresis (SDS-PAGE) and Western Blotting
Cultures were grown in THY supplemented with 2 mM cysteine to the late exponential phase to support SSA expression, as demonstrated previously [36]. Filter-sterilised culture supernatant was precipitated using 10% trichloroacetic acid (TCA). The precipitate was then resuspended in LDS loading buffer (NuPAGE) containing 100 mM dithiothreitol (DTT) normalised to the OD600 of the culture. Samples were boiled for 5 min, subjected to SDS-PAGE, and then transferred to a PVDF membrane (Sigma Aldrich, # IPFL00010, Saint Louis, MO, USA) by wet transfer for the detection of immune-reactive bands using a LI-COR Odyssey Imaging System (LI-COR Biosciences, Lincoln, NE, USA). The primary antibodies used to detect the SpeA, SpeC, SSA and SpeB protein in GAS culture supernatants were rabbit antibodies to SpeA (#PAI11, Toxin Technology, Sarasota, FL, USA), SpeC (PCI333, Toxin Technology), SSA (produced by Mimotopes [37]), and SpeB (PBI222, Toxin Technology) respectively, all at 1:1000 dilution. The murine primary antibodies to detect Spd1 and SLO protein in GAS culture supernatants were at 1:1000 dilution [36]. Of note, the Spd1 blot was incubated in primary antibody that had first been pre-adsorbed using HKU16Δssa/speC/spd1 [36] to minimise non-specific bands. Briefly, HKU16Δssa/speC/spd1 was subjected to SDS-PAGE, then transferred to a PVDF membrane via wet transfer as previously described. The membrane was then incubated in murine antibody to Spd1 at a 1:1000 dilution. This antibody then served as the primary antibody for the Spd1 blots. Either anti-rabbit IgG ((H + L) (DyLight™800 4X PEG Conjugate, NEB #5151P)) or anti-mouse IgG ((H + L) (Dylight™ 800 4X PEG Conjugate, NEB #5257S)) were used as secondary antibodies at a 1:10,000 dilution. Western blot images were captured using an Odyssey LI-COR instrument and Image Studio (v.5.2) software. Culture supernatants from HKU488 [31] served as a positive control for all toxins.
2.6. SpeB Caseinolytic Activity Assay
All isolates were grown on HBA plates overnight at 37 °C. GAS M1global isolate 5448 was used as a positive control, while isolates 5448AP and 5448ΔspeB [38] were used as negative controls. Cultures were statically grown to an OD600 of 0.4 in THY where 5 μL of undiluted culture was pipetted onto Columbia agar plus 15% skin milk and incubated at 37 °C for 72 h. SpeB activity was determined after 72 h based on the clear zone with an opaque perimeter surrounding the culture growth indicating caseinolytic activity [39].
2.7. SLO Activity Assay
SLO activity was measured by the standard SLO haemolysis assay [40]. All isolates were grown on HBA plates overnight at 37 °C. GAS M1global isolate 5448 was used as a positive control, while isolate 5448Δslo was used as a negative control. All bacterial cultures were statically grown in THY to late exponential phase with the exception of SP1494, which was grown in THY supplemented with 20 mM NaHCO3 to compensate for its in vitro growth defect characteristic of emergent, chimeric emm4 GAS [41]. Fresh whole human blood was centrifuged at 500 × g for 10 min to separate red blood cells (RBCs), buffy coat and plasma. The buffy coat and plasma were aspirated and discarded. The tube was filled to the original level of plasma with Hanks Balanced Salt Solution (HBSS), inverted gently to mix, and centrifuged at 500 × g for 10 min. The washing step was repeated for a total of two washes. The supernatant was aspirated and replaced with HBSS, inverted to mix, and was considered to contain only RBCs. A 2% RBC solution was prepared in HBSS, and 200 µL was aliquoted into a 96-well plate. Then, 40 µL of filter-sterilised culture supernatant was added to each well. Triton X-100 (final concentration of 1%) was used as a positive control for 100% RBC lysis, while phosphate-buffered saline was used as a negative control. The plate was incubated for 30 min at 37 °C and then centrifuged at 1000 × g for 10 min to pellet any intact RBCs. The amount of extracellular haemoglobin was measured spectrophotometrically at 405 nm using a CLARIOStar Plus microplate reader (BMG LABTECH). Results were graphed using GraphPad Prism version 10.2.0. Blood samples from a minimum of three healthy adult donors were used and performed in triplicate.
2.8. Ethics Approvals
Clinical approval for this project was granted by the Children’s Health Queensland Human Research Ethics Committee HREC/10/QRCH/113. The ethics approval number for the blood collected for the SLO activity assay is 2010/HE001586).
2.9. Statistical Analysis
All statistical analyses were completed using Prism software (GraphPad; version 10.2.0). Significance was calculated using one-way ANOVA and Tukey’s post-test.
2.10. Data Availability
Whole genome sequencing data obtained in this study were submitted to the Sequence Read Archive database (https://www.ncbi.nlm.nih.gov/sra, accessed on 30 September 2024; BioProject accession PRJNA1172375).
3. Results
3.1. Investigation of 17 Contemporary GAS Isolates
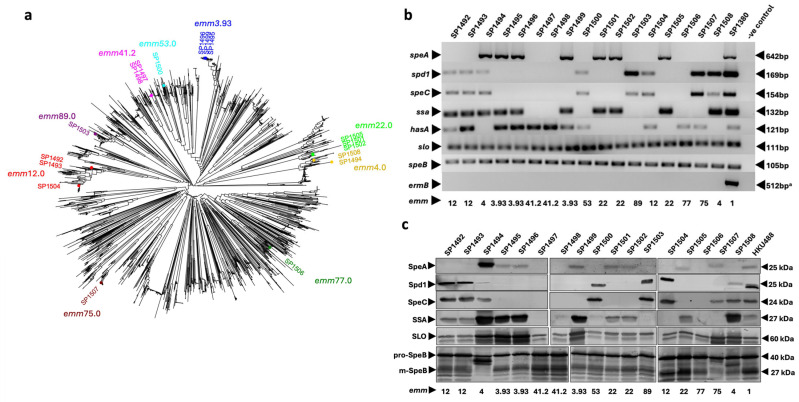
A total of 17 non-emm1 clinical GAS isolates were collected as a clinical snapshot from the Gold Coast University Hospital in subtropical southeast Queensland between 2021 and 2022. Of the 17 clinical isolates, 12 were isolated from invasive infections (iGAS) and 5 were non-invasive (scarlet fever [n = 4] and tonsillitis [n = 1]) (Table 1). A total of nine different emm types were identified following whole genome sequencing (WGS) (Table 1). emm22, emm12 and emm3.93 were the most common emm types (three isolates each), emm41 and emm4 had two isolates each, while emm89, emm77, emm75 and emm53 were each represented by a single isolate. Interestingly, (6/17, 35%) isolates were lacking the hasABC operon genes required for capsule biosynthesis (SP1494 and SP1508 (emm4), SP1501, SP1502 and SP1505 (emm22), and SP1503 (emm89)) [42]. A maximum-likelihood phylogenetic tree was constructed to understand the distribution of the 17 clinical GAS isolates across the global GAS population comprising a geographically and clinically diverse set of 2083 genomes with 150 different emm types [36]. All 17 isolates were found to be represented across the global GAS population (Figure 1a). All emm4, emm12 and emm41.2 isolates were found to be invasive. Of the five non-invasive isolates, three were acapsular.
Figure 1.
Genetic analysis and protein expression of 17 GAS clinical isolates from Gold Coast University hospital. (a) Population structure of 2100 globally distributed GAS genomes [36] with addition of the 17 locally acquired GAS genomes. The tree was generated using Mashtree software v1.4.6 [37]. The 17 isolates from this study are highlighted with colours matching the respective emm type. (b) PCR screening for toxin and antibiotic resistance genes. Genes examined are indicated on the left, and PCR product sizes (bp) are indicated on the right. M1UK GAS isolate SP1380 was used as a positive control for all toxins [31]. a emm12 GAS isolate HKU16 was used as a positive control for ermB [23]. (c) Western blot analysis of toxin expression in culture supernatants. Toxins are indicated on the left, and the protein mass (kDa) is indicated on the right. M1global GAS isolate HKU488 was used as a positive control. The slightly lower molecular weight band detected for SLO likely represents an SLO isoform or breakdown product [43].
3.2. Detection of Diverse Toxin Profiles in GAS Clinical Isolates
To study the toxin profile of the 17 clinical isolates, we performed PCR screening to confirm the presence of scarlet fever-associated superantigen toxins speA, speC, and ssa and the DNase gene spd1 [11,44] (Figure 1b). Of the superantigens, ssa was identified in 59% (10/17) of isolates, speA was detected in 7/17 isolates (42%), while speC and spd1 were detected in 8/17 isolates (47%). Two of the three emm12 isolates (SP1492 and SP1493) were found to carry ssa, speC, and spd1, while SP1504 carried speC and spd1 but lacked ssa. This toxin profile was reported previously in Asian emm1 and emm12 scarlet fever isolates [20,37]. The emm22 isolates were speA and ssa positive, but speC negative, which was a feature shared with emm22 isolates from other geographical locations, such as the UK and Europe [45,46,47,48]. The emm41.2 isolates SP1497 and SP1498 and emm75 isolate SP1506 were negative for ssa, speC, ssa and spd1, while emm4 isolate SP1494 was found to possess all four scarlet fever-associated toxins. The carriage of superantigen genes speC and ssa alongside DNase spd1 is commonly reported for emm4 isolates, while the carriage of speA in this emm type is variable [44,47,48,49,50]. As expected, the acapsular phenotypes of all emm4, emm22, and emm89 isolates were accounted for by the absence of the hasA gene (Figure 1b). The SLO encoding gene slo and cysteine protease gene speB are highly conserved across all GAS isolates [51], and as expected, 17/17 isolates carried both genes. Macrolide-resistance gene ermB was also investigated, as macrolide-resistance is observed frequently in clinical scarlet fever isolates from mainland China and Hong Kong [52]. To test for potential macrolide resistance, we screened the 17 isolates for the ermB gene. However, none of the isolates carried the ermB gene (Figure 1b). Full PCR gel images can be found in Supplementary Figure S1.
3.3. Concordance of GAS Virulence Factor Expression and Increased Virulence Factor Expression
To confirm toxin carriage and investigate toxin expression in the 17 isolates, we performed Western blot analysis to detect secreted toxins in bacterial culture supernatants grown to an OD600 of 0.8. Overall, the toxin profile detected by Western blot analysis matched the PCR gene screening and WGS findings (Figure 1c, Table 2). The expression of SpeA, SpeC, Spd1, and SSA was found to reflect the PCR results, except for SP1507, which was found to express SpeC but not Spd1, despite detecting spd1 using WGS and PCR (Figure 1b). Since no mutation was detected in the spd1 gene locus, the reason for the absence of Spd1 in SP1507 culture supernatants remains unclear. All isolates were found to secrete control proteins SLO and SpeB; however, no mature (and therefore active) SpeB was detected for SP1494, with a band detected at a higher molecular level than expected for m-SpeB. A SpeB caseinolytic assay was performed to investigate the proteolytic activity of all 17 isolates. All isolates were found to produce SpeB activity except for SP1494 and negative controls (Supplementary Figure S2, Supplementary Table S2). It should be noted that SP1494 was found to possess an attenuated growth phenotype, delaying the isolate’s ability to achieve the same levels of growth as the other isolates in this study. This may explain the lack of SpeB activity detected in this assay. Full Western blot images can be found in Supplementary Figure S3.
Table 2.
Summary of all identified toxins in the 17 clinical GAS isolates analysed in this study.
| Isolate | emm Type | speA a | speC a | ssa a | spd1 a | hasA b | speB a | slo a | ermB b |
|---|---|---|---|---|---|---|---|---|---|
| SP1492 | 12 | − | + | + | + | + | + | + | − |
| SP1493 | 12 | − | + | + | + | + | + | + | − |
| SP1494 | 4 | + | + | + | + | − | + | + | − |
| SP1495 | 3.93 | + | − | + | − | + | + | + | − |
| SP1496 | 3.93 | + | − | + | − | + | + | + | − |
| SP1497 | 41.2 | − | − | − | − | + | + | + | − |
| SP1498 | 41.2 | − | − | − | − | + | + | + | − |
| SP1499 | 3.93 | + | − | + | − | + | + | + | − |
| SP1500 | 53 | − | + | − | + | + | + | + | − |
| SP1501 | 22 | + | − | + | − | − | + | + | − |
| SP1502 | 22 | + | − | + | − | − | + | + | − |
| SP1503 | 89 | − | + | − | + | − | + | + | − |
| SP1504 | 12 | - | + | − | + | + | + | + | − |
| SP1505 | 22 | + | − | + | − | − | + | + | − |
| SP1506 | 77 | − | − | − | − | + | + | + | − |
| SP1507 | 75 | − | + | − | Ind c | + | + | + | − |
| SP1508 | 4 | − | + | + | + | − | + | + | − |
GAS, Group A Streptococcus; PCR, polymerase chain reaction; WGS, whole genome sequencing; +, positive; −, negative, Ind, indefinite. a identified using WGS, PCR and Western blot analysis. b identified using WGS and PCR analysis only.c spd1 gene positive but Spd1 expression negative.
In addition, emm3.93 isolates SP1495, SP1496, and SP1499 all expressed high levels of SSA and SLO, while emm75 isolate SP1507 only displayed increased expression levels of SLO. emm4 isolates SP1494 and SP1508 both secreted elevated levels of SSA and SLO; however, only SP1494 expressed increased levels of SpeA. It should be noted that SP1494 was found to possess an attenuated growth phenotype. Therefore, toxin expression by this isolate should be interpreted with some caution. Intriguingly, all isolates that showed increased SLO expression also displayed increased expression levels of SSA, suggesting they may respond to similar, yet unidentified, regulatory signals.
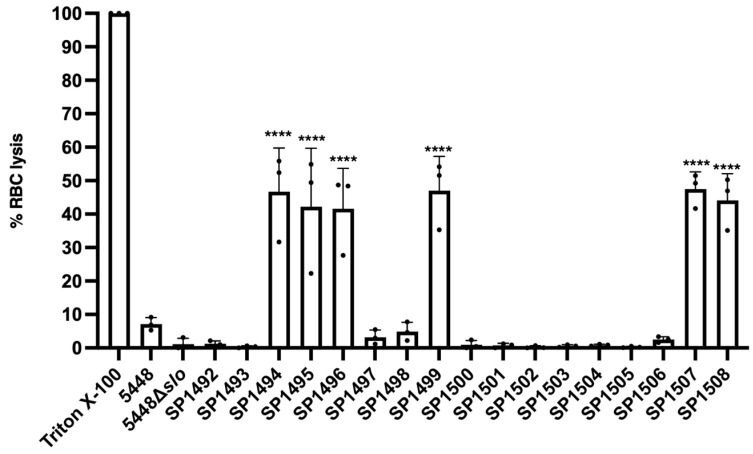
Next, we performed an exploratory phenotypic characterisation of SLO to validate the differential expression of these toxins in the clinical isolates. SLO is a cholesterol- and glycan-dependent cytolysin that perforates the lipid bilayer of various host cells, including epithelial and immune cells [53,54,55,56]. SLO is a major GAS virulence factor, and the emergence and pathogenicity of highly virulent GAS genotypes are often associated with a high-activity promoter recombination event at the slo gene locus, resulting in increased SLO expression [57]. Such genotypes include numerous acapsular emm types, such as emm28, emm87 and emm89 that have all been shown to express increased levels of SLO, potentially compensating for the loss of capsule [58]. RBC lysis and haemoglobin release can be measured spectrophotometrically to quantify SLO activity [40]. Using human RBCs, the haemolytic activity of culture supernatants of all 17 isolates was compared to GAS emm1 strain 5448 [59] to determine SLO expression levels. Significantly increased haemolytic activity was found for emm3.93 (SP1495, SP1496, SP1499), emm4 (SP1494, SP1508) and emm75 (SP1507) isolates, validating the increased SLO expression levels identified by Western blotting (Figure 2).
Figure 2.
SLO haemolytic activity of 17 clinical GAS isolates from Gold Coast University Hospital. Levels of RBC lysis and the release of haemoglobin was measured spectrophotometrically at OD405. SLO activity was assessed in comparison to the control strain 5448. Data are plotted as the mean ± SD from three independent biological experiments, indicated as black dots. Data were analysed by one-way ANOVA and Tukey’s post-test. **** p < 0.0001. Positive control = 1% Triton X-100. ANOVA, analysis of variance; OD, optical density; RBCs, red blood cells; SD, standard deviation; SLO, streptolysin O.
3.4. Mutations Within Major GAS Virulence Factors May Drive Differential Toxin Expression
Toxin expression in GAS is a highly regulated process orchestrated by gene regulators including the control of virulence two-component regulatory system CovRS and the two stand-alone transcriptional regulators RocA and RopB [60]. Non-synonymous mutations in the coding sequences of these regulators frequently arise during infections which often leads to an altered expression of target genes, including several virulence factors [60]. We therefore hypothesised that mutations in these regulatory genes may account for differential expression levels of SpeA, SSA and SLO observed in some of the 17 clinical isolates.
Analysis of the covRS, rocA and ropB gene sequences revealed that both emm4 isolates SP1494 and SP1508 harboured a single nucleotide deletion at position 83 of covS resulting in an early stop codon at position 36 of the amino acid sequence and the premature truncation of CovS (Table 3). However, these isolates maintained production of the protease SpeB (Figure 1c, Figures S2 and S3), the expression of which is commonly lost in CovS truncation mutants [61]. emm75 isolate SP1507 has acquired an 18 base pair insertion at position 86 of the covS gene sequence, encoding the six amino acids IFCIFC at position 30–36 in the CovS protein sequence (Table 3). While these specific mutations have not been documented previously, inactivating mutations in CovS result in the de-repression of CovRS-regulated virulence genes such as slo [62,63], thus providing a possible explanation for the increased SLO expression and activity observed in these isolates. emm3.93 isolates SP1495, SP1496 and SP1499 were found to possess a non-synonymous SNP at position 1228 of the rocA gene sequence, giving rise to a premature stop codon in the RocA protein sequence (Table 3). This SNP was reportedly previously to result in an enhanced expression of SLO [64,65], which may account for an increased expression of SLO in SP1495, SP1496 and SP1499.
Table 3.
GAS master regulators and associated non-synonymous mutations of the 17 clinical GAS isolates analysed in this study.
| Isolate | covR | covS | rocA | ropB | ||||
|---|---|---|---|---|---|---|---|---|
| Carriage | Mutation | Carriage | Mutation | Carriage | Mutation | Carriage | Mutation | |
| SP1492 | + | − | + | − | + | − | + | − |
| SP1493 | + | − | + | − | + | − | + | − |
| SP1494 | + | − | + | truncation | + | − | + | T104I a |
| SP1495 | + | − | + | − | + | insertion | + | − |
| SP1496 | + | − | + | − | + | insertion | + | − |
| SP1497 | + | − | + | − | + | − | + | − |
| SP1498 | + | − | + | − | + | − | + | − |
| SP1499 | + | − | + | − | + | insertion | + | − |
| SP1500 | + | − | + | − | + | − | + | − |
| SP1501 | + | − | + | − | + | − | + | − |
| SP1502 | + | − | + | − | + | − | + | − |
| SP1503 | + | − | + | − | + | − | + | − |
| SP1504 | + | − | + | − | + | − | + | − |
| SP1505 | + | − | + | − | + | − | + | − |
| SP1506 | + | − | + | − | + | − | + | − |
| SP1507 | + | − | + | insertion | + | − | + | − |
| SP1508 | + | − | + | truncation | + | − | + | − |
GAS, Group A Streptococcus; +, gene detected; −, mutation not detected. a Threonine substituted for isoleucine at position 104 of the amino acid sequence.
Furthermore, we detected a non-synonymous mutation in the SpeB regulator ropB in the emm4 isolate SP1494 with a C → T mutation at position 311 in the nucleotide sequence. This resulted in a threonine to isoleucine change at position 104 in RopB (Table 3). This T104I substitution in RopB has been shown previously to drive increased SpeB production in emm4 isolates [49]. However, since SP1494 showed markedly reduced expression levels of mature SpeB and displayed negligible SpeB activity (Supplementary Figure S2, Supplementary Table S2), we speculate there are potentially other yet-unidentified mutations in this isolate that affect SpeB maturation.
4. Discussion
In this study, we conducted a molecular characterisation of 17 invasive and non-invasive clinical GAS isolates from an Australian tertiary hospital in the post-COVID period of 2021–2022, specifically focusing on non-emm1 GAS isolates to capture a small-scale genomic snapshot of the GAS population diversity. An increased expression of pore-forming toxin SLO was noted in isolates SP1494 and SP1508 (emm4), SP1495, SP1496 and SP1499 (emm3.93) and SP1507 (emm75). A further characterisation of SLO activity confirmed increased SLO activity in these isolates. This observed phenotype underscores the importance of molecular characterisation and ongoing surveillance of clinical GAS isolates.
In total, nine different emm types were detected in this clinical strain set. The three most prevalent emm types were emm22, emm12, and emm3.93 (three isolates each). Overall, 6/17 (35%) strains were acapsular. Capsule-negative emm types, such as emm4, emm22 and emm89, represent approximately 30% of the global GAS population and are major contributors to both invasive and non-invasive disease globally [48,66,67,68,69,70,71]. In contrast, emm22, emm12, and emm3.93 were the most common emm types; these emm types are common in geographical regions such as mainland China and Hong Kong or within invasive isolates from Australia [4,20,72,73,74]. The prevalence of scarlet fever-associated toxins within this strain set is of concern. Of the seven isolates found to possess speA, four were non-invasive isolates. While speA has been shown to be more commonly associated with iGAS isolates, it has also been found in association with non-invasive isolates [47,75,76,77]. It was further determined that the carriage of these toxin genes was non-emm type specific.
Of the 17 isolates, only emm4 isolate SP1494 was found to possess all tested toxins. The isolate also displayed an increased production of SLO, SpeA, and SSA in comparison to other isolates. Recent research from the United States has identified a novel fusion event between the emm and enn genes in several clinical emm4 isolates, resulting in the creation of a chimeric emm protein (designated emm4C) [78]. The authors found that the emm4C isolates had a marked growth defect in vitro but showed increased survival in human blood and increased virulence in a murine model when compared to emm4 isolates [49]. The identification of the growth defect of SP1494, like that observed elsewhere [49], prompted further investigation into this isolate. It was determined that SP1494 possessed a SNP in the putative carbonic anhydrase gene saca responsible for the observed growth impairment as well as the chimeric emm gene [41]. SP1494 was also shown to possess other emm4C clade-defining mutations, including the previously mentioned ropB and saca SNPs, confirming its position within the emergent emm4C population, and that this hypervirulent clone has now been detected in Australia [49]. The changes in M protein sequence observed in emm4C pose a potential difficulty to current vaccines targeting the M protein [79]. However, the development of non-M protein-based vaccines may circumvent this issue.
We also detected the presence of several mutations in key regulatory genes in this strain cohort. The invasive isolates SP1494 and SP1508, both emm4, and the emm75 isolate SP1507 carried non-synonymous mutations in covS. Although the repression of SpeB is a predictive marker for identifying CovS-inactivated isolates [61], none of these mutations affected SpeB expression levels. However, these isolates demonstrated high levels of SLO expression and activity, which is generally observed in CovS-inactivated isolates [61]. The emm3.93 isolates SP1495, SP1496 and SP1499 were all found to possess the same rocA mutation, which has been reported previously to contribute to emm3-associated GAS invasive propensity [64,65]. As mutations resulting in increased toxin production may result in the switch from colonisation to invasive disease, vaccination, which aims to prevent colonisation, would not be impacted by these mutations [80].
We wish to acknowledge that this study has several limitations. Firstly, the small size of the strain set (17 isolates) hinders the ability to make broader conclusions about emm type prevalence and toxin carriage in the general population. Secondly, the study is restricted to a single hospital in southeast Queensland, therefore only providing a regional snapshot of clinical GAS isolates circulating currently in Australia. Future research, incorporating larger and more diverse strain sets from multiple regions and healthcare institutions, is needed to provide a better understanding of emm type distribution and toxin carriage.
5. Conclusions
In this study, we performed the molecular characterisation of 17 non-emm1 clinical GAS isolates from a single tertiary hospital in southeast Queensland, Australia. The detection of the hypervirulent emm4C clone in Australia, alongside the increased SLO production observed in several isolates, highlight the need for the continuous molecular surveillance of clinical GAS isolates. Despite our study’s limitations, these observations emphasize the importance of expanding genomic and phenotypic monitoring to detect emerging strains with increased virulence or resistance characteristics that may pose a broader public health threat.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/pathogens13110956/s1, Figure S1: Images of agarose gels following polymerase chain reaction amplification of toxin and antibiotic resistance genes from 17 clinical Group A Streptococcus isolates from Gold Coast University Hospital. SP1380 and HKU16 served as positive controls and distilled H2O was used as a negative control. HyperLadder™ 1kb (Meridian Bioscience #BIO-33053) was used as molecular weight marker; Figure S2: Results from the streptococcal pyrogenic exotoxin B (SpeB) caseinolytic activity assay for 17 clinical isolates from Gold Coast University Hospital. SpeB activity is indicated by a clear zone with an opaque halo surrounding the culture on the plate. Positive control: 5448; negative controls: 5448ΔspeB and 5448AP; Figure S3: Images of Western blots for the detection of Group A Streptococcus toxins in supernatant from the 17 clinical isolates from the Gold Coast University Hospital. Isolate HKU488 was used as positive control. PageRuler™ pre-stained protein ladder (ThermoScientific #26616) was used as molecular weight marker; Table S1: Primers for polymerase chain reaction screening of Group A streptococcal scarlet fever toxins and antibiotic resistance genes; Table S2: Summary of SpeB caseinolytic assay results.
Author Contributions
Conceptualization, P.K.S., M.J.W. and S.B.; methodology, P.K.S., A.J.H., M.R.D., M.J.W. and S.B.; software, A.J.H. and M.R.D.; validation, P.K.S., A.J.H., M.R.D., M.J.W. and S.B.; formal analysis, P.K.S., A.J.H. and M.R.D.; investigation, P.K.S. and A.J.H.; resources, A.J.H., M.L., A.B., K.G., M.R.D. and M.J.W.; writing—original draft preparation, P.K.S., M.J.W. and S.B.; writing—review and editing, P.K.S., A.J.H., M.L., A.B., K.G., M.R.D., M.J.W. and S.B.; visualization, P.K.S. and A.J.H.; supervision, K.G., M.J.W. and S.B.; funding acquisition, M.J.W. and S.B. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
This study was approved by the Queensland Children’s Health Services Human Research Ethics Committee (HREC/10/QRCH/113, 14 December 2010).
Informed Consent Statement
Informed consent was obtained from all subjects involved in this study.
Data Availability Statement
Whole genome sequencing data obtained in this study were submitted to the Sequence Read Archive database (https://www.ncbi.nlm.nih.gov/sra (accessed on 30 September 2024); BioProject accession PRJNA1172375).
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This research was funded by National Health and Medical Research Council of Australia, grant number 1194130. P.K.S. is funded by the Australian Government under a University of Queensland Graduate Support Scholarship.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Brouwer S., Rivera-Hernandez T., Curren B.F., Harbison-Price N., De Oliveira D.M.P., Jespersen M.G., Davies M.R., Walker M.J. Pathogenesis, epidemiology and control of Group A Streptococcus infection. Nat. Rev. Microbiol. 2023;21:431–447. doi: 10.1038/s41579-023-00865-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cunningham M.W. Pathogenesis of Group A Streptococcal infections. Clin. Microbiol. Rev. 2000;13:470–511. doi: 10.1128/CMR.13.3.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carapetis J.R., Steer A.C., Mulholland E.K., Weber M. The global burden of Group A Streptococcal diseases. Lancet Infect. Dis. 2005;5:685–694. doi: 10.1016/S1473-3099(05)70267-X. [DOI] [PubMed] [Google Scholar]
- 4.Abo Y.N., Oliver J., McMinn A., Osowicki J., Baker C., Clark J.E., Blyth C.C., Francis J.R., Carr J., Smeesters P.R., et al. Increase in invasive Group A Streptococcal disease among Australian children coinciding with northern hemisphere surges. Lancet Reg. Health West. Pac. 2023;41:100873. doi: 10.1016/j.lanwpc.2023.100873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bennett J., Zhang J., Leung W., Jack S., Oliver J., Webb R., Wilson N., Sika-Paotonu D., Harwood M., Baker M.G. Rising ethnic inequalities in acute rheumatic fever and rheumatic heart disease, New Zealand, 2000–2018. Emerg. Infect. Dis. 2021;27:36–46. doi: 10.3201/eid2701.191791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sims Sanyahumbi A., Colquhoun S., Wyber R., Carapetis J.R. Global disease burden of Group A Streptococcus. In: Ferretti J.J., Stevens D.L., Fischetti V.A., editors. Streptococcus pyogenes: Basic Biology to Clinical Manifestations. University of Oklahoma Health Sciences Center; Oklahoma City, OK, USA: 2016. [PubMed] [Google Scholar]
- 7.Yu D., Guo D., Zheng Y., Yang Y. A review of penicillin binding protein and Group A Streptococcus with reduced-beta-lactam susceptibility. Front. Cell. Infect. Microbiol. 2023;13:1117160. doi: 10.3389/fcimb.2023.1117160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ajay Castro S., Dorfmueller H.C. Update on the development of Group A Streptococcus vaccines. NPJ Vaccines. 2023;8:135. doi: 10.1038/s41541-023-00730-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guy R., Williams C., Irvine N., Reynolds A., Coelho J., Saliba V., Thomas D., Doherty L., Chalker V., von Wissmann B., et al. Increase in scarlet fever notifications in the United Kingdom, 2013/2014. Eurosurveillance. 2014;19:20749. doi: 10.2807/1560-7917.ES2014.19.12.20749. [DOI] [PubMed] [Google Scholar]
- 10.Hsieh Y.C., Huang Y.C. Scarlet fever outbreak in Hong Kong, 2011. J. Microbiol. Immunol. Infect. 2011;44:409–411. doi: 10.1016/j.jmii.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 11.Chen M., Yao W., Wang X., Li Y., Chen M., Wang G., Zhang X., Pan H., Hu J., Zeng M. Outbreak of scarlet fever associated with emm12 type Group A Streptococcus in 2011 in Shanghai, China. Pediatr. Infect. Dis. J. 2012;31:e158–e162. doi: 10.1097/INF.0b013e31825874f3. [DOI] [PubMed] [Google Scholar]
- 12.Sika-Paotonu D., Beaton A., Raghu A., Steer A., Carapetis J. Acute rheumatic fever and rheumatic heart disease. In: Ferretti J.J., Stevens D.L., Fischetti V.A., editors. Streptococcus pyogenes: Basic Biology to Clinical Manifestations. University of Oklahoma Health Sciences Center; Oklahoma City, OK, USA: 2016. [Google Scholar]
- 13.Carapetis J.R., Beaton A., Cunningham M.W., Guilherme L., Karthikeyan G., Mayosi B.M., Sable C., Steer A., Wilson N., Wyber R., et al. Acute rheumatic fever and rheumatic heart disease. Nat. Rev. Dis. Primers. 2016;2:15084. doi: 10.1038/nrdp.2015.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.World Health Organisation Increased Incidence of Scarlet Fever and Invasive Group A Streptococcus Infection-Multi-Country. 15 December 2022. [(accessed on 20 May 2024)]. Available online: https://www.who.int/emergencies/disease-outbreak-news/item/2022-DON429.
- 15.Rampersadh K., Salie M.T., Engel K.C., Moodley C., Zuhlke L.J., Engel M.E. Presence of Group A Streptococcus frequently assayed virulence genes in invasive disease: A systematic review and meta-analysis. Front. Cell. Infect. Microbiol. 2024;14:1337861. doi: 10.3389/fcimb.2024.1337861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cole J.N., Barnett T.C., Nizet V., Walker M.J. Molecular insight into invasive Group A Streptococcal disease. Nat. Rev. Microbiol. 2011;9:724–736. doi: 10.1038/nrmicro2648. [DOI] [PubMed] [Google Scholar]
- 17.Sanderson-Smith M., De Oliveira D.M., Guglielmini J., McMillan D.J., Vu T., Holien J.K., Henningham A., Steer A.C., Bessen D.E., Dale J.B., et al. A systematic and functional classification of Streptococcus pyogenes that serves as a new tool for molecular typing and vaccine development. J. Infect. Dis. 2014;210:1325–1338. doi: 10.1093/infdis/jiu260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.US Centers for Disease Control and Prevention emm Typing Overview and Guidelines. 8 April 2024. [(accessed on 18 September 2024)]; Available online: https://www.cdc.gov/strep-lab/php/group-a-strep/emm-typing.html.
- 19.Yang P., Peng X., Zhang D., Wu S., Liu Y., Cui S., Lu G., Duan W., Shi W., Liu S., et al. Characteristics of Group A Streptococcus strains circulating during scarlet fever epidemic, Beijing, China, 2011. Emerg. Infect. Dis. 2013;19:909–915. doi: 10.3201/eid1906.121020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.You Y., Davies M.R., Protani M., McIntyre L., Walker M.J., Zhang J. Scarlet fever epidemic in China caused by Streptococcus pyogenes serotype M12: Epidemiologic and molecular analysis. eBioMedicine. 2018;28:128–135. doi: 10.1016/j.ebiom.2018.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turner C.E., Pyzio M., Song B., Lamagni T., Meltzer M., Chow J.Y., Efstratiou A., Curtis S., Sriskandan S. Scarlet fever upsurge in England and molecular-genetic analysis in north-west London, 2014. Emerg. Infect. Dis. 2016;22:1075–1078. doi: 10.3201/eid2206.151726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chalker V., Jironkin A., Coelho J., Al-Shahib A., Platt S., Kapatai G., Daniel R., Dhami C., Laranjeira M., Chambers T., et al. Genome analysis following a national increase in Scarlet Fever in England 2014. BMC Genom. 2017;18:224. doi: 10.1186/s12864-017-3603-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tse H., Bao J.Y., Davies M.R., Maamary P., Tsoi H.W., Tong A.H., Ho T.C., Lin C.H., Gillen C.M., Barnett T.C., et al. Molecular characterization of the 2011 Hong Kong scarlet fever outbreak. J. Infect. Dis. 2012;206:341–351. doi: 10.1093/infdis/jis362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanchez-Encinales V., Ludwig G., Tamayo E., Garcia-Arenzana J.M., Munoz-Almagro C., Montes M. Molecular characterization of Streptococcus pyogenes causing invasive disease in pediatric population in Spain: A 12-year study. Pediatr. Infect. Dis. J. 2019;38:1168–1172. doi: 10.1097/INF.0000000000002471. [DOI] [PubMed] [Google Scholar]
- 25.Venkatesan P. Rise in Group A Streptococcal infections in England. Lancet Respir. Med. 2023;11:e16. doi: 10.1016/S2213-2600(22)00507-0. [DOI] [PubMed] [Google Scholar]
- 26.Chiang-Ni C., Hsu C.Y., Yeh Y.H., Chi C.Y., Wang S., Tsai P.J., Chiu C.H. Detection of toxigenic M1(UK) lineage Group A Streptococcus clones in Taiwan. J. Microbiol. Immunol. Infect. 2024;57:269–277. doi: 10.1016/j.jmii.2024.01.004. [DOI] [PubMed] [Google Scholar]
- 27.Vieira A., Wan Y., Ryan Y., Li H.K., Guy R.L., Papangeli M., Huse K.K., Reeves L.C., Soo V.W.C., Daniel R., et al. Rapid expansion and international spread of M1(UK) in the post-pandemic UK upsurge of Streptococcus pyogenes. Nat. Commun. 2024;15:3916. doi: 10.1038/s41467-024-47929-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vesty A., Ren X., Sharma P., Lorenz N., Proft T., Hardaker A., Straub C., Morgan J., Tiong A., Anderson A., et al. The Emergence and Impact of the M1(UK) Lineage on Invasive Group A Streptococcus Disease in Aotearoa New Zealand. Open Forum Infect. Dis. 2024;11:ofae457. doi: 10.1093/ofid/ofae457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gouveia C., Bajanca-Lavado M.P., Mamede R., Araujo Carvalho A., Rodrigues F., Melo-Cristino J., Ramirez M., Friaes A., Portuguese Group for the Study of Streptococcal Infections. Portuguese Study Group of Pediatric Invasive Streptococcal Disease et al. Sustained increase of paediatric invasive Streptococcus pyogenes infections dominated by M1(UK) and diverse emm12 isolates, Portugal, September 2022 to May 2023. Eurosurveillance. 2023;28:2300427. doi: 10.2807/1560-7917.ES.2023.28.36.2300427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vrenna G., Rossitto M., Agosta M., Cortazzo V., Fox V., De Luca M., Lancella L., Gargiullo L., Granaglia A., Fini V., et al. First Evidence of Streptococcus pyogenes M1UK Clone in Pediatric Invasive Infections in Italy by Molecular Surveillance. Pediatr. Infect. Dis. J. 2024;43:e421–e424. doi: 10.1097/INF.0000000000004455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davies M.R., Keller N., Brouwer S., Jespersen M.G., Cork A.J., Hayes A.J., Pitt M.E., De Oliveira D.M.P., Harbison-Price N., Bertolla O.M., et al. Detection of Streptococcus pyogenes M1(UK) in Australia and characterization of the mutation driving enhanced expression of superantigen SpeA. Nat. Commun. 2023;14:1051. doi: 10.1038/s41467-023-36717-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brouwer S., Barnett T.C., Ly D., Kasper K.J., De Oliveira D.M.P., Rivera-Hernandez T., Cork A.J., McIntyre L., Jespersen M.G., Richter J., et al. Prophage exotoxins enhance colonization fitness in epidemic scarlet fever-causing Streptococcus pyogenes. Nat. Commun. 2020;11:5018. doi: 10.1038/s41467-020-18700-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davies M.R., Holden M.T., Coupland P., Chen J.H., Venturini C., Barnett T.C., Zakour N.L., Tse H., Dougan G., Yuen K.Y., et al. Emergence of scarlet fever Streptococcus pyogenes emm12 clones in Hong Kong is associated with toxin acquisition and multidrug resistance. Nat. Genet. 2015;47:84–87. doi: 10.1038/ng.3147. [DOI] [PubMed] [Google Scholar]
- 34.Prjibelski A., Antipov D., Meleshko D., Lapidus A., Korobeynikov A. Using SPAdes De Novo Assembler. Curr. Protoc. Bioinform. 2020;70:e102. doi: 10.1002/cpbi.102. [DOI] [PubMed] [Google Scholar]
- 35.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 36.Davies M.R., McIntyre L., Mutreja A., Lacey J.A., Lees J.A., Towers R.J., Duchene S., Smeesters P.R., Frost H.R., Price D.J., et al. Atlas of Group A Streptococcal vaccine candidates compiled using large-scale comparative genomics. Nat. Genet. 2019;51:1035–1043. doi: 10.1038/s41588-019-0417-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katz L.S., Griswold T., Morrison S.S., Caravas J.A., Zhang S., den Bakker H.C., Deng X., Carleton H.A. Mashtree: A rapid comparison of whole genome sequence files. J. Open Source Softw. 2019;4:1762. doi: 10.21105/joss.01762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aziz R.K., Pabst M.J., Jeng A., Kansal R., Low D.E., Nizet V., Kotb M. Invasive M1T1 Group A Streptococcus undergoes a phase-shift in vivo to prevent proteolytic degradation of multiple virulence factors by SpeB. Mol. Microbiol. 2004;51:123–134. doi: 10.1046/j.1365-2958.2003.03797.x. [DOI] [PubMed] [Google Scholar]
- 39.Ly A.T., Noto J.P., Walwyn O.L., Tanz R.R., Shulman S.T., Kabat W., Bessen D.E. Differences in SpeB protease activity among Group A Streptococci associated with superficial, invasive, and autoimmune disease. PLoS ONE. 2017;12:e0177784. doi: 10.1371/journal.pone.0177784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lamb C.L., Price E., Field K.P., Dayton C., McIndoo E.R., Katahira E.J., Stevens D.L., Hobdey S.E. Enrichment of antigen-specific class-switched B cells from individuals naturally immunized by infection with Group A Streptococcus. mSphere. 2019;4 doi: 10.1128/mSphere.00598-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Odo C.M., Vega L.A., Mukherjee P., DebRoy S., Flores A.R., Shelburne S.A. Emergent emm4 Goup A Streptococcus evidences a survival strategy during interaction with immune effector cells. Infect. Immun. 2024;92:e0015224. doi: 10.1128/iai.00152-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wessels M.R. Cell wall and surface molecules of Streptococcus pyogenes: Capsule. In: Ferretti J.J., Stevens D.L., Fischetti V.A., editors. Streptococcus pyogenes: Basic Biology to Clinical Manifestations. 2nd ed. University of Oklahoma Health Sciences Center; Oklahoma City, OK, USA: 2022. [PubMed] [Google Scholar]
- 43.Bhakdi S., Roth M., Sziegoleit A., Tranum-Jensen J. Isolation and identification of two hemolytic forms of streptolysin-O. Infect. Immun. 1984;46:394–400. doi: 10.1128/iai.46.2.394-400.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H., Zhou L., Zhao Y., Ma L., Xu J., Liu Y., Qin Q., Hu J., Liu X. Epidemiological analysis of Group A Streptococcus infections in a hospital in Beijing, China. Eur. J. Clin. Microbiol. Infect. Dis. 2020;39:2361–2371. doi: 10.1007/s10096-020-03987-5. [DOI] [PubMed] [Google Scholar]
- 45.Minko A.G., Danilova T.A., Danilina G.A., Adzhieva A.A., Tikhomirov E.E., Zhukhovitsky V.G. Molecular genetic characterization of Streptococcus pyogenes strains isolated from patients with various manifestations of Streptococcal infection. Bull. Exp. Biol. Med. 2023;175:662–666. doi: 10.1007/s10517-023-05922-y. [DOI] [PubMed] [Google Scholar]
- 46.Meehan M., Murchan S., Gavin P.J., Drew R.J., Cunney R. Epidemiology of an upsurge of invasive Group A Streptococcal infections in Ireland, 2012–2015. J. Infect. 2018;77:183–190. doi: 10.1016/j.jinf.2018.05.010. [DOI] [PubMed] [Google Scholar]
- 47.Villalon P., Saez-Nieto J.A., Rubio-Lopez V., Medina-Pascual M.J., Garrido N., Carrasco G., Pino-Rosa S., Valdezate S. Invasive Streptococcus pyogenes disease in Spain: A microbiological and epidemiological study covering the period 2007-2019. Eur. J. Clin. Microbiol. Infect. Dis. 2021;40:2295–2303. doi: 10.1007/s10096-021-04279-2. [DOI] [PubMed] [Google Scholar]
- 48.Hall J.N., Bah S.Y., Khalid H., Brailey A., Coleman S., Kirk T., Hussain N., Tovey M., Chaudhuri R.R., Davies S., et al. Molecular characterization of Streptococcus pyogenes (StrepA) non-invasive isolates during the 2022–2023 UK upsurge. Microb. Genom. 2024;10:001277. doi: 10.1099/mgen.0.001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DebRoy S., Sanson M., Shah B., Regmi S., Vega L.A., Odo C., Sahasrabhojane P., McGeer A., Tyrrell G.J., Fittipaldi N., et al. Population genomics of emm4 Group A Streptococcus reveals progressive replacement with a hypervirulent clone in North America. mSystems. 2021;6:e0049521. doi: 10.1128/msystems.00495-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gergova R., Muhtarova A., Mitov I., Setchanova L., Mihova K., Kaneva R., Markovska R. Relation between emm types and virulence gene profiles among Bulgarian Streptococcus pyogenes clinical isolates. Infect. Dis. 2019;51:668–675. doi: 10.1080/23744235.2019.1638964. [DOI] [PubMed] [Google Scholar]
- 51.Olsen R.J., Raghuram A., Cantu C., Hartman M.H., Jimenez F.E., Lee S., Ngo A., Rice K.A., Saddington D., Spillman H., et al. The majority of 9729 Group A Streptococcus strains causing disease secrete SpeB cysteine protease: Pathogenesis implications. Infect. Immun. 2015;83:4750–4758. doi: 10.1128/IAI.00989-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liang Y., Shen X., Huang G., Wang C., Shen Y., Yang Y. Characteristics of Streptococcus pyogenes strains isolated from Chinese children with scarlet fever. Acta Paediatr. 2008;97:1681–1685. doi: 10.1111/j.1651-2227.2008.00983.x. [DOI] [PubMed] [Google Scholar]
- 53.Bhakdi S., Tranum-Jensen J., Sziegoleit A. Mechanism of membrane damage by streptolysin-O. Infect. Immun. 1985;47:52–60. doi: 10.1128/iai.47.1.52-60.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Palmer M. The family of thiol-activated, cholesterol-binding cytolysins. Toxicon. 2001;39:1681–1689. doi: 10.1016/S0041-0101(01)00155-6. [DOI] [PubMed] [Google Scholar]
- 55.Shewell L.K., Harvey R.M., Higgins M.A., Day C.J., Hartley-Tassell L.E., Chen A.Y., Gillen C.M., James D.B., Alonzo F., 3rd, Torres V.J., et al. The cholesterol-dependent cytolysins pneumolysin and streptolysin O require binding to red blood cell glycans for hemolytic activity. Proc. Natl. Acad. Sci. USA. 2014;111:E5312–E5320. doi: 10.1073/pnas.1412703111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shewell L.K., Day C.J., Jen F.E., Haselhorst T., Atack J.M., Reijneveld J.F., Everest-Dass A., James D.B.A., Boguslawski K.M., Brouwer S., et al. All major cholesterol-dependent cytolysins use glycans as cellular receptors. Sci. Adv. 2020;6:eaaz4926. doi: 10.1126/sciadv.aaz4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nasser W., Beres S.B., Olsen R.J., Dean M.A., Rice K.A., Long S.W., Kristinsson K.G., Gottfredsson M., Vuopio J., Raisanen K., et al. Evolutionary pathway to increased virulence and epidemic Group A Streptococcus disease derived from 3,615 genome sequences. Proc. Natl. Acad. Sci. USA. 2014;111:E1768–E1776. doi: 10.1073/pnas.1403138111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Turner C.E., Holden M.T.G., Blane B., Horner C., Peacock S.J., Sriskandan S. The emergence of successful Streptococcus pyogenes lineages through convergent pathways of capsule loss and recombination directing high toxin expression. mBio. 2019;10 doi: 10.1128/mBio.02521-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sumby P., Porcella S.F., Madrigal A.G., Barbian K.D., Virtaneva K., Ricklefs S.M., Sturdevant D.E., Graham M.R., Vuopio-Varkila J., Hoe N.P., et al. Evolutionary origin and emergence of a highly successful clone of serotype M1 Group A Streptococcus involved multiple horizontal gene transfer events. J. Infect. Dis. 2005;192:771–782. doi: 10.1086/432514. [DOI] [PubMed] [Google Scholar]
- 60.Vega L.A., Malke H., McIver K.S. Virulence-related transcriptional regulators of Streptococcus pyogenes. In: Ferretti J.J., Stevens D.L., Fischetti V.A., editors. Streptococcus pyogenes: Basic Biology to Clinical Manifestations. 2nd ed. University of Oklahoma Health Sciences Center; Oklahoma City, OK, USA: 2022. [PubMed] [Google Scholar]
- 61.Shi Y.A., Chen T.C., Chen Y.W., Liu Y.S., Chen Y.M., Lai C.H., Chiu C.H., Chiang-Ni C. The bacterial markers of identification of invasive CovR/CovS-inactivated Group A Streptococcus. Microbiol. Spectr. 2022;10:e0203322. doi: 10.1128/spectrum.02033-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Plainvert C., Rosinski-Chupin I., Weckel A., Lambert C., Touak G., Sauvage E., Poyart C., Glaser P., Fouet A. A novel CovS variant harbored by a colonization strain reduces Streptococcus pyogenes virulence. J. Bacteriol. 2023;205:e0003923. doi: 10.1128/jb.00039-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Langshaw E.L., Reynolds S., Ozberk V., Dooley J., Calcutt A., Zaman M., Walker M.J., Batzloff M.R., Davies M.R., Good M.F., et al. Streptolysin O deficiency in Streptococcus pyogenes M1T1 covR/S mutant strain attenuates virulence in in vitro and in vivo infection models. mBio. 2023;14:e0348822. doi: 10.1128/mbio.03488-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miller E.W., Danger J.L., Ramalinga A.B., Horstmann N., Shelburne S.A., Sumby P. Regulatory rewiring confers serotype-specific hyper-virulence in the human pathogen Group A Streptococcus. Mol. Microbiol. 2015;98:473–489. doi: 10.1111/mmi.13136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lynskey N.N., Turner C.E., Heng L.S., Sriskandan S. A truncation in the regulator RocA underlies heightened capsule expression in serotype M3 Group A Streptococci. Infect. Immun. 2015;83:1732–1733. doi: 10.1128/IAI.02892-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Flores A.R., Jewell B.E., Fittipaldi N., Beres S.B., Musser J.M. Human disease isolates of serotype M4 and M22 Group A Streptococcus lack genes required for hyaluronic acid capsule biosynthesis. mBio. 2012;3:e00413-12. doi: 10.1128/mBio.00413-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Turner C.E., Abbott J., Lamagni T., Holden M.T., David S., Jones M.D., Game L., Efstratiou A., Sriskandan S. Emergence of a new highly successful acapsular Group A Streptococcus clade of genotype emm89 in the United Kingdom. mBio. 2015;6:e00622. doi: 10.1128/mBio.00622-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Flores A.R., Chase McNeil J., Shah B., Van Beneden C., Shelburne S.A. Capsule-negative emm types are an increasing cause of pediatric Group A Streptococcal infections at a large pediatric hospital in Texas. J. Pediatr. Infect. Dis. Soc. 2019;8:244–250. doi: 10.1093/jpids/piy053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rumke L.W., Davies M.A., Vestjens S.M.T., van der Putten B.C.L., Bril-Keijzers W.C.M., van Houten M.A., Rots N.Y., Wijmenga-Monsuur A.J., van der Ende A., de Gier B., et al. Nationwide upsurge in invasive disease in the context of longitudinal surveillance of carriage and invasive Streptococcus pyogenes 2009–2023, the Netherlands: A molecular epidemiological study. J. Clin. Microbiol. 2024;62:e0076624. doi: 10.1128/jcm.00766-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.de Crombrugghe G., Botteaux A., Osowicki J., Steer A.C., Smeesters P.R. Global epidemiological comparison of Streptococcus pyogenes emm-types associated with pharyngitis and pharyngeal carriage. Clin. Microbiol. Infect. 2024;30:1074.e1–1074.e4. doi: 10.1016/j.cmi.2024.05.007. [DOI] [PubMed] [Google Scholar]
- 71.Virolainen M., Grondahl-Yli-Hannuksela K., Rantakokko-Jalava K., Seiskari T., Lonnqvist E., Kolari T., Rissanen T., Hyyrylainen H.L., DICAR Study Group. Vuopio J. Epidemiology and emm types among Group A Streptococcal pharyngitis in Finland: A prospective laboratory-based study. Eur. J. Clin. Microbiol. Infect. Dis. 2024;43:233–241. doi: 10.1007/s10096-023-04714-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.You Y.H., Song Y.Y., Yan X.M., Wang H.B., Zhang M.H., Tao X.X., Li L.L., Zhang Y.X., Jiang X.H., Zhang B.H., et al. Molecular epidemiological characteristics of Streptococcus pyogenes strains involved in an outbreak of scarlet fever in China, 2011. Biomed. Environ. Sci. 2013;26:877–885. doi: 10.3967/bes2013.016. [DOI] [PubMed] [Google Scholar]
- 73.Butler T.A.J., Story C., Green E., Williamson K.M., Newton P., Jenkins F., Varadhan H., van Hal S. Insights gained from sequencing Australian non-invasive and invasive Streptococcus pyogenes isolates. Microb. Genom. 2024;10:001152. doi: 10.1099/mgen.0.001152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Oliver J., Wilmot M., Strachan J., St George S., Lane C.R., Ballard S.A., Sait M., Gibney K., Howden B.P., Williamson D.A. Recent trends in invasive Group A Streptococcus disease in Victoria. Commun. Dis. Intell. 2019;43 doi: 10.33321/cdi.2019.43.8. [DOI] [PubMed] [Google Scholar]
- 75.Alcolea-Medina A., Snell L.B., Alder C., Charalampous T., Williams T.G.S., Synnovis Microbiology Laboratory G., Tan M.K.I., Al-Yaakoubi N., Humayun G., Newsholme W., et al. The ongoing Streptococcus pyogenes (Group A Streptococcus) outbreak in London, United Kingdom, in December 2022: A molecular epidemiology study. Clin. Microbiol. Infect. 2023;29:887–890. doi: 10.1016/j.cmi.2023.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hamzah S.N.A., Mohd Desa M.N., Jasni A.S., Mohd Taib N., Masri S.N., Hamat R.A. Distribution of virulence genes and the molecular epidemiology of Streptococcus pyogenes clinical isolates by emm and multilocus sequence typing methods. Med. J. Malays. 2021;76:164–170. [PubMed] [Google Scholar]
- 77.Li H., Zhou L., Zhao Y., Ma L., Liu X., Hu J. Molecular epidemiology and antimicrobial resistance of Group A Streptococcus recovered from patients in Beijing, China. BMC Infect. Dis. 2020;20:507. doi: 10.1186/s12879-020-05241-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.DebRoy S., Li X., Kalia A., Galloway-Pena J., Shah B.J., Fowler V.G., Flores A.R., Shelburne S.A. Identification of a chimeric emm gene and novel emm pattern in currently circulating strains of emm4 Group A Streptococcus. Microb. Genom. 2018;4:e000235. doi: 10.1099/mgen.0.000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Walkinshaw D.R., Wright M.E.E., Mullin A.E., Excler J.L., Kim J.H., Steer A.C. The Streptococcus pyogenes vaccine landscape. NPJ Vaccines. 2023;8:16. doi: 10.1038/s41541-023-00609-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sumby P., Whitney A.R., Graviss E.A., DeLeo F.R., Musser J.M. Genome-wide analysis of Group A Streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog. 2006;2:e5. doi: 10.1371/journal.ppat.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Whole genome sequencing data obtained in this study were submitted to the Sequence Read Archive database (https://www.ncbi.nlm.nih.gov/sra (accessed on 30 September 2024); BioProject accession PRJNA1172375).