Abstract
The complete genome of the gram-negative bacterial pathogen Helicobacter pylori, an important etiological agent of gastroduodenal disease in humans, has recently been published. This sequence revealed that the putative products of roughly one-third of the open reading frames (ORFs) have no significant homology to any known proteins. To be able to analyze the functions of all ORFs, we constructed an integration plasmid for H. pylori and used it to generate a random mutant library in this organism. This integration plasmid, designated pBCα3, integrated randomly into the chromosome of H. pylori. To test the capacity of this library to identify virulence genes, subsets of this library were screened for urease-negative mutants and for nonmotile mutants. Three urease-negative mutants in a subset of 1,251 mutants (0.25%) and 5 nonmotile mutants in a subset of 180 mutants (2.7%) were identified. Analysis of the disrupted ORFs in the urease-negative mutants revealed that two had disruptions of genes of the urease locus, ureB and ureI, and the third had a disruption of a unrelated gene; a homologue of deaD, which encodes an RNA helicase. Analysis of the disrupted ORFs in the nonmotile mutants revealed one ORF encoding a homologue of the paralyzed flagellar protein, previously shown to be involved in motility in Campylobacter jejuni. The other four ORFs have not been implicated in motility before. Based on these data, we concluded that we have generated a random insertion library in H. pylori that allows for the functional identification of genes in H. pylori.
Helicobacter pylori is now regarded as one of the most common human pathogens. Infection with this organism leads to type B chronic gastritis, which may progress to duodenal and gastric ulceration. Infection with H. pylori is also a major risk factor for the development of gastric adenocarcinoma and mucosa-associated lymphoid tissue lymphoma (9). Numerous virulence factors, such as urease, are involved in the pathogenesis of H. pylori infections, but still little is known about the actual proteins mediating the infection and the genes encoding these proteins. The sequence of the whole genome of H. pylori 26695 has recently been published (37), and this greatly facilitates research into the virulence genes. However, analysis of the genome also reveals that the putative products of roughly one-third of the open reading frames (ORFs) have no significant homology to known proteins. In contrast to this abundance of ORFs with unknown functions, several key regulatory proteins seem to be absent in H. pylori (3, 37). Detailed functional studies are required to unravel the functions of all ORFs and to establish their putative roles in pathogenesis.
For many pathogens, the generation of random mutant libraries has been a powerful tool for the identification of virulence genes. The creation of a random insertion library in H. pylori, followed by a functional screen, will be able to confirm the proposed functions of the homologous ORFs. This is necessary because homology alone does not prove that the protein encoded has the same properties as its counterpart. In addition, it will allow the establishment of the functions of those ORFs whose products have no homology with known proteins. Currently, molecular genetic studies of H. pylori are severely hampered by the paucity of useful genetic tools. To date, H. pylori mutagenesis has been limited to insertional inactivation of selected genes and transposon shuttle mutagenesis (14, 17). A severe limitation of the latter method is that when selection for phenotypes is performed in Escherichia coli, many H. pylori genes may be toxic to, or may not be expressed in, E. coli (24). We developed a mutagenesis system based on the chromosomal integration of a suicide plasmid for H. pylori, since a similar setup has recently been shown to be effective for the closely related organism Campylobacter coli (7).
In order to generate a random insertion library, we constructed an integration plasmid specific for H. pylori. After it was shown that this plasmid integrates randomly into the chromosome of H. pylori, we used it to generate a mutant library. The quality of the library was tested by screening subsets for urease-negative mutants and for nonmotile mutants. This screen yielded three urease-negative and five nonmotile mutants. The ORFs disrupted in these mutants were identified by a plasmid rescue strategy. Analysis of the disrupted genes in these mutants revealed both genes known to be involved in these phenotypes and novel genes. This work demonstrates the feasibility of performing random mutagenesis in H. pylori to identify the genes underlying its virulence.
MATERIALS AND METHODS
Bacterial strains, plasmids, and cultivation conditions.
The vectors used in this study were pBC SK(−) (Stratagene, La Jolla, Calif.), a phagemid derived from pUC19; pHel 3 (19), an E. coli-H. pylori shuttle vector, and pBCα3 (this work), an H. pylori integration plasmid. H. pylori 1061 (15), a clinical isolate, was used as the parental strain in this study. E. coli 1793 was used as a host for recombinant plasmids (22). E. coli strains were routinely cultured either in liquid or on solid Luria-Bertani (LB) medium. For selection, E. coli media were supplemented with either 50 mg of kanamycin/liter or 20 mg of chloramphenicol/liter. Recombinant plasmids were transformed and maintained in E. coli ER 1793.
H. pylori was routinely cultured on solid Columbia agar medium plates supplemented with 7% lysed horse blood and Dent H. pylori selective supplement (Oxoid, Basingstoke, United Kingdom), referred to below as Dent plates. Bacteria were routinely grown on plates for 48 h at 37°C in an atmosphere of 5% O2–10% CO2–85% N2. When appropriate, Dent plates were supplemented with 10 mg of kanamycin/liter.
General genetic manipulations.
H. pylori was naturally transformed essentially as described in reference 40, according to the following protocol. Bacteria were inoculated as quarter-size patches on Dent plates and grown for 8 h under microaerobic conditions. Subsequently, 8 μl of TE (10 mM Tris-HCl–1 mM EDTA [pH 8.0]), containing approximately 2 μg of plasmid DNA, was added to each patch. After overnight incubation, the bacteria were harvested and resuspended in 1 ml of brucella broth (Oxoid). The bacteria were pelleted, resuspended in 100 μl of brucella broth, and plated on selective plates.
All standard DNA manipulations, heat shock transformation of E. coli, and Southern blot procedures were performed as described by Sambrook et al. (32). Probes for the Southern blot were labeled radioactively with random primers by using the Stratagene Prime-It kit. Plasmid DNA was isolated with the miniprep spin kit of Qiagen Gmbh (Hilden, Germany). All enzymes were obtained from New England Biolabs Inc. (Beverly, Mass.) and were used according to the manufacturer’s instructions.
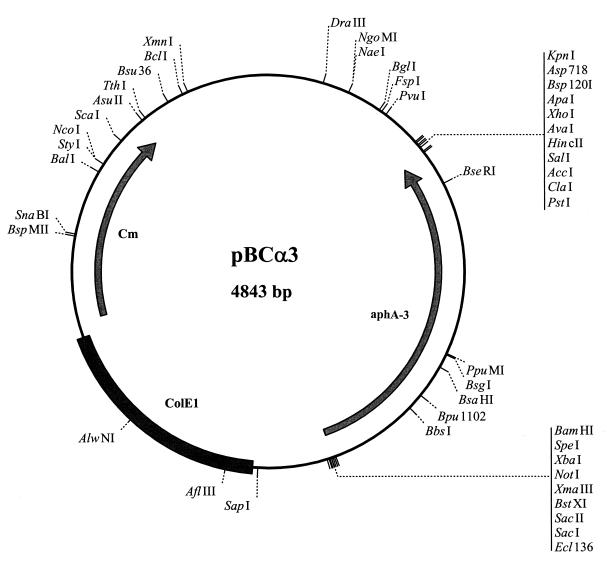
Construction of pBCα3.
The aphA-3 cassette was excised from pHel 3 with SmaI and ligated in the unique SmaI site of the pBC backbone (Fig. 1). The resulting plasmid was called pBCα3. Recombinant plasmids were obtained by transforming the ligation mixture to E. coli ER 1793 with selection on LB-kanamycin agar.
FIG. 1.
Schematic representation of pBCα3. ColE1, origin of replication (not functional in H. pylori); aphA-3, kanamycin cassette (functional in H. pylori and E. coli); Cm, chloramphenicol cassette (not functional in H. pylori). Only unique restriction sites are indicated. The AluI sites used in the plasmid rescue strategy are not indicated because there are 29 AluI sites in the plasmid.
Mutagenesis of H. pylori.
Chromosomal DNA of the parental strain was isolated and digested with Sau3AI. The resulting fragments were size fractionated with a sucrose gradient, after which the average size of the DNA in the fractions was checked on an agarose gel. The fractions containing DNA with an average length around 500 bp were pooled. These fragments were ligated into the dephosphorylated unique BamHI site of pBCα3 (Fig. 1). To obtain recombinant plasmids, the ligation mixture was transformed to E. coli ER 1793 with selection on LB kanamycin. Plasmid DNA was isolated from the pooled transformants. To confirm that inserts were present in the vector, an aliquot was restricted with HincII and NotI, which revealed a smear between 1,500 and 2,000 bp, indicating the presence of inserts of varying lengths (100 to 500 bp). Subsequently, aliquots of uncut plasmids were naturally transformed to H. pylori, with selection on Dent kanamycin plates. After incubation, single colonies were picked and grown in 1 ml of brucella broth supplemented with 7% newborn calf serum (NBCS; Gibco, Paisely, Scotland), Dent supplement, and kanamycin in 24-well plates. After 4 days of growth, 250 μl of glycerol was added to each well, and the library was stored at −80°C.
Screening for urease-negative mutants.
The library was inoculated directly from the −80°C stocks onto Dent kanamycin plates, and after 72 h the colonies were transferred with a grid to 96-well plates with 100 μl of urea broth (2% urea, 0.86% urea broth base; Oxoid) per well. Colonies from wells that did not turn red after 30 min were considered urease negative and were picked, cultured separately, and stored. After incubation the urease activity of these colonies were measured quantitatively with a coupled enzyme assay (8).
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblot analysis of H. pylori urease.
Bacteria were grown for 24 h under standard conditions on Dent plates with kanamycin added when appropriate. Cells were harvested in 1 ml of phosphate-buffered saline (PBS) and washed twice. Whole-cell samples were diluted in sample buffer (4% sodium dodecyl sulfate, 2% 2-mercaptoethanol, 20% glycerol, 125 mM Tris-HCl [pH 6.8], 0.1 mg of bromphenol blue/ml), heated for 5 min at 100°C and fractionated on a 12.5% acrylamide gel by using the Phast-gel minisystem (Pharmacia Biotech), and transferred to a nitrocellulose filter by capillary action. The urease subunits were detected on immunoblots (39) after incubation with a 1:1,000 dilution of antiserum against urease A and B (31).
Ultrastructural localization of urease by electron microscopy (EM).
Bacteria were grown for 2 days on Dent plates, and urease was localized as described elsewhere (31). In brief, the cultures were fixed in 2% paraformaldehyde–0.2% glutaraldehyde-PBS (pH 7.2) for 2 h at room temperature. Bacteria were then embedded in 10% gelatin but without fixation of the gelatin. After the bacteria were pelleted, the gelatin was solidified on ice. Blocks for ultracryotomy were prepared and immunolabeled with 10% goat serum in the blocking buffer. Immunolabeling with primary antibodies (diluted 1:400 to 1:800) was carried out for 2 h. Incubation with secondary antibodies (a 1:25 dilution of 12 nM goat anti- rabbit immunoglobulin G-colloidal gold [Jackson ImmunoResearch Laboratories, West Grove, Pa.]) was carried out for 1 h. Sections were stained with uranyl acetate and embedded in methylcellulose by a modification of the Tokuyasu method introduced by Griffiths et al. (16).
Screening for nonmotile mutants.
The library was inoculated directly from the −80°C stocks onto Dent kanamycin plates. After 48 h the colonies were transferred to soft agar (brucella broth, solidified with 0.4% Bacto agar, supplemented with 7% NBCS and Dent supplement), incubated for 5 days, and screened essentially as described in reference 36. Colonies that seemed nonmotile were picked, cultured separately, and stored. After incubation these mutants were retested in the stab agar assay. Mutants were considered nonmotile when there was a complete absence of swarming in the soft agar.
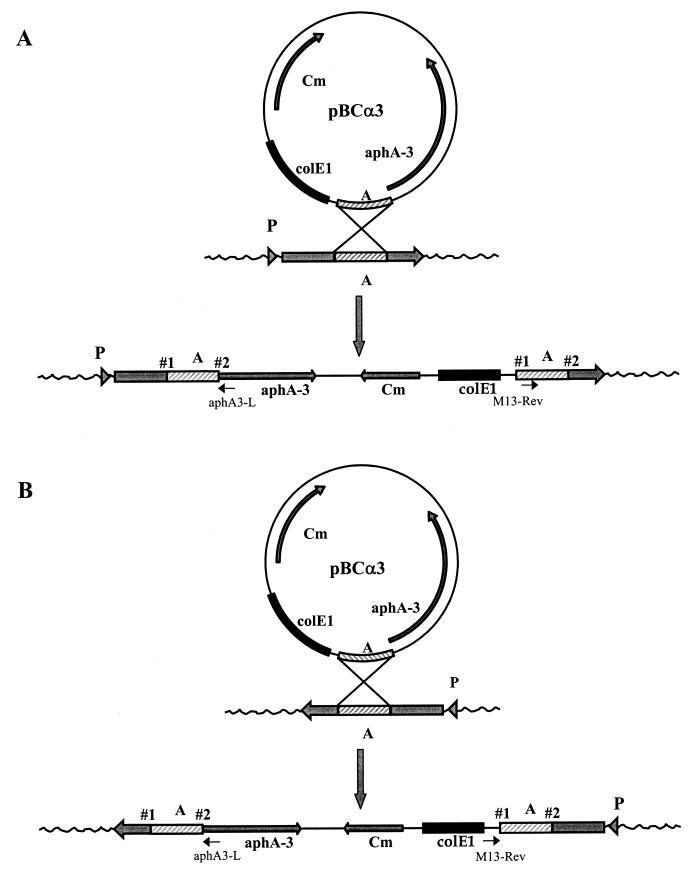
Plasmid rescue, sequencing, and sequence analysis.
Chromosomal DNA of the mutants was isolated and restricted with HindIII to determine insertion point 1 (Fig. 2B). For the determination of insertion point 2 (Fig. 2A), no convenient single cutters were available. Therefore, a partial restriction with AluI was performed; there are 29 AluI sites in pBCα3. Chromosomal fragments were ligated and transformed to E. coli with selection on chloramphenicol and kanamycin. Single colonies were grown overnight in LB medium with chloramphenicol and kanamycin, and plasmid DNA was isolated. These plasmids were used as a template in a sequence reaction with the Thermo-Sequenase premixed cycle sequence kit (Amersham). Either the standard M13 reverse primer or the aphA3-L primer (5′ TCTTACCTATCACTCAAATGG) was used; both primers were labeled with Texas red. Sequencing was performed on an Amersham Vistra 725 sequencer, and data were analyzed with Lasergene software (DNAstar Inc., Madison, Wis.).
FIG. 2.
Schematic representation of the integration of pBCα3 in the chromosome. A hypothetical DNA fragment (fragment A) cloned into the plasmid mediates the integration of the plasmid into the H. pylori chromosome via a Campbell-like mechanism. Integration of pBCα3 results in a duplication of fragment A at both sites of insertion and a subsequent disruption of the gene in which fragment A is present. (A) Integration of pBCα3 is in the same direction as the ORF. In this case, point 2 is the actual insertion site. (B) Integration of pBCα3 is in the direction opposite that of the ORF. In this case, point 1 is the actual insertion site. P, hypothetical promoter. The positions of the primers used to sequence the rescued plasmids, the M13 reverse primer (M13-Rev) and aphA3-L, are indicated.
The sequences obtained were compared to the sequences in the NCBI database with the BlastN program (National Center for Biotechnology Information, Los Alamos, N. Mex.). To determine the insertion point, the sequences were aligned with the complete H. pylori genome as provided by the TIGR database (The Institute for Genomic Research, Rockville, Md.).
EM.
Bacteria were grown for 2 days on Dent plates. They were harvested and resuspended in 1 ml of brucella broth; 50 μl of this suspension was used to inoculate 10 ml of brucella broth. After 72 h of incubation without shaking, the bacteria were spun down, negatively stained, and analyzed by EM as described by Kusters et al. (23).
RESULTS
Construction of the integration vector pBCα3 and the generation of a mutant library.
As a backbone for the construction of the integration plasmid pBCα3, the phagemid pBC SK was used. The aphA-3 kanamycin resistance cassette contains a promoter and a Shine-Dalgarno site that are fully functional in H. pylori (19). This aphA-3 cassette was ligated into pBC, and the resulting plasmid was designated pBCα3 (Fig. 1). Transformation of the parental strain, 1061, with 2 μg of pBCα3, devoid of any chromosomal fragments, yielded no kanamycin-resistant colonies. Fragments of chromosomal DNA of the parental strain, with an average length of 500 bp, were ligated into pBCα3. The resulting pool of plasmids was transformed to H. pylori, and this transformation yielded 500 kanamycin-resistant colonies (250 CFU/μg of DNA). Thus, efficient integration of pBCα3 into the H. pylori chromosome depends on the presence of H. pylori chromosomal fragments. Several independent transformations were performed, each resulting in an average of around 500 colonies per transformation. From three of these transformations, 1,251 single colonies were picked and stored individually in 24-well plates.
Integration occurs randomly throughout the chromosome of H. pylori.
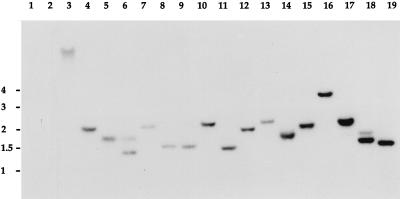
To verify that the integration of pBCα3 into the chromosome of H. pylori 1061 occurred randomly, Southern blotting was performed on 16 mutants arbitrarily selected from the 1,251 kanamycin-resistant colonies. As judged from the hybridization pattern, obtained with the kanamycin cassette as a probe, the cassette was present in all mutants but appeared to be located in different regions of the chromosome (Fig. 3). This suggests that pBCα3 integrates into the chromosome in a random fashion, thereby randomly disrupting different, distinct target genes in the individual mutant strains. The Southern blot data from these 16 arbitrarily selected mutants indicate that the 1,251 kanamycin-resistant colonies indeed represent a more or less random mutant library.
FIG. 3.
Southern blot of 16 randomly selected mutants probed with the kanamycin cassette. Lane 1, plasmid DNA without the kanamycin cassette; lane 2, wild-type chromosomal DNA; lane 3, positive control; lanes 4 to 19, chromosomal DNA of the 16 mutants. Chromosomal DNA was digested with HinfI in all cases. The positions of marker DNA fragments, in kilobase pairs, are given to the left of the autoradiogram.
Screening the library for urease-negative mutants.
To test the library for its suitability for the identification of virulence genes, the 1,251 mutants of the library were phenotypically screened for urease-negative colonies. Three urease-negative mutants were found. In a quantitative assay of urease activity (Table 1), these three mutants were devoid of any detectable urease activity. To confirm that the observed phenotype resulted from the insertion of pBCα3 into the chromosome and not from another, unrelated mutation, total DNA was isolated from the three urease-negative mutants and used to retransform the wild-type parental strain, 1061. Forty independent transformants from each backcross, selected only for Kmr, were subsequently tested for urease activity in the urea broth assay. In all cases, the acquisition of Kmr correlated with the acquisition of the urease-negative phenotype.
TABLE 1.
Analysis of genes disrupted in urease-negative mutants, as determined by plasmid rescue and subsequent sequencing
| Mutant | Site of insertion (bp)a | H. pylori gene no.a | Sp act of urease (U/mg of protein)b | Homologue of interrupted ORF |
|---|---|---|---|---|
| 1 | 76,612 | 0072 | 0.042 ± 0.001 | Urease B gene |
| 2 | 75,258 | 0071 | 0.074 ± 0.003 | Urease accessory gene I |
| 3 | 256,171 | 0247 | 0.050 ± 0.002 | ATP-dependent RNA helicase gene |
According to the sequence of Tomb et al. (37).
Mean from three independent experiments ± standard error of the mean. The urease activity of the parental strain was 34.91 ± 0.043 U/mg of protein.
Characterization of urease-negative mutants.
To identify the disrupted ORFs in the three urease-negative mutants, a plasmid rescue strategy was used, taking advantage of the chloramphenicol and kanamycin resistance cassettes and the origin of replication (ColE1) present on the integrated plasmids. Digestion of the chromosomal DNA with HindIII or AluI, followed by recircularization with T4 DNA ligase and transformation to E. coli ER 1793, resulted in the desired recombinant clones (Fig. 2). The correctness of the rescued plasmid was confirmed by the finding that in all cases the sequence adjacent to the chromosomal DNA consisted of the part of pBCα3 located before or after the insertion. For each integration mutant it was possible to sequence at least 300 bp of flanking chromosomal DNA from the rescued plasmids. Analysis of the chromosomal DNA sequences with the BlastN program revealed that in all cases the rescued plasmids contained H. pylori chromosomal DNA.
Generally, when an integrative plasmid that contains a chromosomal fragment, which only represents a small part of a gene, is used for transformation, a single recombination event will result in two truncated copies of the gene (Fig. 2), and this leads to loss of the function of the affected gene. The duplicated part in the chromosome represents the original fragment present in the integration plasmid. Due to this duplication, the disrupted ORF will not be affected at the actual crossover point but will be correctly transcribed up to the point where the duplication ends. Thus, the mapping of the transcription breakpoint (point of insertion) depends on the relative orientation of the partial ORF present on pBCα3. When integration was in the same direction as the ORF, point 2 (Fig. 2A) was designated as the insertion point. When integration of the plasmid was in the opposite direction, point 1 (Fig. 2B) was designated the insertion point. The site of insertion (Table 1) was determined by aligning the fragments adjacent to pBCα3 with the complete H. pylori sequence, available through the TIGR database. After determination of the interrupted ORF in each mutant, the translated sequence of each ORF was analyzed with the advanced BlastP program of the NCBI database (Table 1).
Mutant 1 had a disruption of ureB, ORF 0072, the gene that encodes one of the structural subunits of the enzyme urease. Mutants constructed by allelic replacement of this gene have been proven urease negative (14). Mutant 2 had a disruption of ureI, ORF 0071, the first gene in the supposed operon of the urease accessory genes, ureIEFGH. To date, no function has been attributed to any of these genes (6). The third mutant had a disruption of ORF 0247, an ORF homologous to deaD, which encodes an ATP-dependent RNA helicase. Homologues of the product of this ORF belong to the DEAD family of proteins (38). This family is thought to be involved in posttranscriptional processes and might play a role in the regulation of gene expression. The two nonhomologous ORFs, 0248 and 0249, located directly behind deaD are transcribed in the same direction, so their function might be impaired by the presence of pBCα3. The gene upstream of deaD is in the reverse direction; therefore, it is unlikely to be affected by the insertion.
Analysis of the urease-negative mutants by Western blotting.
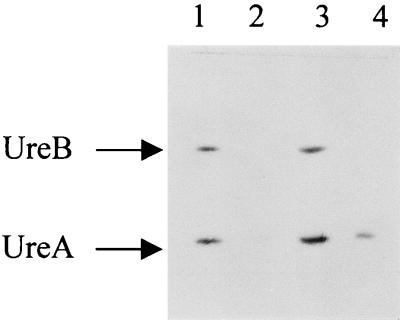
To determine whether the urease-negative phenotype of the mutants was due to the absence of one or both of the structural urease subunits, Western blot analysis was performed with a polyclonal antiserum against UreA and UreB. This analysis (Fig. 4) revealed that both UreA and UreB are still present in the ureI mutant at levels comparable to those in the parental strain. In contrast, in the ureB and deaD mutants, no UreB subunit was present. However, UreA was still present in the deaD mutant, although in significantly reduced amounts compared to those in the wild type. There also was some residual UreA present in the ureB mutant. The near-loss of UreA in the ureB mutant might be due to an instability of the ureAB mRNA or a problem in the translation of the mRNA due to the integration of pBCα3 into the chromosome.
FIG. 4.
Immunoblot of whole-cell preparations with anti-urease (1:1,000). Lane 1, parental strain 1061; lanes 2 through 4, mutants with disruptions of ureB, ureI, and deaD, respectively. Approximately 11 μg of protein was loaded in each lane.
Localization of urease.
To investigate whether the urease-negative phenotype of the ureI mutant, where both subunits were still present, might be due to a mislocalization of the urease enzyme, EM immunolocalization was performed on this mutant and the wild type. EM immunolocalization was also performed on the other two urease-negative mutants. In the wild type, the amount of gold particles and their surface-to-cytoplasm distribution pattern were identical to those in the localization described previously (31) for strain 84-183 (data not shown) (10). In the ureB and deaD mutants, no immunolabeling was visible. This corresponds with the minute amount (for the deaD mutant) or lack (for the ureB mutant) of the UreAB subunits observed in the Western blot analysis of these mutants. The localization of the immunogold particles for the ureI mutant was comparable to that in the parental strain, suggesting that in spite of the urease-negative phenotype, the surface-to-cytoplasm distribution (data not shown) was not altered in this mutant.
Screening the library for nonmotile mutants.
To further test the library for its usefulness for the identification of virulence genes, a subset was tested for the absence of motility, determined by an absence of swarming. Of 180 mutants screened, five displayed a nonmotile phenotype in the stab agar assay. Confirmation that the observed phenotype was due to the insertion of the vector into the chromosome was obtained by retransformation of the wild-type parental strain, 1061, with the isolated total chromosomal DNA of each mutant. From each backcross, 10 independent transformants, selected only for Kmr, were subsequently tested for absence of motility activity. In all cases, the acquisition of Kmr correlated with the acquisition of the nonmotile phenotype.
Characterization of nonmotile mutants.
To identify the disrupted ORFs in the nonmotile mutants, a plasmid rescue strategy was performed and the flanking sequence was determined (Table 2). In mutant 1, ORF 0080, an ORF whose product is not homologous to any known protein and that does not seem to be part of an operon-like structure, was disrupted. Interestingly, however, ORF 0082 is homologous to the methyl-accepting chemotaxis protein (34). Mutant 2 was disrupted 5′ of ORF 0876, which encodes a protein homologous to an iron-regulated outer membrane protein of Neisseria meningitidis (30). Integration of pBCα3 at this position apparently results in an interruption of the presumed promoter of this ORF. It seems unlikely that integration resulted in the disruption of the presumed promoter of ORF 0875, which is transcribed in the other direction, since ORF 0875 encodes catalase and this mutant is still catalase positive. The orientation of ORF 0877 is opposite to that of the disrupted frpB; thus, the phenotype of this mutant is probably caused solely by the disruption of frpB. The ability to sense essential cofactors, such as iron, and the subsequent expression of virulence genes, has been described in various bacteria (25). In mutant 3, the middle of ORF 0904, an ORF whose product is homologous to the phosphotransacetylase of Clostridium acetobutylicum (4), was disrupted. Since ORF 0904 and ORF 0903 are in the same orientation, they might form an operon; thus, the motility-negative phenotype could also result from a polar effect on ORF 0903, which encodes an acetate kinase homologue. In mutant 4, the middle of ORF 1274, an ORF whose product is homologous to the paralyzed flagellar protein of Campylobacter jejuni (41), was disrupted. As a C. jejuni mutant with an insertion in this gene was also nonmotile, it is likely that the phenotype is due to the insertion in ORF 1274, although a polar effect on ORF 1275 cannot be excluded. In mutant 5, ORF 1465, an ORF whose product is homologous to the histidine ATP-binding cassette transporter of Salmonella typhimurium (20), was disrupted. This ORF appears to be the second in an operon consisting of ORFs 1462 to 1466; thus, the phenotype might be due either to the inactivation of ORF 1465 or to the inactivation of ORFs 1465 to 1462.
TABLE 2.
Analysis of genes disrupted in nonmotile mutants, as determined by plasmid rescue and subsequent sequencing
| Mutant | Site of insertion (bp)a | H. pylori gene no.a | Homologue of interrupted ORF product |
|---|---|---|---|
| 1 | 85,141 | 0080 | None |
| 2 | 927,110 | 5′ end of 0876 | Iron-regulated outer membrane protein |
| 3 | 954,971 | 0904 | Phosphotransacetylase |
| 4 | 1,349,381 | 1274 | Paralyzed flagellar protein |
| 5 | 1,536,794 | 1465 | ABC transporter |
According to the sequence of Tomb et al. (37).
Analysis of the nonmotile mutants by EM.
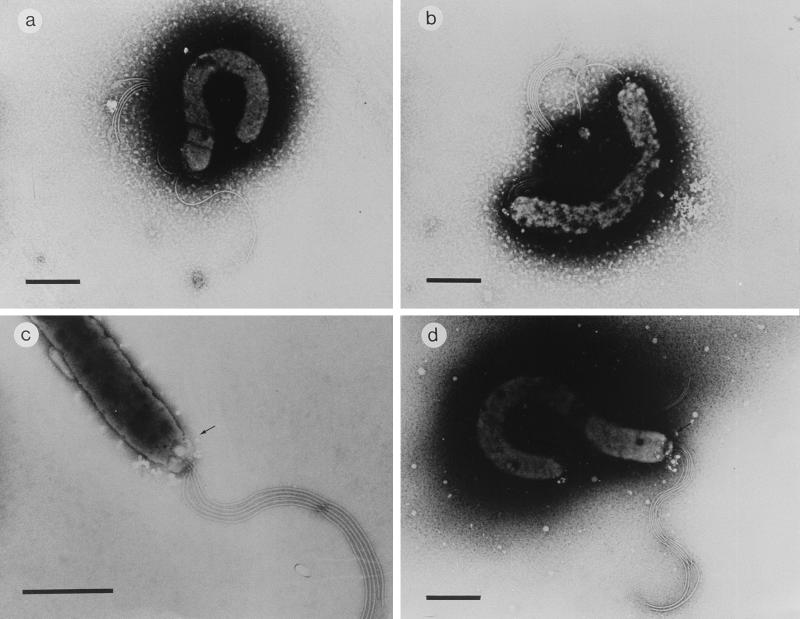
As none of the ORFs disrupted in these five mutants had previously been shown to be involved in the motility of H. pylori, we further characterized the mutants by EM (Fig. 5). They were compared to the parental strain (Fig. 5a). All mutants contained apparently normal flagella, and all of them possessed the terminal bulbs normally seen in H. pylori. However, some aberrations were observed. The most remarkable aberration was noted in mutant 3, where an extra flagellum at the other pole was visible in most of the bacteria (Fig. 5b). In most of the mutant 1 (Fig. 5c) and mutant 4 (Fig. 5d) bacteria, a cap-like structure was present at the site where the flagella protrude from the bacterium. Mutants 2 and 5 had a wild-type appearance in EM (data not shown).
FIG. 5.
Negatively stained electron micrographs of H. pylori nonmotile mutants. Representative results are shown for the wild type (a), mutant 3 (b), mutant 4 (c), and mutant 1 (d). Bar in panel a, 500 nm; bars in panels b through d, 1 μm. Arrow in panel c points to the cap-like structure at the site where the flagella protrude from the bacterium.
DISCUSSION
In this study, the construction of a direct random insertion library in H. pylori is described. The suitability of this library for the identification of putative virulence genes was demonstrated by the isolation of three urease-negative and five nonmotile mutants in part of this library. The mutants had disruptions of genes expected to be involved in these phenotypes (ureB, ureI, and pflA) as well as additional genes.
An integration plasmid, designated pBCα3 (Fig. 1), was constructed for H. pylori by the introduction of a kanamycin cassette into a plasmid that does not replicate in H. pylori. Introduction of pBCα3 containing random H. pylori chromosomal fragments into the parental strain, 1061, resulted in 250 kanamycin-resistant colonies per μg of DNA. A Southern blot of 16 arbitrarily selected mutants was probed with the 32P-labeled aphA-3 cassette (Fig. 3). The data obtained indicate that for each mutant, pBCα3 had integrated in a different part of the chromosome of strain 1061, but we cannot exclude the possibility that some parts of the chromosome are less likely to be hit.
Further indications for the randomness of the mutant library were obtained by two screens of the library. A subset of 1,251 single transformants was screened for the absence of urease activity, and a second subset of the library was tested for mutants with a total lack of motility. Three of 1,251 mutants were urease negative (0.25%), and 5 of 180 mutants (2.7%) were identified as nonmotile. For all mutants, the acquisition of the selected phenotypes correlated with the acquisition of kanamycin resistance when they were recreated in the parental strain, H. pylori 1061.
All fresh clinical isolates of H. pylori express significant amounts of urease, and this enzyme is regarded as one of the essential virulence factors of H. pylori. Isogenic urease-negative mutants of H. pylori and Helicobacter mustelae fail to colonize the gastric mucosae of gnotobiotic piglets and ferrets, respectively (1, 11). In vitro, urease activity in the presence of urea protects H. pylori from severe acid shocks (5, 27). Many genes have been shown to be involved in the urease activity of H. pylori, among them ureA and ureB, which encode the two subunits of the urease enzyme, the accessory genes ureE through ureI (6, 24), and nixA and abcABCD, genes involved in the transport of nickel, an essential cofactor for urease activity (2, 18). Of all these genes, only the disruption of ureB and ureG resulted in a completely urease-negative phenotype in H. pylori (12, 14).
Analysis of the urease-negative mutants revealed three disrupted ORFs. Of these three genes, only ureB has been reported to be essential for urease activity. The ureI gene is one of the five urease accessory genes that were identified as essential for urease activity in E. coli (6). Recently, a nonpolar ureI mutant of H. pylori was constructed, and this mutant is urease positive (35). Western blot analysis of our ureI mutant shows that both UreA and UreB are present at wild-type levels, indicating that the integration of pBCα3 has no effects on the expression of ureA and ureB. Therefore, it seems likely that the urease-negative phenotype of the ureI mutant is due to polar effects of the integration of the plasmid on the downstream urease accessory genes ureE, ureF, ureG, and ureH. It is unlikely that these genes are involved in the transport of the urease enzyme, because in this mutant no differences in urease localization were observed. Further experiments are required to establish which gene(s) is involved in the urease-negative phenotype and its exact function. The finding that in the third mutant a deaD homologue was disrupted indicates that urease is regulated by a putative RNA helicase, but it remains unclear how this would result in the complete absence of the UreB subunit while UreA is present in this mutant. These findings indicate that the DeaD protein affects the stability of the ureAB mRNA, as we did not find any other changes in the protein profile of this mutant (data not shown). Detailed analysis of this mutant will be required to elucidate the precise role of this helicase in the regulation of urease in H. pylori.
Motility is essential for full virulence of H. pylori, since mutants without flagella are unable to efficiently colonize gnotobiotic piglets (13). Several genes involved in the motility of H. pylori have been described; among them are two flagellar subunits, flaA and flaB (21), the hook protein, fliE (29), and a membrane protein involved in the expression of both subunits, flbA (33). In addition, 30 of the 1,590 putative ORFs identified in the H. pylori chromosome (≈2%) are homologous to genes known to be involved in motility in, e.g., E. coli and Salmonella spp. (26). A study of the exported proteins of H. pylori showed that 18 of the 185 (13.3%) secretion-deficient mutants generated were affected in motility, and 8 of those 18 mutants (4.3%) were completely nonmotile (28).
Analysis of the genes disrupted in the five nonmotile mutants found in this study revealed that in four of these mutants genes that had not been implicated in motility before were affected (Table 2). None of the five mutants had disruptions of a homologue of the ORFs known to be involved in the motility of E. coli and Salmonella, but we think this is because only a small subset of the library was tested for motility. EM analysis showed that all mutants still possessed flagella.
However, one of our mutants, mutant 4, had a disruption of ORF 1274 (Table 2), an ORF whose product is homologous to the paralyzed flagellar (plfA) protein of the closely related species C. jejuni (41). This gene was shown to be responsible for the nonmotile phenotype of a C. jejuni insertion mutant. In spite of their nonmotile phenotype, both our H. pylori mutant and the C. jejuni mutant still contained flagella in EM but showed an aberration in the region where the flagella protrude from the bacteria (Fig. 5c). Although nothing is known about the precise function of this gene, our data suggest that the H. pylori homologue has the same function as its C. jejuni counterpart.
Motility mutant 5 has a disruption of ORF 1465, located 2 ORFs upstream of ORF 1462, which is presumed to play a role in motility based on the work of Odenbreit et al. (28). This implies that this cluster of ORFs (1465 to 1462) might be arranged in an operon-like structure that is involved in motility.
Mutant 3 possesses flagella at each pole (Fig. 5b) and had a disruption of ORF 0904 (Table 2), which encodes a phosphotransacetylase homologue. This suggests that either this ORF is involved in the process that determines at which pole the flagella will be assembled or this mutation causes a defect in cell separation. However, in all mutants, polar effects of the interruption cannot be excluded at this stage.
The percentages of urease-negative mutants and nonmotile mutants in our study correlate with the percentages predicted by the genome sequence and previous findings. Furthermore, each of the eight mutants had a different ORF disrupted. This indicates that integration was evenly distributed over the chromosome and that theoretically the complexity of the complete library is large enough to cover the whole genome. As the fragments ligated into the integration plasmid have an average length of 500 bp, they will only contain parts of the ORF present in the chromosome of H. pylori. Hence it is not likely that these fragments will be negatively selected in E. coli due to toxic effects.
We conclude that our random mutant library of H. pylori represents a very useful tool for research into the virulence genes of H. pylori. Because mutation and selection take place directly in H. pylori, the only prerequisite is the careful choice of the selection criteria. This easy and simple method can be used with all readily transformable H. pylori strains and is useful for the identification of virulence genes and the analysis of the underlying regulatory mechanisms.
ACKNOWLEDGMENTS
We thank Monique Gerrits and Rene van Vugt for DNA sequence analysis and Wim Voorhout for excellent technical assistance with EM. We thank Michael Lie a Ling for excellent assistance in the urease screening sessions. We thank Paul Hoffman for the gift of H. pylori 1061 and for helpful discussions. We are grateful to Rainer Haas for the gift of pHel 3 prior to publication.
Part of this work was supported by Public Health Service grants CA67527 and DK39045 (to S.H.P.).
REFERENCES
- 1.Andrutis K A, Fox J G, Schauer D B, Marini R P, Li X, Yan L, Josenhans C, Suerbaum S. Infection of the ferret stomach by isogenic flagellar mutant strains of Helicobacter mustelae. Infect Immun. 1997;65:1962–1966. doi: 10.1128/iai.65.5.1962-1966.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauerfeind P, Garner R M, Mobley L T. Allelic exchange mutagenesis of nixA in Helicobacter pylori results in reduced nickel transport and urease activity. Infect Immun. 1996;64:2877–2880. doi: 10.1128/iai.64.7.2877-2880.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg D E, Hoffman P S, Appelmelk B J, Kusters J G. The Helicobacter pylori genome sequence: genetic factors for long life in the gastric mucosa. Trends Microbiol. 1997;5:468–474. doi: 10.1016/s0966-842x(97)01164-5. [DOI] [PubMed] [Google Scholar]
- 4.Boynton Z L, Bennett G N, Rudolph F B. Cloning, sequencing, and expression of genes encoding phosphotransacetylase and acetate kinase from Clostridium acetobutylicum ATCC 824. Appl Environ Microbiol. 1996;62:2758–2766. doi: 10.1128/aem.62.8.2758-2766.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clyne M, Labigne A, Drumm B. Helicobacter pylori requires an acidic environment to survive in the presence of urea. Infect Immun. 1995;63:1669–1673. doi: 10.1128/iai.63.5.1669-1673.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cussac V, Ferrero R L, Labigne A. Expression of Helicobacter pylori urease genes in Escherichia coli grown under nitrogen-limiting conditions. J Bacteriol. 1992;174:2466–2473. doi: 10.1128/jb.174.8.2466-2473.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dickinson J H, Grant K A, Park S F. Targeted and random mutagenesis of the Campylobacter coli chromosome with integrational plasmid vectors. Curr Microbiol. 1995;31:92–96. doi: 10.1007/BF00294282. [DOI] [PubMed] [Google Scholar]
- 8.Dunn B E, Campbell G P, Perez-Perez G I, Blaser M J. Purification and characterization of urease from Helicobacter pylori. J Biol Chem. 1990;265:9464–9469. [PubMed] [Google Scholar]
- 9.Dunn B E, Cohen H, Blaser M J. Helicobacter pylori. Clin Microbiol Rev. 1997;10:720–741. doi: 10.1128/cmr.10.4.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunn B E, Vakil N B, Schneider B G, Miller M M, Zitzer J B, Peutz T, Phadnis S H. Localization of Helicobacter pylori urease and heat shock protein in human gastric biopsies. Infect Immun. 1997;65:1181–1188. doi: 10.1128/iai.65.4.1181-1188.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eaton K A, Brooks C L, Morgan D R, Krakowka S. Essential role of urease in pathogenesis of gastritis induced by Helicobacter pylori in gnotobiotic piglets. Infect Immun. 1991;59:2470–2475. doi: 10.1128/iai.59.7.2470-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eaton K A, Krakowka S. Effect of gastric pH on urease-dependent colonization of gnotobiotic piglets by Helicobacter pylori. Infect Immun. 1994;62:3604–3607. doi: 10.1128/iai.62.9.3604-3607.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eaton K A, Morgan D R, Krakowka S. Motility as a factor in the colonization of gnotobiotic piglets by Helicobacter pylori. J Med Microbiol. 1992;37:123–127. doi: 10.1099/00222615-37-2-123. [DOI] [PubMed] [Google Scholar]
- 14.Ferrero R L, Cussac V, Courcoux P, Labigne A. Construction of isogenic urease-negative mutants of Helicobacter pylori by allelic exchange. J Bacteriol. 1992;174:4212–4217. doi: 10.1128/jb.174.13.4212-4217.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodwin A, Kersulyte D, Sisson G, Vanzanten S V, Berg D E, Hoffman P S. Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene (rdxa) that encodes an oxygen-insensitive NAD(P)H nitroreductase. Mol Microbiol. 1998;28:383–393. doi: 10.1046/j.1365-2958.1998.00806.x. [DOI] [PubMed] [Google Scholar]
- 16.Griffiths G, McDowall A, Back R, Dubochet J. On the preparation of cryosections for immunocytochemistry. J Ultrastruct Res. 1984;89:65–78. doi: 10.1016/s0022-5320(84)80024-6. [DOI] [PubMed] [Google Scholar]
- 17.Haas R, Meyer T F, Van Putten J. Aflagellated mutants of Helicobacter pylori generated by genetic transformation of naturally competent strains using transposon shuttle mutagenesis. Mol Microbiol. 1993;8:753–760. doi: 10.1111/j.1365-2958.1993.tb01618.x. [DOI] [PubMed] [Google Scholar]
- 18.Hendricks J K, Mobley H L. Helicobacter pylori ABC transporter: effect of allelic exchange mutagenesis on urease activity. J Bacteriol. 1997;179:5892–5902. doi: 10.1128/jb.179.18.5892-5902.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heuermann D, Haas R. A stable shuttle vector system for efficient genetic complementation of Helicobacter pylori strains by transformation and conjugation. Mol Gen Genet. 1998;257:519–528. doi: 10.1007/s004380050677. [DOI] [PubMed] [Google Scholar]
- 20.Higgins C F, Haag P D, Nikaido K, Ardeshir F, Garcia G, Ames G F. Complete nucleotide sequence and identification of membrane components of the histidine transport operon of S. typhimurium. Nature. 1982;298:723–727. doi: 10.1038/298723a0. [DOI] [PubMed] [Google Scholar]
- 21.Josenhans C, Labigne A, Suerbaum S. Comparative ultrastructural and functional studies of Helicobacter pylori and Helicobacter mustelae flagellin mutants: both flagellin subunits, FlaA and FlaB, are necessary for full motility in Helicobacter species. J Bacteriol. 1995;177:3010–3020. doi: 10.1128/jb.177.11.3010-3020.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelleher J E, Raleigh E A. A novel activity in Escherichia coli K-12 that directs restriction of DNA modified at CG dinucleotides. J Bacteriol. 1991;173:5220–5223. doi: 10.1128/jb.173.16.5220-5223.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kusters J G, Gerrits M M, Van Strijp J A, Vandenbroucke-Grauls C M. Coccoid forms of Helicobacter pylori are the morphologic manifestation of cell death. Infect Immun. 1997;65:3672–3679. doi: 10.1128/iai.65.9.3672-3679.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Labigne A, Cussac V, Courcoux P. Shuttle cloning and nucleotide sequences of Helicobacter pylori genes responsible for urease activity. J Bacteriol. 1991;173:1920–1931. doi: 10.1128/jb.173.6.1920-1931.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Litwin C M, Calderwood S B. Role of iron in regulation of virulence genes. Clin Microbiol Rev. 1993;6:137–149. doi: 10.1128/cmr.6.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Macnab R M. Genetics and biogenesis of bacterial flagella. Annu Rev Genet. 1992;26:131–158. doi: 10.1146/annurev.ge.26.120192.001023. [DOI] [PubMed] [Google Scholar]
- 27.Marshall B J, Barrett L J, Prakash C, McCallum R W, Guerrant R L. Urea protects Helicobacter (Campylobacter) pylori from the bactericidal effect of acid. Gastroenterology. 1990;99:697–702. doi: 10.1016/0016-5085(90)90957-3. [DOI] [PubMed] [Google Scholar]
- 28.Oderbreit S, Till M, Haas R. Optimized blam-transposon shuttle mutagenesis of Helicobacter pylori allows the identification of novel genetic loci involved in bacterial virulence. Mol Microbiol. 1996;20:361–373. doi: 10.1111/j.1365-2958.1996.tb02623.x. [DOI] [PubMed] [Google Scholar]
- 29.O’Toole P W, Kostrzynska M, Trust T J. Non-motile mutants of Helicobacter pylori and Helicobacter mustelae defective in flagellar hook production. Mol Microbiol. 1994;14:691–703. doi: 10.1111/j.1365-2958.1994.tb01307.x. [DOI] [PubMed] [Google Scholar]
- 30.Pettersson A, Maas A, van Wassenaar D, van der Ley P, Tommassen J. Molecular characterization of FrpB, the 70-kilodalton iron-regulated outer membrane protein of Neisseria meningitidis. Infect Immun. 1995;63:4181–4184. doi: 10.1128/iai.63.10.4181-4184.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phadnis S H, Parlow M H, Levy M, Ilver D, Caulkins C M, Connors J B, Dunn B E. Surface localization of Helicobacter pylori urease and a heat shock protein homolog requires bacterial autolysis. Infect Immun. 1996;64:905–912. doi: 10.1128/iai.64.3.905-912.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Schmitz A, Josenhans C, Suerbaum S. Cloning and characterization of the Helicobacter pylori flbA gene, which codes for a membrane protein involved in coordinated expression of flagellar genes. J Bacteriol. 1997;179:987–997. doi: 10.1128/jb.179.4.987-997.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seidel R, Scharf B, Gautel M, Kleine K, Oesterhelt D, Engelhard M. The primary structure of sensory rhodopsin II: a member of an additional retinal protein subgroup is coexpressed with its transducer, the halobacterial transducer of rhodopsin II. Proc Natl Acad Sci USA. 1995;92:3036–3040. doi: 10.1073/pnas.92.7.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skouloubris S, Thiberge J M, Labigne A, Dereuse H. The Helicobacter pylori UreI protein is not involved in urease activity but is essential for bacterial survival in vivo. Infect Immun. 1998;66:4517–4521. doi: 10.1128/iai.66.9.4517-4521.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Solnick J V, Josenhans C, Suerbaum S, Tompkins L S, Labigne A. Construction and characterization of an isogenic urease-negative mutant of Helicobacter mustelae. Infect Immun. 1995;63:3718–3721. doi: 10.1128/iai.63.9.3718-3721.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzegerald L M, Lee N, Adams M D, Venter J C. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 38.Toone W M, Rudd K E, Friesen J D. deaD, a new Escherichia coli gene encoding a presumed ATP-dependent RNA helicase, can suppress a mutation in rpsB, the gene encoding ribosomal protein S2. J Bacteriol. 1991;173:3291–3302. doi: 10.1128/jb.173.11.3291-3302.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y, Roos K P, Taylor D E. Transformation of Helicobacter pylori by chromosomal metronidazole resistance and by a plasmid with a selectable chloramphenicol resistance marker. J Gen Microbiol. 1993;139:2485–2493. doi: 10.1099/00221287-139-10-2485. [DOI] [PubMed] [Google Scholar]
- 41.Yao R, Burr D H, Doig P, Trust T J, Niu H, Guerry P. Isolation of motile and non-motile insertional mutants of Campylobacter jejuni: the role of motility in adherence and invasion of eukaryotic cells. Mol Microbiol. 1994;14:883–893. doi: 10.1111/j.1365-2958.1994.tb01324.x. [DOI] [PubMed] [Google Scholar]