Abstract
Marine resources have been important for the survival and economic development of coastal human communities across northern Europe for millennia. Knowledge of the origin of such historic resources can provide key insights into fishing practices and the spatial extent of trade networks. Here, we combine ancient DNA and stable isotopes (δ13C, δ15N, non-exchangeable δ2H and δ34S) to investigate the geographical origin of archaeological cod remains in Oslo from the eleventh to seventeenth centuries CE. Our findings provide genetic evidence that Atlantic cod was obtained from different geographical populations, including a variety of distant-water populations like northern Norway and possibly Iceland. Evidence for such long-distance cod trade is already observed from the eleventh century, contrasting with archaeological and historical evidence from Britain and other areas of Continental Europe around the North and Baltic Seas, where such trade increased during the thirteenth to fourteenth centuries. The genomic assignments of specimens to different populations coincide with significantly different δ13C values between those same specimens, indicating that multiple Atlantic cod populations living in different environments were exploited. This research provides novel information about the exploitation timeline of specific Atlantic cod stocks and highlights the utility of combining ancient DNA (aDNA) methods and stable isotope analysis to describe the development of medieval and post-medieval marine fisheries.
Keywords: cod trade, historical fisheries, aDNA, stable isotopes, long-distance fish trade
1. Introduction
The demand for marine fish as a food source or as trading assets from growing global communities has increased in magnitude over the past millennia [1]. Understanding the extent of historical fish-trading networks is therefore key to identifying locations with long-term exploitation and ensuring their current sustainable management. In the Atlantic Ocean, marine fisheries significantly influenced the development of medieval and post-medieval European societies [1]. Long-distance fish trade from production sites in the north (e.g. northern Norway and/or Iceland) to urban centres in Britain and mainland Europe is well documented by historical and archaeological sources for medieval and early modern times, being especially evident by the thirteenth and fourteenth centuries [2–4]. Nonetheless, for the earlier Middle Ages, temporal and spatial patterns of exploitation and long-distance trade remain poorly documented. While the earliest known example of long-distance trade of Atlantic cod (Gadus morhua) presently has a terminus ante quem of ca 1066 CE (by which date northern Norwegian cod was brought to Haithabu in what is now Schleswig-Holstein) [5], the species was predominantly locally acquired in England and Flanders during the tenth to twelfth centuries [6–8]. Thereafter, an increasingly commercialized long-range trade of air-dried Atlantic cod (stockfish) only appeared from the thirteenth to fourteenth centuries onwards, around the southern North Sea and the eastern Baltic Sea [6–8]. This dried fish was likely traded via Bergen to medieval centres across Europe (Germany, Sweden, Poland, Estonia and England) [3,6,7,9–12]. Nonetheless, the development of the early medieval Atlantic cod trade remains to be discovered, between the early outlier of ca 1066 CE at Haithabu and the more widespread boom of the thirteenth and fourteenth centuries. A promising location to do so is within the milieu of medieval Scandinavia, where processed fish—in particular stockfish—was produced and found a ready cultural reception within local foodways [13]. One such location is Oslo, Norway, which emerged as a prominent town during the tenth to eleventh centuries [14,15]. Large numbers of fish remains (especially of Atlantic cod) [4] have been found during the archaeological excavation of Oslo’s urban settlement layers [16,17]. By the fourteenth century, Oslo had become an important town and centre of consumption, although not a major hub for the transshipment of processed fish [18]. By that time, participation in long-range fish trade is likely to be present. However, earlier patterns and trends of trade through time remain poorly understood.
Recent advances in biomolecular analyses, including ancient DNA (aDNA) and stable isotope approaches, have made it possible to help assign individual fish bones to specific geographical populations or geographical areas. For instance, genetic assignments can be used to detect spatiotemporal changes in the distribution of historical fish populations [19,20] or detect historical fish trade in those cases where local catches can be excluded [5,8]. The application of genetic methodology has also contributed to our understanding of fishing strategies [21,22]. Similarly, stable isotope values measured on bone collagen can also contribute to the assignment of fish to specific geographic regions as these values are influenced by the length of the food web, water temperature and salinity [6,23,24]. Considering that stable isotope values are incorporated into bone collagen predominantly via diet, they can provide information about migratory and habitat use of different species, including fish [6,25–28]. However, the differential geographical resolution of these approaches [6,8], the lack of sufficient DNA [8] or collagen preservation [29] may constrain the number of specimens that can be successfully analysed. Thus, combining these approaches in determining the provenance of economically important species like Atlantic cod can provide complementary evidence to investigate the expansion of medieval marine fisheries and the potential increase in the exploitation of targeted populations.
Here, we used such a multidisciplinary approach, combining genome-wide aDNA and stable isotopes, to describe the development of long-distance trade to Oslo over a period of approximately 600 years during the medieval and post-medieval periods, starting ca 1000 CE. We analysed a total of 106 archaeological specimens using low-coverage whole-genome aDNA approaches (35 out of the 106 specimens) and/or stable carbon (δ13C), nitrogen (δ15N), non-exchangeable hydrogen (δ2H) and sulphur (δ34S) isotope values (86 or 64—after quality thresholds—out of 100 specimens). We aimed to identify ecological differences between these specimens (given their isotopic values, cranial or postcranial bone elements and estimates of body size) and to determine their geographical population source based on genome-wide sequencing. Integrating both approaches, our observations support a diverse origin of Atlantic cod specimens in this assemblage, including specimens obtained through long-distance trade since ca 1000 CE.
2. Material and methods
(a). Sample collection
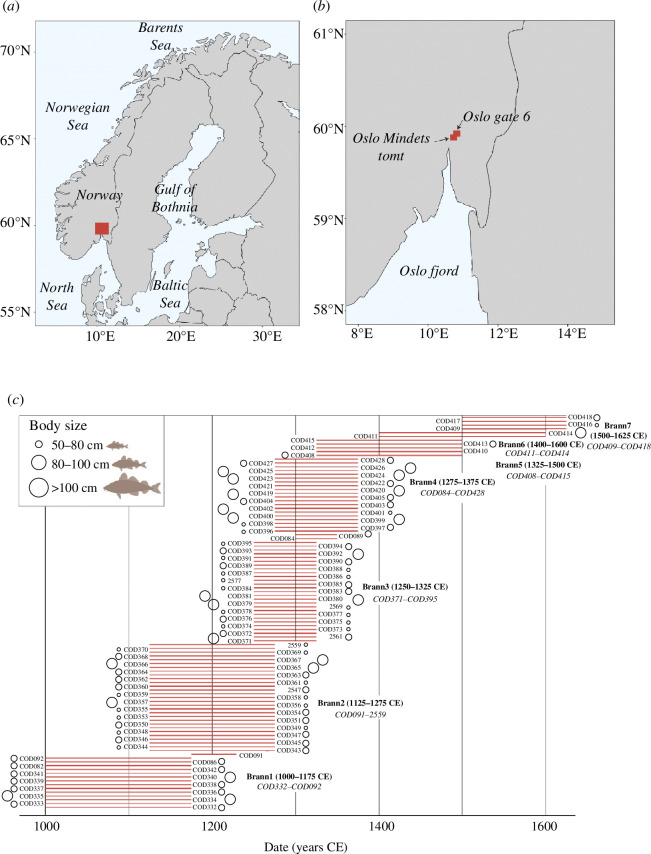
We have analysed 106 Atlantic cod bone samples collected from two archaeological sites: Oslogate 6 (n = 100) and Oslo Mindets tomt (n = 6; electronic supplementary material, table S1). The zooarchaeological assemblages (bones) from both sites are stored in the osteological collections at the University Museum, University of Bergen, under the museum numbers JS-784 (Oslogate 6) and 537 (Oslo Mindets tomt). Oslogate 6 (59.91°N, 10.77°E) was located in the northern part of medieval Oslo [17]. This site was excavated during 1987–1989, while Oslo Mindets tomt (59.90°N, 10.76°E) was excavated in 1973 (figure 1). Remains of barley, fish bones and animal dung (likely from livestock) have been found in the area, while increases in large leather deposits by the late twelfth century in Oslogate 6 are indicative of the development of trading activities (i.e. shoemaking) in the town [17,30].
Figure 1.
Geographical location of the archaeological Atlantic cod specimens collected from two archaeological sites in (a) southeast Norway and (b) Oslo (Oslogate 6, n = 100 and Oslo Mindets tomt, n = 6; electronic supplementary material, table S1). (c) Date estimation range (in red) and body size estimation (in circles) for archaeological samples. Specimens derive from layers placed between distinct fire layers (Branntrinn) that have been dated using archaeological methods (across the eleventh to seventeenth centuries). A total of eight fire stages (Brann1 to Brann8) have been identified and are used to describe the stratigraphy and constructions in urban development [17]. Specimens COD406 and COD407 (from Brann8) do not have a confident date estimation and are excluded from this figure. Body size is significantly different between fire layers, with fish from Brann4 significantly larger than those from Brann2 and Brann3.
Samples—cranial and postcranial elements—have been stored dry and unfrozen after field collection. All specimens were morphologically (see Archive: Osteological collections, University Museum, University of Bergen) and genetically identified as Atlantic cod. Samples have been dated based on the archaeological context (e.g. fire layer) including carbon-14 dating, dendrochronology and typology (for more information, see [17]). Specifically, samples at Oslogate 6 are distributed between seven fire stages (Brann1-7) dated using archaeological methods (across the eleventh to seventeenth centuries) and are used to describe the stratigraphy and constructions in urban development (i.e. waste layers) (for details about the fire layers, see [17]). An eighth layer (Brann8) has been described for Oslogate 6, which includes an uppermost layer that is associated with modern times (i.e. archaeological remains younger than 1624 CE [17]). Samples across the fire layers at Oslogate 6 are distributed as follows (electronic supplementary material, table S1): Brann1 (oldest, where 12 out of 14 samples were processed for genomic analysis), Brann2 (n = 7/30), Brann3 (n = 8/27), Brann4 (n = 10/22), Brann5 (n = 5/5), Brann6 (n = 2/2), Brann7 (n = 4/4) and Brann8 (n = 2/2). Oslo Mindets tomt samples are dated by the archaeological context [31] and are analysed together with those specimens from Oslogate 6 with which they co-occur in time [32]. Three specimens (COD082, COD086 and COD092) co-occur in time with Brann1, one specimen (COD091) co-occurs with Brann2 and two specimens (COD084, COD089) co-occur with Brann4.
(b). aDNA extraction, library preparation and sequencing
A total of 50 samples were processed in the aDNA laboratory at the University of Oslo [33,34] (electronic supplementary material, tables S1 and S2). Samples were treated as per Ferrari et al. [35] and Martínez-García et al. [36] before DNA extraction. Genomic DNA was extracted using a mild bleach treatment and pre-digestion step protocol [37]. Double-indexed blunt-end sequencing libraries were built using either the Meyer–Kircher protocol [38,39] with modifications by Schroeder et al. [40] or the single-stranded Santa Cruz reaction protocol (tier 4) ([41]; electronic supplementary material, table S1). Library quality and concentration were examined with a High Sensitivity NGS Fragment Analysis Kit on the Fragment AnalyzerTM (Advanced Analytical). Libraries were sequenced on the Illumina HiSeq 4000 or on the Novaseq 6000 platform at the Norwegian Sequencing Centre (electronic supplementary material, table S1). Sequencing reads were processed using PALEOMIX v. 1.2.13 [42] and AdapterRemoval v. 2.1.7 [43] to trim residual adapter contamination, to collapse overlapping paired reads, and to filter reads with excessive missing nucleotides. Filtered reads were aligned using the gadMor2 genome as reference [44,45] using BWA v. 0.7.12 [46] with the backtrack algorithm, disabled seeding and minimum quality score of 25. aDNA deamination patterns were characterized using mapDamage v. 2.0.9 [47].
(c). Genomic analysis
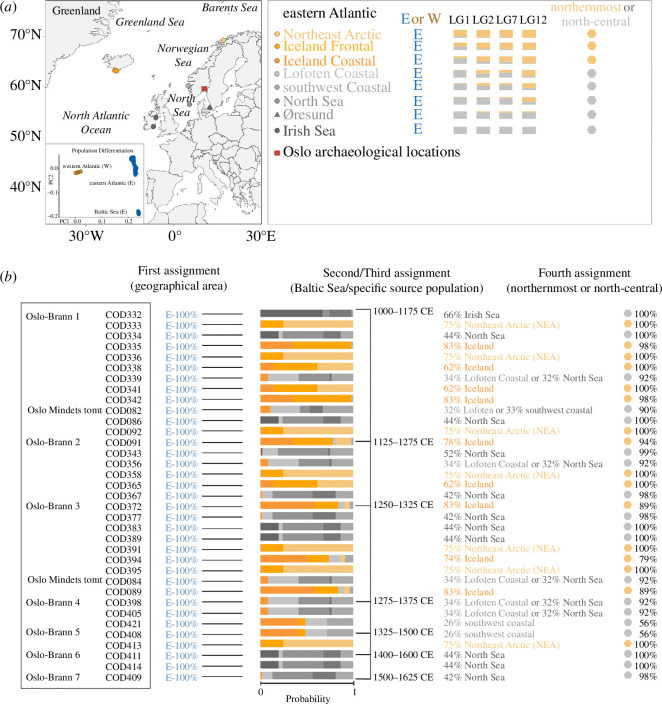
To determine the biological origin of 50 Atlantic cod specimens, we followed the hierarchical approach described in Martínez-García et al. [8]. First, we used the genome-wide approach in the BAMscorer pipeline [48] to assign ancient cod specimens to the eastern or western Atlantic Ocean. Second, among those specimens with an eastern Atlantic origin, we used a similar approach to identify any specimen with a Baltic Sea origin. Third, we used the chromosomal inversion approach to determine the individual haplotypes of the four major chromosomal inversions in Atlantic cod (LG1, LG2, LG7 and LG12) [49] associated with migratory behaviour and temperature clines [49–53]. These combined genotype distributions can indicate an affinity towards a particular ecotype [5,8]. We included reference data on (modern) inversion frequencies from the following populations: the Northeast Arctic (NEA), Iceland (frontal and coastal ecotypes, which differ in their tendency for long-distance migration), the Norwegian Coast (Lofoten and southwest), the North Sea, the Irish Sea and Øresund (figure 2a) [5,53,54]. Samples with >0.1% endogenous DNA and an average of >50 single nucleotide polymorphisms across all four chromosomal inversions have been included for specific genomic assignments (electronic supplementary material, table S1). For each individual specimen, the source population with the highest percentage was considered the likely geographical population source (electronic supplementary material, tables S2 and S3). Specimens with equal or similar assignment probabilities for two populations have both populations as their putative origin (electronic supplementary material, tables S2 and S3). Finally, we recognized two spatially larger scale distinct groups (northernmost and north-central) as described in Martínez-García et al. [8]. The northernmost group includes NEA and Iceland (adding probabilities of both Icelandic ecotypes), while the north-central group includes the Norwegian Coast (coastal Atlantic cod from Lofoten and southwest Norway), the North Sea, the Irish Sea and Øresund [8]. While our ability to obtain high affinities of ancient specimens to specific populations within these two distinct regions is often low, we consider those that fall with high probability within the northernmost group (see §3, figure 1b) as specimens that must have been obtained through long-distance trade following Martínez-García et al. [8]. In contrast, we cannot exclude a putatively local geographical population source (i.e. near Oslo) for those specimens belonging to the north-central group.
Figure 2.
Genetic analyses of the archaeological Atlantic cod specimens from Oslo. (a) Geographical distribution of modern inversion frequencies of chromosomal inversions in Atlantic cod (LG1, LG2, LG7 and LG12) from reference populations across the North Atlantic Ocean. The map is modified from Martínez-García et al. [8]. Alleles associated with a northernmost (Northeast Arctic and Iceland) composite genotype distribution are assigned in orange. Alleles associated with a north-central (Norwegian Coast, the North Sea, the Irish Sea, Øresund) genotype distribution are assigned in grey. Oslo’s archaeological sites are indicated with a red square. (b) Genomic assignments to a geographical area, a source population and a genotypic group are based on the frequencies of chromosomal inversions of Atlantic cod (LG1, LG2, LG7 and LG12) as per Star et al. [5]. Percentages (%) indicate the highest probability of being from one population and area.
We investigated possible associations between the origin of 27 specimens assigned with a >70% probability to a northernmost or north-central group (electronic supplementary material, table S2) and their fish bone element (cranial (premaxilla, articular, dentary, maxilla) or postcranial (vertebra and cleithrum)) [8] using the Fisher’s exact test. Full details of the criteria used to exclude samples for statistical analyses are provided in the electronic supplementary material (table S2).
Additionally, we used the Fisher’s exact test to evaluate whether the binary genetic assignment to the north-central or northernmost groups changed over time (>70% probability). We also evaluated whether the probability—scaled from 0 to 1—of having an NEA or Icelandic origin (adding probabilities of both Icelandic ecotypes) differs over time (i.e. fire layers), body size or bone element. Here, we used logistically transformed probabilities (logit) of NEA or Icelandic origin after testing for normality with a Shapiro–Wilk test. Thereafter, we performed non-parametric Kruskal–Wallis tests between such variables. Only fire layers with sufficient sample sizes (n ≥ 4 in Brann1 (1000–1175 CE) to Brann4 (1275–1375 CE)) were used in these analyses (n = 29). Finally, it is possible that fish coming from distinct geographical population sources may originally have differed in size, or that different fishing practices may themselves select for a particular size range in their catches. Given their often-fragmented condition, archaeological bones were assigned to total (length) body-size categories based on comparison with 1 : 1 scale scans of reference specimens from the known size of Atlantic cod. Using the Fisher’s exact test, we further investigated whether there were significant differences between the estimated total (length) size categories (50–80, 80–100 and >100 cm) in specimens that were genetically assigned to the north-central or northernmost group, or to northern Norway or Iceland.
(d). Stable isotope analysis
Stable isotope analysis was performed on 100 Atlantic cod specimens (50 of which were also processed for genomic sequencing as noted above; electronic supplementary material, table S1). We measured stable carbon (δ13C), nitrogen (δ15N), non-exchangeable hydrogen (hereafter described as δ2H for simplicity, which represents in vivo values rather than the exchange with atmospheric water vapour [28]) and sulphur (δ34S) on purified bone collagen. We processed a cross section of the bone material—for an approximate lifetime average for the isotopic values and thus reducing the impact of changing trophic level associated with age [55]—following the protocols of Barrett et al. [56] and references therein, with the exclusion of a lipid removal step. For δ13C values and δ15N values, the extracted collagen was analysed in triplicate at the University of Cambridge using a Costech elemental analyser coupled to a Thermo Finnigan Delta V Isotope Ratio Mass Spectrometer (IRMS). δ2H and δ34S values (measured by Iso-Analytical Limited, by elemental analyser IRMS) were analysed routinely in duplicate, although some samples yielded only enough collagen for single measurements. All isotope data are reported using international scales: δ13C values are reported relative to Vienna Peedee Belemnite (VPDB), δ15N values to AIR, δ2H values to Vienna Standard Mean Ocean Water (VSMOW) and δ34S to Vienna-Cañon Diablo Troilite (VCDT). The reported non-exchangeable δ2H values are corrected for exchangeable hydrogen by three-point linear calibration using standards. Overall, 64 samples passed appropriate quality-control thresholds for all four measured isotope values (atomic C/N ratio of 2.9–3.6, atomic C/S ratio of 125–225, atomic N/S ratio of 40–80) [57]. An additional 22 samples only passed appropriate C/N ratio quality-control thresholds. Therefore, the δ13C, δ15N and δ2H values are probably reliable for these 22 specimens, but potentially not the δ34S values. For this reason, analyses using only δ13C, δ15N and/or δ2H values (individually and interacting) are based on 86 specimens, while analyses including δ34S values (individually and in combination with other data) are based on 64 specimens.
To evaluate the impact of post-mortem processes on the isotopic values we investigated correlations between C : N ratios and δ13C values. To investigate potential interactions between isotopes, we evaluated the normality of our data with a Shapiro–Wilk test, followed by correlations between δ13C–δ15N, δ15N–δ2H and δ13C–δ34S values. We implemented a Spearman’s correlation between C : N ratios and δ13C values (n = 86), and between δ13C and δ34S values (n = 64), while Pearson’s correlations were used between δ13C and δ15N values (n = 86) and δ15N and δ2H values (n = 86). Based on age, growth and environmental and ecological shifts over time, we investigated the variability of isotope values over time and across body size groups. We performed ANOVA analysis for δ13C, δ15N (n = 86) and δ2H values (n = 83, excluding individuals with missing δ2H values) followed by a Tukey HSD post hoc test; while a non-parametric Kruskal–Wallis test was computed for δ34S values (n = 64) followed by a Dunn post hoc test with a Bonferroni correction (<3 groups).
To identify potentially different trophic levels and/or exposure to ecological conditions among specimens, we examined the distribution of isotope values (δ13C, δ15N and δ2H) within each of the three body size categories. We further evaluated the differences between the body sizes of all archaeological specimens distributed across Brann1 to Brann4 (n = 86) using a Kruskal–Wallis test, followed by a post hoc Dunn test with a Holm correction (>3 groups).
(e). Combined genome–isotope analyses
A summary of isotope values, in relation to all body size categories and the putatively genomic assignment of individuals to either a NEA or Iceland or a north-central or northernmost origin, was represented by a principal component analysis (PCA) on the 64 relevant δ13C, δ15N, δ2H and δ34S individual values. We performed an ANOVA analysis to describe the contribution of size and time (Brann1 to Brann4) to each principal component (PC) in the PCA followed by a Tukey HSD post hoc test.
To investigate whether the isotopic values differ when individuals are genomically assigned to either NEA or Iceland (adding the probabilities of both Icelandic ecotypes), we computed a multivariate linear regression with logistically (logit) transformed probabilities (scaled from 0 to 1) of NEA or Icelandic origin (n = 20) as dependent variables and δ13C, δ15N, δ2H and δ34S values as predictors. We tested the regression for homoscedasticity with a Breusch–Pagan test and validated the model by assessing residuals and symmetry. We further evaluated for multicollinearity by removing δ15N from our regressions (after significant relationships with other isotope values; see §3). We did not observe major differences when removing δ15N; therefore, the final regression model includes all four isotope values. Additionally, considering that stockfish is primarily produced from the Atlantic cod migratory ecotype in Norway, we selected those specimens genomically assigned to the Iceland frontal migratory ecotype (another source of stockfish production) to assess the differences in isotopic values between migratory ecotypes. Multivariate linear regressions with specimens with an Icelandic frontal affinity follow the same procedure previously explained.
Finally, we assessed the direct relation between a binary assignment to NEA or Iceland (adding both Icelandic ecotypes) and the δ13C, δ15N, δ2H and δ34S values. We used one-way ANOVAs (for δ13C, δ15N and δ34S) or Welch’s ANOVAs (for δ2H) after testing for equal variances with a Bartlett test. Only specimens with a >70% probability NEA or overall >70% probability Icelandic assignment were included (electronic supplementary material, table S2). Therefore, 10 specimens were tested against δ13C, δ15N and δ2H values and nine specimens were tested against δ34S values. Icelandic frontal (migratory ecotype) assignments are usually shared with an Icelandic coastal (stationary ecotype) assignment (electronic supplementary material, table S3). Consequently, an Icelandic frontal assignment alone cannot be binary assigned with >70% probability of origin to a population. Thus, we only included samples with a confident (>70% probability) overall Icelandic origin. All statistical analyses have been performed in R [58]. Full details of the packages are provided in the electronic supplementary material.
3. Results
(a). Genomic analysis
We successfully sequenced 35 out of the initial 50 Atlantic cod specimens with a total of ca 770 million paired reads, of which approximately 111 million reads aligned with a range of 0.1% to 47% of endogenous DNA content per specimen (electronic supplementary material, table S2). As expected, patterns of DNA fragmentation and deamination rates are consistent with those of authentic aDNA (electronic supplementary material, figure S1). We found that all 35 specimens can be assigned to an eastern Atlantic origin (100% assignment probability; figure 2b and electronic supplementary material, tables S1 and S2), of which 19 specimens had the highest affinity to the north-central group (56–100% assignment probability). No specimens were identified with a Baltic Sea origin. Within the north-central group, specific population assignments are statistically uncertain, with 10 specimens putatively assigned to the North Sea (42–52% assignment probability), one specimen to the Irish Sea (66% assignment probability), five specimens to both the North Sea (32% assignment probability) and the Norwegian Coast (Lofoten, 34% assignment probability), one specimen to the Norwegian Coast (Lofoten and southwest, 32 and 33% assignment probability respectively) and two specimens to the southwest coast of Norway (26% assignment probability; figure 2b and electronic supplementary material, table S3). Sixteen specimens had the highest affinity to the northernmost group (79–100% assignment probability), of which seven specimens were assigned to the Northeast Arctic (75% assignment probability) and nine were likely assigned to Iceland, with 62–83% assignment probability (adding the probabilities of both migratory and stationary ecotypes; figure 2b and electronic supplementary material, table S3).
While postcranial bones have been associated with long-distance sources (i.e. NEA and Iceland) [8], we did not find a statistically significant association between the bone element (cranial or postcranial) and specimens with a local (north-central group) or traded origin (northernmost group; p‐value = 0.37; n = 27; electronic supplementary material, figure S2a, table S2). In addition, we did not find statistically significant differences over time between specimens genetically assigned to either a north-central or northernmost group (p‐value = 0.77; n = 29; electronic supplementary material, figure S2b, table S1). We did not find any statistical difference between the probability of having an NEA or Icelandic origin in bone element (NEA: Kruskal–Wallis χ2 = 2.28, d.f. = 1, p‐value = 0.13 and Iceland: Kruskal–Wallis χ2 = 0.13, d.f. = 1, p‐value = 0.72; n = 33; electronic supplementary material, figure S3a,b), in body size (NEA: Kruskal–Wallis χ2 = 2.00, d.f. = 2, p‐value = 0.37 and Iceland: Kruskal–Wallis χ2 = 0.25, d.f. = 2, p‐value = 0.88; electronic supplementary material, figure S3c,d) or across time (Brann1 to Brann4; NEA: Kruskal–Wallis χ2 = 0.74, d.f. = 3, p‐value = 0.86 and Iceland: Kruskal–Wallis χ2 = 0.04, d.f. = 3, p‐value = 0.99; electronic supplementary material, figure S3e,f). Furthermore, we did not find statistical differences between the size categories of specimens genetically assigned to Iceland and NEA (p = 0.09, n = 16), the north-central group and Iceland (p = 0.68, n = 25), the north-central group and NEA (p = 0.46, n = 23), the north-central group, NEA and Iceland (p = 0.26, n = 32) and the north-central group and northernmost group (p = 1.0, n = 32; electronic supplementary material, table S4).
(b). Isotope analysis
We successfully extracted collagen from 93 out of 100 Atlantic cod specimens from Oslogate 6, with 86 passing quality thresholds for δ13C, δ15N and δ2H values (electronic supplementary material, table S1). For δ34S values, 64 specimens passed quality control thresholds. Collagen yields of all 93 extractions ranged from 1.5 to 10.6%. Isotope values that passed initial quality control thresholds ranged from −15.9 to −11.9‰ (mean −13.9‰, variance 0.8, s.d. = 0.9) for carbon (δ13C), from +12.7 to +16.8‰ (mean +14.9‰, variance 0.8, s.d. = 0.9) for nitrogen (δ15N), from −13.8 to +38.1‰ (mean +12.9‰, variance 116.9, s.d. = 10.8) for hydrogen (δ2H) and from +6.0 to +17.5‰ (mean +14.0‰, variance 5.3, s.d. = 2.3) for sulphur (δ34S; electronic supplementary material, table S5).
The C : N ratios for the included data ranged from 3.0 to 3.6 (electronic supplementary material, figure S4, table S1) and did not have a significant correlation with δ13C values (Spearman’s ρ = −0.19, p‐value = 0.07). Therefore, we assume that post-mortem processes did not significantly impact our isotopic data [29]. Isotope values were strongly correlated, with a significant relation between δ15N and δ13C values (Pearson’s correlation = 0.46, p‐value ≤0.01; electronic supplementary material, figure S5a) and between δ2H and δ15N values (Pearson’s correlation = 0.51, p‐value ≤0.01; electronic supplementary material, figure S5b). No significant relationship was observed between δ13C and δ34S values (Spearman’s correlation ρ = 0.18, p‐value = 0.15; electronic supplementary material, figure S5c).
Body size significantly influenced three isotope values (electronic supplementary material, table S6a): δ13C (ANOVA: F-value = 6.70, p‐value ≤ 0.01, n = 86; electronic supplementary material, figure S6a), δ15N (ANOVA: F-value = 5.47, p‐value ≤ 0.01, n = 86; electronic supplementary material, figure S6b) and δ2H (ANOVA: F-value = 7.38, p‐value ≤ 0.01, n = 83; electronic supplementary material, figure S6c). Such significant differences in δ13C, δ15N and δ2H values were found between the smallest (50–80 cm) compared to the largest (>100 cm) body size categories (Tukey HSD post hoc test p‐value ≤ 0.01; electronic supplementary material, table S6a). Interestingly, we observed a bimodal distribution within medium-sized fish (80–100 cm) for δ13C and δ15N values but not in δ2H values or smaller and larger body size categories (electronic supplementary material, figure S7). Furthermore, we found significant differences of δ13C (ANOVA: F-value = 5.91, p‐value ≤ 0.01, n = 81; electronic supplementary material, figure S8a), δ15N (ANOVA: F-value = 3.94, p‐value ≤ 0.01, n = 81; electronic supplementary material, figure S8b) and δ2H values (ANOVA: F-value = 3.67, p‐value ≤ 0.01, n = 79; electronic supplementary material, figure S8c) across time (Brann1 to Brann4; electronic supplementary material, table S6b). Specifically, Brann4 (1275–1375 CE) had significantly higher isotope values (δ13C p‐value ≤ 0.01; δ15N p‐value = 0.03 and δ2H p‐value = 0.01) compared to Brann2 (1125–1275 CE). Brann4 also had significantly higher δ13C values (p‐value ≤ 0.01) compared to Brann3 (1250–1325 CE) and significantly higher δ15N values (p‐value = 0.01) compared to Brann1 (1000–1175 CE). No significant correlations were obtained between δ34S values and body size (Kruskal–Wallis χ2 = 0.11, d.f. = 2, p‐value = 0.95, n = 64; electronic supplementary material, figure S6d) or across time (Kruskal–Wallis χ2 = 4.69, d.f. = 3, p‐value = 0.20, n = 60; electronic supplementary material, figure S8d). We found significant differences among the body size of our archaeological specimens across time (Kruskal–Wallis χ2 = 11.93, d.f. = 3, p‐value ≤ 0.01). These differences can be observed between larger fish from Brann4 against Brann2 (TukeyHSD post hoc test p‐value = 0.04) and Brann3 (Tukey HSD post hoc test p‐value = 0.04).
(c). Combined genome–isotope analyses
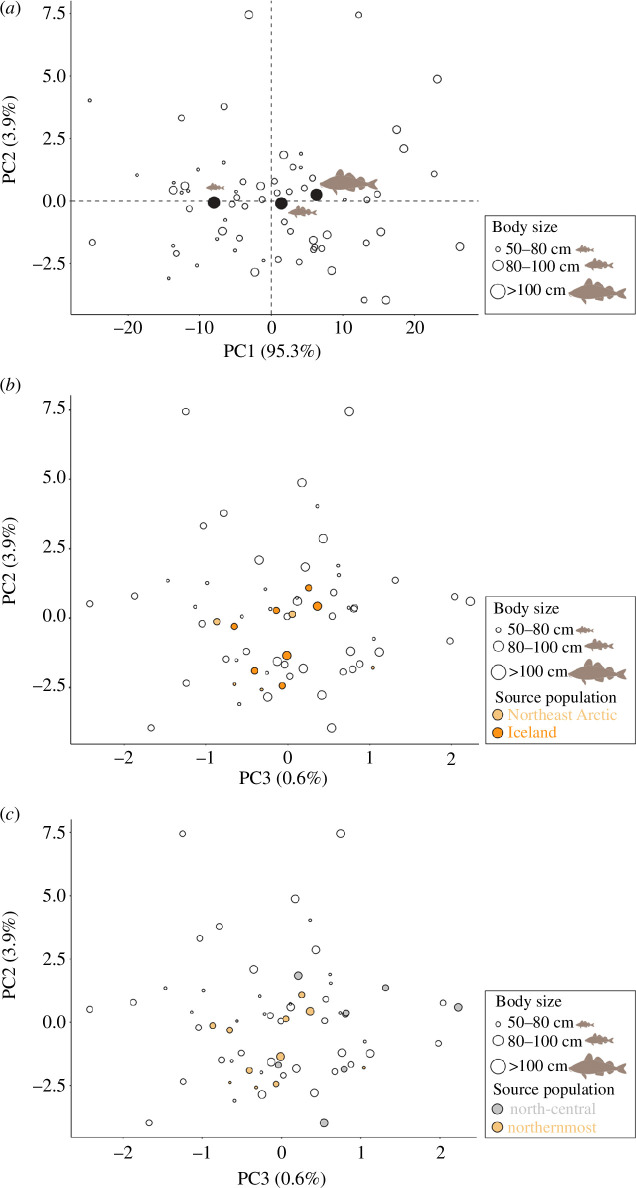
PCA was employed to investigate the relationship between isotope data and genomic assignments while assessing for the contribution of body size. The first two principal component axes (PC1 and PC2) explained 99.18% of the observed variation (figure 3a). PC1 values significantly increased with body size (ANOVA: F-value = 8.85, p‐value ≤ 0.01, n = 64, figure 3a). Such significant differences were found between the smaller (50–80 cm) compared to the medium (80–100 cm; Tukey HSD post hoc test p‐value ≤ 0.01) and larger (>100 cm; Tukey HSD post hoc test p‐value =<0.01) body size categories. We found that the highest contribution to the observed variation in PC1 was that of δ2H values (PC1 contribution = 99.53%), despite similarly strong correlations with body size and δ13C and δ15N values (figure 3a, electronic supplementary material, table S5a,b). Importantly, no body size changes were observed across PC2 (ANOVA: F-value = 0.12, p‐value = 0.89, n = 64, figure 3). PC2 presented the highest contribution and a negative relation with δ34S values according to its loading values (PC2 contribution = 98%, electronic supplementary material, table S5a,b). Furthermore, values in PC3 increased with δ15N and δ13C values (electronic supplementary material, table S5a,b), however, no particular body size changes were observed across PC3 (ANOVA: F-value = 2.25, p‐value = 0.11, n = 64). In the PCAs of isotope values highlighting the genomic assignment of individuals, the second and third principal component axes (PC2 and PC3) explained 4.5% of the observed variation (figure 3b,c). The north-central versus northernmost genomic groups are partially separated with a noticeable overlap between regions on PC3, whereas NEA versus Iceland do not separate on PC2 or PC3 (figure 3b,c).
Figure 3.
Principal component analysis (PCA) for carbon (δ13C), nitrogen (δ15N), non-exchangeable hydrogen (δ2H) and sulphur (δ34S) isotope values, highlighting (a) body size and (b) putative genetically inferred source population assignments of individuals to the Northeast Arctic (NEA) or Iceland populations and (c) north-central or northernmost genomic groups. Values distributed across the PC1–PC2 axes explain 99.18% of the observed variations, while values across the PC2–PC3 axes explain 4.5% of the observed variation. The means (black dots) of each particular body size class (grey cod silhouette) are sorted smallest to largest along PC1s.
Multivariate linear regressions showed a significant relationship between δ13C values and the (genomic) probability of having a northern Norway origin (NEA, p‐value = 0.01, adjusted R2 = 0.50; electronic supplementary material, table S7a), but not for the (genomic) probability of having an Icelandic (p‐value = 0.36; electronic supplementary material, table S7b) or Icelandic frontal origin (p‐value = 0.20; electronic supplementary material, table S7c). We found a significant difference between the δ13C values depending on the binary population assignment between an NEA or Icelandic biological origin (ANOVA: F-value = 5.99, p‐value = 0.04, figure 4). Such differences were not observed for δ15N, δ2H and δ34S values (electronic supplementary material, figure S9a–c). Other isotope values did not show any significant influence on the biological origin of Atlantic cod specimens (electronic supplementary material, table S7).
Figure 4.
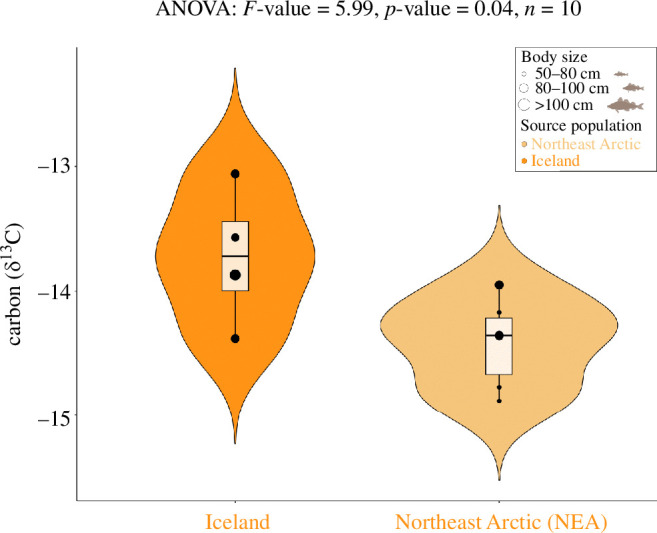
Differences in carbon (δ13C) values across specimens with a binary assignment to genetically inferred source populations: Iceland or NEA. Only specimens with a 70% or higher probability of being assigned to either origin were used in this analysis (see, electronic supplementary material, tables S2, S3).
4. Discussion
We have identified different geographical population sources and ecological differences for ancient Atlantic cod bones obtained from an archaeological assemblage from medieval Oslo over a period of 600 years, by implementing a multidisciplinary approach using aDNA and stable isotopes. We have found an overall diverse provenance of Atlantic cod specimens among the eleventh to seventeenth centuries in Oslo. Atlantic cod obtained through trade (e.g. either the NEA or possibly Iceland) are present throughout time and are found in the earliest deposits. Using multivariate linear regression, we found a significant relationship between carbon (δ13C) values and the (genomic) probability of being assigned to NEA as the original source. Moreover, by assessing for the contribution of Atlantic cod size, PCA analysis supports a separation between the north-central and northernmost genomic groups based on stable isotope values. Our ANOVA results suggest that δ13C values are also significantly different between those specimens with different chromosomal inversion LG1 genotypes (NEA and Iceland). Considering that this inversion is associated with different behaviour patterns and habitat preferences in Atlantic cod [49–53], our observation suggests a diverse geographical population source of Atlantic cod specimens in medieval Oslo. These observations are consistent with known isoscape patterns in the δ13C values of collagen from archaeological Atlantic cod bones, with specimens from Arctic Norway having on average more negative ratios than both Icelandic specimens and Norwegian Sea/North Sea specimens from lower latitudes [6,59]. Below we describe the implications of these findings.
Previous zooarchaeological, isotopic and genetic evidence indicates an increase in long-distance trading of Atlantic cod specimens obtained from remote northern fisheries (e.g. NEA or possibly Iceland) to other locations in Europe, such as England, Flanders, Poland and Estonia, from the thirteenth to fourteenth centuries onwards [6–8,60]. Our observations show a consistent long-distance trading of Atlantic cod specimens, likely obtained from the Lofoten or Vesterålen archipelagos, since the eleventh century (ca 1000 CE). Notably, the stockfish transport from northern Norway to Haithabu—now in northern Germany—has previously been described during the eleventh century (i.e. before ca 1066 CE [5]). Considering that Oslo experienced a pronounced royal and ecclesiastical presence during the eleventh century [14,15,61], elite and ecclesiastical networks may have facilitated access to trading assets (e.g. food) from northern Norway. Nonetheless, Oslo was not a major hub of stockfish trade; therefore, it can be inferred that the Atlantic cod of northern origin present since the eleventh century were primarily for local consumption.
Following Martínez-García et al. [8], we genetically identified a presumed Icelandic origin of processed fish, possibly stockfish [8,62], with specimens assigned to Icelandic frontal (migratory behaviour) or coastal (stationary behaviour) ecotypes. The genomic differences between Icelandic and NEA cod can be found in the chromosomal inversion LG1, where a higher frequency of north-central genotypes can be found in Iceland [53]. However, there are similarities between inversion frequencies (for LG1) between deep water Iceland (migratory ecotype) and NEA cod [8,63]. Considering such similarities, and possible biological complexity, the genetic assignments in this study based on inversion frequencies to either Iceland (either frontal and coastal ecotype) or NEA remain uncertain. Nonetheless, NEA cod has distinct migratory behaviour by feeding in the Barents Sea before spawning along the Norwegian coast [64], while the Icelandic populations consist of a combination of two ecotypes with different spawning, migratory and feeding behaviours [65]. Moreover, modern otolith increment growth indicates that Icelandic cod grow faster than NEA cod during their early stages of life (up to 6 years), whereas NEA cod appears to grow faster during older years [66]. Consequently, previous studies have associated significantly higher δ13C values in Icelandic cod otoliths (δ13Coto) in comparison to NEA cod, with differences in fish growth and also metabolism [66]. The significant differences that we found in δ13C values for specimens assigned to NEA (based on multivariate linear regression) and among specimens assigned to NEA and Iceland are, therefore, consistent with these biological and ecological differences. Our observations then suggest that these specimens may have been exposed to different oceanographic and ecological conditions [67,68] and/or different diet compositions [6,64].
No overall differences in body size categories between genomic assignments to Iceland and NEA specimens, or northernmost and north-central genomic groups, were observed; hence the isotopic differentiation between these genetic assignments is probably not driven by these fish feeding at different trophic levels. In fact, considering the significant differences between body size categories across isotopic values (δ13C, δ15N and δ2H) and across time (fish from Brann4 (1275–1375 CE) are larger compared with those from Brann2 (1125–1275 CE) and Brann3 (1250–1325 CE)), our results reflect the expected ecological complexity during an individual’s lifetime (related to size-specific metabolic rates that decrease as fish grow older), sexual maturation (which differs according to geographical latitude) and diet composition (from lower or higher trophic levels) of different ecotypes or individuals [69–71]. Interestingly, we observed bimodal distributions of δ13C and δ15N values within the medium-size specimens (80–100 cm), which are presumed to feed at similar trophic levels given they are classified in the same body size category. These differences might either reflect differences in their environment (perhaps different locations) or distinct feeding strategies amongst individuals of different sizes [72–74]. Overall, our findings, including variability within the stable isotope data, suggest that multiple localities and/or ecotypes provided stockfish to Oslo from the eleventh century onwards.
5. Conclusion
For millennia, people have relied on Atlantic cod as a food source and key income product for developing coastal communities across northern Europe. Here, we identified a continuous occurrence of Atlantic cod obtained from remote geographical population sources like northern Norway or possibly Iceland since the eleventh century in medieval and post-medieval Oslo. Our observations on the extent of the long-distance fish trade can provide valuable information about the exploitation timeline of specific Atlantic cod stocks and provide a rationale for the long-term baseline assessment for the impacts of historic exploitation. While the interpretation of the genomic assignment of ancient fish specimens to Iceland remains uncertain based upon inversion frequencies only, the association of genetic data with differences in isotopic values does provide evidence for the existence of mixed fisheries, targeting either fish at different spatial locations or co-occurring ecotypes that supported the long-distance trade to Oslo. This study highlights the utility of combining ancient DNA methods with isotope analysis to provide complementary insights into geographical and ecological differences among zooarchaeological fish remains to describe the long-term exploitation of economically important marine species during the medieval and post-medieval development of coastal communities.
Acknowledgements
We thank J. Rolfe (Cambridge) who assisted with the isotope ratio mass spectrometry, T. O’Connell (Cambridge) who facilitated the use of the Dorothy Garrod Laboratory for Isotopic Analysis and Katrien Dierickx for giving advice in statistical analysis. The hydrogen and sulphur isotope analyses were conducted by Iso-Analytical Limited (I. Begley). We thank M. Skage, S. Kollias and A. Tooming-Klunderud at the Norwegian Sequencing Centre for sequencing and processing of samples. We also thank K.S. Jakobsen and S. Jentoft for insightful discussions and Eric J. Guiry for commenting on an early version of this manuscript. Sequencing analysis was performed on the SAGA Cluster under project NN9244K using the resources and assistance from the SIGMA2 Metacenter, the Norwegian National Infrastructure for High Performance Computing and Data Storage.
Contributor Information
Lourdes Martínez-García, Email: lourdes.martinez.garcia@umu.se.
Angélica Pulido, Email: angelica.cuevas@evobio.eu.
Giada Ferrari, Email: giada.ferrari@ibv.uio.no.
Anne Karin Hufthammer, Email: anne.hufthammer@uib.no.
Marianne Vedeler, Email: marianne.vedeler@khm.uio.no.
Alex Hirons, Email: ajh120@leicester.ac.uk.
Catherine Kneale, Email: cjk37@cam.ac.uk.
James H. Barrett, Email: james.barrett@ntnu.no.
Bastiaan Star, Email: bastiaan.star@ibv.uio.no.
Ethics
This work did not require ethical approval from a human subject or animal welfare committee.
Data accessibility
The raw reads for the ancient specimens are released under the ENA accession numbers PRJEB37681 and PRJEB71940. This paper is available as a pre-print in BioRxiv [75]. Code is available on Dryad [76]. Full detail materials and methods are provided online in the electronic supplementary material [77].
Declaration of AI use
We have not used AI-assisted technologies in creating this article.
Authors’ contributions
L.M.-G.: data curation, formal analysis, methodology, visualization, writing—original draft, writing—review and editing; A.P.: data curation, formal analysis, methodology, visualization, writing—original draft, writing—review and editing; G.F.: data curation, methodology, writing—review and editing; A.K.H.: resources, writing—review and editing; M.V.: resources, writing—review and editing; A.H.: methodology, writing—review and editing; C.K.: methodology, writing—review and editing; J.H.B.: conceptualization, data curation, funding acquisition, project administration, resources, supervision, writing—review and editing; B.S.: conceptualization, funding acquisition, project administration, supervision, writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
We declare we have no competing interests.
Funding
This work was supported by Research Council of Norway projects 'FOODIMPACT' (NFR300829), the European Union’s Horizon 2020 Research and Innovation Programme under the Marie Skłodowska-Curie grant agreement no. 813383 (SeaChanges) and the 4 OCEANS Synergy grant agreement no. 951649. The European Research Agency is not responsible for any use that may be made of the information this work contains.
References
- 1. Holm P, Nicholls J, Hayes PW, Ivinson J, Allaire B. 2022. Accelerated extractions of North Atlantic cod and herring, 1520–1790. Fish Fish. 23, 54–72. ( 10.1111/faf.12598) [DOI] [Google Scholar]
- 2. Barrett JH, Locker AM, Roberts CM. 2004. 'Dark age economics’ revisited: the English fish bone evidence AD 600-1600. Antiquity 78, 618–636. ( 10.1017/S0003598X00113262) [DOI] [Google Scholar]
- 3. Maltin E, Jonsson L. 2018. Cod heads, stockfish, and dried spurdog: unexpected commodities in Nya Lödöse (1473–1624), Sweden. Int. J. Hist. Archaeol. 22, 343–363. ( 10.1007/s10761-017-0405-6) [DOI] [Google Scholar]
- 4. Hufthammer AK. 2016. Fish trade in Norway AD 800–1400: zooarchaeological evidence. In Cod and herring (eds Barrett JH, Orton DC), pp. 221–230. Oxford, UK: Oxbow Books. [Google Scholar]
- 5. Star B, et al. 2017. Ancient DNA reveals the arctic origin of Viking Age cod from Haithabu, Germany. Proc. Natl Acad. Sci. USA 114, 9152–9157. ( 10.1073/pnas.1710186114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barrett JH, et al. 2011. Interpreting the expansion of sea fishing in medieval Europe using stable isotope analysis of archaeological cod bones. J. Archaeol. Sci. 38, 1516–1524. ( 10.1016/j.jas.2011.02.017) [DOI] [Google Scholar]
- 7. Orton DC, Morris J, Locker A, Barrett JH. 2014. Fish for the city: meta-analysis of archaeological cod remains and the growth of London’s Northern trade. Antiquity 88, 516–530. ( 10.1017/S0003598X00101152) [DOI] [Google Scholar]
- 8. Martínez-García L, et al. 2022. Ancient DNA evidence for the ecological globalization of cod fishing in medieval and post-medieval Europe. Proc. R. Soc. B 289, 20221107. ( 10.1098/rspb.2022.1107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Heinrich D. 1983. Temporal changes in fishery and fish consumption between early medieval Haithabu and its successor, Schleswig. In Animals and archaeology: 2, shell middens, fishes and birds (eds Grigson C, Clutton-Brock J), pp. 151–156. Oxford, UK: BAR Publishing. [Google Scholar]
- 10. Hallström A. 1979. Die fischknochen. In Eketorp: befestigung und siedlung auf öland/schweden, die fauna (ed. Boessneck J), pp. 422–492. Stockholm, Sweden: Almquist & Wiksell. [Google Scholar]
- 11. Hoffmann RC. 2001. Frontier foods for late Medieval consumers: culture, economy, ecology. Environ. Hist. 7, 131–167. ( 10.3197/096734001129342432) [DOI] [Google Scholar]
- 12. Perdikaris S. 1999. From chiefly provisioning to commercial fishery: long‐term economic change in Arctic Norway. World Archaeol. 30, 388–402. ( 10.1080/00438243.1999.9980419) [DOI] [Google Scholar]
- 13. Garnier N, Vedeler M. 2021. Revealing Medieval culinary practices in Norway: a first metabolomic-based approach. J. Archaeol. Sci. 40, 103206. ( 10.1016/j.jasrep.2021.103206) [DOI] [Google Scholar]
- 14. Nordeide SW, Gulliksen S. 2007. First Generation Christians, Second Generation Radiocarbon Dates: The Cemetery at St. Clement’s in Oslo. Norw. Archaeol. Rev. 40, 1–25. ( 10.1080/00293650601054362) [DOI] [Google Scholar]
- 15. Sørheim, H. 1997. The origin of commercial fisheries and the trade of stockfish in the Northern part of Western Norway. In Exchange and trade in medieval europe: papers of the medieval europe brugge 1997 conference (eds de Boe G, Verhaeghe F), pp. 107–117, vol. 3. [Google Scholar]
- 16. Eide OE. 2007. Om muremåter Og Kirkedateringer i 1100-Tallets Oslo. Coll. Mediev. 20, 137–168. https://ojs.novus.no/index.php/CM/article/view/78 [Google Scholar]
- 17. Molaug PM, Skre D, Flodin L. 2018. Oslogate 6: Arkeologiske utgravninger 1987-1989. Oslo, Norway. NIKU OPPDRAGSRAPPORT 4/2018. See https://niku.brage.unit.no/niku-xmlui/handle/11250/2632540.
- 18. Nedkvitne A, Langholm S. 1991. Byen under Eikaberg: fra byens oppkomst til 1536. Oslo, Norway: Cappelen. [Google Scholar]
- 19. Martínez-García L, Ferrari G, Hufthammer AK, Jakobsen KS, Jentoft S, Barrett JH, Star B. 2022. Ancient DNA reveals a southern presence of the Northeast Arctic cod during the Holocene. Biol. Lett. 18, 20220021. ( 10.1098/rsbl.2022.0021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Atmore LM, Martínez-García L, Makowiecki D, André C, Lõugas L, Barrett JH, Star B. 2022. Population dynamics of Baltic herring since the Viking Age revealed by ancient DNA and genomics. Proc. Natl Acad. Sci. USA 119, e2208703119. ( 10.1073/pnas.2208703119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Grier C, Flanigan K, Winters M, Jordan LG, Lukowski S, Kemp BM. 2013. Using ancient DNA identification and osteometric measures of archaeological Pacific salmon vertebrae for reconstructing salmon fisheries and site seasonality at Dionisio Point, British Columbia. J. Archaeol. Sci. 40, 544–555. ( 10.1016/j.jas.2012.07.013) [DOI] [Google Scholar]
- 22. Elliott Smith EA, Rick TC, Hofman CA. 2022. Investigating fishing strategies and habitat differences in late Holocene Oregon coast sturgeon (Acipenser spp.) through coupled genetic and isotope analyses. Quat. Sci. Adv. 7, 100062. ( 10.1016/j.qsa.2022.100062) [DOI] [Google Scholar]
- 23. Barrett J, et al. 2008. Detecting the medieval cod trade: a new method and first results. J. Archaeol. Sci. 35, 850–861. ( 10.1016/j.jas.2007.06.004) [DOI] [Google Scholar]
- 24. Nikulina E, Meadows J. 2013. Ancient DNA and stable isotope analysis – two innovative scientific methods in the archaeology of hunting. In Hunting in northern Europe until 1500 AD. Old traditions and regional developments, continental sources and continental influences. Papers presented at a workshop organized by the Centre for Baltic and Scandinavian Archaeology (ZBSA) Schleswig, Germany, 16 and 17 June 2011 (eds Grimm O, Schmölcke U), pp. 597–604. Neumünster, Germany: Wachholtz Verlag. [Google Scholar]
- 25. Barrett JH, Boessenkool S, Kneale CJ, O’Connell TC, Star B. 2020. Ecological globalisation, serial depletion and the medieval trade of walrus rostra. Quat. Sci. Rev. 229, 106122. ( 10.1016/j.quascirev.2019.106122) [DOI] [Google Scholar]
- 26. Fuller BT, Müldner G, Van Neer W, Ervynck A, Richards MP. 2012. Carbon and nitrogen stable isotope ratio analysis of freshwater, brackish and marine fish from Belgian archaeological sites (1st and 2nd millennium AD). J. Anal. Atomic Spectrom. 27, 807. ( 10.1039/c2ja10366d) [DOI] [Google Scholar]
- 27. Olafsdottir AS, Thorsdottir I, Wagner KH, Elmadfa I. 2006. Polyunsaturated fatty acids in the diet and breast milk of lactating Icelandic women with traditional fish and cod liver oil consumption. Ann. Nutr. Metab. 50, 270–276. ( 10.1159/000091685) [DOI] [PubMed] [Google Scholar]
- 28. Clauzel T, et al. 2022. Hydrogen isotope measurements of bone and dental tissues from archaeological human and animal samples and their use as climatic and diet proxies. J. Archaeol. Sci. 147, 105676. ( 10.1016/j.jas.2022.105676) [DOI] [Google Scholar]
- 29. Guiry EJ, Szpak P. 2021. Improved quality control criteria for stable carbon and nitrogen isotope measurements of ancient bone collagen. J. Archaeol. Sci. 132, 105416. ( 10.1016/j.jas.2021.105416) [DOI] [Google Scholar]
- 30. Molaug PB. 2015. Nordic middle ages - artefacts, landscapes and society. Essays in honour of Ingvild Øye on her 70th birthday, (eds Baug I, Larsen J, Mygland S), pp. 213–226. Bergen: University of Bergen. [Google Scholar]
- 31. Lie R. 1988. Animal bones. In 'Mindets tomt'– 'Søndre felt'. Animal bones, moss-, plant-, insect- and parasite remains. De arkeologiske utgravninger i Gamlebyen, Oslo, pp. 153–196. Alvheim & Eide Akademisk Forlag, Øvre Ervik. [Google Scholar]
- 32. Lidén HE. 1977. Stratigrafisk-topografisk beskrivelse av feltet “mindets tomt.” In Feltet “mindets tomt”. stratigrafi, topografi, daterende funngrupper (eds Høeg HI, Liestøl A, Molaug PB, Schia E, Wiberg C), pp. 11–71. Oslo, Bergen, Tromsø, Sweden: Scandinavian University Press (Universitetsforlaget). [Google Scholar]
- 33. Cooper A, Poinar HN. 2000. Ancient DNA: do it right or not at all. Science 289, 1139. ( 10.1126/science.289.5482.1139b) [DOI] [PubMed] [Google Scholar]
- 34. Gilbert MTP, Bandelt HJ, Hofreiter M, Barnes I. 2005. Assessing ancient DNA studies. Trends Ecol. Evol. 20, 541–544. ( 10.1016/j.tree.2005.07.005) [DOI] [PubMed] [Google Scholar]
- 35. Ferrari G, et al. 2021. The preservation of ancient DNA in archaeological fish bone. J. Archaeol. Sci. 126, 105317. ( 10.1016/j.jas.2020.105317) [DOI] [Google Scholar]
- 36. Martínez-García L, et al. 2021. Historical demographic processes dominate genetic variation in ancient Atlantic cod mitogenomes. Front. Ecol. Evol. 9, 342. ( 10.3389/fevo.2021.671281) [DOI] [Google Scholar]
- 37. Boessenkool S, Hanghøj K, Nistelberger HM, Der Sarkissian C, Gondek AT, Orlando L, Barrett JH, Star B. 2017. Combining bleach and mild predigestion improves ancient DNA recovery from bones. Mol. Ecol. Resour. 17, 742–751. ( 10.1111/1755-0998.12623) [DOI] [PubMed] [Google Scholar]
- 38. Meyer M, Kircher M. 2010. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 2010, pdb.prot5448. ( 10.1101/pdb.prot5448) [DOI] [PubMed] [Google Scholar]
- 39. Kircher M, Sawyer S, Meyer M. 2012. Double indexing overcomes inaccuracies in multiplex sequencing on the illumina platform. Nucleic. Acids. Res. 40, e3. ( 10.1093/nar/gkr771) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schroeder H, et al. 2015. Genome-wide ancestry of 17th-century enslaved Africans from the Caribbean. Proc. Natl Acad. Sci. USA 112, 3669–3673. ( 10.1073/pnas.1421784112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kapp JD, Green RE, Shapiro B. 2021. A fast and efficient single-stranded genomic library preparation method optimized for ancient DNA. J. Hered. 112, 241–249. ( 10.1093/jhered/esab012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schubert M, et al. 2014. Characterization of ancient and modern genomes by SNP detection and phylogenomic and metagenomic analysis using PALEOMIX. Nat. Protoc. 9, 1056–1082. ( 10.1038/nprot.2014.063) [DOI] [PubMed] [Google Scholar]
- 43. Lindgreen S. 2012. AdapterRemoval: easy cleaning of next-generation sequencing reads. BMC Res. Notes 5, 337. ( 10.1186/1756-0500-5-337) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Star B, et al. 2011. The genome sequence of Atlantic cod reveals a unique immune system. Nature 477, 207–210. ( 10.1038/nature10342) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tørresen OK, et al. 2017. An improved genome assembly uncovers prolific tandem repeats in Atlantic cod. BMC Genomics 18, 95. ( 10.1186/s12864-016-3448-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760. ( 10.1093/bioinformatics/btp324) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jónsson H, Ginolhac A, Schubert M, Johnson PLF, Orlando L. 2013. mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 29, 1682–1684. ( 10.1093/bioinformatics/btt193) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ferrari G, Atmore LM, Jentoft S, Jakobsen KS, Makowiecki D, Barrett JH, Star B. 2022. An accurate assignment test for extremely low-coverage whole-genome sequence data. Mol. Ecol. Resour. 22, 1330–1344. ( 10.1111/1755-0998.13551) [DOI] [PubMed] [Google Scholar]
- 49. Matschiner M, et al. 2022. Supergene origin and maintenance in Atlantic cod. Nat. Ecol. Evol. 6, 469–481. ( 10.1038/s41559-022-01661-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Barney BT, Munkholm C, Walt DR, Palumbi SR. 2017. Highly localized divergence within supergenes in Atlantic cod (Gadus morhua) within the Gulf of Maine. BMC Genomics 18, 271. ( 10.1186/s12864-017-3660-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sodeland M, et al. 2016. 'Islands of divergence' in the Atlantic cod genome represent polymorphic chromosomal rearrangements. Genome Biol. Evol. 8, 1012–1022. ( 10.1093/gbe/evw057) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Berg PR, Star B, Pampoulie C, Sodeland M, Barth JMI, Knutsen H, Jakobsen KS, Jentoft S. 2016. Three chromosomal rearrangements promote genomic divergence between migratory and stationary ecotypes of Atlantic cod. Sci. Rep. 6, 23246. ( 10.1038/srep23246) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Berg PR, Star B, Pampoulie C, Bradbury IR, Bentzen P, Hutchings JA, Jentoft S, Jakobsen KS. 2017. Trans-oceanic genomic divergence of Atlantic cod ecotypes is associated with large inversions. Heredity 119, 418–428. ( 10.1038/hdy.2017.54) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Johansen T, Besnier F, Quintela M, Jorde PE, Glover KA, Westgaard JI, Dahle G, Lien S, Kent MP. 2020. Genomic analysis reveals neutral and adaptive patterns that challenge the current management regime for East Atlantic cod Gadus morhua L. Evol. Appl. 13, 2673–2688. ( 10.1111/eva.13070) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Guiry EJ, Hunt BPV. 2020. Integrating fish scale and bone isotopic compositions for 'deep time' retrospective studies. Mar. Environ. Res. 160, 104982. ( 10.1016/j.marenvres.2020.104982) [DOI] [PubMed] [Google Scholar]
- 56. Barrett JH, Khamaiko N, Ferrari G, Cuevas A, Kneale C, Hufthammer AK, Pálsdóttir AH, Star B. 2022. Walruses on the Dnieper: new evidence for the intercontinental trade of Greenlandic ivory in the Middle Ages. Proc. R. Soc. B 289, 20212773. ( 10.1098/rspb.2021.2773) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nehlich O, Richards MP. 2009. Establishing collagen quality criteria for sulphur isotope analysis of archaeological bone collagen. Archaeol. Anthropol. Sci. 1, 59–75. ( 10.1007/s12520-009-0003-6) [DOI] [Google Scholar]
- 58. R Core Team. 2022. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 59. Hutchinson WF, Culling M, Orton DC, Hänfling B, Lawson Handley L, Hamilton-Dyer S, O’Connell TC, Richards MP, Barrett JH. 2015. The globalization of naval provisioning: ancient DNA and stable isotope analyses of stored cod from the wreck of the Mary Rose, AD 1545. R. Soc. Open Sci. 2, 150199. ( 10.1098/rsos.150199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Orton DC, et al. 2011. Stable isotope evidence for Late Medieval (14th-15th C) origins of the Eastern Baltic cod (Gadus morhua) fishery. PLoS One 6, e27568. ( 10.1371/journal.pone.0027568) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sørheim H. 2015. The first Norwegian towns seen on the background of European history. In Nordic middle ages – artefacts, landscapes and society (eds Baug I, Larsen J, Mygland SS), pp. 301–315. Oslo, Norway: University of Bergen Archaeological Series. [Google Scholar]
- 62. Perdikaris S, McGovern T. 2008. Codfish and kings, seals and subsistence: Norse marine resource use in the North Atlantic. In Human impacts on ancient marine ecosystems: a global perspective (eds Rick T, Erlandson J), pp. 187–214. Berkeley, CA: University of California Press. [Google Scholar]
- 63. Pampoulie C, Ruzzante DE, Chosson V, Jörundsdóttir TD, Taylor L, Thorsteinsson V, Daníelsdóttir AK, Marteinsdóttir G. 2006. The genetic structure of atlantic cod (Gadus morhua) around iceland: insight from microsatellites, the Pan I locus, and tagging experiments. Can. J. Fish. Aquat. Sci. 63, 2660–2674. ( 10.1139/f06-150) [DOI] [Google Scholar]
- 64. Sundby S, Nakken O. 2008. Spatial shifts in spawning habitats of Arcto-Norwegian cod related to multidecadal climate oscillations and climate change. ICES J. Mar. Sci. 65, 953–962. ( 10.1093/icesjms/fsn085) [DOI] [Google Scholar]
- 65. Pampoulie C, Berg PR, Jentoft S. 2023. Hidden but revealed: after years of genetic studies behavioural monitoring combined with genomics uncover new insight into the population dynamics of Atlantic cod in Icelandic waters. Evol. Appl. 16, 223–233. ( 10.1111/eva.13471) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Smoliński S, Denechaud C, von Leesen G, Geffen AJ, Grønkjær P, Godiksen JA, Campana SE. 2021. Differences in metabolic rate between two Atlantic cod (Gadus morhua) populations estimated with carbon isotopic composition in otoliths. PLoS One 16, e0248711. ( 10.1371/journal.pone.0248711) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Johnson SP, Schindler DE. 2012. Four decades of foraging history: stock-specific variation in the carbon and nitrogen stable isotope signatures of Alaskan sockeye salmon. Mar. Ecol. Prog. Ser. 460, 155–167. ( 10.3354/meps09772) [DOI] [Google Scholar]
- 68. Trueman CN, MacKenzie KM, Palmer MR. 2012. Identifying migrations in marine fishes through stable‐isotope analysis. J. Fish Biol. 81, 826–847. ( 10.1111/j.1095-8649.2012.03361.x) [DOI] [PubMed] [Google Scholar]
- 69. Jennings S, Pinnegar JK, Polunin NVC, Boon TW. 2001. Weak cross‐species relationships between body size and trophic level belie powerful size‐based trophic structuring in fish communities. J. Anim. Ecol. 70, 934–944. ( 10.1046/j.0021-8790.2001.00552.x) [DOI] [Google Scholar]
- 70. Soto DX, Wassenaar LI, Hobson KA, Catalan J. 2011. Effects of size and diet on stable hydrogen isotope values (δD) in fish: implications for tracing origins of individuals and their food sources. Can. J. Fish. Aquat. Sci. 68, 2011–2019. ( 10.1139/f2011-112) [DOI] [Google Scholar]
- 71. Köster FW, Trippel EA, Tomkiewicz J. 2013. Linking size and age at sexual maturation to body growth, productivity and recruitment of Atlantic cod stocks spanning the North Atlantic. Fish. Res. 138, 52–61. ( 10.1016/j.fishres.2012.07.002) [DOI] [Google Scholar]
- 72. Sherwood GD, Rideout RM, Fudge SB, Rose GA. 2007. Influence of diet on growth, condition and reproductive capacity in Newfoundland and Labrador cod (Gadus morhua): insights from stable carbon isotopes (δ13C). Deep Sea Res. II Top. Stud. Oceanogr. 54, 2794–2809. ( 10.1016/j.dsr2.2007.08.007) [DOI] [Google Scholar]
- 73. Badalamenti F, D’Anna G, Pinnegar J, Polunin N. 2002. Size-related trophodynamic changes in three target fish species recovering from intensive trawling. Mar. Biol. 141, 561–570. ( 10.1007/s00227-002-0844-3) [DOI] [Google Scholar]
- 74. Miller MJ, Capriles JM, Hastorf CA. 2010. The fish of Lake Titicaca: implications for archaeology and changing ecology through stable isotope analysis. J. Archaeol. Sci. 37, 317–327. ( 10.1016/j.jas.2009.09.043) [DOI] [Google Scholar]
- 75. Martínez-García L, Pulido A, Ferrari G, Hufthammer AK, Vedeler M, Hirons A, Kneale C, Barrett JH, Star B. Tracing 600 years of long-distance Atlantic cod trade in medieval and post-medieval Oslo using stable isotopes and ancient DNA. bioRxiv. ( 10.1101/2024.01.25.577044) [DOI] [PubMed]
- 76. Martínez-García L, Pulido A, Ferrari G, Hufthammer AK, Vedeler M, Hirons A, Kneale C, Barrett JH, Star B. 2024. Data from: Tracing 600 years of long-distance Atlantic cod trade in medieval and post-medieval Oslo using stable isotopes and ancient DNA. Dryad Digital Repository. ( 10.5061/dryad.547d7wmj1) [DOI] [PubMed]
- 77. Martínez-García L, Pulido A, Ferrari G, Hufthammer AK, Vedeler M, Hirons A, Kneale C, Barrett JH, Star B. 2024. Supplementary material from: Tracing 600 years of long-distance Atlantic cod trade in medieval and post-medieval Oslo using stable isotopes and ancient DNA. Figshare ( 10.6084/m9.figshare.c.7539164) [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw reads for the ancient specimens are released under the ENA accession numbers PRJEB37681 and PRJEB71940. This paper is available as a pre-print in BioRxiv [75]. Code is available on Dryad [76]. Full detail materials and methods are provided online in the electronic supplementary material [77].