Abstract
Purpose
Hepatocellular carcinoma is the most frequent liver cancer and constitutes one of the main causes of cancer mortality. The combination of targeted therapy drugs, such as selumetinib and perifosine that inhibit cell signaling pathways involved in cell survival and proliferation, with the expression of tumor suppressor transgenes, such as PTEN, may result in an efficient therapeutic approach against HCC. Thus, the main objective of this work was to develop a new lipid-polymer hybrid nanosystem (HNP), composed of a PLGA core coated with a pH-sensitive lipid bilayer functionalized with the targeting ligand GalNAc, in order to specifically and efficiently deliver this novel combination of therapeutic agents in HCC cells.
Methods
Transmission electron microscopy, zeta potential, Fourier transform infrared spectroscopy, and dynamic light scattering were used to determine the physicochemical properties of hybrid nanosystems and their components. The biological activity and specificity of nanosystems were evaluated using luminescence and flow cytometry. A variety of techniques were used to assess the therapeutic activity of hybrid nanosystems, including the Alamar Blue assay for cell viability; flow cytometry for cell death mechanisms, mitochondrial membrane potential and cell cycle; luminescence for caspase activity; flow cytometry and fluorescence microscopy for cell proliferation; and Western blot for molecular targets levels.
Results
The obtained results showed that this new hybrid nanosystem not only has a high loading capacity of both drugs, but also allows for substantial expression of the PTEN transgene. In addition, the developed formulation has high stability, adequate physicochemical properties and high specificity to HCC cells. Moreover, the achieved data revealed that this innovative nanosystem presents a high antitumor effect, demonstrated not only by the enhancement on the programmed cell death, but also by the reduction in cell proliferation capacity.
Conclusion
The generated formulation shows a high anticancer effect, demonstrating a high translational potential for future clinical application in HCC treatment.
Keywords: hybrid nanosystem, gene and drug delivery, chemotherapy, gene therapy, hepatocellular carcinoma
Graphical Abstract
Introduction
About ninety percent of primary liver tumors, which constitute the third main cause of cancer mortality worldwide, are hepatocellular carcinomas (HCC).1–3 The treatment of HCC depends not only on the stage at which the disease is diagnosed, but also on the patient’s baseline clinical conditions. Liver transplantation, surgical resection, chemotherapy, targeted therapy and immunotherapy are the main therapeutic strategies for HCC.4 Over the years, several studies involving immunotherapy, targeted therapy, alternative therapies, gene therapy, among others, have been carried out in order to develop a more effective treatment against HCC.5–8 However, the prognosis for HCC remains poor in all regions of the world. The incidence and mortality rates are approximately equivalent, being the five-year survival rate for HCC around 18%. The development of HCC is influenced by numerous risk factors, such as exposure to toxic compounds (alcohol or aflatoxin B), hepatitis infections (HBV or HCV), as well as metabolic and genetic/epigenetic alterations, that can change several important cell signaling pathways and the expression profile of oncogenes and tumor suppressor genes, leading to HCC.
One of the most critical signaling pathways involved in the initiation and development of HCC is the PI3K/AKT/mTOR pathway, which is essential to the proliferation and survival of tumour cells.9,10 In non-cancerous tissues, this pathway activity is under control of the tumour suppressor protein PTEN. However, alterations in PTEN expression and function are frequently detected in HCC, leading to hyperactivation of the PI3K/AKT/mTOR pathway. In fact, most HCC patients present high levels of AKT phosphorylation, which result in early recurrence and poor prognosis.11
On the other hand, RAF/MEK/ERK pathway is a ubiquitous signal transduction pathway that regulates crucial cellular processes, including differentiation, angiogenesis, proliferation and cell survival.12 It is known that in this cancer disease the RAF/MEK/ERK pathway is constitutively active and that there is a very significant increase in mitogen-activated protein kinase 1 and 2 (MEK1/2) phosphorylation compared to normal liver tissues, suggesting its involvement in tumorigenesis.13,14
Several works have shown that increasing PTEN expression not only significantly reduces the proliferation of cancer cells, but also makes them more sensitive to targeted therapy drugs such as sorafenib.15,16 On the other side, Phase 2 clinical trials are presently testing perifosine, an oral bioactive alkylphospholipid. Perifosine interferes with the pleckstrin homology domain of AKT inhibiting its activity.17,18 Studies have shown that perifosine induces cell cycle arrest and promotes apoptosis of cancer cells by blocking AKT phosphorylation.19 In turn, selumetinib strongly and selectively inhibits MEK1/2. Several studies have shown that this MEK1/2 inhibitor blocks growth and promotes apoptosis of cancer cells, suppressing tumor growth in HCC xenografts.7,20 Since hepatocellular carcinoma is a highly complex and difficult-to-treat disease, the generation of combined and multitarget therapies can result in a superadditive therapeutic activity. In this regard, a combined strategy involving perifosine, selumetinib and the expression of PTEN transgene could induce a high anticancer effect. However, both perifosine and selumetinib present associated toxicity that results in very adverse side effects,21,22 likewise genetic material encoding PTEN is not able to efficiently and selectively target HCC cells on its own.
In this regard, nanotechnology can have a decisive role in circumventing these limitations, as it enables the generation of functionalized nanostructures that allow the simultaneous transport and delivery of one or more therapeutic agents, improving their stability, prolonging their systemic circulation half-life and releasing them in the target tissue at a controlled rate, consequently increasing their therapeutic efficiency and minimizing the associated side effects. The therapeutic use of nucleic acids is challenging, not only because of their unfavorable physicochemical characteristics, which prevent their efficient internalization in target cells by themselves, but also because they are vulnerable to enzymatic degradation in the bloodstream and undergo rapid renal clearance.23,24 In this respect, poly (lactic-co-glycolic acid) (PLGA) nanoparticles and liposomes have been extensively studied as drug and gene delivery systems. Both liposomes and PLGA nanoparticles exhibit long circulation times, ability to efficiently load different types of molecules, biocompatibility, biodegradability and physicochemical versatility.25,26 However, they also present exclusive characteristics. PLGA nanoparticles allow the control of the degradation rate, just by changing the lactic acid/glycolic acid molar ratio, in addition to permit the loading of molecules with different physicochemical properties.27 Regarding the liposomes, the choice of the lipid type and concentration used for their formulation allows them to have different surface chemistries and responsiveness to a diverse range of stimuli, such as pH and temperature, that permits to obtain a controlled release of the encapsulated molecules.28,29 In turn, lipid-polymer hybrid nanosystems are designed to merge the advantageous features of liposomes with those of PLGA nanoparticles.30 The hybrid structure of these formulations allows the simultaneous transport and delivery of different therapeutic molecules, great drug loading capacity, high serum stability, surface functionality, and adjustable release profile.31 Moreover, they have shown high ability to overcome multidrug resistance (MDR), which is increasingly common in cancer, thus promoting an enhanced therapeutic response.7,32 Furthermore, these nanosystems can also induce high therapeutic efficacy with reduced toxicity, since coating by targeting ligands allows them to perform a specific delivery of the therapeutic agents in target cells.
In this context, the functionalization of nanosystems with targeting ligands to the asialoglycoprotein receptor has allowed the specific and efficient delivery of therapeutic molecules to various HCC models.6,7 The asialoglycoprotein receptor, a plasma membrane protein found in hepatocytes, regulates the levels of glycoproteins that expose alpha 2,6-linked sialic acid residues at the terminals. This receptor has an excellent binding capacity to terminal carbohydrate residues of D-galactose and N-acetyl-D-galactosamine, promoting their efficient cellular uptake.7 Practically non-existent in other tissues, it is expressed in the liver and overexpressed in HCC.33,34 We already proved that the incorporation of triantennary N-acetylgalactosamine (GalNAc) cluster, which has a great binding ability to this receptor, in a new lipid-polymer hybrid formulation results in specific and efficient drug delivery in HCC cells.7
In the present study, we developed a new lipid-polymer hybrid nanocarrier constituted of PLGA nanoparticles sequentially coated with genetic material and a pH-sensitive lipid bilayer containing DSPE-PEG(2000)-GalNAc. This targeted formulation was developed to mediate a new therapeutic strategy to HCC, involving the combination of selumetinib, perifosine and the expression of PTEN transgene, allowing a specific and simultaneous drug and gene delivery to target cells, in order to induce high therapeutic efficiency and reduced side effects.
Experimental Section
Materials
Poly(d,l-lactic-co-glycolic acid) (PLGA) 50:50 DL (24–38kDa), polyvinyl alcohol (PVA) (MW 9000–10,000, 80% hydrolyzed), didodecyldimethylammonium bromide (DMAB), ethyl acetate, acetone, JC-1 probe, fluorescein diacetate, propidium iodide, and perifosine were obtained from Sigma-Aldrich. Sulfo-Cyanine3 azide, FAM azide 6-isomer, and 5-ethynyl-2′-deoxyuridine (EdU) were acquired from Lumiprobe. 1-palmitoyl-2-oleoyl-sn-glycero-3-ethylphosphocholine (EPOPC), cholesterol (Chol), cholesteryl hemisuccinate (CHEMS), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), N-palmitoyl homocysteine (PHC), and 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[amino(polyethylene glycol)-2000] (DSPE-PEG2000) were obtained from Avanti Polar Lipids. Triantennary N‐acetylgalactosamine (GalNAc) cluster was gently provided by Ionis Pharmaceuticals and selumetinib was generously given by AstraZeneca.
Nanoparticles Production
In this work, the solvent emulsification and diffusion-evaporation method was used to produce the PLGA nanoparticles.35 Succinctly, 50 mg of PLGA 50:50 block copolymer (24–38 kDa) solution in 2.5 mL ethyl acetate was dispensed in 5 mL of aqueous phase having various quantities of PVA and/or DMAB and stirred for 30 minutes, to get a primary emulsion, using a homogenizer (T 10 basic, Ultra Turrax, IKA). In order to extract the organic solvent, the main emulsion was diluted with water at 50 °C and agitated overnight. For uptake studies and for studies with selumetinib, 50 µg of coumarin-6 or 1 mg of selumetinib, respectively, were dissolved in the organic phase. Nanoparticles were recovered by ultracentrifugation at 17,000 g which allowed the removal of non-encapsulated selumetinib in PLGA nanoparticles.
Large unilamellar liposomes (LUV) were prepared by extrusion of multilamellar liposomes (MLV) composed of: 1:1 (molar ratio) mixtures of 1-palmitoyl-2-oleoyl-sn-glycero-3-ethylphosphocholine (EPOPC) and cholesterol (Chol); 0.6:1:0,4 (molar ratio) mixtures of EPOPC, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) and cholesteryl hemisuccinate (CHEMS); or 0.6:1:0,4 (molar ratio) mixtures of EPOPC, DOPE and N-palmitoyl homocysteine (PHC). Briefly, lipids dissolved in chloroform were mixed and dried under vacuum in a rotator evaporator. Then, deionized water was used to hydrate the dried lipid films and the resultant MLVs were subsequently sonicated for three minutes. For drug studies, different molar ratios of perifosine (relative to the total lipid amount) were added in chloroform.
Liposomes containing the GalNAc ligand were prepared by the postinsertion method.36 The DSPE-PEG2000-amine micelles dissolved at a concentration of 1.1mM in sodium tetraborate buffer at pH 8.5 were coupled to 3 equivalent of GalNAc PFP ester dissolved in acetone. The coupling reaction was performed overnight with stirring. After evaporation of acetone, the GalNAc O-acetyl groups were removed by the addition of aqueous ammonia. Micelles were purified by size exclusion chromatography on a Sephadex G-25 M (GE Healthcare) column using water as running buffer to remove unreacted GalNAc. The lyophilized product was analysed by Jasco FTIR-420 (Jasco, USA), and 32 scans were collected at a resolution of 2 cm−1 in the range of 4000–650 cm−1.
The insertion of DSPE-PEG-GalNAc conjugate or plain DSPE-PEG-amine on the preformed liposomes, at 3 mol % (relative to the total lipid amount), was done through micelles incubation with liposomes in a water bath at 40 °C for 16 hours under agitation.
The PLGA nanoparticles/DNA complexes were prepared by sequentially mixing 1 μg of pCMVluc DNA plasmid encoding luciferase (a gift of Dr. P. Felgner, Vical, CA, USA), or 1 μg of pCMVgfp DNA plasmid encoding green fluorescent protein (GFP) (Clontech, CA, USA), or 1 μg of the DNA plasmid pcDNA3-FLAG PTEN (Addgene) with PLGA nanoparticles (the volume necessary to obtain the desired PLGA nanoparticles/DNA (±) charge ratio). The mixtures were then incubated for 15 min at room temperature. The PLGA nanoparticles/DNA complexes were used immediately after being prepared.
Hybrid nanosystems (HNP) were prepared by adding a dispersion of PLGA nanoparticles/DNA complexes, containing 1 μg of DNA, to preformed liposomes containing DSPE-PEG-GalNAc or DSPE-PEG-amine (the volume required to obtain the desired DNA/lipid mass ratio). The mixture was agitated for 1 hour at 60 °C (T> Tm) and 300 rpm.
In the case of PEI/DNA complexes (polyplexes), they were prepared by mixing 1 μg of pCMVluc or 1 μg of pCMVgfp with polyethylenimine (PEI) (the volume necessary to obtain the desired PEI/DNA (±) charge ratio). The mixtures were then incubated for 15 min at room temperature. The polyplexes were used immediately after being prepared.
Physicochemical Characterization of Nanoparticles
Transmission electron microscopy, zeta potential, hydrodynamic diameter, and polydispersity index were evaluated to accomplish the physicochemical characterization of the HNP and their components, as we have previously described.7 Regarding the hydrodynamic diameter and polydispersity index, a Zetasizer Nano-ZS (Malvern Instruments Ltd., UK) was used to measure the dynamic light scattering (DLS) of nanoparticles and their components and, then, DLS measurements were translated into average hydrodynamic diameter and polydispersity index using the software Zetasizer 7.02. The zeta potential was also evaluated using a Zetasizer Nano-ZS (Malvern Instruments Ltd., UK) coupled to laser Doppler electrophoresis and determined using a Smoluchovski model. The analyses were performed at 25 °C, in water, and at a backward scattering angle of 173°.
Transmission electron microscopy was used to analyze the size and structure of nanosystems. The nanosystems were adsorbed on copper grids covered with formvar film and samples contrast was ensured by using uranyl acetate. A JEM 1400 Transmission Electron Microscope (Jeol, Tokyo, Japan) was employed to examine the grids. Images were digitally recorded using a SC1000 ORIUS CCD camera (Gatan, PA, USA) and analysed with an EDX Microanalysis System (Oxford Instruments, Abingdon, UK).
Ethidium Bromide Intercalation
The accessibility of ethidium bromide to the DNA of PLGA nanoparticles/DNA complexes and HNP was analyzed by fluorescence measurements in a SPECTRAmax GEMINI EM fluorometer (Molecular Devices, CA, USA). PLGA nanoparticles/DNA complexes and HNP, prepared at pH 5 or pH 7.4, containing 0.5 μg of DNA were mixed with EtBr in order to obtain a final EtBr concentration of 400nM. Following 10 min incubation, fluorescence was measured with excitation and emission wavelengths of 518 and 605 nm, respectively. The fluorescence scale was calibrated so that residual fluorescence corresponded to EtBr fluorescence with a final concentration of 400nM, and the fluorescence obtained with 0.5 μg of free DNA (control), in the presence of 400nM EtBr, corresponded to 100%. The amount of DNA available to interact with the probe was calculated by subtracting the value of residual fluorescence from those obtained for the samples and expressed as the percentage of the control.
Agarose Gel Electrophoresis Assay
To evaluate the DNA complexation in the PLGA nanoparticles/DNA complexes, agarose gel electrophoresis was performed. Nanosystems were produced as referred above and then mixed with loading buffer (15% v/v Ficoll® 400, 0.05% w/v bromophenol blue, 1% w/v SDS, 0.1 M EDTA, pH 7.8). The samples were then transferred to a 1% agarose gel prepared in TBE solution (89 mm Tris-buffer (pH 8.6), 89 mm boric acid, and 2.5 mm EDTA) and containing 1 μg/mL of EtBr. Electrophoresis was then performed at 100 mV for 60 minutes. Visualization of samples on the gel was performed in a GelDoc® system (BioRad®, USA) using QuantityOne® software.
In vitro Release Studies
The in vitro release studies were performed using a dialysis membrane, with a molecular weight cut-off of 14,000 Da, against phosphate buffer solutions with two different pH values (5 and 7.4). Briefly, 50 mg of HNP loaded with coumarin-6 were added to a dialysis bag which was then introduced in 50 mL of release medium at 37 °C and stirred continuously at 250 rpm. Small aliquots (1 mL) of release medium were removed over time and replaced with the same volume of fresh medium. Then, the aliquots were lyophilized, dissolved in DMSO and the fluorescence intensity was measured in a SPECTRAmax GEMINI EM fluorometer (Molecular Devices, CA, USA). The concentration of the probes was determined by comparing the fluorescence intensity of the samples to that of the standards (calibration curves) submitted to the same conditions.
Cell Cultures
HepG2 and HuH-7 human hepatocellular carcinoma cell lines were cultured in Dulbecco’s modified Eagle’s medium-high glucose (DMEM-HG) (Sigma–Aldrich, MO, USA) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS) (Alfagene), 100 U/mL penicillin, and 100 mg/mL streptomycin. The cells were maintained at 37 °C and 5% CO2. The HepG2 and HuH-7 cells, were seeded at a density of 60 × 103 cells/cm2 and 65 × 103 cells/cm2, respectively. The cell lines were obtained from ATCC.
Spheroid Cultures
The cells suspended in complete culture medium were seeded at a density of 6×103 cells/well in 96-well round-bottomed ultra-low attachment microplates (Corning® Costar®) to create spheroids. The plates were then incubated at 37 °C for a minimum of three days in a humidified atmosphere with 5% CO2.
Transfection Activity
For luminescence evaluation of luciferase expression, twenty-four hours before incubation with the nanosystems, HepG2 and HuH-7 cells were seeded in 48-well culture plates. The different formulations of nanosystems containing the pCMVluc plasmid were added to HepG2 or HuH-7 cells, previously covered with DMEM-HG medium. In the competition studies, 30 min after incubating cells with DMEM-HG medium, containing or not 1.5 mg/mL of asialofetuin, the nanosystems were added to each well. After 48 hours incubation (5% CO2 at 37 °C), cells were washed twice with phosphate-buffered saline solution (PBS) and lysed with lysis buffer (1 mM dithiothreitol; 1 mM ethylenediaminetetraacetic acid; 25 mM Tris-phosphate (pH 7.8); 8 mM MgCl2; 15% glycerol; 1% (v/v) Triton X-100). The levels of luciferase expression in the lysates were quantified through measurement of light production by luciferase in a Synergy HT luminometer (Biotek, USA). The protein content of the lysates was measured by the DC Protein Assay reagent (Bio-Rad CA, USA) using bovine serum albumin (BSA) as the standard. The data were expressed as relative light units of luciferase per milligram of total cell protein.
For flow cytometry analysis of GFP expression, twenty-four hours before incubation with the nanosystems, HepG2 and HuH-7 cells were seeded in 12-well culture plates. Nanosystems containing 4 μg of pCMVgfp plasmid were added to HepG2 or HuH-7 cells, previously covered with DMEM-HG medium. After 48 h incubation (5% CO2 at 37 °C), cells were washed twice with PBS and detached with trypsin (5 min at 37 °C). After that, cells were washed and resuspended in PBS, and immediately analysed in a FACSCalibur flow cytometer (Becton Dickinson, NJ, USA). Live cells were gated by forward/side scattering from a total of 20000 events and data was analysed using CellQuest software.
Cellular Uptake
Twenty-four hours before incubation with HNP, HepG2 and HuH-7 cells were seeded in 12-well culture plates. In the competition studies, 30 min after incubating cells with 1.7 mL of DMEM-HG medium containing or not 1.5 mg/mL of asialofetuin, the hybrid nanosystems, prepared with coumarin-6, were added to each well. Following 4 hours incubation at 37 °C, cells were washed twice with PBS, detached, and resuspended in PBS. Then, the samples were evaluated by flow cytometry (FACSCalibur flow cytometer, Becton Dickinson, NJ, USA). Live cells were gated by forward/side scattering from a total of 20,000 events and data was analysed using CellQuest software.
Cell Viability
The Alamar Blue assay was selected to assess the cytotoxicity of the different formulations as we previously performed.7 This assay measures the redox capacity of the cells due to the production of metabolites as a result of cell growth. Briefly, after incubation with nanosystems for 48 or 72 hours, DMEM-HG medium containing 10% (v/v) alamar blue dye was added to each well. This medium was prepared from a 0.1 mg/mL stock solution of alamar blue. Then, after one hour of incubation at 37 °C, the absorbance was determined at 570 and 600 nm in a SPECTRAmax PLUS 384 spectrophotometer (Molecular Devices, CA, USA). Cell viability (as a percentage of untreated control cells) was calculated according to the formula (A570 - A600) of treated cells x 100 / (A570 - A600) of control cells.
Cell Death
To evaluate the cell death levels promoted by the various therapeutic approaches, flow cytometry was employed by using the probes FITC-Annexin V (Immunostep, Salamanca, Spain) and propidium iodide (PI, Sigma) in agreement with our established procedure.7 After a 72-hour HNP incubation period, the cell media and detached cells were harvested, washed with ice-cold PBS, and resuspended in 100 μL of binding buffer (10 mm HEPES (pH 7.4), 2.5 mm CaCl2, 140 mm NaCl), to which 1 µL of PI (0.05 mg/mL) and 2 µL of FITC-annexin V (0.05 mg/mL) were added. The samples were analysed in a FACSCalibur flow cytometer (Becton Dickinson, NJ, USA) after being incubated for five minutes at room temperature in the dark. Twenty thousand events were collected, propidium iodide was evaluated in the FL-3 channel, and FITC fluorescence was assessed in the FL-1 channel. The data were analysed using CellQuest software.
Caspases Activity
The activities of caspase-3/7, −8 and −9 were measured by a luminescence assay, using Caspase-Glo 3/7, Caspase-Glo 8 and Caspase-Glo 9 assay kit (Promega, USA).7 The evaluation was carried out in compliance with the manufacturer’s protocol. Twenty-four hours after HepG2 and HuH-7 cells have been plated on 96-well plates, they were incubated with the different nanosystems for 72 hours. In the case of spheroids, after 3 days of spheroid formation, they were incubated (5% CO2 at 37 °C) with the various therapeutic strategies for 12 days. Following that, each well received 100 µL of assay reagent, being then incubated for one hour at room temperature. Finally, a Synergy HT luminometer (Biotek, USA) was used to measure the luminescence.
Mitochondrial Membrane Potential
JC-1 fluorescent dye was used to study the mitochondrial membrane potential through flow cytometry as described previously.7 Briefly, 24 hours after HepG2 and HuH-7 cells have been plated on 12-well plates, they were incubated with the different nanosystems for 48 hours. Then, the cells were resuspended and incubated with fresh culture medium having JC-1 dye (2.5 µg/mL), for 40 minutes at 37 °C in the dark. Following incubation, the cells were washed with PBS and examined using a FACSCalibur flow cytometer (Becton Dickinson, NJ, USA). The decrease in mitochondrial membrane potential was indicated by the shift from red (JC-1 oligomer) to green (JC-1 monomer) fluorescence.
Cell Proliferation
Cell proliferation was measured by incorporation of 5-ethynyl-2’-deoxyuridine (EdU).37,38 Four hours before the end of the 48 hours treatment with the different therapeutic strategies, HepG2 and HuH-7 cells were incubated with 10 µM EdU (5% CO2 at 37 °C) for 4 hours. After this incubation the cells were detached with trypsin and washed with PBS, and then fixed with 70% (v/v) ethanol and incubated overnight at 4 °C. Posteriorly, the cells were washed with PBS containing 2% (m/v) BSA. Cell pellets were collected by centrifugation, resuspended and incubated in the dark for 30 minutes with a reaction cocktail containing 5 μM FAM azide 6-isomer, 2 mm copper sulfate pentahydrate and 20 mg/mL ascorbic acid. Finally, the cells were washed and resuspended in PSB and analysed in a FACSCalibur flow cytometer (Becton Dickinson, NJ, USA). Emitted FAM fluorescence was measured at FL1 and 15000 events were collected for each sample. The data were analysed using CellQuest software.
To obtain fluorescence microscopy images, cells were incubated with Edu for 4 hours, then washed with PBS and fixed with 4% paraformaldehyde for 15 minutes. Posteriorly, they were washed with PBS and permeabilized with PBS containing 0.2% Triton X-100 for 10 minutes. The cells were then washed again with PBS and incubated for 30 minutes in the dark with a reaction cocktail containing 5 μM Sulfo-Cyanine3 azide, 2 mm copper sulfate pentahydrate and 20 mg/mL ascorbic acid. Nuclei were stained with 2 μg/mL of Hoechst 33258 dye (Molecular Probes) in PBS for 5 min. Cells were then washed, maintained in PBS and analysed under a widefield fluorescence microscope (Zeiss Axio Observer Z1; Carl Zeiss, Oberkochen, Germany).
Cell Cycle Analysis
After 72 hours of incubation with the nanosystems, HepG2 and Huh-7 cells were detached with trypsin and washed with PBS. Posteriorly, the cells were fixed with 70% (v/v) ethanol and incubated overnight at 4 °C. After washing with PBS containing 2% (m/v) BSA, cellular pellets were collected by centrifugation, resuspended and incubated for 15 minutes in the dark with a FxCycle™ PI/RNase Staining Solution (Thermo Fisher). Finally, the cells were analysed in a FACSCalibur flow cytometer (Becton Dickinson, NJ, USA). Emitted PI fluorescence was measured at FL2 and 10000 events were collected for each sample. The percentages of cell populations in different phases of the cell cycle were determined using ModFit LT software.
Western Blotting
Total protein extracts were obtained from pellets of HepG2 and HuH-7 cells treated with the different nanosystems for 48 hours. Cells were lysed for 30 minutes in RIPA buffer (150 mm NaCl, 50 mm Tris-base, 5 mm EGTA, 1% Triton X-100, 0.5% DOC, and 0.1% SDS) that contained a protease inhibitor cocktail (Sigma-Aldrich) and phosphatase inhibitors. The concentration of protein lysates was determined using the BC protein assay kit (Biorad, CA, USA). Samples with 30 μg of protein each were resuspended in loading buffer (20% glycerol, 10% SDS, 0.1% bromophenol blue) and incubated at 95 °C for 5 minutes. Then, they were resolved on SDS-PAGE in a 12% polyacrylamide gel. After that, proteins were transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore). The membrane was then blocked with 5% BSA for 1 hour at room temperature and incubated, overnight at 4 °C, with anti-PTEN (#9552, 1:1000) anti-ERK1/2 (#9107, 1:2000), anti-pERK1/2 (Thr202/Tyr204) (#4370, 1:2000), anti-AKT (#4685, 1:1000), anti-pATK (Ser473) (#4058, 1:1000) (Cell Signaling), anti-FLAG (F1804, 1:1000) (Sigma-Aldrich), and anti-GAPDH (#MAB1667, 1:1000) (Millipore) antibodies, and with the appropriate alkaline phosphatase labelled-secondary antibodies (1:10,000) (Thermo Fisher) for 2 hours at room temperature. Following the antibody incubation period, the membranes underwent four TBS-T washes before being incubated for five minutes at room temperature with the enzyme substrate ECF (Thermo Fisher). A ChemiDoc Touch System (Bio-Rad) was used to detect the chemofluorescence and the Quantity One software (Bio-Rad) was used to analyse the band intensity.
Spheroid Treatment and Growth
After the initial 3 days of formation, spheroids were incubated with different nanosystems for the desired time, at 37 °C and in a humidified atmosphere of 5% CO2. To monitor spheroid diameter and compaction over the course of treatment, phase contrast images (Analysis imaging system CELENA® S, Logos Biosystems) were obtained. Spheroids were stained with fluorescein diacetate (FDA) and propidium iodide for the live-dead analysis. Briefly, 100μL of a solution containing 2 mg/mL propidium iodide in PBS and 2 µL of 5 mg/mL FDA in acetone was added to the spheroids and incubated for 5 minutes at room temperature. Then, after being washed with PBS, spheroids were visualized using CELENA® S Digital Imaging System by Logos Biosystems.
Statistical Analysis
Statistical significance of the differences between the data was assessed with Prism software (version 7.04) using one-way ANOVA and Student’s t-tests. A value of p < 0.05 was considered significant.
Results and Discussion
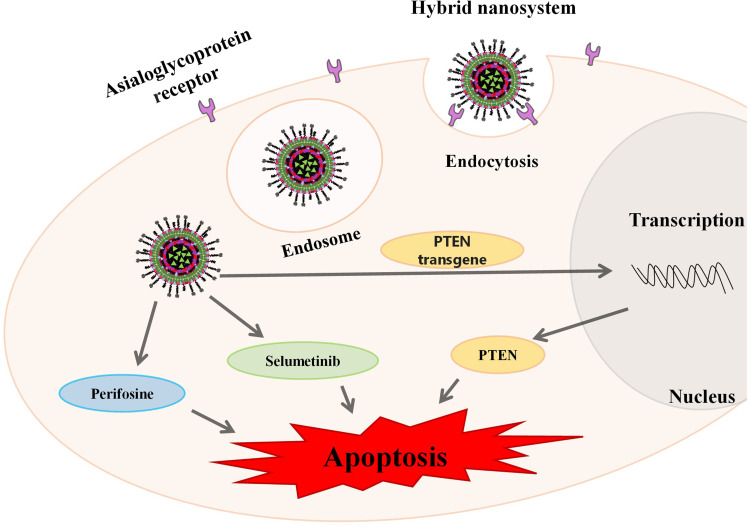
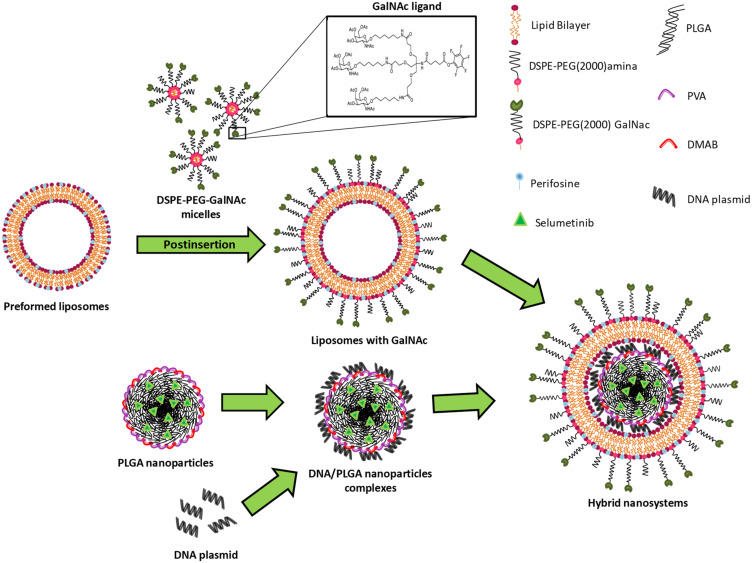
In this work, a new nanosystem formulation with the ability to specifically co-deliver two targeted therapy drugs and a DNA plasmid containing a tumor suppressor gene into HCC cells was developed. As illustrated in Figure 1, these nanosystems consisted of a core-shell nanostructure comprising a PLGA core complexed with DNA, through electrostatic interactions, coated with a lipid bilayer composed of EPOPC, DOPE, PHC, DSPE-PEG2000-amine and DSPE -PEG2000-GalNac.
Figure 1.
Schematic representation of the hybrid nanosystems preparation.
The production of these lipid/polymer hybrid nanoparticles (HNP) was carried out in two steps. In the first step, the polymeric core was produced with the biodegradable and biocompatible PLGA copolymer using the solvent emulsification diffusion-evaporation method.39 In order to produce PLGA nanoparticles with a positive surface charge and reduced size, the influence of different combinations of DMAB and PVA surfactants on these parameters was evaluated. As illustrated in Figure S1A and B, it was found that as the quantity of DMAB increases, there is a reduction in the size and an enhancement in the surface charge of the PLGA nanoparticles, that is in line with what was expected, since the DMAB surfactant, in addition to be a positively charged surfactant, allows the formation of small nanoparticles.39 Based on the obtained data, the PLGA nanoparticles produced with 30 mg of DMAB and 175 mg of PVA were selected to develop the HNP, since in these conditions it was attained nanoparticles smaller than 100 nm (≅ 92.3 nm), with moderately positive zeta potential (≅ +19.2 mV). The PLGA nanoparticles/DNA complexes were prepared, by electrostatic interactions, at different (±) charge ratios, by maintaining constant the genetic material amount and varying the PLGA nanoparticles quantity.
The PLGA nanoparticles showed the ability to complex the genetic material at higher charge ratios (Figure S1C), however none of the produced complexes showed transfection activity in the HCC cell lines used (data not shown). The absence of biological activity may be due, in the case of high charge ratios, to the reduced ability to release the genetic material or, in the case of low charge ratios, to the reduced capability to complex and protect nucleic acids from enzymatic and/or mechanical degradation, as well as to a decreased ability to interact and be internalized by the target cells.40,41 In order to circumvent this problem, PLGA nanoparticle/DNA complexes with lower charge ratios were coated with three distinct HCC-targeted lipid bilayers (Figure 2A–C). Thus, in the second stage of hybrid nanoparticle production, the PLGA nanoparticles/DNA complexes were mixed with preformed liposomes, as previously described.7,42 Four charge ratios of PLGA nanoparticles/DNA complexes and three mass ratios of DNA/lipid, for the three distinct lipid bilayers, were tested. To confer specificity to HNPs for HCC cells, we conjugated the triantennary N-acetylgalactosamine (GalNAc) cluster to the DSPE-PEG2000-amine. This cluster has high affinity for the asialoglycoprotein receptor (ASGP-R), which is overexpressed in HCC cells.7 The conjugation of the cluster to the DSPE-PEG2000-amine was evaluated by FITIR. As observed in Figure S2, DSPE-PEG2000-amine does not present bands above 3000 cm−1 that are characteristic of secondary amines. On the other hand, DSPE-PEG-GalNac as well as GalNac cluster present bands above 3000 cm−1, indicating the successful conjugation of the GalNAc cluster to DSPE-PEG2000-amine.
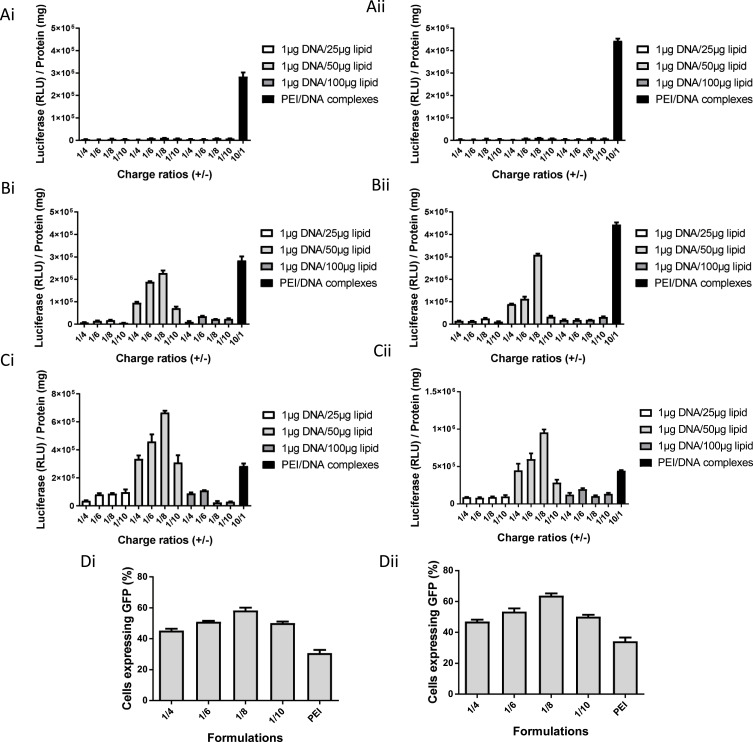
Figure 2.
Transfection activity of HNP. Luciferase gene expression (RLU/mg of total cell protein) in HepG2 (Ai, Bi and Ci) and HuH-7 (Aii, Bii and Cii) cells treated, for 48 hours, with HNP produced with the lipid bilayer composed of: EPOPC/Chol/DSPE-PEG2000-amine/DSPE-PEG2000-GalNac (Ai and Aii); EPOPC/DOPE/CHEMS/DSPE-PEG2000-amine/DSPE-PEG2000-GalNac (Bi and Bii); EPOPC/DOPE/PHC /DSPE-PEG2000-amine /DSPE -PEG2000-GalNac (Ci and Cii). Different charge ratios (PLGA nanoparticles/DNA) and mass ratios (DNA/lipid) were tested, keeping the amount of DNA fixed and varying the quantity of PLGA nanoparticles and lipid, respectively. Cells were transfected in 48-well plates with different HNP formulations containing 1 μg of DNA. Transfection efficiency in HepG2 (Di) and HuH-7 (Dii) cells treated, for 48 hours, with HNP composed of EPOPC/DOPE/PHC/DSPE-PEG2000-amine/DSPE-PEG2000-GalNac, prepared at different charge ratios (PLGA nanoparticles/DNA) and at 1/50 DNA/lipid mass ratio. Cells were transfected in 12-well plates with different hybrid formulations containing 4 μg of DNA. Transfection efficiency (Percentage of cells expressing GFP) was evaluated by flow cytometry. PEI/DNA polyplexes prepared at the charge ratio 10/1 (±) were used as a positive control. Data are expressed as mean ± SD (n=3) obtained from three independent experiments.
The biological activity of the different HNP formulations was evaluated and, as illustrated in Figure 2, there was only significant biological activity, in both HCC cell lines, with HNP prepared with the lipid bilayers constituted by EPOPC/DOPE/CHEMS/DSPE-PEG2000-amine/DSPE-PEG-GalNac and EPOPC/DOPE/PHC/DSPE-PEG2000-amine/DSPE-PEG-GalNac, at the mass ratio of 1µg DNA to 50 µg of lipid. This biological activity was higher for nanosystems prepared with the lipid bilayer composed of EPOPC/DOPE/PHC/DSPE-PEG2000-amine/DSPE-PEG-GalNac (Figure 2C), being the best HNP formulation constituted by PLGA nanoparticle/DNA complexes prepared at a charge ratio of 1/8 (±) and coated by the lipid bilayer at a mass ratio of 1 µg DNA to 50 µg of lipid. The biological activity of these best hybrid nanoparticles was about twice as high as the biological activity obtained with PEI-based polyplexes (positive control). This new hybrid nanoformulation also showed the highest transfection efficiency, with approximately 60% of transfected cells, in both HCC cell lines, a value that once again was twice of that obtained with the positive control (Figure 2D). In subsequent studies, this best HNP formulation was used.
The stability of these new hybrid nanosystems was evaluated, and they did not exhibit major changes in their transfection activity and their physicochemical properties (mean diameter, polydispersity index, and zeta potential), over 30 days, when kept at room temperature or at 4 °C, the same being not observed when they were stored at −20 °C (Figure S3 and Table S1). The reduction of transfection activity after storage at −20 °C was probably due to the fact that the freezing/thawing process promoted changes in their structure (Table S1) and consequently loss of biological activity. Additionally, contrary to what was observed in the PLGA nanoparticles/DNA complexes, these new hybrid nanosystems allowed a greater protection of the genetic material at pH 7.4, observed through a significant decrease on the access of ethidium bromide to the DNA when compared to that observed with the PLGA nanoparticles/DNA complexes at this pH (Figure 3A). These results, together with those observed both in terms of size (no substantial changes in the mean diameter of the formulation throughout the hybrid nanoparticles preparation), and in terms of zeta potential (positive surface charge for the PLGA nanoparticles, negative for the PLGA nanoparticle/DNA complex, and positive for the HNP produced without GalNac) (Table 1), as well as in the TEM images (Figure S4), suggest that the formation of this HNP formulation occurred initially via the coating of the PLGA nanoparticles by a DNA layer, resulting in the formation of the complexes, which were then coated by the lipid bilayer forming the new hybrid nanosystems.
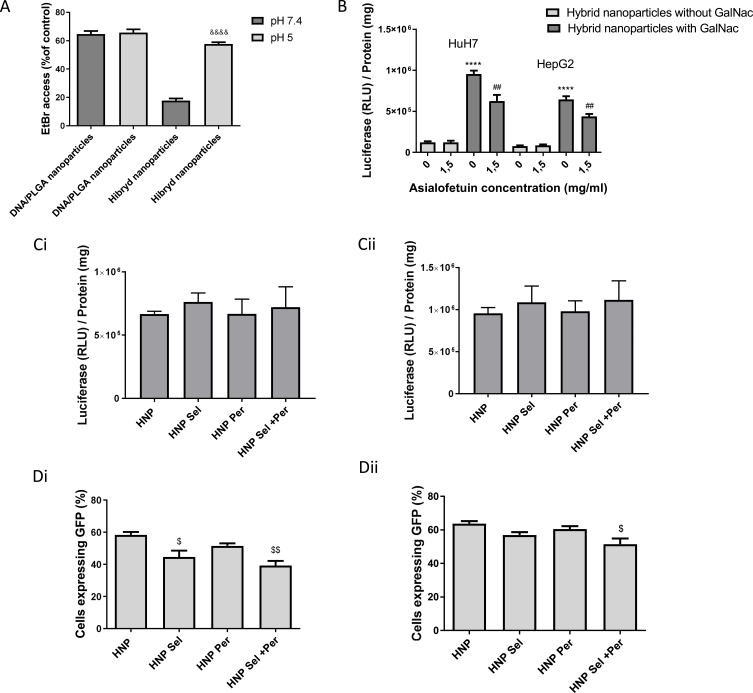
Figure 3.
Properties of HNP and effect of encapsulation of chemotherapeutic drugs on their transfection activity. (A) Access of Ethidium bromide to DNA in HNP and in PLGA nanoparticles/DNA complexes at pH 5 and pH 7.4. The PLGA nanoparticle/DNA complexes were produced at the charge ratio (1/8) (±), and the HNP were prepared at the charge ratio 1/8 (±) and at the DNA/lipid mass ratio of 1/50. (B) Influence of free asialofetuin on biological activity of HNP with and without GalNac on HepG2 and HuH-7 cells. Luciferase expression (RLU/mg of total cell protein) in HCC cells treated, for 48 hours in 48-well plates, with HNP produced at the charge ratio 1/8 (±) and at the DNA/lipid mass ratio of 1/50, containing 1 μg of DNA. (C and D) Effect of selumetinib and/or perifosine on the biological activity of HNP in HepG2 (Ci and Di) and HuH-7 (Cii and Dii) cells. HNP were produced with and without selumetinib (0.73 µg/mg HNP) and/or perifosine (27.5 µg/mg HNP). (C) Luciferase expression (RLU/mg of total cell protein) in cells treated, for 48 hours in 48-well plates, with HNP containing 1 μg of DNA. (D) Percentage of cells expressing GFP, evaluated through flow cytometry, after cells treatment, for 48 hours in 12-well plates, with HNP containing 4 μg of DNA. Results are presented as mean ± SD (n=3) obtained from 3 independent experiments. (&&&&P< 0.0001) Denotes a statistically significant difference when compared to ethidium bromide access at pH 7.4 for the same formulation. (****P< 0.0001) Denotes a statistically significant difference in comparison to the biological activity observed with the HNP prepared without GalNac, for the same cell line. (##P< 0.01) Denotes a statistically significant difference when comparing with the biological activity registered with the same formulation in the lack of asialofetuin, for the same cell line. ($$P< 0.01); ($P< 0.05) Denotes a statistically significant difference in comparison to the percentage of cells expressing GFP obtained with HNP without drugs.
Table 1.
Physicochemical Characterization of Selected Hybrid Nanosystems and Their Components
| Size (nm) | Polydispersity Index (PDI) | Zeta Potential (mV) | |
|---|---|---|---|
| PLGA nanoparticles | 92.3 ± 0.4 | 0.02 ± 0.01 | 21.5 ± 0.61 |
| PLGA nanoparticles/DNA (1/8 ratio) | 112.5 ± 2.3 | 0.20 ± 0.02 | −31.2 ± 7.39 |
| Liposomes without Galnac | 97.9 ± 0.4 | 0.17 ± 0.05 | 25.3 ± 1.28 |
| Liposomes with Galnac | 102 ± 0.1 | 0.18 ± 0.03 | −15.4 ± 2.01 |
| Hybrid nanoparticles without Galnac | 129 ± 0.8 | 0.08 ± 0.07 | 19.5 ± 0.78 |
| Hybrid nanoparticles with GalNac | 135 ± 0.5 | 0.07 ± 0.09 | −17.6 ± 0.93 |
Notes: Results are presented as mean ± SD obtained at least from four independent samples.
As illustrated in Figure 3A, the increase of ethidium bromide access to DNA of the HNP, when the pH was decreased from 7.4 to 5, suggests that the lipid bilayer (EPOPC/DOPE/PHC /DSPE-PEG2000-amine/DSPE-PEG2000-GalNac) of the HNP undergoes destabilization at acidic pH, most probably because of the presence of the lipids PHC and DOPE, which can allow the creation of pH-sensitive lipid bilayers when used simultaneously.43 This pH sensitivity of the HNP may allow the transport and protection of the nucleic acids at pH 7.4, while once within the endocytic pathway, it may facilitate the release of the nucleic acids to the cytoplasm.44,45 In this regard, we also found that this best formulation of nanosystems exhibited a faster release of the coumarin-6 under acid conditions (pH 5) than at pH 7.4, thus corroborating that the developed nanosystems are pH-responsive, and are most likely sensitive to the acidic pH of the endosome (Figure S5).
On the other hand, it was observed that the incorporation of the GalNac cluster increased the transfection activity (Figure 3B) and the cell uptake (Figure S6) of the hybrid nanoparticles, most probably due to the specific interaction of GalNac with the ASGP-R, which is highly expressed in HCC cells, resulting in greater cell binding and uptake when compared to HNP prepared without GalNac (Figure S6), thus leading to a greater biological activity.7,46 To support this hypothesis, we evaluated the influence of pre-incubation with free asialofetuin, a specific ligand for the ASGP-R, on the biological activity (Figure 3B) and cell uptake (Figure S6) of HNP prepared with or without the GalNac cluster. We found that 1.5 mg/mL of free asialofetuin significantly reduced the cellular internalization and the transfection capacity of the hybrid nanoparticles containing the GalNac ligand, but did not change these parameters in the case of the HNP prepared without this cluster (Figures 3B and S6). These results, together with the fact that the hybrid nanoparticles prepared with the GalNac cluster have a negative surface charge (avoiding their nonspecific interaction with the cellular membrane) (Table 1), support the hypothesis that cell binding and internalization of targeted HNP occur through the specific interaction of the GalNac cluster with the ASGP-R.7,46,47 Xunan Li et al and Youngsoo Kim et al also demonstrated that conjugation with N-acetylgalactosamine allows efficient targeting to the asialoglycoprotein receptor, promoting the specific delivery of micelles with therapeutic agents and antisense oligonucleotides, respectively, to HCC cells.48,49
As the aim of our work was to produce a new hybrid nanosystem with the capacity to efficiently deliver a therapeutic gene and two antitumor drugs, the concentration and dosage of the therapeutic agents was optimized based on that objective. In this regard, the selected dosage of DNA (1 µg of DNA/cm2) was chosen taking into account the amount of nanosystems, prepared without drugs, that resulted in a higher expression of the transgene (the reporter gene luciferase) without causing unspecific cytotoxicity (Figure S7). The amount of selumetinib used (0.73 µg/mg HNP, corresponding to a loading efficiency of 67%) was the quantity of drug loaded onto the PLGA nanoparticles that corresponded to the number of positive charges needed to complex DNA and allow the highest transgene (luciferase) expression in the target cells (Figures 2 and 3C and D). On the other hand, the selected amount of perifosine was 3 mol% (in relation to the total amount of lipids) (27.5 µg of perifosine/mg HNP, corresponding to a loading efficiency of 100%) because it was the lowest concentration that showed targeted toxicity without significantly affecting the transfection activity of nanosystems (Figures S7 and 3C and D). As shown in Figure 3C and D, although the presence of these drugs did not promote substantial changes in transfection activity in terms of luciferase gene expression (Figure 3C), a significant decrease in the percentage of transfected cells, with greater evidence in HepG2 cells, namely when both drugs were present, was observed (Figure 3D). This reduction in transfection efficiency was most probably due to the fact that these drugs induce some cell death that was more evident when both drugs were incorporated in the HNP (Figure S8). In the case of the evaluation by luciferase expression the possible reduction on transfection was annulled by the normalization with the total cell protein. It should also be noted that drug-free hybrid nanoparticles did not show cytotoxicity (Figure S8).
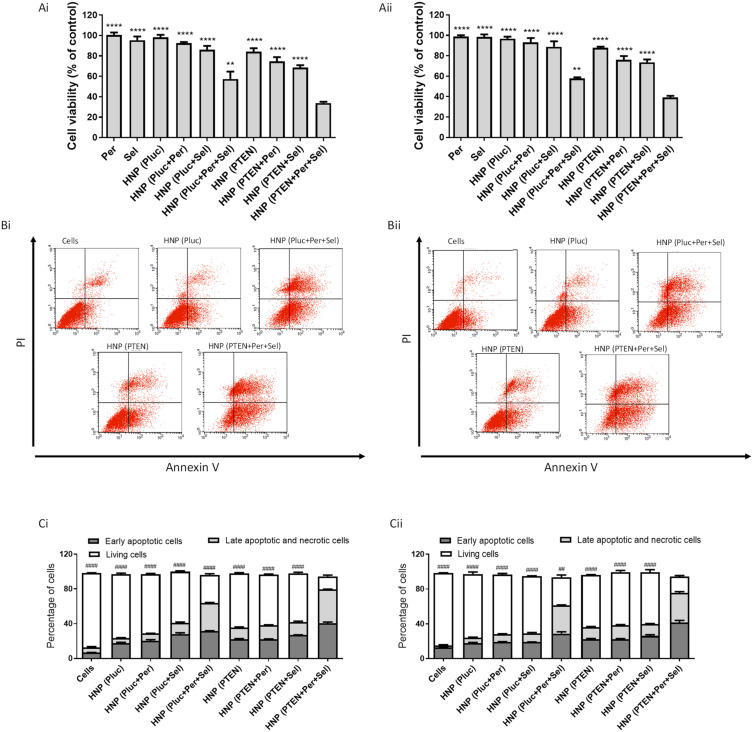
The developed HNP not only allow the simultaneous transport of antitumor drugs and genetic material, but also their targeted delivery into HCC cells. In order to generate an effective combined antitumor approach, comprising chemotherapy and gene therapy, we evaluated the potential of the generated HNP to mediate such therapeutic strategy, involving two targeted therapy drugs (selumetinib loaded in the PLGA nanoparticles, and perifosine incorporated in the lipid bilayer) and a DNA plasmid encoding PTEN, in two HCC cells lines (HepG2 and HuH-7). The selection of these therapeutic agents is justified by the fact that selumetinib acts as an inhibitor of MEK, which is normally overactive in HCC; perifosine acts as an inhibitor of AKT, which is also overactive in HCC; and PTEN is a tumor suppressor protein that normally is underexpressed in HCC.11,14,15 In order to determine the potential of this triple therapeutic approach, the other possible combinations and the individual strategies were also evaluated. Furthermore, the cytotoxicity of the free drugs, in the same concentration as used in nanoparticles, was also assessed. As demonstrated by the data presented in Figure 4A, the cytotoxicity profile of the various therapeutic conditions was identical in both HCC cell lines. The achieved results revealed that the combined strategies were always more toxic than the individual approaches. Moreover, the triple combination demonstrated to be significantly more efficient in promoting cancer cell death than the other approaches, with about 70% of cytotoxicity in HepG2 cells (Figure 4Ai) and about 60% in HuH-7 cell line (Figure 4Aii). On the other hand, the HNP and the different therapeutic conditions did not show significant cytotoxicity in a non-tumor control cell line (Figure S9).
Figure 4.
Therapeutic potential of the different approaches mediated by HNP. HepG2 (Ai) and HuH-7 (Aii) cells were treated with the different therapeutic formulations, corresponding to 1 μg of DNA/well, for 72 hours, and then cell viability was analyzed using the Alamar blue assay. The tested formulations were: empty HNP containing the luciferase plasmid (the control gene); HNP containing perifosine and/or selumetinib and luciferase plasmid; HNP containing only the PTEN plasmid; HNP containing perifosine and/or selumetinib and PTEN plasmid; free drugs used in the same concentration as in the HNP. HepG2 (Bi and Ci) and HuH-7 (Bii and Cii) cells lines were stained with Annexin V and PI double-staining for 15 min after being treated, for 72 hours in 12-well plates, with the different formulations corresponding to 4 μg of DNA. The apoptosis levels were evaluated by flow cytometry. The percentages of live cells, cells in early apoptosis and cells in late apoptosis/necrosis were presented in bar graphs (Ci and Cii). Data are presented as mean ± SD (n=3) obtained from three independent experiments. Asterisks represent significant differences in cell viability. Cardinals represent significant differences in terms of living cells. (**** and ####P < 0.0001); (** and ##P < 0.01) Denotes a statistically significant difference when compared with cells incubated with HNP containing perifosine, selumetinib and PTEN plasmid.
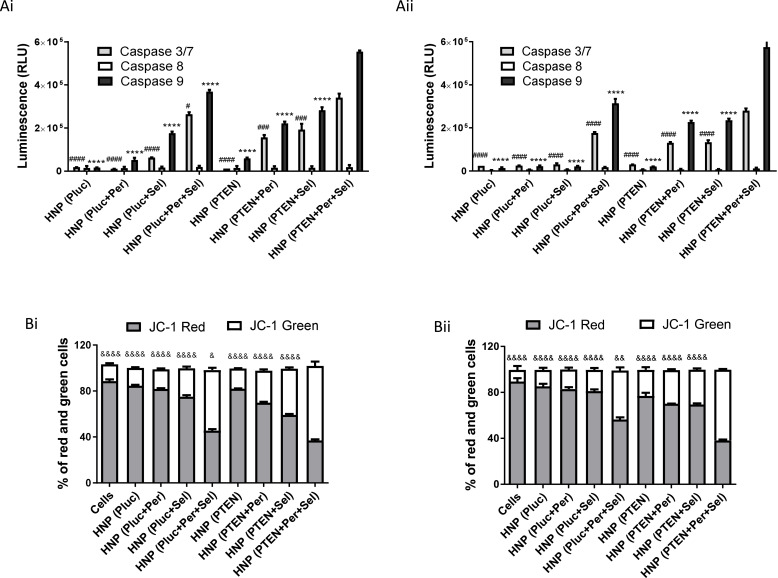
In an attempt to identify the mechanisms involved in the anticancer activity induced by the individual and combined strategies, mediated by the new HNP, we evaluated the extent of early apoptosis and necrosis/late apoptosis using annexin V and PI. As presented in Figure 4B and C, for both HCC cell lines, incubation for 72 hours with the HNP containing the triple combination resulted in a greater number of cells stained with PI and/or annexin V than registered with the other therapeutic conditions, whether in double combinations or individually, which confirmed once again the higher capacity of the triple combination to promote cancer cell death. Although there was a significant percentage of cells labelled only for annexin V, an indicator of initial apoptosis, there was also a significant percentage of cells with double labelling, annexin V and PI, not allowing the distinction between cells in late apoptosis or necrosis. However, we can assume that these latter ones are cells that have evolved from an early apoptosis process to a late apoptosis phase.7,50 In order to corroborate the hypothesis that the triple therapeutic strategy induces cell death through a process of programmed cell death, we proceeded to assess the activity of caspases, since the activation of caspases is a critical event in the induction process of cellular apoptosis. The apoptotic process is carried out through the cleavage of cellular components by effector caspases (caspases-3/7). These caspases are activated by initiator caspases, such as caspase-9 and caspase-8, that in turn are activated by intrinsic and extrinsic apoptotic pathways, respectively.51 In this context, the activity of caspases-3/7, −8 and −9 was evaluated in order to measure the activation of caspases and determine the apoptotic pathway associated with HCC cell death promoted by the different treatments. The results presented in Figure 5A show that treatment of HCC cells with the new HNP, containing the triple combination, resulted in a strong increase in the activity of caspases-3/7 and −9, this activation being significantly higher than that obtained for the remaining therapeutic strategies. On the other side, activation of caspase-8 was shown to be residual for both HCC cell lines, exhibiting no significant differences between the different therapeutic conditions. Thus, the increased activation of caspases-3/7 and caspase-9 demonstrated that the intrinsic apoptotic pathway is involved in the apoptotic process induced by the treatment comprising the triple therapeutic combination. These facts indicate that most probably the cytochrome c is released from the mitochondria to cytosol, triggering the activation of caspase-9 that in turn activated caspases-3/7. This cytochrome c release involves mitochondrial changes, such as the collapse of the mitochondrial membrane potential, that in turn originate alterations in mitochondrial permeability and consequent release of cytochrome c.52,53 Considering this, we proceeded to analyze the mitochondrial membrane potential, after treatment of HCC cells with the different therapeutic approaches, by using the JC-1 fluorescent cation probe. This probe, in case of high mitochondrial membrane potential, accumulates in the mitochondria forming aggregates that produce a red fluorescence, and in case of mitochondrial membrane potential collapse, it remains in the cytosol in low concentrations not allowing the creation of aggregates and, consequently, exhibiting green fluorescence.54,55 As can be seen in Figure 5B, the treatment of both HCC cell lines with the HNP mediating the triple therapeutic approach caused an enhancement in the percentage of cells that exhibited green fluorescence. This increase was significantly higher comparing with those percentages registered with the other tested conditions. These data revealed that the new HNP mediating the triple therapeutic strategy were capable of inducing a substantial reduction in mitochondrial membrane potential, which normally occurs during the apoptotic process. These results once again prove that the triple combination approach, mediated by our HNP, has a greater therapeutic potential than the dual combinations or the individual strategies.
Figure 5.
Caspase activation levels and mitochondrial membrane potential of treated cells. Luminescence generated by the activity of caspase −3/7, −8 and −9 in (Ai) HepG2 and (Aii) HuH-7 cells incubated, for 72 hours in 96-well plates, with HNP containing the luciferase plasmid (the control gene); HNP containing perifosine and/or selumetinib and the luciferase plasmid; HNP containing the PTEN plasmid; and HNP containing perifosine and/or selumetinib and PTEN plasmid (nanoparticles corresponding to 0.5 μg of DNA). Percentage of HepG2 (Bi) and HuH-7 (Bii) cells with JC-1 green or red fluorescence, quantified by flow cytometry, after treatment with the different therapeutic approaches. The percentage of green and red cells are expressed as mean ± SD obtained at least from three independent experiments (n=3). Asterisks represent significant differences in caspase−9 activity. Cardinals represent significant differences regarding the activity of caspase−3/7. Ampersands represent significant differences regarding the percentage of cells with JC-1 green fluorescence. (****, #### and &&&&P < 0.0001); (###P< 0.001); (&&P < 0.01) (& and #P< 0.05) Denotes a statistically significant difference when compared with cells treated with HNP containing perifosine, selumetinib and PTEN plasmid (triple combination).
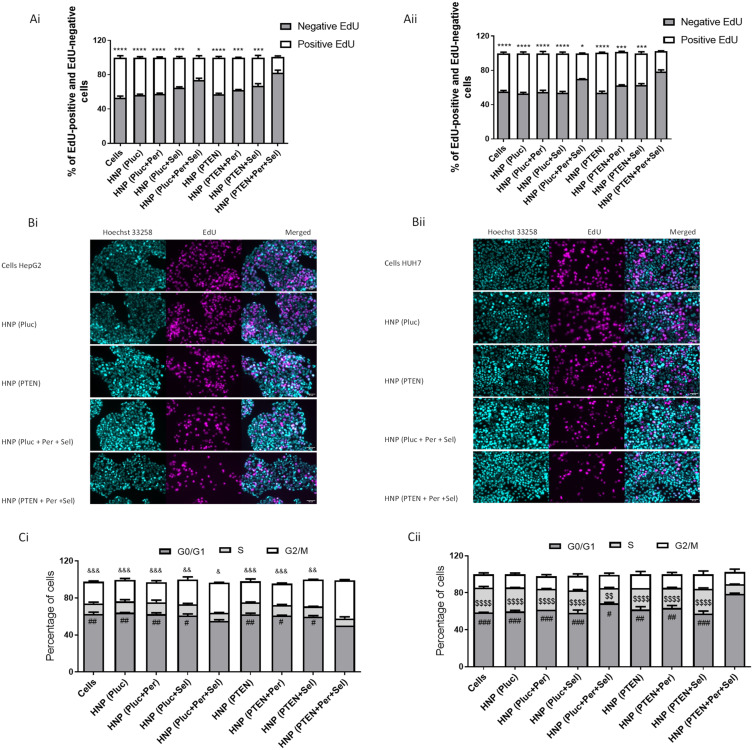
In addition to assessing the death mechanisms involved in the antineoplastic effect of individual and combined strategies, mediated by the new HNP, we also evaluated their influence on cell proliferation through the EdU incorporation assay. EdU is a thymidine nucleotide analogue and can be used to measure the synthesis of new DNA, since during the S phase of the cell cycle it is integrated into the newly synthesized DNA of replicating cells.38 The EdU assay provides reliable and direct measurements of cellular multiplication in actively dividing cells.56 As shown in Figure 6A and B, treatment of HepG2 and HuH-7 cells with the new HNP, containing the triple combination of therapeutic agents, promoted a significant decrease in the percentage of EdU positive cells. This decrease was greater than that achieved for the other therapeutic conditions, as demonstrated by the results obtained not only through flow cytometry (Figure 6A) but also by fluorescence microscopy (Figure 6B). The obtained results demonstrated that HCC cells treatment with HNP, mediating the triple therapeutic strategy, promoted the highest decrease in cell proliferation. In an attempt to verify whether the effect observed in cell proliferation was also associated to changes in the cell cycle, this latter parameter was evaluated after treatment with the different approaches. As elucidated in Figure 6C, the HNP delivering the triple combination resulted in significant alterations in cell cycle phases when comparing with those registered with the other treatments either combined or individual. For HepG2 cells, incubation with the best therapeutic formulation resulted in a significant increase in cells in the G2/M phase, while for HuH-7 cells, it promoted a significant enhancement in the G0/G1 phase. As previously reported by other authors, these differences found between the two cell lines are probably due to cellular heterogeneity, and genetic and epigenetic factors.57,58
Figure 6.
Cell proliferation and cell cycle of treated cells. Evaluation of cell proliferation, through the incorporation of EdU, in HepG2 (Ai and Bi) and HuH-7 (Aii and Bii) cells treated, for 48 hours in 12-well plates, with HNP containing the luciferase plasmid (the control gene); HNP containing perifosine and/or selumetinib and luciferase plasmid; HNP containing the PTEN plasmid; and HNP containing perifosine and/or selumetinib and PTEN plasmid (nanoparticles corresponding to 4 μg of DNA). Percentage of EdU positive and EdU negative HepG2 (Ai) and HuH-7 (Aii) cells quantified by flow cytometry. Representative microscopy imagens of EdU positive HepG2 (Bi) and HuH-7 (Bii) cells (magenta, Sulfo-Cyanine3), nuclear staining (cyan, Hoechst 33258) and the merged imagens (scale bars, 50 μm). Percentages of the various phases of the cell cycle for HepG2 (Ci) and Huh-7 (Cii) cells treated, for 72 hours in 6-well plates, with the different therapeutic formulations (nanosystems corresponding to 8 μg of DNA). Data are expressed as mean ± SD obtained from three independent experiments (n=3). Asterisks represent significant differences in percentage of EdU positive cells. Cardinals represent significant differences in the G0/G1 phase of the cell cycle. Ampersands represent significant differences in the G2/M phase of the cell cycle. Dollar sign represent significant differences in the S phase of the cell cycle. (**** and $$$$P< 0.0001); (***, ### and &&&P< 0.001); (##, && and $$P< 0.01); (*, # and &P< 0.05) Denotes a statistically significant difference when compared with cells treated with HNP containing perifosine, selumetinib and PTEN plasmid (triple combination).
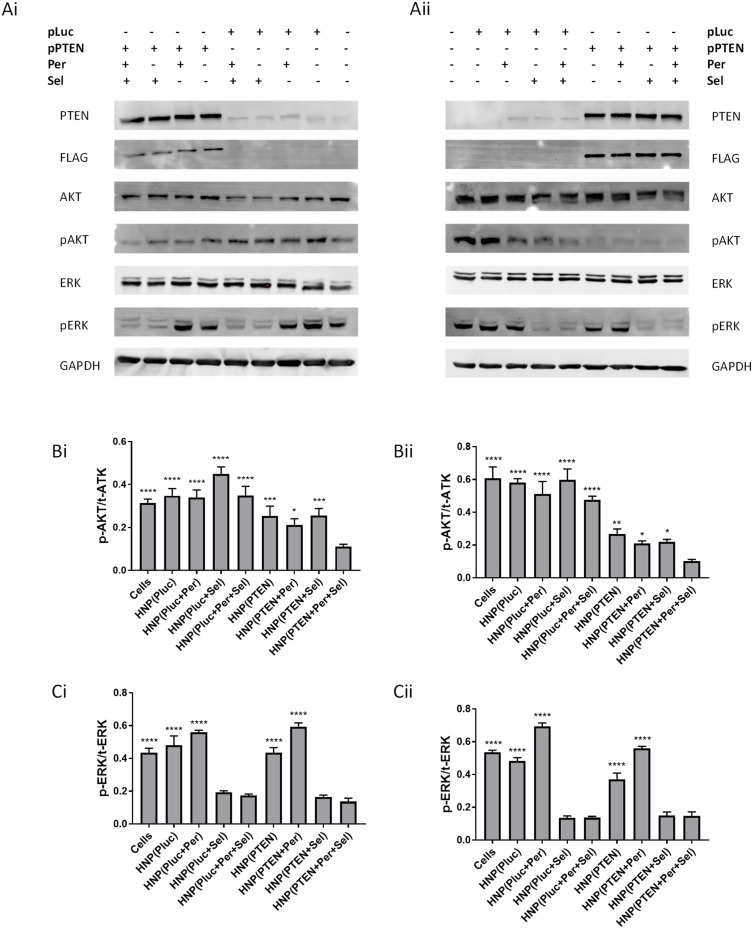
The increase in apoptotic cells and the decrease in cell proliferation, promoted by the new HNP containing the triple combination, is possibly due to a high ability of this combined approach to inhibit the signaling pathways involved in both cell survival and proliferation, namely the RAS/RAF/MEK/ERK and PI3K/AKT/mTOR pathways.59,60 Possibly, the inhibition of MEK and AKT proteins induced by the targeted therapy drugs selumetinib and perifosine, respectively, in combination with the PTEN transgene expression, which acts as a negative regulator of the activation of the PI3K/AKT pathway and has nuclear functions regulating and maintaining the integrity of the genome, resulted in an enhancement in the antitumor activity, observed through increased programmed cell death and decreased cell proliferation capacity. In an attempt to elucidate the process and pathways involved in the anticancer strategies, we evaluated not only the levels of pERK and pATK but also the levels of PTEN transgene expression in HCC cells, through immunoblotting, after treatment with the HNP mediating the combined and individual approaches. As demonstrated by the results presented in Figure 7A, the incubation of HCC cells with the HNP containing the plasmid encoding PTEN promoted a substantial increase in the levels of PTEN protein in both cell lines, which was not obtained after treatment with nanosystems prepared with the plasmid containing the luciferase transgene. Indeed, the increase observed in PTEN protein levels was a result of the PTEN transgene expression, since the protein expressed by the used PTEN plasmid has a FLAG (polypeptide protein tag) labelling.61 Treatment of both cells with HNP containing the PTEN plasmid promoted a decrease in the p-AKT levels, this reduction being more evident in the HuH-7 cell line, quite possibly because of the superior transgene expression observed in these cells (Figure 2C and D). Therefore, the higher levels of PTEN promoted a greater inhibition of AKT protein and, consequently, lower p-AKT levels.62 The decrease in p-AKT levels was further accentuated when cells were incubated with HNP containing not only the plasmid encoding PTEN but also the drug perifosine (Figure 7B). In contrast, cells treated with HNP containing the drug selumetinib presented lower p-ERK levels. However, the lowest levels of p-AKT and p-ERK were only achieved concomitantly when cells were incubated with nanosystems mediating the triple combination strategy (Figure 7A–C). In fact, cells treatment with nanoparticles simultaneously loaded with selumetinib (MEK inhibitor), perifosine (AKT inhibitor) and the plasmid encoding PTEN, caused a robust inhibition of target proteins that most probably promoted a lower activity of the signalling pathways involved in cell proliferation and survival (RAS/RAF/MEK/ERK and PI3K/Akt/mTOR pathways), consequently inducing a greater antitumor activity than the other therapeutic approaches.
Figure 7.
Influence of the therapeutic strategies on PTEN, ATK and ERK protein levels. Western blot analysis of phosphorylated (p) and total (t) AKT and ERK proteins in HepG2 (Ai, Bi and Ci) and HuH-7 (Ai, Bi and Ci) cells treated, for 48 hours in 6-well plates, with HNP containing the luciferase plasmid; HNP containing perifosine and/or selumetinib and luciferase plasmid; HNPcontaining the PTEN plasmid; and HNP containing perifosine and/or selumetinib and PTEN plasmid (nanosystems corresponding to 8 μg of DNA). Representative Western blot images displaying the levels of PTEN, FLAG, p/t-AKT and p/t-ERK in HepG2 (Ai) and HuH-7 (Aii) cells. Graphical representation of the p-AKT/t-AKT (Bi and Bii) and p-ERK/t-ERK (Ci and Cii) ratios in HepG2 and HuH-7 cells. Data are expressed as mean ± SD obtained from three independent experiments (n=3). (**** P < 0.0001); (*** P< 0.001); (** P< 0.01); (* P< 0.05) Denotes a statistically significant difference when compared with cells treated with HNP containing perifosine, selumetinib and PTEN plasmid.
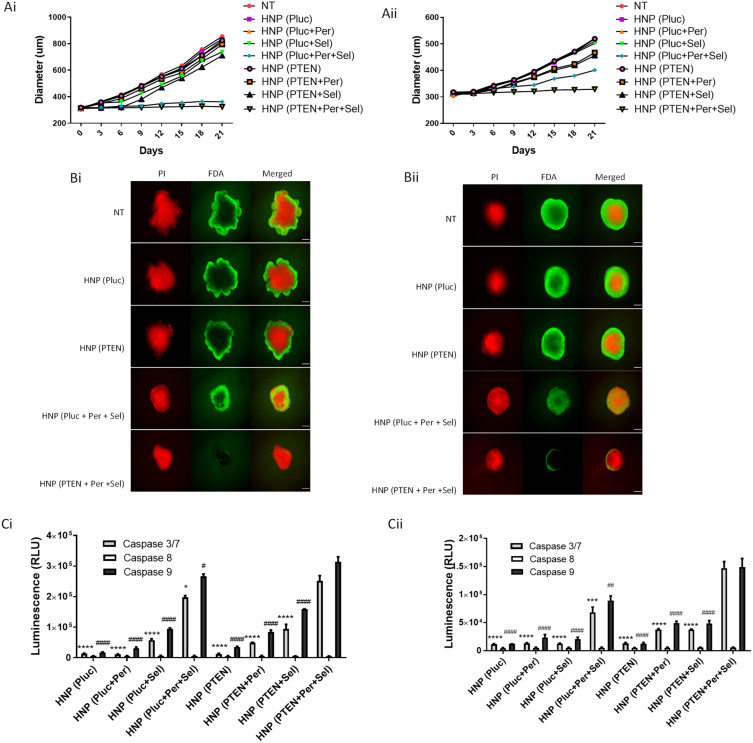
In an attempt to obtain more physiologically relevant data and overcome some limitations associated with 2D cell cultures, the use of three-dimensional (3D) cell culture systems has gained prominence.63 In this way, the anticancer effect of the individual and combined therapeutic approaches, mediated by the new HNP, was assessed in HCC-tumor spheroid, obtained with HepG2 and HuH-7 cells, with an initial average diameter of approximately 300 µm. As elucidated in Figure 8A, the spheroids of both cell lines did not show an enhancement in their sizes over 21 days of incubation with the HNP containing the triple therapeutic combination. Although a similar size was observed in spheroids treated with nanoparticles containing the combination of the two drugs, this being particularly evidence in HepG2 spheroids (Figure 8Ai), the number of live cells at the end of the 21 days of treatment was significantly lower in spheroids incubated with nanosystems delivering the triple therapeutic combination (Figure 8B). In fact, the spheroids of both cell lines, at the end of 21 days of treatment with the HNP containing the triple combination, not only exhibited the smaller size but also the smaller number of living cells, when compared with the other therapeutic strategies. Regarding the activation of caspases, as shown in Figure 8C, the treatment with nanosystems containing the triple therapeutic combination promoted a robust activation of caspases-3/7 and −9 in both HepG2 and HuH-7 spheroids. This activation was significantly higher than that achieved with the other strategies. In contrast, as in monolayer cultures, caspase-8 activation was practically non-existent and no substantial differences were observed. These data indicate that the programmed cell death occurred through the participation of the intrinsic apoptotic pathway. Thus, these results demonstrated that also in 3D HCC cell cultures hybrid nanoparticles containing the triple therapeutic combination exhibited greater antitumor activity than the other tested approaches.
Figure 8.
Impact of the treatment approaches mediated by the HNP in 3D cultures of HCC cells. Growth curves of (Ai) HepG2- and (Aii) HuH-7-derived spheroids after treatment, over 21 days, with HNP containing the luciferase plasmid; HNP containing perifosine and/or selumetinib and luciferase plasmid; HNP containing the PTEN plasmid; and HNP containing perifosine and/or selumetinib and PTEN plasmid (corresponding to 5 μg of DNA). Representative fluorescence images of (Bi) HepG2- and (Bii) HuH-7-derived spheroids, at the end of 21 days of incubation with the different therapeutic approaches, using FDA-PI as live-dead staining (scale bar = 150 µm). Relative luminescence produced by the activity of caspase-3/7, −8 and −9 in (Ci) HepG2- and (Cii) HuH-7-derived spheroids treated, for 12 days, with the different therapeutic formulations. Data are expressed as mean ± SD obtained from three independent experiments (n=3). Cardinals represent significant differences regarding the activity of caspase 9. Asterisks represent significant differences in caspase 3/7 activity. (**** and ####P< 0.0001); (***P< 0.001); (##P< 0.01); (* and #P< 0.05) Denotes a statistically significant difference when compared with cells treated with HNP containing perifosine, selumetinib and PTEN plasmid (triple combination).
Conclusion
In the present study, we generated a new hybrid nanosystem with adequate physicochemical properties, namely, negative zeta potential and sizes below 200 nm, which is biocompatible and biodegradable. This new nanosystem allows the simultaneous transport of different therapeutic agents, namely antitumor drugs and nucleic acids, and their specific delivery in HCC cells, due to its functionalization with the GalNAc ligand. In fact, this formulation has the capability to mediate combined chemotherapy and gene therapy, as it efficiently loaded and delivered, in target cells, the drugs selumetinib and perifosine, and the PTEN transgene. The treatment of liver cancer cells with this new nanosystem, containing these three therapeutic agents, promoted a high antitumor effect involving the inhibition of the signalling pathways associated with both HCC cell proliferation and cell survival. This antitumor effect occurred not only through the enhancement in programmed cell death, demonstrated by the increase in the number of cells in initial apoptosis, by the activation of initiator and effector caspases and by the loss of mitochondrial membrane potential, but also through the reduction of the ability of HCC cells to proliferate, observed by the decreased ability to incorporate EdU and by the cell cycle changes. Moreover, the therapeutic potential of this new HNP was also demonstrated in 3D HCC cell cultures, where it was verified not only a smaller growth capacity of the spheroids tumor mass, but also an increased programmed cell death. Therefore, this new formulation allows a concerted action of chemotherapy and gene therapy, promoting a high therapeutic effect, and has the capacity to reduce the side effects associated to the therapeutic drugs because of its specific to HCC cells. To conclude, our data revealed that this new therapeutic approach, mediated by the generated nanosystems, can be considered an auspicious strategy for future clinical application in hepatocellular carcinoma treatment.
Acknowledgments
This work was financed by the European Regional Development Fund (ERDF) through the COMPETE 2020 program (Operational Program for Competitiveness and Internationalization) and Portuguese national funds via FCT – Fundação para a Ciência e a Tecnologia, under projects: IF/01007/2015, POCI-01-0145-FEDER-30916, LA/P/0058/2020, UIDB/04539/2020 and UIDP/04539/2020. The authors would like to thank Dr. Michael Migawa and Ionis Pharmaceuticals Inc. for kindly providing the Triantennary N‐acetylgalactosamine (GalNAc) cluster. Selumetinib was kindly provided by AstraZeneca PLC. Dina Farinha acknowledges FCT for the Grant: SFRH/BD/104894/2014.
Funding Statement
This work was financed by the European Regional Development Fund (ERDF) through the COMPETE 2020 program (Operational Program for Competitiveness and Internationalization) and Portuguese national funds via FCT – Fundação para a Ciência e a Tecnologia, under projects: IF/01007/2015, POCI-01-0145-FEDER-30916, LA/P/0058/2020, UIDB/04539/2020 and UIDP/04539/2020.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Refolo MG, Messa C, Guerra V, Carr BI, D’alessandro R. Inflammatory mechanisms of HCC development. Cancers. 2020;12(3):641. doi: 10.3390/cancers12030641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca a Cancer J Clinicians. 2021;71(3):209–249. doi: 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 3.McGlynn KA, Petrick JL, El‐Serag HB. Epidemiology of hepatocellular carcinoma. Hepatology. 2021;73(S1):4–13. doi: 10.1002/hep.31288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suddle A, Reeves H, Hubner R, et al. British society of gastroenterology guidelines for the management of hepatocellular carcinoma in adults. Gut. 2024;73(8):1235–1268. doi: 10.1136/gutjnl-2023-331695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morazán-Fernández D, Mora J, Molina-Mora JA. In silico pipeline to identify tumor-specific antigens for cancer immunotherapy using exome sequencing data. Phenomics. 2023;3(2):130–137. doi: 10.1007/s43657-022-00084-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farinha D, de Lima MCP, Faneca H. Specific and efficient gene delivery mediated by an asialofetuin-associated nanosystem. Int J Pharm. 2014;473(1–2):366–374. doi: 10.1016/j.ijpharm.2014.07.019 [DOI] [PubMed] [Google Scholar]
- 7.Farinha D, Migawa M, Sarmento-Ribeiro A, Faneca H. A combined antitumor strategy mediated by a new targeted nanosystem to hepatocellular carcinoma. Int J Nanomed. 2021;16:3385. doi: 10.2147/IJN.S302288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo B-J, Ruan Y, Wang Y-J, et al. Jiedu recipe, a compound Chinese herbal medicine, inhibits cancer stemness in hepatocellular carcinoma via Wnt/β-catenin pathway under hypoxia. J Integr Med. 2023;21(5):474–486. doi: 10.1016/j.joim.2023.06.008 [DOI] [PubMed] [Google Scholar]
- 9.Whittaker S, Marais R, Zhu A. The role of signaling pathways in the development and treatment of hepatocellular carcinoma. Oncogene. 2010;29(36):4989–5005. doi: 10.1038/onc.2010.236 [DOI] [PubMed] [Google Scholar]
- 10.Kahraman DC, Kahraman T, Cetin-Atalay R. Targeting PI3K/Akt/mTOR pathway identifies differential expression and functional role of IL8 in liver cancer stem cell enrichment. Mol Cancer Ther. 2019;18(11):2146–2157. doi: 10.1158/1535-7163.MCT-19-0004 [DOI] [PubMed] [Google Scholar]
- 11.Nakanishi K, Sakamoto M, Yamasaki S, Todo S, Hirohashi S. Akt phosphorylation is a risk factor for early disease recurrence and poor prognosis in hepatocellular carcinoma. Cancer. 2005;103(2):307–312. doi: 10.1002/cncr.20774 [DOI] [PubMed] [Google Scholar]
- 12.Yang S, Liu G. Targeting the Ras/Raf/MEK/ERK pathway in hepatocellular carcinoma. Oncol Lett. 2017;13(3):1041–1047. doi: 10.3892/ol.2017.5557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hwang YH, Choi JY, Kim S, et al. Over-expression of c-raf-1 proto-oncogene in liver cirrhosis and hepatocellular carcinoma. Hepatol Res. 2004;29(2):113–121. doi: 10.1016/j.hepres.2004.02.009 [DOI] [PubMed] [Google Scholar]
- 14.Huynh H, Nguyen TTT, Chow K-HK-P, Tan PH, Soo KC, Tran E. Over-expression of the mitogen-activated protein kinase (MAPK) kinase (MEK)-MAPK in hepatocellular carcinoma: its role in tumor progression and apoptosis. BMC Gastroenterol. 2003;3(1):1–21. doi: 10.1186/1471-230X-3-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruan Z-P, Xu R, Lv Y, et al. PTEN enhances the sensitivity of human hepatocellular carcinoma cells to sorafenib. Oncol Res Featuring Preclinical Clin Cancer Ther. 2012;20(2–3):113–121. doi: 10.3727/096504012X13477145152995 [DOI] [PubMed] [Google Scholar]
- 16.Ha SE, Kim SM, Vetrivel P, et al. Inhibition of cell proliferation and metastasis by scutellarein regulating PI3K/Akt/NF-κB signaling through PTEN activation in hepatocellular carcinoma. Int J Mol Sci. 2021;22(16):8841. doi: 10.3390/ijms22168841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holohan B, Hagiopian MM, Lai T-P, et al. Perifosine as a potential novel anti-telomerase therapy. Oncotarget. 2015;6(26):21816. doi: 10.18632/oncotarget.5200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gills JJ, Dennis PA. Perifosine: update on a novel Akt inhibitor. Curr Oncol Reports. 2009;11(2):102–110. doi: 10.1007/s11912-009-0016-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fei H-R, Chen G, Wang J-M, Wang F-Z. Perifosine induces cell cycle arrest and apoptosis in human hepatocellular carcinoma cell lines by blockade of Akt phosphorylation. Cytotechnology. 2010;62(5):449–460. doi: 10.1007/s10616-010-9299-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huynh H, Soo KC, Chow PK, Tran E. Targeted inhibition of the extracellular signal-regulated kinase kinase pathway with AZD6244 (ARRY-142886) in the treatment of hepatocellular carcinoma. Mol Cancer Ther. 2007;6(1):138–146. doi: 10.1158/1535-7163.MCT-06-0436 [DOI] [PubMed] [Google Scholar]
- 21.Leijen S, Soetekouw PM, Jeffry Evans T, et al. A Phase I, open-label, randomized crossover study to assess the effect of dosing of the MEK 1/2 inhibitor Selumetinib (AZD6244; ARRY-142866) in the presence and absence of food in patients with advanced solid tumors. Cancer Chemother Pharmacol. 2011;68(6):1619–1628. doi: 10.1007/s00280-011-1732-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Campos L, Nemunaitis J, Stephenson J, et al. Phase II study of single agent perifosine in patients with hepatocellular carcinoma (HCC). J Clin Oncol. 2009;27(15_suppl):e15505–e15505. doi: 10.1200/jco.2009.27.15_suppl.e15505 [DOI] [Google Scholar]
- 23.Dowdy SF. Overcoming cellular barriers for RNA therapeutics. Nature Biotechnol. 2017;35(3):222–229. doi: 10.1038/nbt.3802 [DOI] [PubMed] [Google Scholar]
- 24.Witzigmann D, Kulkarni JA, Leung J, Chen S, Cullis PR, van der Meel R. Lipid nanoparticle technology for therapeutic gene regulation in the liver. Adv Drug Delivery Rev. 2020;159:344–363. doi: 10.1016/j.addr.2020.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.E Grottkau B, Cai X, Wang J, Yang X, Lin Y. Polymeric nanoparticles for a drug delivery system. Curr Drug Metabol. 2013;14(8):840–846. doi: 10.2174/138920021131400105 [DOI] [PubMed] [Google Scholar]
- 26.Mallick S, Choi JS. Liposomes: versatile and biocompatible nanovesicles for efficient biomolecules delivery. J Nanosci Nanotechnol. 2014;14(1):755–765. doi: 10.1166/jnn.2014.9080 [DOI] [PubMed] [Google Scholar]
- 27.Danhier F, Ansorena E, Silva JM, Coco R, Le Breton A, Préat V. PLGA-based nanoparticles: an overview of biomedical applications. J Control Release. 2012;161(2):505–522. doi: 10.1016/j.jconrel.2012.01.043 [DOI] [PubMed] [Google Scholar]
- 28.Ahmed KS, Hussein SA, Ali AH, Korma SA, Lipeng Q, Jinghua C. Liposome: composition, characterisation, preparation, and recent innovation in clinical applications. J Drug Targeting. 2019;27(7):742–761. doi: 10.1080/1061186X.2018.1527337 [DOI] [PubMed] [Google Scholar]
- 29.Rayamajhi S, Marchitto J, Nguyen TDT, Marasini R, Celia C, Aryal S. pH-responsive cationic liposome for endosomal escape mediated drug delivery. Colloids Surf B. 2020;188:110804. doi: 10.1016/j.colsurfb.2020.110804 [DOI] [PubMed] [Google Scholar]
- 30.Zhang L, Chan JM, Gu FX, et al. Self-assembled lipid− polymer hybrid nanoparticles: a robust drug delivery platform. ACS nano. 2008;2(8):1696–1702. doi: 10.1021/nn800275r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mandal B, Bhattacharjee H, Mittal N, et al. Core–shell-type lipid–polymer hybrid nanoparticles as a drug delivery platform. Nanomed Nanotechnol Biol Med. 2013;9(4):474–491. doi: 10.1016/j.nano.2012.11.010 [DOI] [PubMed] [Google Scholar]
- 32.Fraix A, Conte C, Gazzano E, Riganti C, Quaglia F, Sortino S. Overcoming doxorubicin resistance with lipid–polymer hybrid nanoparticles photoreleasing nitric oxide. Mol Pharmaceut. 2020;17(6):2135–2144. doi: 10.1021/acs.molpharmaceut.0c00290 [DOI] [PubMed] [Google Scholar]
- 33.Steirer LM, Park EI, Townsend RR, Baenziger JU. The asialoglycoprotein receptor regulates levels of plasma glycoproteins terminating with sialic acid α2, 6-galactose. J Biol Chem. 2009;284(6):3777–3783. doi: 10.1074/jbc.M808689200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pranatharthiharan S, Patel MD, Malshe VC, et al. Asialoglycoprotein receptor targeted delivery of doxorubicin nanoparticles for hepatocellular carcinoma. Drug Delivery. 2017;24(1):20–29. doi: 10.1080/10717544.2016.1225856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwon H-Y, Lee J-Y, Choi S-W, Jang Y, Kim J-H. Preparation of PLGA nanoparticles containing estrogen by emulsification–diffusion method. Colloids Surf A. 2001;182(1–3):123–130. doi: 10.1016/S0927-7757(00)00825-6 [DOI] [Google Scholar]
- 36.Costa PM, Cardoso AL, Mendonça LS, et al. Tumor-targeted chlorotoxin-coupled nanoparticles for nucleic acid delivery to glioblastoma cells: a promising system for glioblastoma treatment. Mol Ther Nucleic Acids. 2013;2:e100. doi: 10.1038/mtna.2013.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morais CM, Cunha PP, Melo T, et al. Glucosylceramide synthase silencing combined with the receptor tyrosine kinase inhibitor axitinib as a new multimodal strategy for glioblastoma. Human Molecular Genetics. 2019;28(21):3664–3679. doi: 10.1093/hmg/ddz152 [DOI] [PubMed] [Google Scholar]
- 38.Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci. 2008;105(7):2415–2420. doi: 10.1073/pnas.0712168105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhardwaj V, Ankola D, Gupta S, Schneider M, Lehr C-M, Kumar MR. PLGA nanoparticles stabilized with cationic surfactant: safety studies and application in oral delivery of paclitaxel to treat chemical-induced breast cancer in rat. Pharm Res. 2009;26(11):2495–2503. doi: 10.1007/s11095-009-9965-4 [DOI] [PubMed] [Google Scholar]
- 40.Ogris M, Steinlein P, Kursa M, Mechtler K, Kircheis R, Wagner E. The size of DNA/transferrin-PEI complexes is an important factor for gene expression in cultured cells. Genet Ther. 1998;5(10):1425–1433. doi: 10.1038/sj.gt.3300745 [DOI] [PubMed] [Google Scholar]
- 41.Godbey W, Barry MA, Saggau P, Wu KK, Mikos AG. Poly (ethylenimine)‐mediated transfection: a new paradigm for gene delivery. J Biomed Mater Res. 2000;51(3):321–328. doi: [DOI] [PubMed] [Google Scholar]
- 42.Troutier A-L, Delair T, Pichot C, Ladavière C. Physicochemical and interfacial investigation of lipid/polymer particle assemblies. Langmuir. 2005;21(4):1305–1313. doi: 10.1021/la047659t [DOI] [PubMed] [Google Scholar]
- 43.Won SH, Lee JU, Sim SJ. Fluorogenic pH-sensitive polydiacetylene (PDA) liposomes as a drug carrier. J Nanosci Nanotechnol. 2013;13(6):3792–3800. doi: 10.1166/jnn.2013.7205 [DOI] [PubMed] [Google Scholar]
- 44.Lim E-K, Chung BH, Chung SJ. Recent advances in pH-sensitive polymeric nanoparticles for smart drug delivery in cancer therapy. Curr Drug Targets. 2018;19(4):300–317. doi: 10.2174/1389450117666160602202339 [DOI] [PubMed] [Google Scholar]
- 45.Elkin SR, Lakoduk AM, Schmid SL. Endocytic pathways and endosomal trafficking: a primer. Wiener Medizinische Wochenschrift. 2016;166(7–8):196–204. doi: 10.1007/s10354-016-0432-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prakash TP, Yu J, Migawa MT, et al. Comprehensive structure–activity relationship of triantennary N-acetylgalactosamine conjugated antisense oligonucleotides for targeted delivery to hepatocytes. J Med Chem. 2016;59(6):2718–2733. doi: 10.1021/acs.jmedchem.5b01948 [DOI] [PubMed] [Google Scholar]
- 47.Tripathi P, Dwivedi P, Khatik R, et al. Development of 4-sulfated N-acetyl galactosamine anchored chitosan nanoparticles: a dual strategy for effective management of Leishmaniasis. Colloids Surf B. 2015;136:150–159. doi: 10.1016/j.colsurfb.2015.08.037 [DOI] [PubMed] [Google Scholar]
- 48.Li X, Wang X, Liu N, Wang Q, Hu J. Inhibition of metastatic hepatocarcinoma by combined chemotherapy with silencing VEGF/VEGFR2 genes through a GalNAc-modified integrated therapeutic system. Molecules. 2022;27(7). doi: 10.3390/molecules27072082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim Y, Jo M, Schmidt J, et al. Enhanced potency of GalNAc-conjugated antisense oligonucleotides in hepatocellular cancer models. Mol Ther. 2019;27(9):1547–1557. doi: 10.1016/j.ymthe.2019.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crowley LC, Marfell BJ, Scott AP, Waterhouse NJ. Quantitation of apoptosis and necrosis by annexin V binding, propidium iodide uptake, and flow cytometry. Cold Spring Harbor Protocols. 2016;2016(11):pdb.prot087288. doi: 10.1101/pdb.prot087288 [DOI] [PubMed] [Google Scholar]
- 51.Shi Y. Mechanisms of caspase activation and inhibition during apoptosis. Molecular Cell. 2002;9(3):459–470. doi: 10.1016/S1097-2765(02)00482-3 [DOI] [PubMed] [Google Scholar]
- 52.Brentnall M, Rodriguez-Menocal L, De Guevara RL, Cepero E, Boise LH. Caspase-9, caspase-3 and caspase-7 have distinct roles during intrinsic apoptosis. BMC Cell Biol. 2013;14(1):1–9. doi: 10.1186/1471-2121-14-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kashyap D, Garg VK, Goel N. Intrinsic and extrinsic pathways of apoptosis: role in cancer development and prognosis. Adv Protein Chem Struct Biol. 2021;125:73–120. [DOI] [PubMed] [Google Scholar]
- 54.Perelman A, Wachtel C, Cohen M, Haupt S, Shapiro H, Tzur A. JC-1: alternative excitation wavelengths facilitate mitochondrial membrane potential cytometry. Cell Death Dis. 2012;3(11):e430–e430. doi: 10.1038/cddis.2012.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cossarizza A, Baccaranicontri M, Kalashnikova G, Franceschi C. A new method for the cytofluorometric analysis of mitochondrial membrane potential using the J-aggregate forming lipophilic cation 5, 5′, 6, 6′-tetrachloro-1, 1′, 3, 3′-tetraethylbenzimidazolcarbocyanine iodide (JC-1). Biochem Biophys Res Commun. 1993;197(1):40–45. doi: 10.1006/bbrc.1993.2438 [DOI] [PubMed] [Google Scholar]
- 56.Menyhárt O, Harami-Papp H, Sukumar S, et al. Guidelines for the selection of functional assays to evaluate the hallmarks of cancer. Biochimica Et Biophysica Acta (BBA)-Rev Cancer. 2016;1866(2):300–319. [DOI] [PubMed] [Google Scholar]
- 57.De Falco M, De Luca A. Cell cycle as a target of antineoplastic drugs. Curr Pharm Des. 2010;16(12):1417–1426. doi: 10.2174/138161210791033914 [DOI] [PubMed] [Google Scholar]
- 58.Wang X, Chen Z, Mishra AK, et al. Chemotherapy-induced differential cell cycle arrest in B-cell lymphomas affects their sensitivity to Wee1 inhibition. Haematologica. 2018;103(3):466. doi: 10.3324/haematol.2017.175992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Asati V, Mahapatra DK, Bharti SK. PI3K/Akt/mTOR and Ras/Raf/MEK/ERK signaling pathways inhibitors as anticancer agents: structural and pharmacological perspectives. Eur J Med Chem. 2016;109:314–341. doi: 10.1016/j.ejmech.2016.01.012 [DOI] [PubMed] [Google Scholar]
- 60.Weng L-P, Smith WM, Dahia PL, et al. PTEN suppresses breast cancer cell growth by phosphatase activity-dependent G1 arrest followed by cell death. Cancer Res. 1999;59(22):5808–5814. [PubMed] [Google Scholar]
- 61.Lee M-S, Jeong M-H, Lee H-W, et al. PI3K/AKT activation induces PTEN ubiquitination and destabilization accelerating tumourigenesis. Nat Commun. 2015;6(1):7769. doi: 10.1038/ncomms8769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen C-Y, Chen J, He L, Stiles BL. PTEN: tumor suppressor and metabolic regulator. Front Endocrinol. 2018;9:338. doi: 10.3389/fendo.2018.00338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu X, Su J, Wei J, Jiang N, Ge X. Recent advances in three-dimensional stem cell culture systems and applications. Stem Cells Int. 2021;2021:1–13. doi: 10.1155/2021/9477332 [DOI] [PMC free article] [PubMed] [Google Scholar]