Abstract
The porin proteins of the pathogenic Neisseria species, Neisseria gonorrhoeae and Neisseria meningitidis, are important as serotyping antigens, putative vaccine components, and for their proposed role in the intracellular colonization of humans. A three-dimensional structural homology model for Neisseria porins was generated from Escherichia coli porin structures and N. meningitidis PorA and PorB sequences. The Neisseria sequences were readily assembled into the 16-strand β-barrel fold characteristic of porins, despite relatively low sequence identity with the Escherichia proteins. The model provided information on the spatial relationships of variable regions of peptide sequences in the PorA and PorB trimers and insights relevant to the use of these proteins in vaccines. The nucleotide sequences of the porin genes from a number of other Neisseria species were obtained by PCR direct sequencing and from GenBank. Alignment and analysis of all available Neisseria porin sequences by use of the structurally conserved regions derived from the PorA and PorB structural models resulted in the recovery of an improved phylogenetic signal. Phylogenetic analyses were consistent with an important role for horizontal genetic exchange in the emergence of different porin classes and confirmed the close evolutionary relationships of the porins from N. meningitidis, N. gonorrhoeae, Neisseria lactamica, and Neisseria polysaccharea. Only members of this group contained three conserved lysine residues which form a potential GTP binding site implicated in pathogenesis. The model placed these residues on the inside of the pore, in close proximity, consistent with their role in regulating pore function when inserted into host cells.
The genus Neisseria comprises a number of species associated with mucosal carriage and disease in animals and humans (3, 26); these species include Neisseria meningitidis, a major cause of meningitis and septicemia worldwide (6), and Neisseria gonorrhoeae, the etiological agent of gonorrhoea (31). Comparative studies of these two pathogenic species with others not frequently associated with disease, some of which are closely related (16), are helpful in understanding both the pathogenesis of meningococcal and gonococcal infections and the evolutionary events that led to their emergence. Such studies are particularly valuable when they target cell components potentially involved in immunity or other host-pathogen interactions, such as outer membrane proteins (OMPs) (35–37).
The Neisseria porins, a distinct class within the porin superfamily (20), are major components of the outer membranes of all members of the genus Neisseria (11, 35, 37). Most Neisseria species express only one, referred to as Por, the meningococcus exceptionally expressing two, PorA and PorB (17). The gonococcus is the only other Neisseria species known to have a porA gene, which is not expressed due to frameshift and promoter mutations (11). The meningococcal and gonococcal porins are targets for serological typing schemes (14, 15), candidates for inclusion in vaccines (13), and have been implicated in pathogenesis (29).
Interest in the antigenic variability of the Neisseria porins, from both typing and vaccination perspectives, has resulted in the generation of many sequences of their genes. Detailed interpretation of this database of sequences has been hindered by the lack of a molecular structure for the gene products, although a two-dimensional model has been proposed on the basis of sequence similarity (23, 36). This model comprises a β-barrel porin structure, similar to that of the Escherichia coli porins (9), with regions that are relatively conserved among Neisseria sequences forming the β barrel. Other regions, which are less well conserved among and within species, form surface-exposed loops which protrude from the bacterial surface into the surrounding environment (23, 36). While this model was useful, it was not three-dimensional, being proposed before the molecular structures of the E. coli porins were established; the assignment of residues to membrane-embedded or surface-exposed environments was somewhat uncertain.
In this work, we have used molecular modelling techniques to generate a three-dimensional homology model of the Neisseria porins using E. coli porin structures, enabling us to identify more precisely the structural roles of individual residues. Phylogenetic analyses of the porins from different Neisseria species were enhanced when the model was used to assist sequence alignment and to identify structurally conserved parts of the proteins, identifying features likely to be important in the emergence of the pathogenic Neisseria species.
MATERIALS AND METHODS
Neisseria strains and sequences from databases.
The Neisseria porin sequences were obtained by translation of nucleotide sequences obtained by direct nucleotide sequence determination in this study or from GenBank, as detailed in Table 1. The porin of Neisseria polysaccharea was excluded from the analysis, as it was essentially identical to the Neisseria lactamica porin. Multiple examples of the porins were not included, with the exception of five additional N. gonorrhoeae porins classified elsewhere as “intermediate” and “hybrid ”(8) and both Neisseria sicca porins, which were not closely similar.
TABLE 1.
Porin sequences analyzeda
| Designation used in this work | Description | GenBank nucleotide accession no. | Source or reference(s)b |
|---|---|---|---|
| Eco OmpF | E. coli OmpF porin | J01655 | 19 |
| Eco PhoE | E. coli PhoE porin | V00316 | 28 |
| Nme PorA | N. meningitidis PorA porin | X12899 | 2 |
| Ngo PorA | N. gonorrhoeae hypothetical translation of porA pseudogene | AJ223447 | 11 |
| Nel Por | N. elongata porin | AF121870 | ATCC 25295; this work |
| Nca Por | N. canis porin | AF121871 | NCTC 10296-H6; this work |
| Nmu Por | N. mucosa porin | AF121872 | NCTC 10777-M3; this work |
| Nfl Por2 | N. flavescens locus 2 | Y09311 | 34 |
| Nsi Por-2 | N. sicca porin allele 2 | Y09307 | 34, 35 |
| Nsi Por-1 | N. sicca porin allele 1 | X65461 | 37 |
| Nflava Por | N. flava porin | AF121873 | ATCC 14221; this work |
| Nfl Por1 | N. flavescens locus 1 | Y09310 | 34 |
| Nde Por | N. denitrificans porin | AF121874 | ATCC 14686; this work |
| Nan Por | N. animalis porin | AF121875 | NCTC 10212-NA10; this work |
| Nci Por | N. cinerea porin | AF121876 | NCTC Z10294-194; this work |
| Nme PorB2 | N. meningitidis class 2 porin | X67940 | 12; this work |
| Ngo PorB1b | N. gonorrhoeae PIB porin | X52823 | 5 |
| Nla Por | N. lactamica porin | X65533 | 37 |
| Ngo PorB1a | N. gonorrhoeae PIA porin | X58073 | Strain JSK 412 (serovar P1A18); unpublished data |
| Nme PorB3 | N. meningitidis class 3 porin | X59992 | 38 |
| AF015117 to AF015122 | N. gonorrhoeae porins including intermediate porin class and hybrid porins | AF015117 to AF015122 | 8 |
The porin sequences analyzed were obtained by translation of the appropriate nucleotide sequences.
Accession numbers for bacterial strains from the American Type Culture Collection (ATCC) or the National Collection of Type Cultures (NCTC) are given.
For the purposes of this work, a nomenclature system suggested previously for Neisseria porins (11) has been adopted and extended. As there is a porA pseudogene in the gonococcus, its expressed porin, previously identified as Por, has been renamed PorB for consistency with the meningococcal nomenclature. Within meningococcal and gonococcal PorB porins, distinct allele classes are distinguished with numbers: the meningococcal class 2 OMP becomes PorB2, and the class 3 OMP becomes PorB3. The gonococcal PIA porin becomes PorB1a, and the PIB protein becomes PorB1b. These designations were intended to maintain as much consistency with previous nomenclature as possible while indicating more accurately the relationships among the proteins and the genes that encode them. The porins of other species have been labelled Por, as there remains no evidence for a porA gene in these species. The two distinct porins present in Neisseria flavescens were identified as Por1 and Por2, as neither bore a close relationship to the meningococcal and gonococcal PorA and PorB proteins. For N. sicca, the designations Por-1 and Por-2 indicated the two alleles identified in this species, which were insufficiently diverse to be regarded as representative of distinct allele classes. Final designation of the N. flavescens and N. sicca porins requires further study.
Nucleotide sequence determination of Neisseria porin genes.
Chromosomal DNA was extracted from each of the Neisseria strains by the Isoquick DNA extraction procedure (Orca Research). The porin genes were amplified with PCR primers 27 and 28 (12), purified as described previously (10), and their sequences were determined on both strands with Big Dye terminators (PE Applied Biosystems). The reaction products were separated with an Applied Biosystems Prism 377 automated sequencer, and the sequences were assembled with the Staden sequence analysis package (33).
The nucleotide sequences were determined with the following oligonucleotide primers: Neisseria animalis, 27, 28, 8U, and 8L (12), ani-S1 (5′-ACGCGTAAAAGCCACCATCG-3′), ani-S2 (5′-AGAAGTCGTAATCAGCACC-3′), ani-S3 (5′-AGTACATCGCTTGACTCTGG-3′), and ani-S4 (5′-CAGCAGGTTGTTGGCATCG-3′); Neisseria canis, 27, 28, can-S1 (5′-GCTGGTAAACTGAGCACCC-3′), can-S2 (5′-GGGTGCTCAGTTTACCAGC-3′), can-S3 (5′-GGCTACCAATACACCAATGG-3′), can-S4 (5′-CCATTGGTGTATTGGTAGCC-3′), and ani-S2; Neisseria cinerea, 27, 28, 8U, 8L, cin-S1 (5′-GTACTAAACACACCTATGCC-3′), cin-S2 (5′-AGAAGTCGTAATCCGCACCG-3′), cin-S3 (5′-GGCTTCTTCGGCCAATATGC-3′), cin-S4 (5′-GCATATTGGCCGAAGAAGCC-3′), and cin-S5 (5′-CATACCGGCAGTGGTAACGG-3′); Neisseria denitrificans, 27, 28, 8U, and den-S1 (5′-AGAACCGTCAGTCAGGTCG-3′); Neisseria elongata, 27, 28, 8U, and 8L; Neisseria flava, 27, 28, fla-S1 (5′-CTTCTTCGGTCGCTACGC-3′) and fla-S2 (5′-AATGTATCCAAGTAGCCAGC-3′); and Neisseria mucosa, 27, 28, muc-S1 (5′-GGTAAACTGAATACCCAACTG-3′), muc-S2 (5′-CAGTTGGGTATTCAGTTTACC-3′), muc-S3 (5′-GGATACTGTCGGTACTTACCG-3′), and muc-S4 (5′-CGGTAAGTACCGACAGTATCC-3′).
Generation of PorA and PorB structural models.
The initial model of the PorA monomer was generated by MODELLER (release 3) (30), a sophisticated software package that calculates protein three-dimensional structure based on sequence alignments, spatial restraints, and stereochemical considerations. The models for the Neisseria PorA and PorB sequences were constructed with two E. coli structures, OmpF (Protein Database [PDB] accession no. 2omf) and PhoE (PDB accession no. 1pho), as templates. All parameters within MODELLER were set to their default values and, in order to generate the trimer, the loop regions corresponding to Gly 23 to Ile 59, Ala 188 to Ser 221, and His 248 to Asn 281 (numbers as in Fig. 1) were removed from the model. The model was subjected to 50 steps of conjugate gradient energy refinement, as implemented within X-plor version 3.1 (4), to remove poor contacts. This step was performed by inserting the model for the PorA monomer into the OmpF unit cell (P321 a,b = 118.50 Å; c = 52.70Å), effectively generating the full PorA trimer by crystallographic symmetry. After refinement, coordinates for the full trimer were assembled with PDBSET, as implemented with the CCP4 suite of crystallographic programs (7).
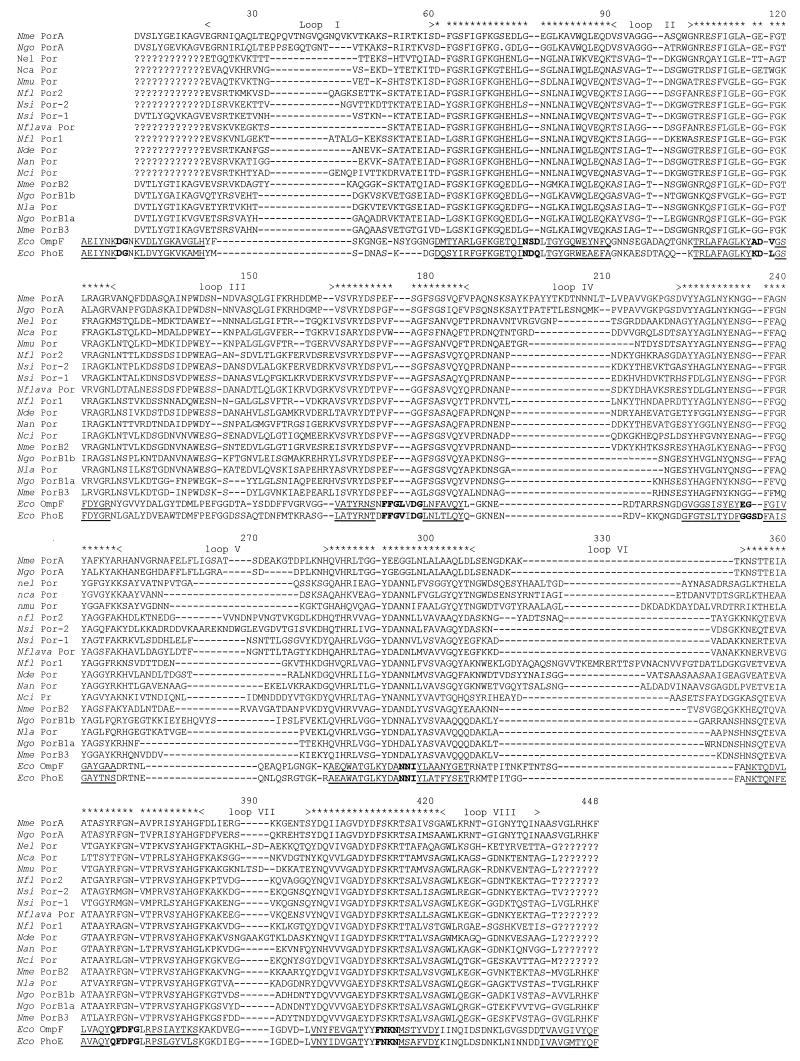
FIG. 1.
Alignment of Neisseria and E. coli porins included in this study. Eco OmpF, E. coli OmpF protein; Eco PhoE, E. coli PhoE protein; the designations of Neisseria porin sequences are as given in Table 1. A question mark indicates a region of the protein for which the sequence is unknown, and a dash indicates an alignment gap. The regions of the E. coli porins known from structural data to form the β strands and β turns of the β barrel are shown by underlining and bold text, respectively. < and >, putative loop regions (I to VIII) of the Neisseria porins; ∗, regions corresponding to parts of the β barrel for which data were available for all of the Neisseria porins and were used in phylogenetic analysis of only structural regions.
The PorB model was generated in the same way, from a PorB3 representative sequence, with the exception that only one loop region was deleted before the full trimer was built (Glu 22 to Val 60 inclusive; numbers as in Fig. 1). In this case, 30 cycles, rather than 50 cycles, of conjugate gradient energy minimization were performed.
Alignment of Neisseria porin gene sequences.
The PorA and PorB models were used in the alignment of the translated porin genes (Fig. 1) with the program SeqLab, part of the GCG sequence analysis software package (Wisconsin Package version 9.1; Genetics Computer Group, Madison, Wis.). Alignments were performed manually so as to maintain the reading frame and to ensure that the β strands identified in the models of PorA and PorB were minimally disrupted. Many alignment gaps were required to accommodate the differences in the lengths of the porins, the majority of these occurring in regions of the proteins predicted by the model to form surface loops. As a consequence of high sequence diversity, many of the loop region alignments were essentially arbitrary.
Phylogenetic analyses.
Nucleotide sequence alignments derived from the peptide sequence alignments shown in Fig. 1 were analyzed with MEGA (22), which was used to generate nonsynonymous distances, estimated by the method of Nei and Gojobori (27), with deletions excluded in pairwise comparisons. In some analyses, only sites within regions predicted to form the β barrel of the protein were included to minimize the effect of the large diversity present in the loop regions. The distance matrices obtained were used to reconstruct phylogenetic trees by the neighbor-joining method; these phylogenies were represented as unrooted radial trees with the program TREEVIEW (data not shown). The distance data were also visualized as split graphs generated by use of the split decomposition analysis technique with the program SPLITSTREE version 2.4 (18).
Nucleotide sequence accession numbers.
The novel sequences determined in this study have been deposited in GenBank under accession no. AF121870 to AF121876 (Table 1).
RESULTS
Novel porin sequences and alignments.
The novel porin sequences obtained were consistent with those previously described for various Neisseria porins and are shown in Fig. 1, aligned with the sequences for the two E. coli porins with known three-dimensional structures and one example of each previously sequenced porin gene. The alignment was based on that of Jeanteur et al. (20), modified to take account of the additional structural information available (9). The initial alignment made was that of the E. coli porins and the meningococcal PorA and PorB proteins; this alignment was refined and the other Neisseria porins were added after construction of the structural homology models. Correct alignment of meningococcal PorA and PorB with the E. coli porins was critical for the generation of accurate models, and each putatively conserved residue in the E. coli crystal structures was examined to verify that there was a sound structural basis for its conservation between Neisseria and E. coli porins. The most striking feature of the alignment was that the Neisseria sequences began at the second β strand, within the β-barrel framework, effectively truncating the first β strand, unlike the situation for E. coli. This alignment provided more convincing homology models and was also justified from sequence variations within the Neisseria porins that would otherwise have occurred within the β-barrel framework.
Homology models.
Given the high degree of sequence divergence between the Neisseria and E. coli porins, the β-barrel framework of the modelled porins was constructed remarkably well. Both the PorA and the PorB models formed a well-defined main-chain hydrogen-bonding pattern characteristic of the β-sheet motif, although no hydrogen-bonding or backbone dihedral-angle restraints were used in the calculation. Larger loop regions were less well modelled by this procedure and showed a variety of conformations, depending on the starting alignment and other restraints. Loop regions within proteins are notoriously difficult to model, and a more reliable calculation of their conformations requires further experimental data. The PorA monomer model is illustrated in Fig. 2.
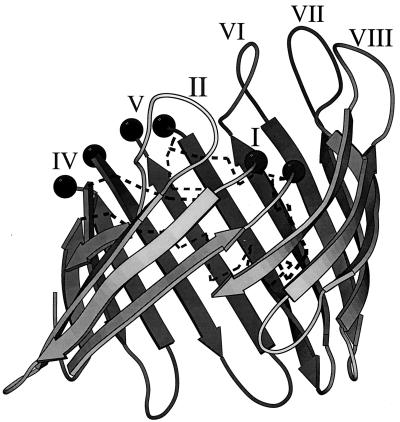
FIG. 2.
Molecular surface of a porin eyelet from the PorA model. The PorA model was calculated as described in Materials and Methods, and the diagram was generated with MOLSCRIPT (21). The positions of the external loop regions are indicated as I, II, and so forth, with the exception of loop III, which lies inside the protein and is shown by the broken line. The positions in space of the beginning and end residues of the longer loops (I, IV, and V), which were too long to be modelled reliably and were excluded from the final model, are marked with a solid sphere at the position of the Cα atom in the residue.
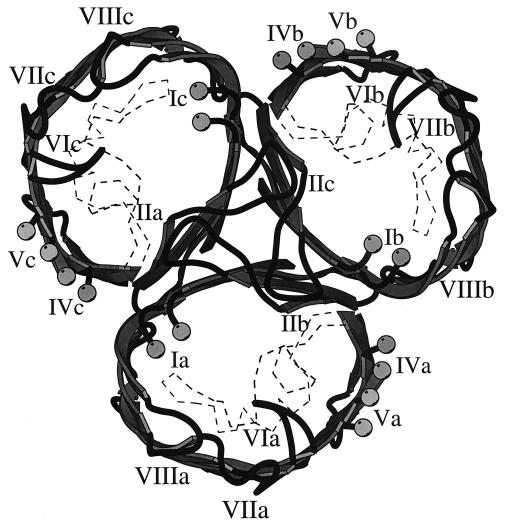
The limitations of the MODELLER package required that the porin models be calculated as monomers, although they exist in a trimeric state within the outer membrane (25). A more realistic model, which constructed the trimer from its constituent monomers, was produced with X-plor (4) by placing the monomers within the original trigonal crystallographic unit cell that was used for the E. coli OmpF porin. Several cycles of conjugate gradient energy refinement provided the final model, which was readily accommodated into a trimer without substantial modifications. The PorA trimer model is shown in Fig. 3, viewed from the top. A similar trimer was assembled within the OmpF crystal graphic unit cell with the PorB monomer model. The fact that the modelled PorA and PorB monomers fit well into the trigonal space group of the OmpF trimer with no major steric clashes indicated that these models were likely to be correct and, further, that the other Neisseria porins were likely to conform to these models.
FIG. 3.
Model of a PorA trimer, viewed from above. The positions of the external loop regions are marked as for Fig. 2, with the subunits designated a, b, and c. Other details are also as for Fig. 2.
The models had several structural features that are common to porins with known three-dimensional structures. Each model had a well-defined pore that was lined with hydrophilic and hydrophobic side chains of appropriate dimensions for the passage of low-molecular-weight solutes. The distribution of aromatic amino acid side chains within the PorA and PorB models was similar to that established for other porins: the majority of the Phe, Tyr, and Trp side chains point outward from the barrel and presumably would play an important role in interactions with phospholipids. A subset of these residues interact around the threefold symmetry axis of the PorA trimer and may play a role in stabilizing the quaternary structure of the porin.
Phylogenetic relationships of Neisseria porin genes.
A number of phylogenetic reconstructions were made with nucleotide sequence alignments based on the peptide sequence alignments shown in Fig. 1. The final phylogeny, shown in Fig. 4, was reconstructed with the following considerations. Due to the strategy used in sequencing, some of the porin genes were truncated; regions corresponding to the first 12 and last 8 amino acids were missing (12, 23). These regions were therefore excluded from all sequences for the phylogenetic analyses. As these regions were relatively small and located in parts of the protein that are relatively conserved, the consequent loss of phylogenetic resolution was minimal. Given the large distances among sequences, nonsynonymous distances, estimated as described previously (27), were used. Further, only those regions of the gene encoding the β barrel were included to remove any distortions of the phylogeny introduced by signals from the rapidly evolving antigenically variable loops (32). Finally, split decomposition, which does not assume a tree-like phylogenetic structure (1), was used to ensure that any signal implying that recombination had occurred in the evolution of the Neisseria porin genes was identified.
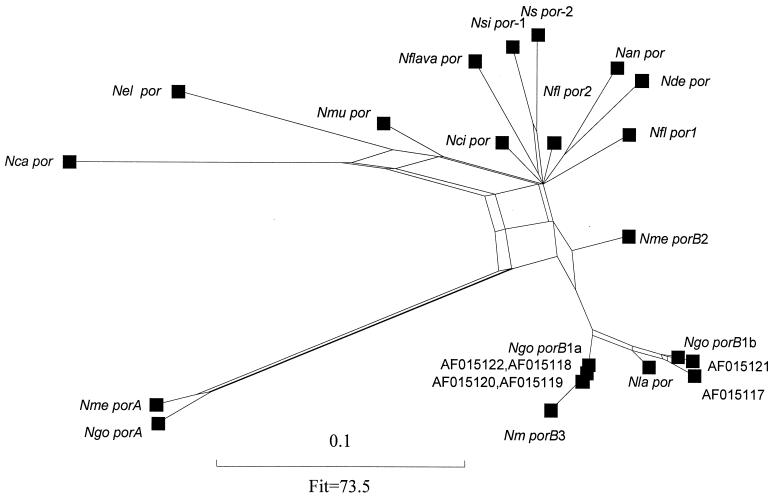
FIG. 4.
Phylogenetic analyses of the Neisseria porins. A refined split graph was generated with a distance matrix obtained by the method of Nei and Gojobori (27) from nucleotide sequence alignments derived from Fig. 1, including only residues corresponding to the amino acids marked with an asterisk. Branch lengths are drawn to scale, and the fit parameter is shown.
DISCUSSION
The antigenic variation of human pathogens remains incompletely understood, impeding vaccine development and complicating molecular typing, which relies on the characterization of such variation to distinguish isolates. An improved understanding of the complex interactions of the host immune system with variable antigens requires a multidisciplinary approach that integrates the inferences available from structural, immunological, and evolutionary data. At present, this integration is most readily achieved for protein antigens, where the relationships among genetic change in the pathogen, antigen structure, and interaction with the host immune system are relatively easily investigated. The porins of the genus Neisseria are an instructive example for a number of reasons, including their intrinsic interest, the large number of sequences available, and the fact that the genus includes both pathogenic and commensal species, whose lifestyle imposes constant selection pressure from the host immune response.
Neisseria porin antigens comprise regions of relatively conserved sequence, which are predicted to form the β-barrel structure of the proteins, interspersed with more variable regions, which form the putative surface-exposed loops. With two exceptions, these loops are highly variable within and among the Neisseria species. These exceptions, putative loops II and III, have structural roles in the three-dimensional model. Putative loop II is important in monomer-monomer interactions within the porin trimer, while putative loop III is sequestered in the pore of each monomer, potentially influencing pore function (Fig. 3). The other loops, which exhibit diversity in both length and sequence, are the determinants of antigenic variability in species that have been extensively studied. Loops I, IV, V, and VI (Fig. 1) show the greatest length variation. The hypervariable regions of meningococcal PorA, VR1 and VR2 (24), correspond to loop regions I and IV of the model (36). In the monomer PorA model, these loops appear to be widely spaced (Fig. 2), making it unlikely that the hypervariable regions would come into direct contact across a single monomer. However, the view of the PorA trimer from above (Fig. 3) indicates that the proposed locations of VR1 and VR2 (loops I and IV) are in close proximity between adjacent monomers, raising the possibility that these regions may interact to some extent across the monomer-monomer interface. In principle, therefore, a PorA epitope could extend across more than one polypeptide chain in a PorA trimer.
Comparison of the pairing of β strands and the positioning of the loop regions present in the three-dimensional PorA model with those predicted in the two-dimensional model of van der Ley et al. (36) showed good agreement for the locations of loops I, IV, V, VIII and for the β strands, with the exception of strands 1, 2, 4, 5, and 14. The deviations observed were due at least in part to the fact that the lengths of the β strands vary in the OmpF crystal structure, illustrating the additional information that a three-dimensional structure can add to a model based on sequence alignments.
The variability of the loop sequences, which is likely to be the result of strong positive selection (32), may distort the phylogenetic signal present in the sequences. One result would be to make closely related porins appear more diverse. For example, a recent study proposed hybrid and intermediate porin classes for the gonococcal porins (8). Including porins from other species, limiting the analysis only to those parts of the protein that form the β barrel, and applying more sensitive phylogenetic methods to the alignments generated here show that these proteins cluster closely with other members of their respective porin classes, the apparent differences being introduced mainly by the highly variable loop sequences (Fig. 4).
The phylogenetic relationships of the porin genes from most of the commensal and animal Neisseria species (N. animalis, N. canis, N. cinerea, N. dentirificans, N. flavescens, N. flava, and N. sicca) were reconstructed as a “star” phylogeny (Fig. 4), with many lineages emerging near the root. This configuration may be due to rapid population growth during the divergence of the different Neisseria species, positive selection as they specialized into different niches, high rates of intraspecies recombination, or simply a lack of phylogenetically informative changes for the appropriate branching events. High rates of recombination among species are unlikely, given the sequence diversity of the porin genes. Selection is also an unlikely explanation given that the star phylogeny is still present when only the structural regions of the proteins are included. Furthermore, as the lack of resolution is present when all sites are included (data not shown) or just those from the β-barrel-encoding regions (Fig. 4), it is unlikely that the star phylogeny is caused by the lack of a phylogenetic signal. We therefore propose that rapid population growth is the most likely explanation for this observation.
Whatever the cause of the star phylogeny, the porins of N. canis, N. elongata, and N. mucosa appear to share a common ancestor, occupying one branch of the star, as do those of N. denitrificans and N. animalis. The two N. sicca variants are diverse, perhaps indicating difficulties in the definition of this species. The two separate porin genes present in one N. flavescens isolate are sufficiently diverse that they may have been assembled in the same species relatively recently by horizontal genetic exchange.
Meningococcal PorA and gonococcal PorA are the most diverse members of the family of Neisseria porins and might constitute an outgroup relative to the other porins. The PorB porins of N. meningitidis and N. gonorrhoeae and the Por porin of N. lactamica (and the closely related N. polysaccharea), with the exception of meningococcal PorB2, form a well-resolved and supported bifurcating tree. The interrelationships of these porins indicate that interspecies genetic exchange of porin genes has occurred during the emergence of the present-day species. It is possible that the common ancestor of the meningococcus and the gonococcus possessed a gene ancestral to the porB1b and porB3 genes. The porB1b gonococcal porin gene appears to have arisen by the replacement of a porB1a allele with a gene sharing a common ancestor with the N. lactamica porin gene.
The meningococcal porB2 gene is a special case which appears to have arisen as a result of intragenic recombination during the evolution of the pathogenic Neisseria species. The split graph shows that PorB2 occupies an intermediate position between N. gonorrhoeae and N. meningitidis PorB and the N. lactamica porin on the one hand and the commensal-animal Neisseria porins on the other, with a network of relationships suggesting a phylogenetic signal that is inconsistent with a bifurcating tree. The data contributing to these relationships are illustrated in Fig. 5, which shows the amino acid identities within the β-barrel-forming regions among PorB2, PorB3, and the N. flavescens Por2 protein (similar results were obtained in comparisons of other representative porins from the commensal-animal and pathogenic groups) (data not shown). PorB2 was most similar to the N. flavescens porin (19 amino acid identities in structural regions) but was also related to PorB3 (17 amino acid identities). However, while the identities between PorB2 and the N. flavescens porin were distributed throughout the sequence, those between PorB2 and PorB3 were mainly localized in β sheets on either side of loop II. This region includes the three lysine residues (positions 66, 82, and 113 in Fig. 1) putatively identified as being involved in the binding of ATP and GTP. This binding provides a gating mechanism that down-regulates pore size and changes voltage dependence and ion selectivity when PorB proteins are inserted into mammalian cells (29). In the PorB model, the side chains of these three residues protrude into the center of the pore, providing a plausible steric explanation of the role of GTP in obstructing the passage of solutes and changing the selectivity of the porin. As this gating mechanism, which is only observed in PorB2, PorB3, PorB1a, PorB1b, and the N. lactamica and N. polysaccharea porins, is conserved, it is tempting to suppose that PorB proteins with this characteristic were important in the evolution of the pathogenic Neisseria species, perhaps by promoting intracellular invasion and thereby improving carriage in humans. The fact that there is a network of interrelationships among PorB2, the porins of the pathogenic Neisseria species, and the porins of the commensal-animal group suggests that PorB2 may have evolved by recombination of two proteins ancestral to the pathogenic and commensal porins, resulting in an antigenically different porin that retained the GTP binding site.
FIG. 5.
Comparison of variable amino acids present in the β-barrel-forming regions of the N. meningitidis PorB2 (Nme PorB2) and PorB3 (Nme PorB3) and N. flavescens Por2 (Nfl Por2) proteins. The numbers printed vertically above each residue in the alignment correspond to the position in the alignment shown in Fig. 1. The 18 residues which are identical between Nfl Por2 and Nme PorB2 are shown by black text on a grey background, the 16 residues which are identical between the two N. meningitidis porins are shown by white text on a black background, and the 11 residues which are identical between Nfl Por2 and Nme PorB3 are shown by white text on a grey background. The three lysine residues which form the PorB2 and PorB3 GTP binding sites are marked by asterisks.
While advances in automated sequence technology have enabled large numbers of nucleotide sequences of variable antigens to be sequenced rapidly, interpretation of these data continues to be limited by the lack of structural data, which remain comparatively difficult to obtain. The insights available from structural biology can enhance understanding of the evolutionary, immunological, and pathological significance of the gene sequence data. The approach taken in the present work, of combining experimental observations, structural modelling, and phylogenetic analyses, is valid, notwithstanding the wide sequence variation between the Neisseria and E. coli porins, as the β-barrel porin structure is well conserved in a wide variety of species. The porin model presented here not only extends the two-dimensional models proposed earlier, enabling the design of further structural and immunological analyses, but also permits more accurate phylogenetic analyses, providing insights into the biochemical mechanisms that may have contributed to the evolution of the pathogenic Neisseria species.
ACKNOWLEDGMENTS
J.P.D. is a fellow of the Lister Institute of Preventive Medicine. M.C.J.M. is a Wellcome Trust senior fellow in biodiversity.
We thank Eddie Holmes for comments on the manuscript and advice on the phylogenetic analyses.
REFERENCES
- 1.Bandelt H J, Dress A W. Split decomposition: a new and useful approach to phylogenetic analysis of distance data. Mol Phylogenet Evol. 1992;1:242–252. doi: 10.1016/1055-7903(92)90021-8. [DOI] [PubMed] [Google Scholar]
- 2.Barlow A K, Heckels J E, Clarke I N. The class 1 outer membrane protein of Neisseria meningitidis: gene sequence and structural and immunological similarities to gonococcal porins. Mol Microbiol. 1989;3:131–139. doi: 10.1111/j.1365-2958.1989.tb01802.x. [DOI] [PubMed] [Google Scholar]
- 3.Barrett S J, Sneath P H A. A numerical phenotypic taxonomic study of the genus Neisseria. Microbiology. 1994;140:2867–2891. doi: 10.1099/00221287-140-10-2867. [DOI] [PubMed] [Google Scholar]
- 4.Brunger A T. X-plor version 3.1: a system for X-ray crystallography and NMR. New Haven, Conn: Yale University Press; 1992. [Google Scholar]
- 5.Butt N J, Lambden P R, Heckels J E. The nucleotide sequence of the por gene from Neisseria gonorrhoeae strain P9 encoding outer membrane protein PIB. Nucleic Acids Res. 1990;18:4258. doi: 10.1093/nar/18.14.4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cartwright K A V. Meningococcal disease. Chichester, England: John Wiley & Sons, Inc.; 1995. [Google Scholar]
- 7.CCP4. The SERC (U.K.) Collaborative Computing Project No. 4, a suite of programmes for protein crystallography. Warrington, United Kingdom: Daresbury Laboratory; 1979. [Google Scholar]
- 8.Cooke S J, Jolley K, Ison C A, Young H, Heckels J E. Naturally occurring isolates of Neisseria gonorrhoeae, which display anomalous serovar properties, express PIA/PIB hybrid porins, deletions in PIB or novel PIA molecules. FEMS Microbiol Lett. 1998;162:75–82. doi: 10.1111/j.1574-6968.1998.tb12981.x. [DOI] [PubMed] [Google Scholar]
- 9.Cowan S W, Schirmer T, Rummel G, Steiert M, Ghosh R, Paupit R A, Jansonius J N, Rosenbusch J P. Crystal structures explain functional properties of two E. coli porins. Nature. 1992;358:727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- 10.Embley T M. The linear PCR reaction: a simple and robust method for sequencing amplified rRNA genes. Lett Appl Microbiol. 1991;13:171–174. doi: 10.1111/j.1472-765x.1991.tb00600.x. [DOI] [PubMed] [Google Scholar]
- 11.Feavers I M, Maiden M C J. A gonococcal porA pseudogene: implications for understanding the evolution and pathogenicity of Neisseria gonorrhoeae. Mol Microbiol. 1998;30:647–656. doi: 10.1046/j.1365-2958.1998.01101.x. [DOI] [PubMed] [Google Scholar]
- 12.Feavers I M, Suker J, McKenna A J, Heath A B, Maiden M C J. Molecular analysis of the serotyping antigens of Neisseria meningitidis. Infect Immun. 1992;60:3620–3629. doi: 10.1128/iai.60.9.3620-3629.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frasch C E. Meningococcal vaccines: past, present, and future. In: Cartwright K A V, editor. Meningococcal disease. Chichester, England: John Wiley & Sons, Inc.; 1995. pp. 246–283. [Google Scholar]
- 14.Frasch C E, Zollinger W D, Poolman J T. Serotype antigens of Neisseria meningitidis and a proposed scheme for designation of serotypes. Rev Infect Dis. 1985;7:504–510. doi: 10.1093/clinids/7.4.504. [DOI] [PubMed] [Google Scholar]
- 15.Gill M J. Serotyping Neisseria gonorrhoeae: a report of the Fourth International Workshop. Genitourin Med. 1991;67:53–57. doi: 10.1136/sti.67.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guibourdenche M, Popoff M Y, Riou J Y. Deoxyribonucleic acid relatedness among Neisseria gonorrhoeae, N. meningitidis, N. lactamica, N. cinerea, and “N. polysaccharea.”. Ann Inst Pasteur/Microbiol (Paris) 1994;137B:177–185. doi: 10.1016/s0769-2609(86)80106-5. [DOI] [PubMed] [Google Scholar]
- 17.Hitchcock P J. Unified nomenclature for pathogenic Neisseria species. Clin Microbiol Rev. 1989;2:S64–S65. doi: 10.1128/cmr.2.suppl.s64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huson D H. SplitsTree: a program for analysing and visualising evolutionary data. Bioinformatics. 1998;14:68–73. doi: 10.1093/bioinformatics/14.1.68. [DOI] [PubMed] [Google Scholar]
- 19.Inokuchi K, Mutoh N, Matsuyama S, Mizushima S. Primary structure of the ompF gene that codes for a major outer membrane protein of Escherichia coli K-12. Nucleic Acids Res. 1982;10:6957–6968. doi: 10.1093/nar/10.21.6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeanteur D, Lakey J H, Pattus F. The bacterial porin superfamily: sequence alignment and structure prediction. Mol Microbiol. 1991;5:2153–2164. doi: 10.1111/j.1365-2958.1991.tb02145.x. [DOI] [PubMed] [Google Scholar]
- 21.Kraulis P J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 22.Kumar S, Tamura K, Nei M. MEGA: molecular evolutionary genetics analysis software for microcomputers. Comput Appl Biosci. 1994;10:189–191. doi: 10.1093/bioinformatics/10.2.189. [DOI] [PubMed] [Google Scholar]
- 23.Maiden M C J, Suker J, McKenna A J, Bygraves J A, Feavers I M. Comparison of the class 1 outer membrane proteins of eight serological reference strains of Neisseria meningitidis. Mol Microbiol. 1991;5:727–736. doi: 10.1111/j.1365-2958.1991.tb00743.x. [DOI] [PubMed] [Google Scholar]
- 24.McGuinness B T, Barlow A K, Clarke I N, Farley J E, Anilionis A, Poolman J T, Heckels J E. Deduced amino acid sequences of class 1 protein PorA from three strains of Neisseria meningitidis. Synthetic peptides define the epitopes responsible for serosubtype specificity. J Exp Med. 1990;171:1871–1882. doi: 10.1084/jem.171.6.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minetti C A S A, Tai J Y, Blake M S, Pullen J K, Liang S M, Remeta D P. Structural and functional characterization of a recombinant PorB class 2 protein from Neisseria meningitidis. Conformational stability and porin activity. J Biol Chem. 1997;272:10710–10720. doi: 10.1074/jbc.272.16.10710. [DOI] [PubMed] [Google Scholar]
- 26.Morse S A, Knapp J S. The Genus Neisseria. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. 2nd ed. New York, N.Y: Springer-Verlag; 1992. pp. 2495–2559. [Google Scholar]
- 27.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 28.Overbeeke N, Bergmans H, van Mansfeld F, Lugtenberg B. Complete nucleotide sequence of phoE, the structural gene for the phosphate limitation inducible outer membrane pore protein of Escherichia coli K12. J Mol Biol. 1983;163:513–532. doi: 10.1016/0022-2836(83)90110-9. [DOI] [PubMed] [Google Scholar]
- 29.Rudel T, Schmid A, Benz R, Kolb H A, Lang F, Meyer T F. Modulation of Neisseria porin (PorB) by cytosolic ATP/GTP of target cells: parallels between pathogen accommodation and mitochondrial endosymbiosis. Cell. 1996;85:391–402. doi: 10.1016/s0092-8674(00)81117-4. [DOI] [PubMed] [Google Scholar]
- 30.Sali A, Potterton L, Yuan F, van Vlijmen H, Karplus M. Evaluation of comparative protein modeling by MODELLER. Proteins. 1995;23:318–326. doi: 10.1002/prot.340230306. [DOI] [PubMed] [Google Scholar]
- 31.Sherrard J S, Bingham J S. Gonorrhoea now. Int J STDs AIDS. 1995;6:162–166. doi: 10.1177/095646249500600304. [DOI] [PubMed] [Google Scholar]
- 32.Smith N H, Maynard Smith J, Spratt B G. Sequence evolution of the porB gene of Neisseria gonorrhoeae and Neisseria meningitidis: evidence of positive Darwinian selection. Mol Biol Evol. 1995;12:363–370. doi: 10.1093/oxfordjournals.molbev.a040212. [DOI] [PubMed] [Google Scholar]
- 33.Staden R. The Staden sequence analysis package. Mol Biotechnol. 1996;5:233–241. doi: 10.1007/BF02900361. [DOI] [PubMed] [Google Scholar]
- 34.Suker J. Variation of meningococcal porin antigens. Ph.D. thesis. London, England: University of London; 1997. [Google Scholar]
- 35.Suker J, Feavers I M, Maiden M C J. Structural analysis of the variation in the major outer membrane proteins of Neisseria meningitidis and related species. Biochem Soc Trans. 1993;21:304–306. doi: 10.1042/bst0210304. [DOI] [PubMed] [Google Scholar]
- 36.van der Ley P, Heckels J E, Virji M, Hoogerhout P, Poolman J T. Topology of outer membrane porins in pathogenic Neisseria spp. Infect Immun. 1991;59:2963–2971. doi: 10.1128/iai.59.9.2963-2971.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ward M J, Lambden P R, Heckels J E. Sequence analysis and relationships between meningococcal class 3 serotype proteins and other porins of pathogenic and non-pathogenic Neisseria species. FEMS Microbiol Lett. 1992;94:283–290. doi: 10.1016/0378-1097(92)90644-4. [DOI] [PubMed] [Google Scholar]
- 38.Wolff K, Stern A. The class 3 outer membrane protein (PorB) of Neisseria meningitidis: gene sequence and homology to the gonococcal porin PIA. FEMS Microbiol Lett. 1991;83:179–185. doi: 10.1016/0378-1097(91)90351-a. [DOI] [PubMed] [Google Scholar]