Abstract
Fimbriae of Porphyromonas gingivalis are thought to play an important role in the colonization and invasion of periodontal tissues. In this study, we analyzed the interactions of P. gingivalis fimbriae with human hemoglobin, fibrinogen, and salivary components (i.e., proline-rich protein [PRP], proline-rich glycoprotein [PRG], and statherin) based on surface plasmon resonance (SPR) spectroscopy with a biomolecular interaction analyzing system (BIAcore). The real-time observation showed that the fimbriae interacted more quickly with hemoglobin and PRG than with other proteins and more intensely with fibrinogen. The significant association constant (ka) values obtained by BIAcore demonstrated that the interactions between fimbriae and these host proteins are specific. These estimated Ka values were not too different; however, the Ka values for hemoglobin (2.43 × 106) and fibrinogen (2.16 × 106) were statistically greater than those for the salivary proteins (1.48 × 106 to 1.63 × 106). The Ka value of anti-fimbriae immunoglobulin G for fimbriae was estimated to be 1.22 × 107, which was 6.55-fold higher than the mean Ka value of the host proteins. Peptide PRP-C, a potent inhibitor of PRP-fimbriae interaction, dramatically inhibited fimbrial association to PRP and PRG and was also inhibitory against other host proteins by BIAcore. The binding of fimbriae to these proteins was also evaluated by other methods with hydroxyapatite beads or polystyrene microtiter plates. The estimated binding abilities differed considerably, depending on the assay method that was used. It was noted that the binding capacity of PRP was strongly diminished by immobilization on a polystyrene surface. Taken together, these findings suggest that P. gingivalis fimbriae possess a strong ability to interact with the host proteins which promote bacterial adherence to the oral cavity and that SPR spectroscopy is a useful method for analyzing specific protein-fimbriae interactions.
Porphyromonas gingivalis, a gram-negative anaerobic rod, is well recognized as a major etiologic agent of periodontal diseases (27). This putative periodontopathogen can adhere to a variety of surface components lining the oral cavity, and the adherence is thought to be mediated by the bacterial components such as fimbriae, vesicles, hemagglutinin, and proteases (25). Fimbriae are thought to play a major role in the interaction of the organism with host proteins such as saliva and plasma components, extracellular matrix proteins, epithelial cells, erythrocytes, fibroblasts, and other bacteria (11, 23). One of these proteins, saliva, interacts with the surface components of P. gingivalis, including fimbriae, in the very early phase of its infection of the oral cavity.
A search for salivary components that specifically interact with P. gingivalis fimbriae indicates that fimbriae strongly bind to acidic proline-rich proteins (PRP), basic proline-rich glycoproteins (PRG), and statherin immobilized onto nitrocellulose membranes or hydroxyapatite (HA) beads (2, 5). These bindings occur via protein-protein interactions through definitive domains of fimbriae (4) and salivary proteins (3, 14). The minimum active domain of PRP1 (a major variant of acidic PRP) for the binding to P. gingivalis fimbriae was found to be Pro-Gln-Gly-Pro-Pro-Gln (PQGPPQ), a typical repeating sequence common to various salivary proline-rich (glyco-) protein variants (2, 14). The synthetic PRP peptide (i.e., peptide PRP-C) analogous to the carboxyl-terminal 21-amino-acid sequence containing PQGPPQ and PQGPPPQ showed significant inhibition in the binding of fimbriae to PRP and PRG on HA beads (14). Peptide PRP-C also inhibited fimbrial binding to PRP, PRG, and their size variants in whole saliva transferred onto a nitrocellulose membrane (2).
The recently developed biomolecular interaction analysis (BIAcore) system involves the use of surface plasmon resonance (SPR) to measure the binding of test samples to ligandary protein (6, 8, 10, 12, 19, 26). In this system, one interactant (ligand) is covalently immobilized onto a sensory chip surface via amino-terminal and ɛ-amino groups of the ligandary protein (6). The other interactant, referred as the analyte, flows over the sensory chip surface in solution. This miniaturized flow system can detect small changes on or near the chip surface by measuring refractive index and can specify which ligands are immobilized. The benefits of SPR assay are (i) direct and real-time observation of the interactions without any labeling of the proteins, (ii) kinetic analysis to provide rate and affinity constants of one-to-one interactions, (iii) comparison of the binding properties of different interactants such as other proteins and mutated recombinant proteins by a point mutation or deletion, and (iv) screening of unknown interactants in crude samples (13, 22).
Several host proteins have been reported to bind to fimbriae; however, their binding specificities and the underlying mechanisms are still unknown. In this study, the binding of fimbriae to the host proteins, including PRP, PRG, statherin, hemoglobin, and fibrinogen, was analyzed by the BIAcore system. The inhibitory effects of peptide PRP-C on these interactions were also investigated. The binding profiles of the BIAcore analyses were compared with those of other assay methods involving HA beads or polystyrene microtiter plates.
MATERIALS AND METHODS
Purification of fimbriae.
Fimbriae were mechanically detached from P. gingivalis ATCC 33277 cells grown anaerobically and purified chromatographically as previously described (28).
Preparation of host proteins.
The salivary proteins PRP, low-molecular-weight proline-rich glycoprotein (L-PRG), and statherin were prepared as outlined in our previous study (2, 5). Hemoglobin was isolated from human blood in our previous study (17), and fibrinogen was purchased (Kabi Vitrum, Stockholm, Sweden). Lipid-free bovine serum albumin (BSA; A-7030; Sigma Chemical Co., St. Louis, Mo.) was used as a negative control. The protein content of samples was determined with bicinchoninic acid protein assay reagent (Pierce, Rockford, Ill.), with BSA as a standard, according to the manufacturer’s manual.
Antibodies.
The preparation of rabbit antifimbriae immunoglobulins was described previously (9), and immunoglobulin G was fractionated with Protein G affinity column chromatography (HiTrap Protein G; Amersham Pharmacia Biotech, Uppsala, Sweden).
Preparation of peptide PRP-C.
Peptide PRP-C, corresponding to the carboxyl-terminal segment composed of 21-amino-acid residues of PRP1, was synthesized and purified in our previous study (14). The amino acid sequence of the peptide is PQGPPPQGGRPQGPPQGQSPQ. Two synthetic peptides, as follows, which showed no effects in the binding of fimbriae to salivary components were used as negative controls: peptide SM15, corresponding to residues 15 to 29 of statherin (GYGYGPYQPVPEQPL) (3), and peptide A1, corresponding to residues 22 to 41 of P. gingivalis fimbrillin (EQQEAIKSAENATKVEDIKC) (5).
Measurement of molecular interactions by the BIAcore method.
The interactions between fimbriae and the host proteins were analyzed with a model 1000 system from BIAcore (Uppsala, Sweden) as described in our previous study (16). The BIAcore system is equipped with the sensory chip CM5, a small metal chip with a carboxymethyldextran surface, to allow ligand immobilization via native NH2 (12). An amine coupling kit containing N-hydroxysuccinimide (NHS), N-ethyl-N′-[(3-dimethylamino)-propyl]-carbodiimide hydrochloride (EDC), and ethanolamine-HCl (Amersham Pharmacia Biotech) was used to immobilize the ligand to the chip. The host proteins were immobilized on the sensory chip CM5 to measure their interaction with fimbriae. A mixture of NHS and EDC (1:1) was injected into the dextran matrix on the sensory chip to activate it at the flow rate of 5 μl/ml at 25°C, and each intact protein (100 μg/ml) in 10 mM sodium acetate buffer (pH 4.0 or 4.8) was immobilized on the matrix. To equalize the amount (mol) of the immobilized proteins, the increase in resonance units (RU) produced by the immobilization was manually set at 400× (molecular mass of immobilized protein/molecular mass of fimbrillin [41 kDa]) RU according to the manufacturer’s manual (BIAcore operations manual). The excess active sites of the matrix were blocked with ethanolamine-HCl (1 M) (12, 19) and washed with regeneration buffer (1 M NaCl in PBS). All materials were dissolved in PBS, which was also used as a running buffer in the experiments. Fimbriae were injected at a flow rate of 10 μl/min at 25°C, and the binding of fimbriae was monitored and presented in a sensorgram (a plot of RU versus time). A 0.1° shift in the SPR angle, corresponding to 1,000 RU, corresponds to a change in the surface concentration of 1 ng/mm2 (8). For kinetic studies, fimbriae in increasing concentrations were injected over the sensory chip, and continuous response of RU per second (dRU/dt) versus RU values were plotted as a slope from the sensorgram of each interaction. The slopes at different fimbrial concentrations were replotted, and then the rate constant was obtained with equation 1 as follows: slope (dRU/dt versus RU) = kas × C + kdis, where kas is the association rate constant (1/M/s), kdis is the dissociation rate constant (1/s), and C is the concentration of fimbriae. The first-order kinetics were obtained according to equation 2 as follows: ln(Rt1/Rtn) = kdis × t, where Rt1 is the RU at the time of the initial phase of the dissociation (t1), Rtn is the RU at time tn, and t = tn − t1. The association constant (Ka) was calculated from equation 3 as follows: Ka = kas/kdis. The analyses of these kinetic parameters were performed by using BIA EVALUATION 3.0, a software designed to analyze experimental sensorgram data for kinetics and affinity of interactions, according to the manufacturer’s manual.
Binding of fimbriae to salivary protein-coated HA.
The binding assays of 125I-labeled fimbriae to salivary protein-coated HA beads were carried out as described previously (2). The specific activity of iodinated protein was 1.9 mCi/μmol of fimbrillin. The HA beads (3 mg of spherical HA beads; BDH Chemicals, Poole, England) in a tube were coated with PRP, L-PRG, statherin, hemoglobin, fibrinogen, or BSA (100 μl of 0.1 mg/ml solution), respectively. Aliquots (100 μl each) of 125I-labeled fimbriae (5 nmol/ml) and, if necessary, peptide PRP-C (50 nmol) as an inhibitor were added to tubes containing the host protein-coated HA beads and incubated at room temperature (RT) for 1 h. The value for specific binding was calculated by subtracting that for nonspecific binding, which was obtained by the preincubation of protein-coated HA beads with nonlabeled fimbriae (500 μl of 50 nmol/ml solution) at RT for 1 h. All assays were performed in triplicate, on three separate occasions.
Binding of fimbriae to the host proteins immobilized on a polystyrene surface.
Binding of fimbriae to the host proteins was investigated by using 96-well microtiter plates (Maxisorp; Nalge Nunc International, Roskilde, Denmark). The plates were coated with the host proteins or BSA (100 μl of 0.1 mg/ml solution in phosphate buffer [PB] containing 150 mM sodium chloride, pH 7.4 [PBS]) at 37°C for 2 h. After being washed with 300 mM NaCl in PB, the wells were blocked with 100 μl of 1% casein solution (Block Ace; Snow Brand Co. Ltd., Sapporo, Japan) in PBS at RT for 30 min. Following a wash with 300 mM NaCl in PB, aliquots of fimbriae (5 μg/ml in PBS) and, if necessary, peptide PRP-C as an inhibitor were added to the wells, followed by incubation at RT for 1 h. The wells were washed with 300 mM NaCl in PB three times, and then rabbit anti-fimbriae immunoglobulin G (1:1,000) was added and incubated at RT for 1 h. The amount of fimbriae bound to the wells was measured at A405 following the incubation of alkaline-phosphatase-conjugated goat anti-rabbit immunoglobulin G (1:1,000) at RT for 1 h and the subsequent addition of diethanolamin buffer containing p-nitrophenylphosphate disodium salt (1 mg/ml). The background absorbance was set on fimbriae-bound wells coated only with blocking agents, i.e., Block Ace. All assays were performed in triplicate, on three separate occasions.
Statistical analysis.
The data, expressed as means ± standard deviations, were averaged, and a t test was used for comparison. P values of <0.01 were considered statistically significant.
RESULTS
Estimation of fimbrial binding affinity to the host proteins by using the BIAcore system.
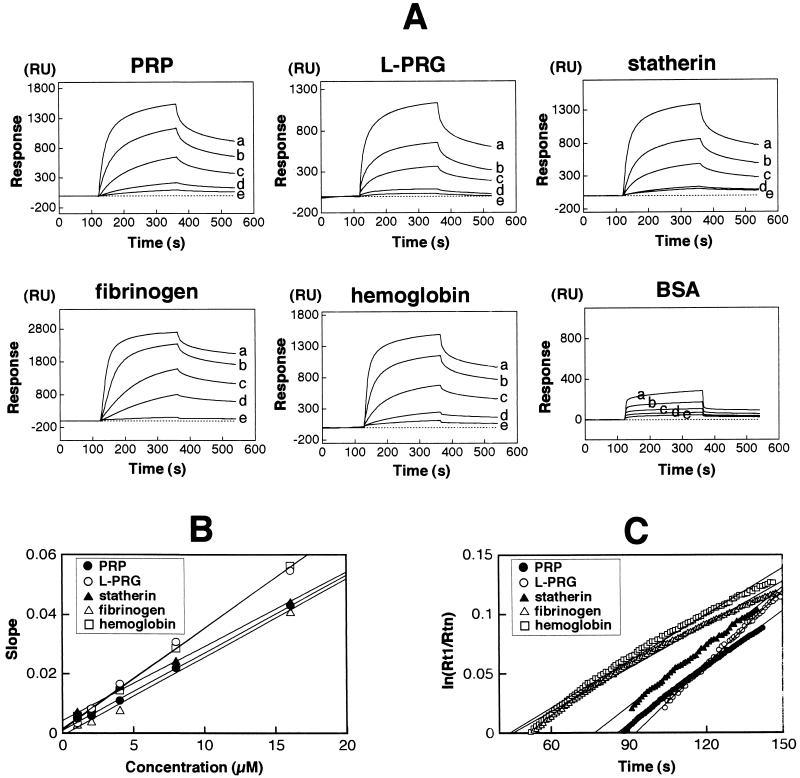
After given amounts of the host proteins (10 fmol/mm2) were immobilized on a dextran matrix of the sensory chip, various concentrations of fimbriae (1, 2, 4, 8, and 16 μM) were repeatedly injected over the immobilized proteins for the kinetic studies. The sensograms of the fimbrial bindings to the host proteins were monitored as shown in Fig. 1A. Following injection of fimbriae (time, 120 s), the very first linear increase of RU was observed due to the mass change of fimbriae diffusion, which is called mass transport limitation (21, 22). The following curve showed an RU increase in complex concentration. The logarithmic decrease after the sample pulse had passed (time, 360 s) indicated the dissociation of fimbriae from the immobilized protein. In the interaction with BSA as a negative control, the RU was increased by mass transport limitation and nonspecific interaction, followed by a decrease to the baseline at the end of the pulse (Fig. 1A). A dRU/dt versus RU plot (slope) was calculated from the sensograms, and then the slopes at different concentrations were plotted against the concentrations of fimbriae (Fig. 1B). Different angles of the obtained linear lines represent the variation of affinities, which gave the association rate constants (kas) from equation 1. The dissociation rate constant (kdis) was directly determined from the linear lines of plots at the dissociation phase (see Fig. 3C). The angles of regression linear lines represent the stabilities of complexes. First-order kinetics were obtained by equation 2, and the calculated specific constants are shown in Table 1. L-PRG and hemoglobin showed significantly higher kas values than the other proteins, i.e., 3.38 × 103 and 3.42 × 103, respectively, indicating quick association with fimbriae. The kas values of PRP, statherin, and fibrinogen were within similar ranges. In the analysis of dissociation phase, significant differences were observed among all kdis values of the proteins. L-PRG showed the biggest kdis value, representing the lowest stability, while fibrinogen exhibited a kdis value of 1.22 × 10−3 s−1, indicating the highest stability. Total affinities are presented as Ka. The affinities of fimbriae with hemoglobin and fibrinogen (Ka = 2.43 × 106 M−1 and 2.16 × 106 M−1) were found to be statistically higher than those of the salivary proteins. For comparison, the affinity of fimbriae to rabbit anti-fimbriae immunoglobulin G was also measured (data not shown). The interaction showed 2.1-fold-higher kas (6.11 × 103 M−1/s−1) and 3.1-fold-lower kdis (5.00 × 10−4 s−1) than the mean value of the host proteins (Table 1). The Ka was 6.55-fold higher than the mean Ka value of the host proteins.
FIG. 1.
Estimation of fimbrial binding affinity to the host proteins by the BIAcore system. The same molar amount of each host protein was immobilized on the matrix of the chip. Fimbriae were injected at flow rate 10 μl/min for 240 s. (A) The binding ability of fimbriae to each host protein was monitored and presented as a sensogram (plotted as RU versus time). For kinetic studies, fimbriae with increasing concentrations (a, 16 μM; b, 8 μM; c, 4 μM; d, 2 μM; e, 1 μM) were injected over the sensor chip. (B) Kinetic analysis of fimbrial binding to host proteins. A plot of dRU/dt versus RU (slope) was calculated from the sensograms, and then the slopes at different concentrations were replotted against the concentrations of fimbriae. The different angles of the obtained linear lines represent the variation of the affinities, giving the kas from equation 1. (C) kdis (1/s) was determined directly from the linear lines of the plots at the dissociation phase. A ln(Rt1/Rtn) plot was calculated [Rt1 is the RU at the initial phase of dissociation (t1), and Rtn is the RU at time tn] and replotted against the times. The angles of the regression linear lines represent resistibility to dissociation.
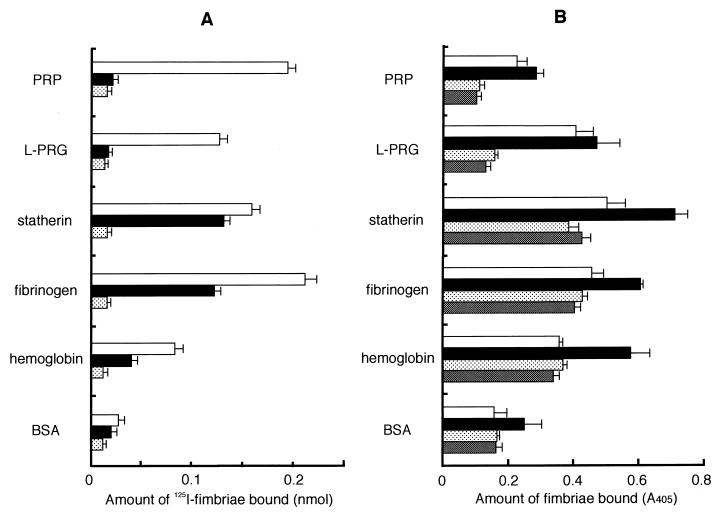
FIG. 3.
Binding of P. gingivalis fimbriae to the host proteins on an apatitic or a polystyrene surface. (A) Binding abilities of fimbriae to host proteins were assayed by using HA beads. 125I-labeled fimbriae (0.5 nmol) was added to a tube containing host protein-coated HA beads and incubated at RT for 1 h. 125I-labeled fimbriae (0.5 nmol) added (□); 125I-labeled fimbriae (0.5 nmol) plus peptide PRP-C (50 nmol) added (■); fimbriae (0.5 nmol) plus nonlabeled fimbriae (25 nmol) added to obtain the nonspecific binding level ( ). The specific binding level was calculated by subtracting the nonspecific binding level. Significant differences (P < 0.01) were observed among fimbrial binding levels (□) to host proteins in the absence of peptide PRP-C. (B) Binding assay performed with polystyrene surface. The wells of 96-well microtiter plates were coated with host proteins or BSA (100 μl of 0.1 mg/ml in PBS) at 37°C for 2 h. An aliquot of fimbriae and, if necessary, peptide PRP-C as an inhibitor was added to the wells and then incubated at RT for 1 h. Rabbit anti-fimbriae immunoglobulin G (1:1,000) was added to detect the amount of bound fimbriae. □, 12.2 pmol of added fimbriae; ■, 24.4 pmol of added fimbriae; , 12.2 pmol of fimbriae plus 100 pmol of added peptide PRP-C; ▩, 12.2 pmol of fimbriae plus 1 nmol of added peptide PRP-C. Significant differences (P < 0.01) were observed among fimbrial binding levels (■) to host proteins in the absence of peptide PRP-C and between PRP and BSA and between fibrinogen and hemoglobin. All assays were performed in triplicate, on three separate occasions.
TABLE 1.
Binding constants of P. gingivalis fimbriae to host proteinsa
| Protein | kas (1/M/s) | kdis (1/s) | Ka (1/M) |
|---|---|---|---|
| PRP | 2.61 × 103a | 1.60 × 10−3 | 1.63 × 106c |
| L-PRG | 3.38 × 103b | 2.08 × 10−3 | 1.62 × 106c |
| Statherin | 2.49 × 103a | 1.68 × 10−3 | 1.48 × 106c |
| Fibrinogen | 2.63 × 103a | 1.22 × 10−3 | 2.16 × 106d |
| Hemoglobin | 3.42 × 103b | 1.41 × 10−3 | 2.43 × 106d |
| Anti-fimbriae immunoglobulin G | 6.11 × 103 | 5.00 × 10−4 | 1.22 × 107 |
Differences between values indicated by roman superscripts are not significant (P ≥ 0.01). Differences among constant values are significant (P < 0.01).
Demonstration of the effect of peptide PRP-C by using the BIAcore system.
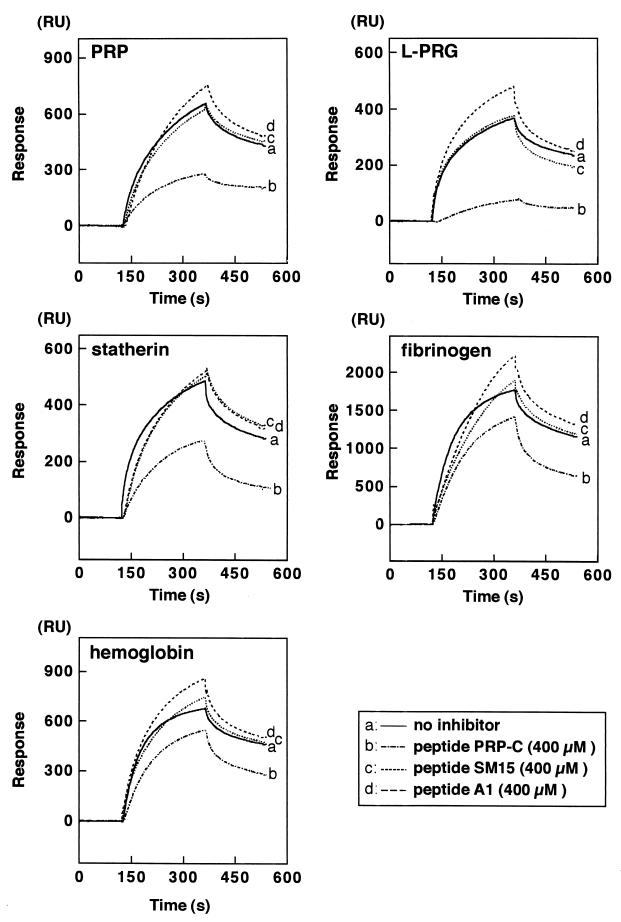
Peptide PRP-C (400 μM) was simultaneously injected with fimbriae (4 μM) to the sensory chip of the BIAcore system. As shown in Fig. 2, the peptide significantly inhibited the fimbrial association with all immobilized proteins (P < 0.01), especially PRP and L-PRG. On the other hand, peptides SM15 and A1 (400 μM) showed no inhibitory effects.
FIG. 2.
Demonstration of the effect of peptide PRP-C by the BIAcore system. Fimbriae (4 μM) were injected with or without simultaneous addition of each peptide (400 μM of peptide PRP-C, SM 15, or A1) to the sensory chip of the BIAcore system as described in the text.
Binding of fimbriae to the host proteins on solid surfaces.
The binding ability of fimbriae to the proteins was evaluated by other methods with HA beads or a polystyrene microtiter plate in addition to BIAcore. Although fimbriae significantly bound to the proteins on HA beads, the relative levels of fimbrial binding were different from those of BIAcore (Fig. 3A). The binding ability of hemoglobin measured markedly lower than that of BIAcore. The effects of peptide PRP-C by BIAcore and HA beads were similar, but the effect of peptide PRP-C on statherin was not as significant by the HA beads assay as by BIAcore. Results obtained by the microtiter plate assay did not agree with those obtained by BIAcore or HA beads assay (Fig. 3B). The fimbriae bound to statherin most effectively, while PRP showed a minimum binding by the microtiter plate assay. The levels of the inhibitory effects of peptide PRP-C were similar to those obtained by the other assays.
DISCUSSION
We employed the SPR technology with BIAcore for kinetic analysis of the fimbrial interactions with the host proteins. The affinity constant (Ka) was estimated based on two kinetic constants, kas, which determines the speed of the association, and kdis, reflecting the stability of the complexes of fimbriae and the host protein. Interestingly, different kinetic constants were obtained among these interactions. For example, fimbriae interacted more quickly with hemoglobin and PRG than with other proteins, while the fimbrial binding to fibrinogen was very tight. The Ka values obtained by BIAcore analyses demonstrated that the interactions between fimbriae and the host proteins are specific. These estimated Ka values were not too different; however, hemoglobin and fibrinogen possess statistically greater affinities for fimbriae than do the salivary proteins. The fimbrial interaction with anti-fimbriae immunoglobulin G showed significant kinetic constants. Compared with these constants for host proteins and antibodies, the kdis value of the antibodies was remarkably lower (3.1-fold) than the mean of the host proteins, indicating significant stability of the associated molecules. In other words, the negligible dissociation of the complex should reflect a tight interaction between antigen and antibody. The binding of fimbriae with the host proteins leads to subsequent invasion by P. gingivalis of periodontal tissues (11, 25), and it is expected that the strong association of fimbriae with the host proteins mediates bacterial adherence to the oral surface and that high dissociation helps P. gingivalis leave for other sites for subsequent colonization and invasion.
The BIAcore system has been used to assay various interactions between antigens and antibodies. The reported constants of many antibody-protein antigen interactions by BIAcore analyses are as follows: kas = ∼104 to 106 (M−1 s−1) kdis = ∼10−2 to 10−4 (s−1), and Ka = ∼106 to 1010 (M−1) (13, 15). For example, the reported Ka values are as follows: 3.73 × 109 M−1 (cyclosporin and Fab fragment [20]), 3.10 × 109 M−1 (epithelial A33 antigen and monoclonal antibody [6]), and 1.9 × 1010 M−1 (carcinoembryonic antigen and polyclonal immunoglobulin G [1]). In the present study, the fimbriae-polyclonal immunoglobulin G interaction gave a Ka of 1.22 × 107 M−1, indicating that the interaction was moderate compared with the above-reported interactions. It should be noted that the constant value was calculated here based on the molecular mass of fimbrillin (a 41-kDa subunit protein of fimbriae), because the exact molecular mass of P. gingivalis native fimbriae is difficult to determine. In our previous study, it was calculated to be ∼106 to 104 kDa from the elution profile of gel filtration of the purified fimbrial preparation (16). Given that the molecular mass of fimbriae is 1 × 104 kDa, the Ka value of the fimbriae-polyclonal immunoglobulin G interaction will be more than 103 times higher, >1.22 × 109 M−1, a level similar to those of other reported antibodies with high affinities. In addition, the Ka of fimbrial binding to the host proteins will be >1.62 × 108 M−1, which is as high as those of moderate interactions between antibodies and protein antigens. These results suggest that the interaction of fimbriae with the host proteins is fairly strong and thus important in the establishment of infection by P. gingivalis in vitro.
The inhibitory effects of peptide PRP-C on fimbrial binding to all host proteins, especially to PRP and L-PRG, were shown to be significant by BIAcore. The BIAcore assay also indicates that the inhibition is based on a competitive effect in the association phase of these molecules. In addition, peptide PRP-C was inhibitory in the other assays, suggesting that fimbriae interact with the proteins through a similar mechanism. However, why effects inhibitory to fibrinogen and hemoglobin were weaker than those to salivary proteins by BIAcore is not known. This finding indicates that there might be additional binding site(s) on these two proteins.
In this study, estimated binding capacities varied by assay method. The proteins are fixed onto the polystyrene surfaces by hydrophobic interaction (7, 18, 24), and the assay might be impeded by several factors, such as poor adsorption, random orientation, alteration of protein conformation, steric hindrance, and altered kinetics, leading to a partial or complete loss of binding capacities (24). The bindings of P. gingivalis fimbriae to PRP, L-PRG, and statherin are mediated by unique hidden receptors termed cryptitopes (2–5). Therefore, the occurrence of the binding to fimbriae depends on the conformation of the immobilized salivary proteins. The hydrophilic environment in the matrix of BIAcore was reported to be valuable, as it prevents conformational changes attributable to adsorption to polystyrene surfaces (21). The binding of fimbriae to PRP was negligible by the present microtiter plate assay, suggesting that the binding capacity of PRP is diminished by the nature of the polystyrene surface.
In the HA beads assay, the proteins were immobilized onto HA beads via ionic and other interactions (7), and fimbriae were radiolabeled. The protein labeling could affect the biological activity of the proteins, including binding ability (29). In the BIAcore system, the ligand protein is covalently immobilized onto the chip surface via amino-terminal and ɛ-amino groups of the protein without any labeling (6, 12). This mechanism seems to minimize the above-mentioned events leading to a loss of binding capacities (6, 8, 12, 19). The fimbrial binding to hemoglobin might be interpreted by the 125I labeling.
Peptide PRP-C exhibited a marked inhibition in the fimbrial binding to the host proteins by three different assay methods employed in this study. This finding suggests that the fimbrial binding to the host proteins occurs by similar molecular mechanisms. Of these assays, BIAcore most clearly demonstrated the specific interactions between fimbriae and the host proteins, including PRP. On the basis of the present results, we conclude that SPR spectroscopy is a useful method to analyze the specific interactions of bacterial surface components with host (glyco)proteins.
ACKNOWLEDGMENTS
We thank S. Hase (Osaka University Institute for Protein Research) for generously allowing us to use the BIAcore system. We are also grateful to Masanori Kontani for his valuable suggestions.
REFERENCES
- 1.Abraham R, Buxbaum S, Link J, Smith R, Venti C, Darsley M. Screening and kinetic analysis of recombinant anti-CEA antibody fragments. J Immunol Methods. 1995;183:119–125. doi: 10.1016/0022-1759(95)00039-d. [DOI] [PubMed] [Google Scholar]
- 2.Amano A, Shizukuishi S, Horie H, Kimura S, Morisaki I, Hamada S. Binding of Porphyromonas gingivalisfimbriae to proline-rich glycoproteins in parotid saliva via a domain shared by major salivary components. Infect Immun. 1998;66:2072–2077. doi: 10.1128/iai.66.5.2072-2077.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amano A, Kataoka K, Raj P A, Genco R J, Shizukuishi S. Binding sites of salivary statherin to Porphyromonas gingivalisrecombinant fimbrillin. Infect Immun. 1996;64:4249–4254. doi: 10.1128/iai.64.10.4249-4254.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amano A, Sharma A, Lee J-Y, Sojar H T, Raj P A, Genco R J. Structural domains of Porphyromonas gingivalisrecombinant fimbrillin that mediate binding to salivary proline-rich protein and statherin. Infect Immun. 1996;64:1631–1637. doi: 10.1128/iai.64.5.1631-1637.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amano A, Sojar H T, Lee J-Y, Sharma A, Levine M J, Genco R J. Salivary receptors for recombinant fimbrillin of Porphyromonas gingivalis. Infect Immun. 1994;62:3372–3380. doi: 10.1128/iai.62.8.3372-3380.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Catimel B, Nerrie M, Lee F T, Scott A M, Ritter G, Welt S, Old L J, Burgess A W, Nice E C. Kinetic analysis of the interaction between the monoclonal antibody A33 and its colonic epithelial antigen by the use of an optical biosensor: a comparison of immobilization strategies. J Chromatogr. 1997;776:15–30. doi: 10.1016/s0021-9673(97)00087-3. [DOI] [PubMed] [Google Scholar]
- 7.Doyle R J, Rosenberg M. Measurement of microbial adhesion to hydrophobic substrata. Methods Enzymol. 1995;253:542–550. doi: 10.1016/s0076-6879(95)53046-0. [DOI] [PubMed] [Google Scholar]
- 8.Fägerstam L G, Frostell-Karlsson Å, Karlsson R, Persson B, Rönnberg I. Biospecific interaction analysis using surface plasmon resonance detection applied to kinetic, binding site and concentration analysis. J Chromatogr. 1992;597:397–410. doi: 10.1016/0021-9673(92)80137-j. [DOI] [PubMed] [Google Scholar]
- 9.Fujiwara T, Morishima S, Takahashi I, Hamada S. Molecular cloning and sequencing of the fimbrilin gene of Porphyromonas gingivalisstrains and characterization of recombinant proteins. Biochem Biophys Res Commun. 1993;197:241–247. doi: 10.1006/bbrc.1993.2467. [DOI] [PubMed] [Google Scholar]
- 10.Granzow R, Reed R. Interactions in the fourth dimension. Bio/Technology. 1992;10:390–393. doi: 10.1038/nbt0492-390. [DOI] [PubMed] [Google Scholar]
- 11.Hamada S, Amano A, Kimura S, Nakagawa I, Kawabata S, Morisaki I. The importance of fimbriae in the virulence and ecology of some oral bacteria. Oral Microbiol Immunol. 1998;13:129–138. doi: 10.1111/j.1399-302x.1998.tb00724.x. [DOI] [PubMed] [Google Scholar]
- 12.Johnsson B, Lofås S, Lindquist G. Immobilization of proteins to a carboxymethyldextran-modified gold surface for biospecific interaction analysis in surface plasmon resonance sensors. Anal Biochem. 1991;198:268–277. doi: 10.1016/0003-2697(91)90424-r. [DOI] [PubMed] [Google Scholar]
- 13.Karlsson R, Fält A. Experimental design for kinetic analysis of protein-protein interactions with surface plasmon resonance biosensors. J Immunol Methods. 1997;200:121–133. doi: 10.1016/s0022-1759(96)00195-0. [DOI] [PubMed] [Google Scholar]
- 14.Kataoka K, Amano A, Kuboniwa M, Horie H, Nagata N, Shizukuishi S. Active sites of salivary proline-rich protein for binding to Porphyromonas gingivalisfimbriae. Infect Immun. 1997;65:3159–3164. doi: 10.1128/iai.65.8.3159-3164.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khilko S N, Jelonek M T, Corr M, Boyd L F, Bothwell A L, Margulies D H. Measuring interactions of MHC class I molecules using surface plasmon resonance. J Immunol Methods. 1995;183:77–94. doi: 10.1016/0022-1759(95)00033-7. [DOI] [PubMed] [Google Scholar]
- 16.Kontani M, Kimura S, Nakagawa I, Hamada S. Adherence of Porphyromonas gingivalisto matrix proteins via a fimbrial cryptic receptor exposed by its own arginine-specific protease. Mol Microbiol. 1997;24:1179–1187. doi: 10.1046/j.1365-2958.1997.4321788.x. [DOI] [PubMed] [Google Scholar]
- 17.Kuboniwa M, Amano A, Shizukuishi S. Hemoglobin-binding protein purified from Porphyromonas gingivalisis identical to lysine-specific cysteine proteinase (Lys-Gingipain) Biochem Biophys Res Commun. 1998;249:38–43. doi: 10.1006/bbrc.1998.8958. [DOI] [PubMed] [Google Scholar]
- 18.Ljungh Å, Wadström T. Binding of extracellular matrix proteins by microbes. Methods Enzymol. 1995;253:501–514. doi: 10.1016/s0076-6879(95)53041-x. [DOI] [PubMed] [Google Scholar]
- 19.Löfås S, Johnsson B. A novel hydrogel matrix on gold surfaces in surface plasmon resonance sensors for fast and efficient covalent immobilization of ligands. J Chem Soc Chem Commun. 1990;21:1526–1528. [Google Scholar]
- 20.Lutz Z G, Rauffer N, Altschuh D, van Regenmortel M H V. Analysis of cyclosporin interactions with antibodies and cyclophilin using the BIAcore. J Immunol Methods. 1995;183:131–140. doi: 10.1016/0022-1759(95)00041-8. [DOI] [PubMed] [Google Scholar]
- 21.Malmqvist M. Biospecific interaction analysis using biosensor technology. Nature. 1993;361:186–187. doi: 10.1038/361186a0. [DOI] [PubMed] [Google Scholar]
- 22.Myszka D G. Kinetic analysis of macromolecular interactions using surface plasmon resonance biosensors. Curr Opin Biotechnol. 1997;8:50–57. doi: 10.1016/s0958-1669(97)80157-7. [DOI] [PubMed] [Google Scholar]
- 23.Naito Y, Gibbons R J. Attachment of Bacteroides gingivalisto collagenous substrata. J Dent Res. 1988;67:1075–1080. doi: 10.1177/00220345880670080301. [DOI] [PubMed] [Google Scholar]
- 24.Ofek I, Doyle R J. Methods, models, and analysis of bacterial adhesion. In: Otek I, Doyle R J, editors. Bacterial adhesion to cells and tissues. New York, N.Y: Chapman and Hall; 1994. pp. 16–40. [Google Scholar]
- 25.Okuda K. Attachment mechanisms and colonization. In: Shah H N, Mayrand D, Genco R J, editors. Biology of the species Porphyromonas gingivalis. Boca Raton, Fla: CRC Press Inc.; 1993. pp. 139–158. [Google Scholar]
- 26.Stenberg E, Persson B, Roos H, Urbaniczky C. Quantitative determination of surface concentration of protein with surface plasmon resonance by using radiolabelled proteins. J Colloid Interface Sci. 1991;143:513–526. [Google Scholar]
- 27.van Steenbergen T J M, van Winkelhoff A J, deGraaff J. Black-pigmented oral anaerobic rods: classification and role in periodontal disease. In: Hamada S, Holt S C, McGhee J R, editors. Periodontal disease: pathogens & host immune responses. Tokyo, Japan: Quintessence Publisher Co., Ltd.; 1991. pp. 41–52. [Google Scholar]
- 28.Yoshimura F, Takahashi K, Nodasaka Y, Suzuki T. Purification and characterization of a novel type of fimbriae from the oral anaerobe Bacteroides gingivalis. J Bacteriol. 1984;160:949–957. doi: 10.1128/jb.160.3.949-957.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilder R L, Yuen C C, Mage R G. Lactoperoxidase catalyzed radioiodination of cell surface immunoglobulin: incorporated radioactivity may not reflect relative cell surface Ig density. J Immunol. 1979;122:459–463. [PubMed] [Google Scholar]