Abstract
Purpose
While immunotherapy has revolutionized the oncology field, variations in therapy responsiveness limit the broad applicability of these therapies. Diagnostic imaging of immune cell, and specifically CD8+ T cell, dynamics could allow early patient stratification and result in improved therapy efficacy and safety. In this study, we report the development of a nanobody-based immunotracer for non-invasive SPECT and PET imaging of human CD8+ T-cell dynamics.
Methods
Nanobodies targeting human CD8β were generated by llama immunizations and subsequent biopanning. The lead anti-human CD8β nanobody was characterized on binding, specificity, stability and toxicity. The lead nanobody was labeled with technetium-99m, gallium-68 and copper-64 for non-invasive imaging of human T-cell lymphomas and CD8+ T cells in human CD8 transgenic mice and non-human primates by SPECT/CT or PET/CT. Repeated imaging of CD8+ T cells in MC38 tumor-bearing mice allowed visualization of CD8+ T-cell dynamics.
Results
The nanobody-based immunotracer showed high affinity and specific binding to human CD8 without unwanted immune activation. CD8+ T cells were non-invasively visualized by SPECT and PET imaging in naïve and tumor-bearing mice and in naïve non-human primates with high sensitivity. The nanobody-based immunotracer showed enhanced specificity for CD8+ T cells and/or faster in vivo pharmacokinetics compared to previous human CD8-targeting immunotracers, allowing us to follow human CD8+ T-cell dynamics already at early timepoints.
Conclusion
This study describes the development of a more specific human CD8+ T-cell-targeting immunotracer, allowing follow-up of immunotherapy responses by non-invasive imaging of human CD8+ T-cell dynamics.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00259-024-06896-3.
Keywords: Immuno-imaging, Nuclear imaging, Nanobodies, T-cell dynamics
Introduction
In the last decade, immunotherapy has revolutionized the oncology field and has become the standard of care for some cancer types [1]. However, variations in therapy responsiveness and side effects limit the broad applicability and effectiveness of these therapies [2]. Diagnostic imaging of immune cell dynamics during immunotherapy could allow early patient stratification and result in improvements regarding therapy efficacy, safety and therapy costs [3]. Consequently, the development of diagnostic imaging tracers, capable of non-invasively visualizing distinct immune cell populations, has gained a lot of interest over the last years.
Among the key players in immunotherapy responses, cytotoxic CD8+ T cells have received particular attention [4]. Indeed, CD8+ T cell presence and cell density during immune checkpoint therapy has been associated with therapy response [5, 6]. As a result, several PET imaging tracers have been developed to target murine or human CD8+ T cells, facilitating the monitoring and/or prediction of immunotherapy responses [4, 7–10]. However, most of these tracers suffer from suboptimal pharmacokinetics (i.e. long circulation time), necessitating the use of long-lived radionuclides and potentially raising concerns regarding radiotoxicity [3]. Furthermore, all of these tracers bind the CD8α-chain, which is also expressed on other types of immune cells, decreasing the tracer’s specificity for CD8+ T cells [11, 12].
Nanobodies (Nbs), camelidae heavy chain-only antibody fragments, have emerged as interesting scaffolds for the development of diagnostic tracers [13]. Nbs are smaller (15 kDa) than conventional monoclonal antibodies, while still maintaining their ability to bind their target with high affinity and specificity [14]. This results in rapid tumor uptake and blood clearance, allowing fast and specific imaging of immune cells and a lower radiation burden [15].
In this study, we describe the development of a radiolabeled anti-human (h)CD8β Nb, allowing non-invasive imaging of human CD8+ T cells by SPECT and PET. These Nb-based tracers show high affinity and specific binding to CD8+ T cells and ideal in vivo pharmacokinetics. Furthermore, we show the ability of these Nb-based tracers to follow up immune cell dynamics during tumor growth. Overall, this study describes the development of a more specific hCD8+ T-cell-targeting tracer, allowing the follow-up of hCD8+ T-cell dynamics.
Materials and methods
Cell culture
SUP-T1 cells were purchased from ATCC (Wesel, Germany). MC38 cells were provided by Massimiliano Mazzone (VIB-KU Leuven, Belgium). Primary PBMCs of healthy volunteers were provided by Karine Breckpot (VUB, Belgium). Details regarding culturing can be found in the supplementary material and methods section.
Animal models
Male and female C57BL6/J mice, nu(ncr)-foxn1nu and human CD8 transgenic mice (B6;SJL-Tg(CD8αCD8β)57Scr/J) were purchased from Charles River (Ecully, France) or Jackson laboratory (Bar Harbor, USA). In case of tumor-bearing mice, mice were subcutaneously injected with 1 million MC38 or 5 million SUP-T1 cells in the flank. In case of SUP-T1 cells, cells were resuspended in 50% Matrigel (Corning, Somerville, USA). Mice were examined daily and tumor growth was measured using a caliper. Tumor volume was calculated using the formula (length × width2)/2. Two young adult male cynomolgus macaques (Macaca fascicularis), aged 4 and 5 years, F2 generation originating from Mauritian AAALAC certified breeding centers, were used.
Nanobody generation and production
Two llamas were subcutaneously injected 6 times with recombinant human (h)CD8β protein. After immunizations, peripheral blood was collected to generate phage display libraries and perform biopanning [16]. More details can be found in the supplementary material and methods section. Nanobodies were produced and purified as previously described [17].
Surface plasmon resonance
The affinity of purified anti-hCD8β nanobody to recombinant hCD8αβ protein (Sino Biological) was determined using a BIACORE-T200 device (Cytiva, Freiburg, Germany). More details can be found in the supplementary material and methods section.
Affinity determination using flow cytometry
Serial dilution of the anti-hCD8β nanobody was incubated with 500.000 SUP-T1 cells in FACS buffer (HBSS (Gibco) supplemented with 1% FBS and 2mM EDTA (Duchefa Biochemie, Haarlem, The Netherlands)) for 1 h at 4 °C. Cells were washed once with FACS buffer. Next, nanobody binding was detected by incubation of the cells with an Alexa Fluor®-488 tagged anti-HA antibody (1:1000 in FACS buffer, clone 16B12, Biolegend, San Diego, USA) or PE-conjugated rabbit anti-camelid VHH cocktail (1:500 in FACS buffer, Genscript, Piscataway, USA) for 30 min at 4 °C. Cells were washed once with FACS buffer. Nanobody binding was determined using the FACS CANTO II analyser (BD Biosciences, Franklin Lakes, USA). The mean fluorescence intensity of nanobody binding was determined using FlowJo version 10.
Affinity determination using ELISA
A 96 well MicroWell MaxiSorp flat bottom plate (Thermo Fisher Scientific) was coated with 0.2 µg of recombinant hCD8αβ protein, cynomolgus CD8β-Fc protein (Sino Biologicals) or PBS overnight at 4 °C. The next day, wells were washed with PBS-T (PBS + 0.05% Tween20 (Merck-Millipore, Burlington, USA) and blocked with blocking buffer (2% skimmed milk powder (Régilait) in PBS) for 1 h at room temperature (RT). Different concentrations of nanobodies, diluted in blocking buffer, were incubated for 1 h at RT. Nanobody binding was detected using a mouse-anti-HA antibody (1:2000, clone 16B12, Biolegend) and alkaline-phosphatase conjugated goat-anti-mouse antibody (1:2000, clone A90-116AP, Bethyl Laboratories, Montgomery, USA). Wells were washed with PBS-T between all incubation steps. Binding was determined using p-nitrophenyl phosphate (2 mg/mL resuspended in AP blot buffer (12.12 g/L Trizma base, 10.17 g/L MgCl2.6H20, 5.84 g/L NaCl, pH 9.5); Thermo Fisher Scientific). Absorbance at 405 nm was measured using a VersaMax ELISA Microplate Reader (Molecular Devices, San Jose, USA).
Thermal shift assay
The anti-hCD8β nanobody (concentration 0.2 mg/mL) was mixed with 1x SYPRO™ Orange Protein Gel Stain (Thermo Fisher Scientific) in PBS and added to white 96-well PCRs plates (Bio Rad, Pleasanton, CA, USA). Fluorescence signal was measured during increasing temperature steps ranging from 20 to 95 °C, with stepwise increments of 0.5 °C, using CFX connect™ Real-Time PCR (Bio Rad). The melting temperature of the nanobody was calculated using the Boltzmann equation.
Nanobody binding and T-cell activation assay using primary PBMCs
Primary PBMCs of 3 independent donors were co-incubated with different nanobodies to assess binding and T-cell activation. More details can be found in the supplementary material and methods section.
Immunofluorescence staining of non-human primate tissue
Lymph node tissues were stained using Alexa Fluor®-647 conjugated anti-hCD8β or irrelevant nanobody. More details can be found in the supplementary material and methods section.
Dendritic cell/T cell restimulation experiments
Immunogenicity of the nanobodies was determined using a dendritic cell/T cell restimulation assay and was outsourced to Lonza (Basel, Switzerland). The assay was performed using PBMCs of 30 pre-HLA-typed healthy donors as described previously [18].
99mTc-labeling of nanobodies
Nanobodies were labeled with technetium-99m as previously described [19]. More details can be found in the supplementary material and methods section.
SPECT-CT imaging and image analysis
SPECT-CT imaging and image analysis were performed as previously described [20]. More details can be found in the supplementary material and methods section.
Alphafold nanobody binding prediction
Nanobody binding models to human CD8 were generated using Colabfold (patch v1.5.2) [21]. The number of recycles were set to 6 while all other standard parameters were left unchanged. Analysis of the Alphafold model was done using pyMOL.
NOTA-conjugation of nanobodies
The conjugation of the anti-hCD8β nanobody to p-SCN-Bn-NOTA (NOTA-NCS, Macrocyclics, Inc., Plano, USA) was based on the standard protocol previously described with some adaptations [22]. More details can be found in the supplementary material and methods section.
68Ga-labeling of nanobodies
Nanobodies were labeled with gallium-68 as previously described [23]. More details can be found in the supplementary material and methods section.
PET/CT imaging and image analysis in mice
Mice were injected (i.v.) with 5 µg of radiolabeled nanobody (15.5 ± 0.34 MBq; 39.7 MBq/nmol (anti-hCD8β nanobody) or 44.3 MBq/nmol (irrelevant nanobody)). One hour post-injection, mice were anesthetized with 75 mg/kg ketamine and 1 mg/kg medetomidine by intraperitoneal injection or isoflurane (5% induction, 2.5% maintenance, oxygen flow rate between 0.3 and 1.5 L/min; Virbac, Nice, France) by inhalation during PET/CT imaging (MoleCubes, Gent, Belgium). PET scans of 12–20 min were performed, followed by a CT scan. After imaging, mice were euthanized and organs were collected. Radioactivity in each organ was measured using a Wizard2 γ-counter (Perkin-Elmer). PET/CT image analysis was performed using VivoQuant software (Invicro, Needham, MA, USA).
Processing organs and flow cytometry analysis
Single cell preparations of MC38 tumors were prepared as previously described [24]. Antibodies used for staining of single cell preparations can be found in Supplemental Table 1. Delta median fluorescence intensity (ΔMFI) was determined by subtraction of the MFI of the staining and the MFI of the isotype control. Data were acquired using the FACS CANTO II or FACS CELESTA analyser and analyzed using FlowJo software.
64Cu-labeling of nanobodies
Copper-64 (64Cu) in 1 M HCl (1 GBq, 375 µL, ARRONAX, Nantes, France) was concentrated at 90 °C under an argon stream to dryness. NaOAc buffer 0.1 M was prepared and the pH was adjusted to 6.5 using HCl. Then, 365 µL of NaOAc buffer was added to solubilized [64Cu]CuCl2 and this solution was transferred to NOTA-hCD8β Nb. The resulting mixture was stirred at 500 rpm in a thermoshaker at 37 °C. Radio-TLC was performed, using 50 mM citric acid as eluent, to monitor the reaction. Full conversion was observed after 1 h (Rf = 0.05) as no residual free 64Cu was observed (Rf = 0.9). In the meantime, a PD-10 column (GE Healthcare, USA) was rinsed with 20 mL of PBS. The reaction mixture was loaded on the column and the flow-through was discarded. PBS was used as eluent and the flow-through was collected in 500 µL fractions. Radioactivities were measured in a dose calibrator (Capintec®, Berthold, France), fractions showing the highest activities were pooled together and analyzed by SEC chromatography (Alliance e2695 system, Waters, USA). Radiolabeled nanobodies were identified as radioactive peak detected by a gamma detector (Berthold, France).
PET/CT imaging and image analysis of macaques
All imaging acquisitions were performed using the Digital Photon Counting (DPC) PET/CT system (Vereos-Ingenuity, Philips). Animals were first anesthetized with 10 mg/kg ketamine and 0.05 mg/kg medetomidine, intubated, and then maintained under 0.5-1% isoflurane and placed in a supine position on a warming blanket (Bear Hugger, 3 M) on the machine bed with monitoring of the cardiac rate, oxygen saturation, and temperature. The CT detector collimation used was 64 × 0.6 mm, the tube voltage was 120 kV, and the intensity was approximately 150 mA. Whole-body CT images were reconstructed with a slice thickness of 1.5 mm and an interval of 0.75 mm. A whole-body PET scan (5 bed positions, 1 min/bed position) was performed approximately 60 min post-injection of 500 µg of [64Cu]Cu-NOTA-anti-hCD8β Nb via the saphenous vein (230 ± 23 MBq, 5.9 MBq/nmol). PET images were reconstructed onto a 256 × 256 matrix using OSEM (3 iterations, 15 subsets). PET and CT images were analyzed using INTELLISPACE PORTAL 8 (Philips Healthcare) and 3DSlicer (open-source tool) software. For segmentation, various regions of interest were semi-automatically contoured according to anatomical information and PET signal. A 3D volume of interest (VOI) was interpolated from several ROIs in different image slices to cover the entire organ or anatomical structure. Mean radioactive signal in each VOI was expressed in mean standardized uptake value (SUVmean ± S.D.).
Results
The anti-human CD8β nanobody has ideal in vitro characteristics
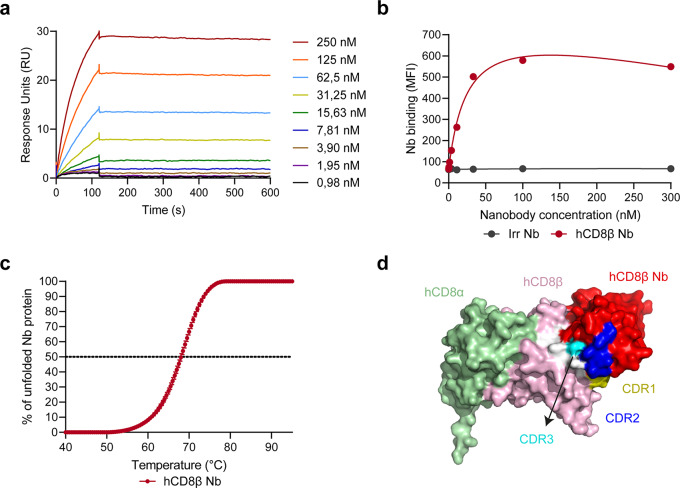
Immunization followed by phage display panning, screening and characterization resulted in the selection of a lead anti-hCD8β Nb. The anti-hCD8β Nb displayed low nanomolar binding affinity to the hCD8αβ protein (Fig. 1a and Supplemental Table 2). In line with this, a similar binding affinity to SUP-T1 cells, a T-cell lymphoma cell line endogenously expressing hCD8αβ, was observed, while no binding was seen with an irrelevant Nb, binding the 5T2 multiple myeloma M protein (Fig. 1b and Supplemental Table 1). Next, we assessed the cross-reactive binding of the anti-hCD8β Nb (Supplemental Fig. 1 and Supplemental Table 2). While no Nb binding to murine CD8β protein was observed (Supplemental Fig. 1a), similar low nanomolar binding affinities to the human and cynomolgus CD8β proteins were seen (Supplemental Fig. 1b). This result is in line with the poor homology between the human and mouse proteins (50%), and the very high homology between human and cynomolgus CD8β proteins (93%). Subsequently, the thermostability of the Nb was determined, considering that the Nb will be incubated at higher temperatures during radiolabeling (Fig. 1c and Supplemental Table 2). The anti-hCD8β Nb displayed a melting temperature of 68.1 ± 0.3 °C. Finally, the binding epitope of the anti-hCD8β Nb was modelled using Alphafold (Fig. 1d). Although the predicted local distance difference test (pLDDT) score of the model indicated that the exact interaction between the CDR regions of the Nb and CD8 could only be estimated (Supplemental Fig. 2), the Alphafold model nicely predicted binding to the β chain of human CD8.
Fig. 1.
The anti-human CD8β nanobody binds to human CD8 with high affinity. (a) Surface plasmon resonance plots of different nanobody concentrations to the human CD8αβ protein. (b) Binding of different human CD8β-targeting nanobody (hCD8β Nb) or irrelevant nanobody (Irr Nb) concentrations to T-cell-lymphoma SUP-T1 cells. Binding was detected as mean fluorescence intensity (MFI) via the C-terminal HA-tag and a fluorescently labeled anti-HA antibody using flow cytometry. (c) Thermostability of the hCD8β Nb was measured via a Thermofluor Assay using Sypro Orange dye. The percentage of unfolded protein was determined at increasing temperatures to calculate the melting temperature (50% unfolded protein). Data is presented as mean ± S.D. (d) Alphafold prediction model of the binding between the anti-hCD8β Nb and human CD8
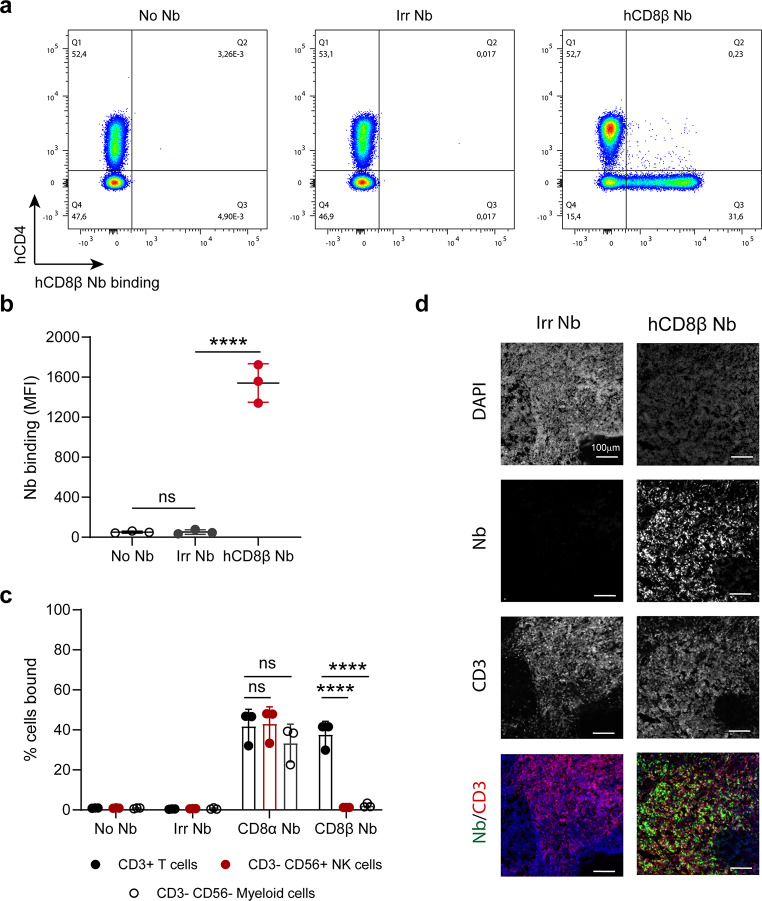
Next, we determined the ability of the anti-hCD8β Nb to bind primary CD8+ T cells ex vivo (Fig. 2a-b). While no binding was seen for the irrelevant Nb, the anti-hCD8β Nb bound the CD8+ T cell population within a pool of primary T cells. Moreover, the anti-hCD8β Nb did not show any binding to NK or myeloid cells, known to express hCD8α but not hCD8β (Fig. 2c and Supplemental Fig. 3). In contrast, binding of an anti-hCD8α Nb (WO2017134306A1) to NK or myeloid cells was observed, illustrating the superior specificity of the anti-hCD8β Nb for CD8+ T cells. Next, binding of the anti-hCD8β Nb to non-human primate lymph node tissue was determined using fluorescence immunohistochemistry (Fig. 2d). While no binding of the irrelevant Nb was observed, the anti-hCD8β Nb bound to CD3+ T cells present in the lymph node tissue.
Fig. 2.
The anti-human CD8β nanobody binds specifically to primary CD8+ T cells. (a) Binding of an irrelevant nanobody (Irr Nb) or the human CD8β-targeting nanobody (hCD8β Nb) to primary T cells obtained from a healthy donor. CD3+ T cells (CD45+, CD11b-, CD19-, CD3+) were gated and anti-CD4 mAb staining was plotted against Nb binding. Nb binding was compared to a sample without any Nb (No Nb) and detected via the C-terminal HA-tag by a fluorescently labeled anti-HA antibody. (b) Quantification of Nb binding to CD8+ T cells. Nanobody binding on primary T cells of 3 independent donors was detected as mean fluorescence intensity (MFI). (c) Binding of an Irr Nb, anti-human CD8α Nb or hCD8β Nb to primary T cells, NK cells and myeloid cells obtained from 3 independent healthy donors. T cells (CD45+, CD11b-, CD19-, CD3+), NK cells (CD45+, CD11b-, CD19-, CD3-, CD56+) or myeloid cells (CD45+, CD19-, CD3-, CD56-) were gated. Nb binding was compared to a sample without any Nb (No Nb) and detected via the C-terminal HA-tag by a fluorescently labeled anti-HA antibody. (d) Fluorescence immunohistochemistry staining of non-human primate lymph node tissue with a fluorescently labeled Irr Nb or hCD8β Nb. Nuclei were stained using DAPI and CD3+ T cells were detected using a fluorescently labeled anti-CD3 antibody. All data are presented as mean ± S.D.. Statistical analyses were performed using one-way ANOVA with Dunnett’s post hoc test or two-way ANOVA with Šídák’s multiple comparisons test. ns, p > 0.05; ****, p < 0.0001
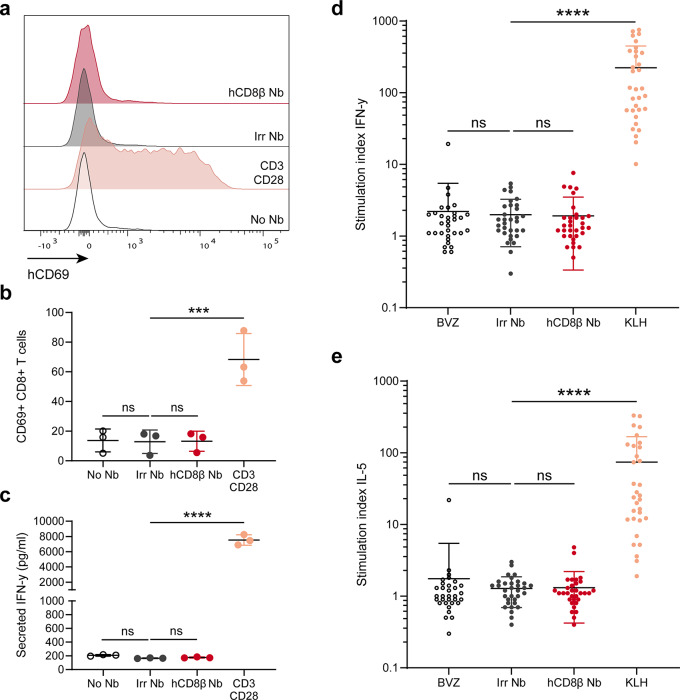
To ensure lack of unwanted cytotoxicity, we assessed the effect of Nb binding on T-cell activation. Hereto, primary human peripheral blood mononuclear cells (PBMCs) were incubated overnight with the anti-hCD8β Nb, irrelevant Nb or anti-CD3/CD28 dynabeads. The next day, T-cell activation was assessed by CD69 expression, an early activation marker (Fig. 3a-b), and secretion of IFN-γ (Fig. 3c). Stimulation with anti-CD3/CD28 dynabeads resulted in a significant increase of CD69 expression and IFN-γ secretion. In contrast, no changes in CD69 expression nor secreted IFN- γ levels were observed upon incubation with the anti-hCD8β Nb or the irrelevant Nb. Finally, the immunogenicity of the Nbs was determined in a dendritic cell-T cell co-culture assay using PBMCs of 30 independent healthy donors (Fig. 3d-e). To this end, monocyte-derived dendritic cells (moDCs) were loaded with the Nbs and matured, followed by CD4+ T cells co-culturing and a rechallenge. Afterwards, IFN-γ (Th1 cytokine) and IL-5 (Th2 cytokine) levels were assessed. As negative and positive controls, we took along clinically-benchmarked Bevacizumab and immunogenic KLH protein. In contrast to KLH, neither of the Nbs showed any sign of immunogenicity, as no significant increase of IFN-γ nor IL-5 levels was observed.
Fig. 3.
The anti-human CD8β nanobody does not induce T-cell activation. (a) Histogram plots of human CD69 expression on CD8+ T cells without treatment (No Nb) or after overnight incubation with anti-CD3/CD28 dynabeads, GLP-grade irrelevant nanobody (Irr Nb) or human CD8β-targeting nanobody (hCD8β Nb) to human primary peripheral blood mononuclear cells. (b) Quantification of human CD69+ CD8+ T cells without treatment (No Nb) or after overnight incubation with anti-CD3/CD28 dynabeads, GLP-grade Irr Nb or hCD8β Nb. (c) ELISA quantification of secreted IFN-γ levels in the supernatant of human primary peripheral blood mononuclear without treatment (No Nb) or after overnight incubation with anti-CD3/CD28 dynabeads, GLP-grade Irr Nb or hCD8β Nb. d-e) Stimulation indices of IFN-γ (d) and IL-5 (e) in the DC/CD4+ T cell (from 30 healthy donors) restimulation assays after incubation with non-immunogenic Bevacizumab (BVZ), immunogenic keyhole limpet hemocyanin (KLH), GLP-grade Irr Nb or hCD8β Nb. Stimulation index indicates the amount of positive cells compared to untreated cells. All data are presented as mean ± S.D.. Statistical analyses were performed using one-way ANOVA with Dunnett’s post hoc test. ns, p > 0.05; ***, p < 0.001; ****, p < 0.0001
[99mTc]Tc-anti-human CD8β nanobody non-invasively images CD8+ T cells in naïve and tumor-bearing mice
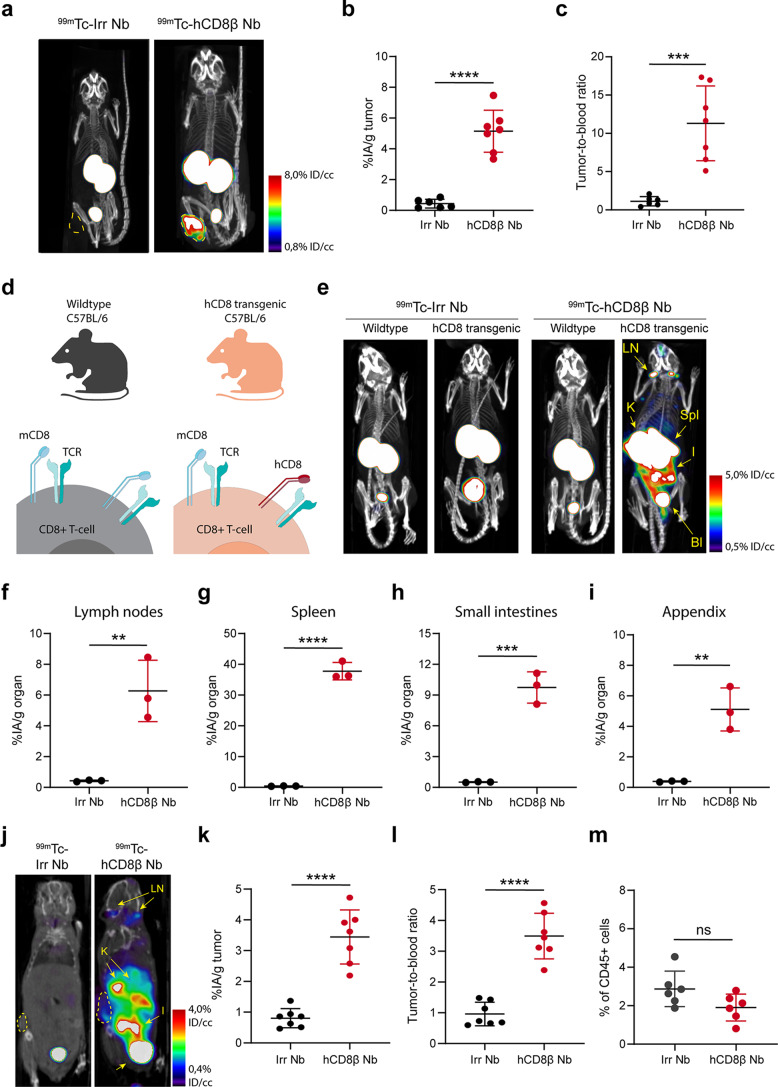
Next, we determined the potential of the anti-hCD8β Nb to visualize hCD8+ T cells in vivo. To this end, the irrelevant and anti-hCD8β Nb were site-specifically radiolabeled with technetium-99m (99mTc) on their C-terminal His-tag. Both Nbs were successfully labeled (Supplemental Table 3). First, the targeting potential of the [99mTc]Tc-Nbs was assessed in CD8+ SUP-T1 tumor-bearing nude mice, which lack endogenous T cells, by SPECT/CT imaging 1 h after intravenous (i.v.) injection (Fig. 4a). Both [99mTc]Tc-Nbs showed a high uptake in the kidneys and the bladder, due to rapid blood clearance [13]. The [99mTc]Tc-anti-hCD8β Nb showed a significantly higher uptake in the SUP-T1 tumors as compared to the [99mTc]Tc-irrelevant Nb, as confirmed by ex vivo γ-counting and tumor-to-blood ratio calculations (Fig. 4b-c and Supplemental Fig. 4). The in vivo specificity of the [99mTc]Tc-anti-hCD8β Nb was further determined in J wild type (WT) and hCD8 transgenic mice (Fig. 4d-e). The [99mTc]Tc-irrelevant Nb did not show any specific accumulation in either mouse. In contrast, the [99mTc]Tc-anti-hCD8β Nb showed uptake in T-cell-rich organs such as lymph nodes, spleen, intestines, lungs and appendix of hCD8 transgenic, but not WT mice [25]. Again, these data were confirmed by ex vivo γ-counting of the isolated organs (Fig. 4f-i and Supplemental Fig. 5). Finally, the ability of the [99mTc]Tc-anti-hCD8β Nb to target intratumoral T cells was assessed in a MC38 tumor model. The [99mTc]Tc-anti-hCD8β Nb showed a significantly higher uptake in the tumor compared to the [99mTc]Tc-irrelevant Nb (Fig. 4j-k and Supplemental Fig. 6a). Moreover, the [99mTc]Tc-anti-hCD8β Nb showed a significantly higher tumor-to-blood ratio compared to the [99mTc]Tc-irrelevant Nb (Fig. 4l and Supplemental Fig. 6b) indicating that the anti-hCD8β Nb binds to intratumoral hCD8+ T cells.
Fig. 4.
SPECT/CT imaging with the 99mTc-labeled anti-human CD8β nanobody. (a) Representative SPECT/CT images of SUP-T1 tumor-bearing mice (tumor outlined in yellow), intravenously injected with 99mTc-labeled irrelevant nanobody (Irr Nb) or human CD8β-targeting nanobody (hCD8β Nb). (b) Ex vivo γ-counting of the isolated tumors 80 min after injection with 99mTc-labeled Nbs. Uptake of the Nbs is expressed as injected activity per gram of tumor (%IA/g). (c) Tumor-to-blood ratios of the 99mTc-labeled Nbs, calculated by dividing the %IA/g tumor by the %IA/g blood. (d) Schematic cartoon of mouse (m) and human (h) CD8 expression on mouse CD8+ T cells of C57BL/6J wild type and human CD8 transgenic mice. (e) Representative SPECT/CT images of wild type and human (h)CD8 transgenic mice intravenously injected with 99mTc-labeled Nbs. Lymph nodes (LN), spleen (Spl), kidneys (K), bladder (Bl) and small intestines (I) are highlighted. f-i) Ex vivo γ-counting of the isolated lymph nodes (f), spleen (g), small intestines (h) and appendix (i) 80 min after injection with 99mTc-labeled Nbs. j) Representative coronal section of a MC38 tumor-bearing hCD8 transgenic mouse intravenously injected with 99mTc-labeled Nbs. The MC38 tumor is delineated. Lymph nodes (LN) and kidneys (K) are highlighted. k) Ex vivo γ-counting of the isolated MC38 tumors 80 min after injection with 99mTc-labeled Nbs. l) Tumor-to-blood ratios of the 99mTc-labeled Nbs in MC38 tumor-bearing mice. All data are presented as mean ± S.D.. Statistical analyses were performed using a two-tailed unpaired t test. ns, p > 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001
The anti-human CD8β PET tracer visualizes human CD8+ T-cell dynamics in vivo
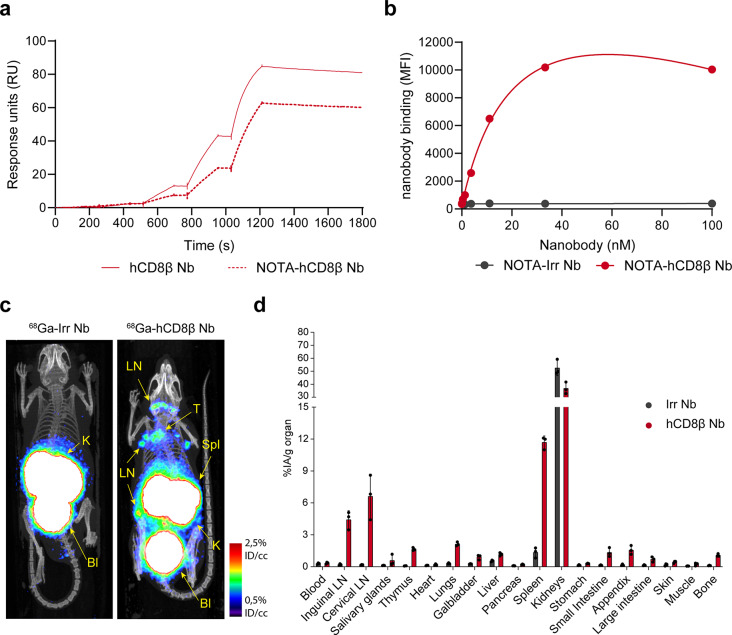
As PET imaging enables a higher spatial resolution and is more commonly used in the clinic, the anti-hCD8β Nb was converted to a PET tracer. To this end, the Nb was first conjugated to NOTA on its lysines and subsequently radiolabeled with gallium-68 (68Ga). A radiochemical purity above 98% was already observed before purification (Supplemental Table 3). In addition, the [68Ga]Ga-NOTA-anti-hCD8β Nb remained stable after incubation in injection buffer and human serum, at room temperature or at 37 °C (Supplemental Fig. 7). Furthermore, NOTA-coupling did not interfere with Nb binding to hCD8 as seen by surface plasmon resonance and flow cytometry (Fig. 5a-b and Supplemental Table 4).
Fig. 5.
PET/CT imaging with 68Ga-labeled anti-human CD8β nanobody. (a) Surface plasmon resonance plots of the kinetic titration of increasing concentrations of human CD8β-targeting nanobody (hCD8β Nb) and NOTA-conjugated hCD8β Nb concentrations to human CD8αβ protein. (b) Binding of different concentrations of the NOTA-conjugated hCD8β Nb or an irrelevant nanobody (Irr Nb) to T-cell-lymphoma SUP-T1 cells. Binding was detected as mean fluorescence intensity (MFI) via a fluorescently-labeled anti-VHH antibody using flow cytometry. (c) Representative PET/CT image of human CD8 transgenic mice intravenously injected with 68Ga-labeled Irr Nb or hCD8β Nb. Lymph nodes (LN), spleen (Spl), thymus (T), kidneys (K) and bladder (Bl) are highlighted. (d) Ex vivo γ-counting of the organs 80 min after injection with 68Ga-labeled Nbs. Biodistribution of the nanobodies in three mice is shown and uptake of the Nbs is expressed as injected activity per gram (%IA/g). All data are presented as mean ± S.D.
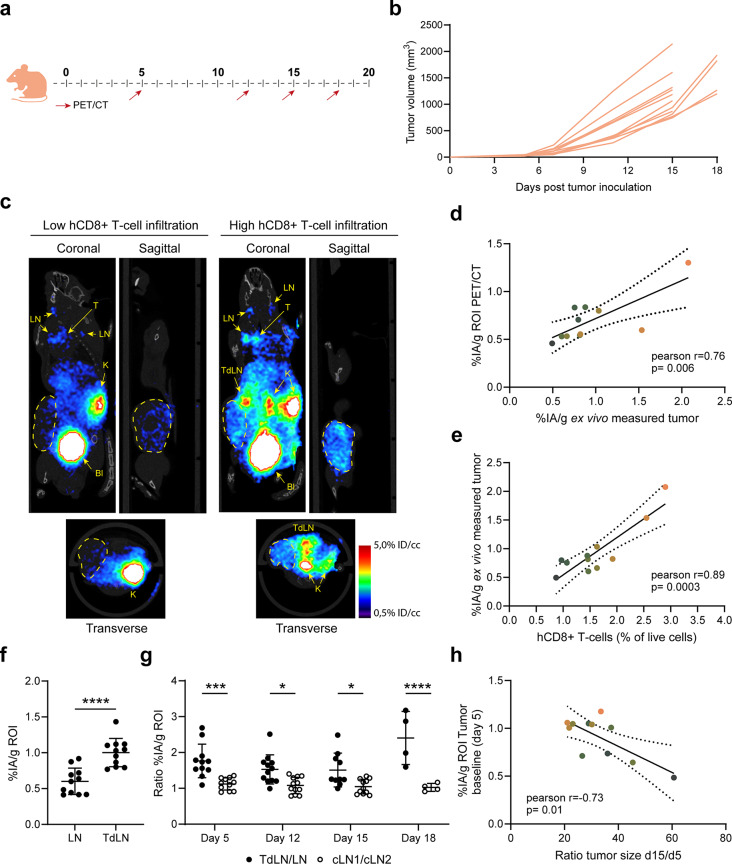
Specific uptake of [68Ga]Ga-NOTA-anti-hCD8β Nb in T-cell-rich organs of naïve hCD8 transgenic mice was observed (Fig. 5c-d) by PET/CT imaging and ex vivo γ-counting. Then, we assessed whether the [68Ga]Ga-NOTA-anti-hCD8β Nb was able to visualize T-cell dynamics in MC38 tumor-bearing mice (Fig. 6a). Tumor growth and CD8+ T-cell dynamics were followed over a period of 18 days. Individual differences in tumor growth could be observed as of day 9 after inoculation (Fig. 6b). Interestingly, PET imaging with the [68Ga]Ga-NOTA-anti-hCD8β Nb allowed, despite overall low levels of T-cell infiltration, the differentiation of mice with lower or higher levels of human CD8+ T-cell infiltration in the MC38 tumors (Fig. 6c and Supplemental Fig. 8). Quantification of radioactive uptake in the tumor on the PET images nicely correlated with the radioactive uptake measured by ex vivo γ-counting of the dissected tumors and flow cytometric analysis of the amount of intratumoral human CD8+ T cells (Fig. 6d-e and Supplemental Fig. 9). Overall, these results show that, despite the low levels of intratumoral human CD8+ T cells, the differences in T-cell tumor infiltration can be visualized using the [68Ga]Ga-NOTA-anti-hCD8β Nb. In addition to visualizing intratumoral T cells, we could also follow up T-cell dynamics in the lymph nodes. Baseline PET scans revealed already a significantly higher level of CD8+ T cells in the tumor-draining lymph node compared to the opposing inguinal lymph node (Fig. 6f). To determine whether this difference was tumor specific, we analyzed the ratio of radioactive uptake between these lymph nodes and compared it to the ratio between opposing cervical lymph nodes (Fig. 6g). Repeated imaging showed that a consistent and significant higher ratio of radioactive uptake was observed in the tumor-draining lymph nodes relative to the contralateral inguinal lymph nodes, compared to that between the cervical lymph nodes during the whole experiment. Finally, we assessed the prognostic value of the [68Ga]Ga-NOTA-anti-hCD8β Nb (Fig. 6h). To this end, we correlated the amount of intratumoral human CD8+ T cells present at baseline, as measured by PET imaging, and the increase of tumor growth, as determined by the ratio of tumor size at day 15 and baseline. First, mice showing higher radioactive uptake levels at baseline also showed higher uptake levels at endpoint (Fig. 6d-e-h). Interestingly, a significant correlation was observed between the radioactive uptake measured at baseline and the tumor growth, indicating that [68Ga]Ga-NOTA-anti-hCD8β Nb may hold prognostic value to differentiate the speed of tumor growth based on the amount of intratumoral hCD8+ T cells at early timepoints.
Fig. 6.
PET/CT imaging of hCD8+ T-cell dynamics with the 68Ga-labeled anti-human CD8β nanobody. (a) Schematic overview of the imaging regimen of MC38 tumor-bearing human (h) CD8 transgenic mice. Mice were inoculated with MC38 cells at day 0. (b) MC38 tumor growth curves in hCD8 transgenic mice. (c) Representative PET/CT image (coronal, sagittal and transverse section) of a MC38 tumor-bearing hCD8 transgenic mouse with low and high hCD8+ intratumoral T-cell infiltration at endpoint (day 15 or 18). The tumor is delineated. Lymph nodes (LN), tumor-draining lymph node (TdLN), thymus (T), kidneys (K) and bladder (Bl) are highlighted. d-e) Correlation plots of ex vivo measured radioactive uptake of the tumor versus the radioactive uptake measured via PET image analysis using a region of interest (ROI) (d) or hCD8+ T cells quantified via flow cytometry (e) at endpoint (day 15 or 18). Each individual mouse is highlighted in a different color. f) Radioactive uptake measured in the inguinal LN or inguinal TdLN via PET image analysis using a ROI at day 5. g) Ratio of radioactive uptake measured in inguinal lymph nodes (TdLN/LN) and cervical lymph nodes (cLN1/cLN2) via PET image analysis using ROIs. h) Correlation plot of the intratumoral hCD8+ T cells at baseline (day 5) versus tumor growth. Intratumoral hCD8+ T cells were determined via the radioactive uptake of 68Ga-hCD8β Nb in the tumor measured via PET image analysis using a ROI. Tumor growth is presented as the ratio of increase of tumor size at day 15 compared to baseline (day 5). Each individual mouse is highlighted in a different color. All data are presented as mean ± S.D.. Correlations were determined via Pearson correlation and 95% confidence bars are presented. Statistical analyses were performed using unpaired two-tailed t-test or one way ANOVA with Dunnett’s post hoc test. *, p < 0.05; ***, p < 0.001, ****p<0.0001
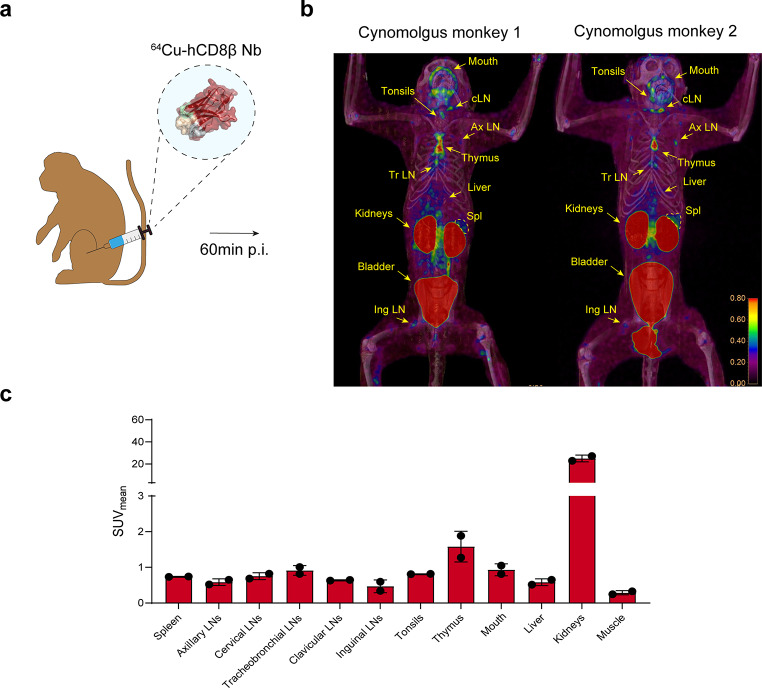
Finally, a proof-of-concept biodistribution study of the anti-hCD8β Nb in non-human primates was performed. 500 µg of [64Cu]Cu-NOTA-anti-hCD8β Nb was injected i.v. in 2 cynomolgus monkeys, followed by PET/CT imaging 1 h post-injection (Fig. 7a). Similar as in mice, a fast renal clearance of the [64Cu]Cu-NOTA-anti-hCD8β Nb was observed with a high uptake in the kidneys and bladder while no signal was observed in the blood (Fig. 7b-c). Furthermore, only minor uptake in the liver was observed. In both animals, the [64Cu]Cu-NOTA-anti-hCD8β Nb showed a similar uptake pattern with the ability to visualize both primary and secondary lymphoid organs, such as thymus, spleen, tonsils and a multitude of lymph nodes. Interestingly, both cynomolgus monkeys showed a high uptake in the mouth area. Finally, the dose of 500 µg of the 64Cu-labeled anti-hCD8β Nb was well-tolerated.
Fig. 7.
PET/CT imaging of CD8+ T cells in naïve cynomolgus monkeys with the 64Cu-labeled anti-human CD8β nanobody. (a) Schematic overview of the imaging experiment. (b) PET/CT images of 2 naïve cynomolgus monkeys intravenously injected with 64Cu-labeled anti-human CD8β Nb. Different regions of uptake are highlighted including cervical (cLN), axillary (Ax LN), tracheobronchial (Tr LN) lymph nodes, spleen (Spl) and thymus. (c) Quantification of the radioactive uptake (SUVmean) in different organs via PET image analysis
Discussion
Here, we report the development of a novel immunotracer for the imaging of hCD8+ T-cell dynamics. In-depth characterization shows high affinity and specificity of this tracer towards human CD8+ T cells and the ability to visualize T-cell dynamics with high sensitivity.
As T-cell-based immunotherapies have been a major point of focus in anti-cancer therapies, non-invasive imaging of T-cell dynamics has gained interest in recent years. To this end, several tracers targeting different T-cell markers have been reported [3]. In particular, imaging of cytotoxic CD8+ T cells has received attention [3, 26].
In this study, we show the ability of our immunotracer to detect human CD8+ T cells using nuclear imaging. Nbs have emerged as an interesting scaffold for imaging due to their in vivo characteristics, enabling same-day imaging of patients with short-lived isotopes [13]. Our study also shows the ability of Nb-based immunotracers to visualize CD8+ T cells already 1 h post-injection using short-lived isotopes such as technetium-99m, gallium-68 or copper-64. Furthermore, the use of these short-lived isotopes results in a lower radiation burden for patients compared to long-lived isotopes, such as zirconium-89, which are used for larger antibody-(fragment-)based tracers [27, 28]. Finally, we visualized human CD8+ T cells using both SPECT and PET. While PET imaging is generally preferred in a clinical setting due to a better contrast and spatial resolution, SPECT imaging of immune cells still holds great value due to an overall lower cost and wide availability of isotopes [29].
In vitro results show high affinity and specific binding of the anti-human CD8β Nb with no unwanted T-cell activation. This is important, since CD8 is an important mediator of T-cell activation and binding of anti-CD8 antibodies may induce T-cell activation [30, 31]. Furthermore, no immunogenic responses towards the Nb were observed. Although Nbs are assumed to be non-immunogenic [32, 33], patients developing anti-drug antibodies after Nb treatments have been reported [34, 35]. In the future, immunogenic responses towards the NOTA-conjugated immunotracers will also need to be assessed during clinical testing. However, immunogenic responses are expected to be low, since diagnostic tracers can be microdosed [27, 36]. This notion is corroborated by previous findings, showing that a HER2-targeted Nb-based tracer used in a phase I clinical trial had a low immunogenicity [32]. Together, these are important parameters for future clinical translatability of this tracer.
To date, a few hCD8-targeting tracers have been reported and are being tested clinically. This includes the 89Zr-labeled minibody Df-IAB22M2C, 89Zr-labeled antibody ZED88082A, 68Ga-labeled Nb SNA006 and 18F-labeled Nb VHH5v2 [6, 7, 9, 10, 37]. While these tracers have shown promising results so far, they still suffer from some disadvantages with respect to pharmacokinetics (in case of the larger tracers) and specificity.
The larger 89Zr-labeled constructs Df-IAB22M2C and ZED88082A are unsuitable for early timepoint and/or fast repeated imaging due to their long in vivo half-life and slower tumor penetration. Nbs could be complementary, or preferred altogether, thanks to the characteristics mentioned before. Indeed, our immunotracer shows similar in vitro and in vivo characteristics as the previously reported Nb-based tracers, including (sub)nanomolar affinity and the ability to detect CD8+ T cells already 1 h post-injection [9, 10, 37]. Importantly, these characteristics could prove to be essential for the prediction and/or follow-up of immunotherapy responses. It has become apparent that both the intratumoral amount and localization of CD8+ T cells may hold predictive value in therapy response [4, 38]. However, a recent clinical study indicated that intratumoral CD8+ T cell dynamics during immunotherapy are quite heterogeneous between different lesions and patients, indicating a potential need for CD8-targeting tracers that allow (early) sequential imaging timepoints in order to track spatio-temporal changes of CD8+ T cells [6]. As such, smaller tracers, such as Nbs, could be more suited to predict immunotherapy outcome. In line with this, we were able to correlate early baseline levels of intratumoral hCD8+ T cells to subsequent tumor growth, suggesting that early timepoint imaging of hCD8+ T cells may indeed hold a prognostic value. In contrast to our immunotracer, imaging of CD8+ T-cell dynamics with the SNA006 and VHH5v2 tracers has not been tested preclinically and the prognostic or predictive value of these tracers remains to be determined.
All reported larger 89Zr-labeled, as well as smaller 68Ga/18F-labeled tracers against CD8+ T cells target the α-chain of the CD8 protein. Several studies have reported the presence of CD8α on other immune cell populations, including NK cells and monocytes [11, 12]. In contrast, the CD8 β-chain is exclusively expressed on CD8+ T cells, making it a more selective target for visualizing T-cell dynamics. Indeed, our results show the higher specificity of our anti-CD8β Nb for CD8+ T cells. Since tumor-associated monocytes can encompass a major immune cell population in the tumor microenvironment [39], which would erroneously also be imaged by CD8α-targeting tracers, it seems reasonable to expect that the CD8β tracer yields an enhanced predictive or prognostic value. One disadvantage that all CD8-targeting tracers, including ours, have is that these tracers do not give any insight in the functional state of CD8+ T cells. As such, additional characterization of the CD8+ T-cell activation state or other tumor microenvironmental parameters may be necessary to provide a more accurate insight in immunotherapy response.
The PET imaging results in cynomolgus monkeys also showed the feasibility to image CD8+ T cells in larger animals. However, it is important to note that this was a proof-of-concept study. In the future, a dose-escalation study and imaging at multiple timepoints could give additional information for the clinical translation. The need for such studies was previously shown for SNA006, where different doses resulted in different uptake levels in T-cell rich organs such as bone marrow. Compared to these results, our immunotracer shows a similar biodistribution as the highest dose of 68Ga-labeled Nb SNA006 in cynomolgus monkeys [9]. To enable future studies, we decided to radiolabel our anti-hCD8β Nb with 64Cu instead of 68Ga. As 64Cu has a half-life of 12.7 h, this would enable us to assess the uptake of the anti-hCD8β Nb for a time period of multiple hours to day(s) and provide additional information on the pharmacokinetics of this Nb. Furthermore, this longer half-life time would allow more flexibility during tracer preparation and transport in a clinical setting, as compared to 68Ga-labeling. While a similar biodistribution profile was observed in cynomolgus monkeys as in our mouse imaging experiments, tracer uptake in the mouth area was also observed. The exact reason for this radioactive signal is still unclear, but it has been reported that CD8+ T cells are present in the oral tissues of non-human primates [40]. Moreover, 68Ga-labeled Nb SNA006 also seems to show an accumulation in the mouth and nasal area [9]. Finally, while no acute inflammation could be observed upon visual inspection, a local inflammation cannot be excluded.
Finally, the non-invasive imaging of CD8+ T-cell dynamics could be interesting for multiple immune-related diseases including viral infections or chronic inflammatory and autoimmune diseases, such as immune thrombocytopenia, rheumatoid arthritis and giant cell arteritis. Furthermore, previous preclinical research has focused on the non-invasive imaging of T-cell populations in graft-versus-host disease and multiple sclerosis, conditions for which imaging of CD8+ T cells might also be interesting [41–46]. Therefore, it is not unlikely that these tracers could be widely applied in multiple disease settings in the future.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Kevin De Jonghe (MITH, VUB), Steve Huvelle (Biomaps Unit, CEA), VIB nanobody core facility, VIB protein core facility, VIB proteomics core facility, VUB In vivo Cellular and Molecular Imaging (ICMI) core facility and Roche for the technical assistance. We also thank ARRONAX (Nantes) for the delivery of copper-64.
Author contributions
Timo W.M. De Groof, Yoline Lauwers, Jo A.Van Ginderachter and Nick Devoogdt designed the study; Timo W.M. De Groof performed the in vitro characterization assays with the help from Yoline Lauwers, Tessa De Pauw and Cécile Vincke; Timo W.M. De Groof and Yoline Lauwers performed the SPECT and PET imaging experiments with the help from Tessa De Pauw and Jolien Van Craenenbroeck. Mohit Saxena, Thibault Naninck, Roger Le Grand and Catherine Chapon performed the immunostaining and PET imaging experiments on non-human primate tissue; Timo W.M. De Groof analyzed the Alphafold binding prediction models; Timo W.M. De Groof, Yoline Lauwers and Mohit Saxena analyzed the experimental data; Geert Raes, Jo A.Van Ginderachter and Nick Devoogdt supervised the study; Timo W.M. De Groof wrote the paper with the help from all the authors
Funding
This work has received funding from the Innovative Medicines Initiative 2 Joint Undertaking (JU) under grant agreement No 831514 (Immune-Image). The JU receives support from the European Union’s Horizon 2020 research and innovation programme and EFPIA. This work was further funded by the Strategic Research Programme and Wetenschappelijk Fonds Willy Gepts from the Vrije Universiteit Brussel. This work is also supported by Kom op Tegen Kanker (Stand up to Cancer, the Flemish cancer society) and Research Foundation Flanders (FWO) research projects G087524N, I005622N and I001618N, and by a Kom op tegen Kanker fellowship grant (projectID: 13022). Timo W.M. De Groof is funded by a post-doctoral fellowship (12ZO723N) from the Research Foundation Flanders (FWO), Belgium. The Infectious Disease Models and Innovative Therapies (IDMIT) research infrastructure is supported by the “Programme Investissements d’Avenir”, managed by the ANR under reference ANR-11-INBS-0008.
Data availability
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
All mouse experiments were approved by the Ethical Committee for laboratory animals of the Vrije Universiteit Brussel and executed in accordance with the European guidelines for animal experimentation (ethical dossier number 21-272-1).Cynomolgus macaques were housed at the IDMIT infrastructure facilities (CEA, Fontenay-aux-roses, France) under BSL-3 containment (Animal facility authorization #D92-032-02, Préfecture des Hauts de Seine, France) and in compliance with European Directive 2010/63/EU, French regulations, and the Standards for Humane Care and Use of Laboratory Animals of the Office for Laboratory Animal Welfare (OLAW, assurance number #A5826-01, US). The protocols were approved by the institutional ethical committee ‘Comité d’Ethique en Expérimentation Animale du Commissariat à l’Energie Atomique et aux Energies Alternatives’ (CEtEA number 44) under statement number A23-057. The study was authorized by the ‘‘Research, Innovation and Education Ministry’’ under registration number APAFIS #46283-202312131546674 v1.
Competing interests
Timo W.M. De Groof, Yoline Lauwers, Cécile Vincke, Geert Raes, Jo A.Van Ginderachter and Nick Devoogdt are co-inventors on a pending patent application (EP23153689.7), which covers the use of the described CD8-targeting immunotracer
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Timo W.M. De Groof, Yoline Lauwers, Jo A. Van Ginderachter and Nick Devoogdt contributed equally to this work.
Contributor Information
Timo W.M. De Groof, Email: timo.de.groof@vub.be
Nick Devoogdt, Email: nick.devoogdt@vub.be.
References
- 1.Esfahani K, Roudaia L, Buhlaiga N, Del Rincon SV, Papneja N, Miller WH. Jr. A review of cancer immunotherapy: from the past, to the present, to the future. Curr Oncol. 2020;27:S87–97. 10.3747/co.27.5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kourie HR, Klastersky J. Immune checkpoint inhibitors side effects and management. Immunotherapy. 2016;8:799–807. 10.2217/imt-2016-0029. [DOI] [PubMed] [Google Scholar]
- 3.Arnouk S, De Groof TWM, Van Ginderachter JA. Imaging and therapeutic targeting of the tumor immune microenvironment with biologics. Adv Drug Deliv Rev. 2022;184:114239. 10.1016/j.addr.2022.114239. [DOI] [PubMed] [Google Scholar]
- 4.Rashidian M, Ingram JR, Dougan M, Dongre A, Whang KA, LeGall C, et al. Predicting the response to CTLA-4 blockade by longitudinal noninvasive monitoring of CD8 T cells. J Exp Med. 2017;214:2243–55. 10.1084/jem.20161950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li F, Li C, Cai X, Xie Z, Zhou L, Cheng B, et al. The association between CD8 + tumor-infiltrating lymphocytes and the clinical outcome of cancer immunotherapy: a systematic review and meta-analysis. EClinicalMedicine. 2021;41:101134. 10.1016/j.eclinm.2021.101134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kist de Ruijter L, van de Donk PP, Hooiveld-Noeken JS, Giesen D, Elias SG, Lub-de Hooge MN, et al. Whole-body CD8(+) T cell visualization before and during cancer immunotherapy: a phase 1/2 trial. Nat Med. 2022;28:2601–10. 10.1038/s41591-022-02084-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pandit-Taskar N, Postow MA, Hellmann MD, Harding JJ, Barker CA, O’Donoghue JA, et al. First-in-humans imaging with (89)Zr-Df-IAB22M2C Anti-CD8 Minibody in patients with solid malignancies: preliminary pharmacokinetics, Biodistribution, and Lesion Targeting. J Nucl Med. 2020;61:512–9. 10.2967/jnumed.119.229781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tavare R, Danton M, Giurleo JT, Makonnen S, Hickey C, Arnold TC, et al. Immuno-PET monitoring of lymphocytes using the CD8-Specific antibody REGN5054. Cancer Immunol Res. 2022;10:1190–209. 10.1158/2326-6066.CIR-21-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Wang C, Huang M, Qin S, Zhao J, Sang S, et al. Pilot study of a novel nanobody (68) Ga-NODAGA-SNA006 for instant PET imaging of CD8(+) T cells. Eur J Nucl Med Mol Imaging. 2022;49:4394–405. 10.1007/s00259-022-05903-9. [DOI] [PubMed] [Google Scholar]
- 10.Zhao H, Wang C, Yang Y, Sun Y, Wei W, Wang C, et al. ImmunoPET imaging of human CD8(+) T cells with novel (68)Ga-labeled nanobody companion diagnostic agents. J Nanobiotechnol. 2021;19:42. 10.1186/s12951-021-00785-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibbings DJ, Marcet-Palacios M, Sekar Y, Ng MC, Befus AD. CD8 alpha is expressed by human monocytes and enhances fc gamma R-dependent responses. BMC Immunol. 2007;8:12. 10.1186/1471-2172-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geng J, Raghavan M. CD8alphaalpha homodimers function as a coreceptor for KIR3DL1. Proc Natl Acad Sci U S A. 2019;116:17951–6. 10.1073/pnas.1905943116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Debie P, Devoogdt N, Hernot S. Targeted nanobody-based Molecular Tracers for Nuclear Imaging and Image-guided surgery. Antibodies (Basel). 2019;8. 10.3390/antib8010012. [DOI] [PMC free article] [PubMed]
- 14.Muyldermans S. Nanobodies: natural single-domain antibodies. Annu Rev Biochem. 2013;82:775–97. 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- 15.De Pauw T, De Mey L, Debacker JM, Raes G, Van Ginderachter JA, De Groof TWM, et al. Current status and future expectations of nanobodies in oncology trials. Expert Opin Investig Drugs. 2023;32:705–21. 10.1080/13543784.2023.2249814. [DOI] [PubMed] [Google Scholar]
- 16.Vincke C, Gutierrez C, Wernery U, Devoogdt N, Hassanzadeh-Ghassabeh G, Muyldermans S. Generation of single domain antibody fragments derived from camelids and generation of manifold constructs. Methods Mol Biol. 2012;907:145–76. 10.1007/978-1-61779-974-7_8. [DOI] [PubMed] [Google Scholar]
- 17.Pardon E, Laeremans T, Triest S, Rasmussen SG, Wohlkonig A, Ruf A, et al. A general protocol for the generation of Nanobodies for structural biology. Nat Protoc. 2014;9:674–93. 10.1038/nprot.2014.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siegel M, Steiner G, Franssen LC, Carratu F, Herron J, Hartman K, et al. Validation of a dendritic cell and CD4 + T cell Restimulation Assay contributing to the Immunogenicity Risk Evaluation of Biotherapeutics. Pharmaceutics. 2022;14. 10.3390/pharmaceutics14122672. [DOI] [PMC free article] [PubMed]
- 19.Xavier C, Devoogdt N, Hernot S, Vaneycken I, D’Huyvetter M, De Vos J, et al. Site-specific labeling of his-tagged nanobodies with (9)(9)mTc: a practical guide. Methods Mol Biol. 2012;911:485–90. 10.1007/978-1-61779-968-6_30. [DOI] [PubMed] [Google Scholar]
- 20.Zeven K, De Groof TWM, Ceuppens H, Awad RM, Ertveldt T, de Mey W, et al. Development and evaluation of nanobody tracers for noninvasive nuclear imaging of the immune-checkpoint TIGIT. Front Immunol. 2023;14:1268900. 10.3389/fimmu.2023.1268900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mirdita M, Schutze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M. ColabFold: making protein folding accessible to all. Nat Methods. 2022;19:679–82. 10.1038/s41592-022-01488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xavier C, Vaneycken I, D’Huyvetter M, Heemskerk J, Keyaerts M, Vincke C, et al. Synthesis, preclinical validation, dosimetry, and toxicity of 68Ga-NOTA-anti-HER2 nanobodies for iPET imaging of HER2 receptor expression in cancer. J Nucl Med. 2013;54:776–84. 10.2967/jnumed.112.111021. [DOI] [PubMed] [Google Scholar]
- 23.Bridoux J, Broos K, Lecocq Q, Debie P, Martin C, Ballet S, et al. Anti-human PD-L1 Nanobody for Immuno-PET imaging: validation of a conjugation strategy for clinical translation. Biomolecules. 2020;10. 10.3390/biom10101388. [DOI] [PMC free article] [PubMed]
- 24.Van Damme H, Dombrecht B, Kiss M, Roose H, Allen E, Van Overmeire E, et al. Therapeutic depletion of CCR8(+) tumor-infiltrating regulatory T cells elicits antitumor immunity and synergizes with anti-PD-1 therapy. J Immunother Cancer. 2021;9. 10.1136/jitc-2020-001749. [DOI] [PMC free article] [PubMed]
- 25.Szabo PA, Miron M, Farber DL. Location, location, location: tissue resident memory T cells in mice and humans. Sci Immunol. 2019;4. 10.1126/sciimmunol.aas9673. [DOI] [PMC free article] [PubMed]
- 26.Krekorian M, Fruhwirth GO, Srinivas M, Figdor CG, Heskamp S, Witney TH, et al. Imaging of T-cells and their responses during anti-cancer immunotherapy. Theranostics. 2019;9:7924–47. 10.7150/thno.37924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berland L, Kim L, Abousaway O, Mines A, Mishra S, Clark L, et al. Nanobodies for medical imaging: about ready for Prime Time? Biomolecules. 2021;11. 10.3390/biom11050637. [DOI] [PMC free article] [PubMed]
- 28.Liu M, Li L, Jin D, Liu Y. Nanobody-A versatile tool for cancer diagnosis and therapeutics. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2021;13:e1697. 10.1002/wnan.1697. [DOI] [PubMed] [Google Scholar]
- 29.Volpe A, Adusumilli PS, Schoder H, Ponomarev V. Imaging cellular immunotherapies and immune cell biomarkers: from preclinical studies to patients. J Immunother Cancer. 2022;10. 10.1136/jitc-2022-004902. [DOI] [PMC free article] [PubMed]
- 30.Li Y, Yin Y, Mariuzza RA. Structural and biophysical insights into the role of CD4 and CD8 in T cell activation. Front Immunol. 2013;4:206. 10.3389/fimmu.2013.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clement M, Ladell K, Ekeruche-Makinde J, Miles JJ, Edwards ES, Dolton G, et al. Anti-CD8 antibodies can trigger CD8 + T cell effector function in the absence of TCR engagement and improve peptide-MHCI tetramer staining. J Immunol. 2011;187:654–63. 10.4049/jimmunol.1003941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ackaert C, Smiejkowska N, Xavier C, Sterckx YGJ, Denies S, Stijlemans B, et al. Immunogenicity Risk Profile of Nanobodies. Front Immunol. 2021;12:632687. 10.3389/fimmu.2021.632687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klarenbeek A, El Mazouari K, Desmyter A, Blanchetot C, Hultberg A, de Jonge N, et al. Camelid Ig V genes reveal significant human homology not seen in therapeutic target genes, providing for a powerful therapeutic antibody platform. MAbs. 2015;7:693–706. 10.1080/19420862.2015.1046648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Papadopoulos KP, Isaacs R, Bilic S, Kentsch K, Huet HA, Hofmann M, et al. Unexpected hepatotoxicity in a phase I study of TAS266, a novel tetravalent agonistic Nanobody(R) targeting the DR5 receptor. Cancer Chemother Pharmacol. 2015;75:887–95. 10.1007/s00280-015-2712-0. [DOI] [PubMed] [Google Scholar]
- 35.Holland MC, Wurthner JU, Morley PJ, Birchler MA, Lambert J, Albayaty M, et al. Autoantibodies to variable heavy (VH) chain ig sequences in humans impact the safety and clinical pharmacology of a VH domain antibody antagonist of TNF-alpha receptor 1. J Clin Immunol. 2013;33:1192–203. 10.1007/s10875-013-9915-0. [DOI] [PubMed] [Google Scholar]
- 36.Barakat S, Berksoz M, Zahedimaram P, Piepoli S, Erman B. Nanobodies as molecular imaging probes. Free Radic Biol Med. 2022;182:260–75. 10.1016/j.freeradbiomed.2022.02.031. [DOI] [PubMed] [Google Scholar]
- 37.Sriraman SK, Davies CW, Gill H, Kiefer JR, Yin J, Ogasawara A, et al. Development of an (18)F-labeled anti-human CD8 VHH for same-day immunoPET imaging. Eur J Nucl Med Mol Imaging. 2023;50:679–91. 10.1007/s00259-022-05998-0. [DOI] [PubMed] [Google Scholar]
- 38.Pages F, Mlecnik B, Marliot F, Bindea G, Ou FS, Bifulco C, et al. International validation of the consensus immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet. 2018;391:2128–39. 10.1016/S0140-6736(18)30789-X. [DOI] [PubMed] [Google Scholar]
- 39.Ugel S, Cane S, De Sanctis F, Bronte V. Monocytes in the Tumor Microenvironment. Annu Rev Pathol. 2021;16:93–122. 10.1146/annurev-pathmechdis-012418-013058. [DOI] [PubMed] [Google Scholar]
- 40.Hernandez JL, Park J, Hughes SM, Hladik F, Woodrow KA. Characterization of Immune cells in oral tissues of non-human Primates. Front Oral Health. 2021;2:821812. 10.3389/froh.2021.821812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rothlauf PW, Li Z, Pishesha N, Xie YJ, Woodham AW, Bousbaine D, et al. Noninvasive Immuno-PET imaging of CD8(+) T cell behavior in Influenza A Virus-infected mice. Front Immunol. 2021;12:777739. 10.3389/fimmu.2021.777739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Audia S, Samson M, Mahevas M, Ferrand C, Trad M, Ciudad M, et al. Preferential splenic CD8(+) T-cell activation in rituximab-nonresponder patients with immune thrombocytopenia. Blood. 2013;122:2477–86. 10.1182/blood-2013-03-491415. [DOI] [PubMed] [Google Scholar]
- 43.Samson M, Ly KH, Tournier B, Janikashvili N, Trad M, Ciudad M, et al. Involvement and prognosis value of CD8(+) T cells in giant cell arteritis. J Autoimmun. 2016;72:73–83. 10.1016/j.jaut.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 44.Carvalheiro H, da Silva JA, Souto-Carneiro MM. Potential roles for CD8(+) T cells in rheumatoid arthritis. Autoimmun Rev. 2013;12:401–9. 10.1016/j.autrev.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 45.Cheng HM, Honda T, Asahina R, Miyake T, Chow Z, Tomura M, et al. In vivo imaging of CD8(+) T-Cell–mediated keratinocyte apoptosis in graft-versus-host disease–like Dermatitis in Involucrin membrane-bound ovalbumin mice. J Invest Dermatol. 2022;142:2827–e313. 10.1016/j.jid.2022.03.010. [DOI] [PubMed] [Google Scholar]
- 46.Salou M, Nicol B, Garcia A, Laplaud DA. Involvement of CD8(+) T cells in multiple sclerosis. Front Immunol. 2015;6:604. 10.3389/fimmu.2015.00604. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.