Abstract
Fluoroquinolone-resistant (FQs-R) microorganisms causing infectious complications after ultrasound-guided needle biopsy of the prostate (TRUS-BP) have become an important challenge in healthcare settings globally, questioning the continued utility of FQ as the preferred prophylactic agent. This study aimed to characterize molecular mechanisms of resistance on FQs-R E. coli isolated from the enteric microbiota of patients undergoing (TRUS-BP) and to highlight their impact on Minimum Inhibitory Concentrations (MICs). From February 2016 to December 2018, the incidence of rectal carriage of Qs-FQs resistant Enterobacterales detected from rectal swabs of patients before undergoing (TRUS-BP) was 61.06% (80/131) all related to E. coli species. Based on the MICs range of Qs (24–256 mg/L) and FQs (0.24–128 mg/L) breakpoint by EUCAST, we categorized these E. coli isolates into three resistance profiles (I, II, and III) associated with the patterns of chromosomal mutations in the quinolone resistance-determining regions (QRDRs) of gyrA and parC and the plasmid-mediated quinolone resistance encoding genes (PMQRs) detected by PCR-based assay and sequencing; MICs increase in an escalation step according to the co-occurrence of multiple molecular mechanisms. The mutation of the gyrA gene was the most frequent on codons (Ser83Leu/Thr/Tyr/Trp and Asp87Asn); mutation on the parC gene was the least on codons (Ser80Iso/Leu and Glu84 Val/Gly/Lys). PMQRs genes (4 qnrB ,7 qnrS, and one aac(6’)-Ib-cr) were determined within 15% of the isolates. Allelic variation allows us to report earliest the qnrB81 determinant in an E. coli isolate. Among isolates (35%) belonged to the notorious ST131 lineage. The phylogenetic group showed a predominance of B2 group (51, 25%), however (PFGE) revealed a high level of clonal variability. Worrying incidence of FQs-R E. coli isolates in the rectal flora of our local population showed the potential to cause post-infection. FQ resistance is a complex interplay between mutations in the QRDRs and PMQR determinants that impact MICs. The importance of intestinal microbiota as a reservoir of resistant strains and pandemic clones encourages driving mitigation challenges to characterize molecular mechanisms of antimicrobial resistance to adapt prophylactic therapy, control infection, and ensure epidemiological monitoring.
Keywords: Fluoroquinolone resistance, Escherichia coli, QRDRs mutation, PMQRs gene, MICs rectal flora, Prostate biopsy
Subject terms: Microbiology, Molecular biology, Health care, Medical research, Pathogenesis, Risk factors
Introduction
Fluoroquinolones (FQs) are among the most widely used antibiotics in the world for the treatment of Gram-negative and Gram-positive bacterial infections; they are classified by the World Health Organisation (WHO) as “Critically Important Antimicrobials” due to their broad spectrum effects and their clinical importance in humans and livestock1 Conversely, a considerable rise in resistance to these compounds has been linked to an increase in the prescription of FQs globally, whether in the community or in hospitals. Due to their remarkable efficiency, the emergence and dissemination of bacteria resistant to these classes of antibiotics are of major concern in the treatment of infectious diseases2. For numerous years, it was thought that FQs resistance was only caused by chromosomal mutations in the quinolone resistance-determining regions (QRDRs) gyrA and parC genes, which encode the subunits A and C of DNA gyrase and topoisomerase, respectively substitutions at these two genes increase ciprofloxacin resistance by more than tenfold; and more rarely in the gyrB or parE genes. FQs resistance is also caused by increased efflux caused by mutations in regulatory genes of chromosomally encoded multidrug resistance pumps and decreased permeability caused by porin loss2. Afterward, several studies conducted over the last two decades have revealed that the acquisition of plasmids containing quinolone resistance genes plasmid-mediated quinolone resistance (PMQRs) plays a critical role in the development of quinolone resistance3,4. Chromosomal mutation confers a high level of resistance to FQs and is transmitted vertically, while (PMQRs) confer a low level of resistance, and are transmitted horizontally among distantly related organisms, making their spread much faster than that of QRDRs mutations and realizing the epidemic spread of resistance to FQs3. These determinants have worldwide dissemination, they are increasingly found in strains isolated from humans, in community, hospital, and carriage infections; To date, at least 3 types of PMQRs determinants have been described in clinical isolates, i) target protection mediated by Qnr protein; (ii) FQs modification through AAC (6’)-Ib-cr, an aminoglycoside acetyltransferase variant; (iii) and plasmid-mediated quinolone efflux pumps (QepA and OqxAB)2. From a historical point of view, the Qnr represents the first determinant of plasmid origin. These determinants are thought to be intercalated between quinolones, topoisomerases, and DNA, thus limiting the subsequent binding of quinolones to their target. Since the first description of QnrA15 more than a hundred other Qnr proteins have been described and classified into QnrA, QnrB, QnrS, QnrC, QnrD, and QnrE. Those variants are frequently found in Enterobacterales, while the seventh variant, QnrVC, is mainly detected in Vibrio cholerae and other Vibrio spp6,7. The Qnr variants are defined by at least 30% nucleic or derived amino acid differences from each other, whereas the Qnr alleles are more closely related and are defined by a maximum difference of 10% between members of the same family4. Numerous alleles (8QnrA, 94 QnrB, 15 QnrS, 1 QnrC, 3 QnrD, and 7 QnrVC) are listed on the Lahey Clinic website (http://www.lahey.org/qnrStudies/). Nonetheless, in the following years, numerous chromosomally encoded Qnr-related proteins were detected, both in Gram-negative and Gram-positive microorganisms8. The AAC (6 ′)-Ib-cr determinant was initially reported in China (2005) contributing independently to resistance to ciprofloxacin by modification of the molecule, and was very widespread in strains of Enterobacterales4. It is an aminoglycoside acetyltransferase with an acetylation spectrum that has been expanded to include ciprofloxacin and norfloxacin9. QepA, an efflux pump belonging to the major facilitator subfamily, is related to a decrease in susceptibility to hydrophilic FQs10 OqxAB, a multidrug efflux pump, is related to reduced FQs susceptibility and resistance to multiple agents11. In terms of preventing Enterobacterales infections, FQs are particularly known to be used in an increased way as the molecule of choice for empirical and targeted therapies, including pre-prostate biopsy prophylaxis, thanks to their broad spectrum of activity, their low toxicity profile, their ease of administration by the oral route, as well as their excellent intra-prostatic penetration, which leads to high tissue concentrations12. Transrectal ultrasound-guided prostate biopsy TRUS-BP is the gold standard for confirming the diagnosis of prostate cancer.13 Knowing that prostate cancer is the second most frequent malignancy (after lung cancer) in men in Tunisia and worldwide https://gco.iarc.fr/today/home, hence the importance of early detection. Although TRUS-BP is a minimally invasive, rapid procedure that can be carried out in the consultation, it is not without potentially serious complications, in particular, infectious ones (acute prostatitis, prostatic abscess, orchi-epididymitis, and bacteremia)12. Thus, antibiotic prophylaxis before TRUS-BP is one of the most effective tools in terms of preventing post-biopsy infections caused by the resident intestinal flora14. Effectively, the intestinal microbiota is made up of more than 500 species of microorganisms that are in equilibrium, this balance can be destabilized by the selection pressure exerted by several factors, such as taking antibiotics, which can lead to the predominance of resistant Enterobacterales15. Especially since FQ-R phenotype is a key characteristic giving fitness advantage common to pandemic E. coli clones which is likely to have been a factor in their overall success18,19. Antimicrobial prophylaxis with a (FQ) or first-, second-,or third-generation cephalosporin is a current guideline recommendation, and currently, FQs are the most used anti-microbial prophylaxis for prostate biopsy. However, numerous reports have documented that FQ-R bacteria have been frequently identified as a cause of significant urinary tract infections; after TRUS-BP, hence the high risk of ineffective antibiotic prophylaxis16,17. The various expert committees therefore agree that it is essential to implement adequate preventive antibiotic therapy to avoid these infectious complications16. Accordingly, this work is included in this context as part of an antibiotic prophylaxis protocol in patients proposed for (TRUS-BP), to evaluate the prevalence of enteric colonization with FQs- R Enterobacterales, their phylogenetic and clonal identification; and an in-depth study on the characterization of chromosome target site mutation and plasmid born mechanisms involved on resistances as well as impact of their co-occurrence on MICs levels.
Materials and methods
Study design
A prospective study was carried out over a period of almost 3 years (February 2016–December 2018) in collaboration between the microbiology and urology departments of the Charles Nicolle Hospital (CNH) in Tunis in order to adapt an antibiotic prophylaxis protocol for patients undergoing TRUS-BP in order to control post-biopsy infections, as well as understand the impact of the genetic basis variation involved on levels of antibiotic resistance.
Ethical approval and consent to participate
for this study protocol, the patient information sheet, and the informed consent form were approved by the Ethics Committee of the Charles Nicolle Hospital: CHN Ethics Committee of the Office for the Protection of Human Research (OHRP) in the United States recognized under the number FWA: FWA00032748/IID: IORG0011243. All procedures performed in this study were following the ethical standards of the institutional research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Sample processing and microbial study
Rectal swabs were received from the urology department on a transport medium (Amies CultureSwab). Then each swab was added to a liquid enrichment medium (brain-heart broth) and incubated at 37 °C for 24 h. Obtained cultures were inoculated on desoxycholate lactose agar plates supplemented with ciprofloxacin (0.05 µg/ml) as selective screening agar for the isolation of Gram-negative bacilli resistant to FQs. After incubation at 37 °C for 18 to 24 h, growing colonies showing a distinct morphotype were identified by classical biochemical methods. The microbial identification was determined by Gram staining, an oxydase test, and API 20E strips (bioMérieux). One ciprofloxacin-resistant isolate per positive sample was further characterized.
Antimicrobial susceptibility testing
Antimicrobial susceptibility tests were determined by the agar disk diffusion method on Mueller-Hinton (MH) agar plates (Bio-Rad, Marnes-la-Coquette, France) according to the CA-SFM guidelines (http://www.sfm-microbiologie.org/). The antibiotics tested were: Ampicillin (10 µg), amoxicillin–clavulanic acid (20 –10 µg), ticarcillin (75 µg), ticarcillin– clavulanic acid (75 –10 µg), piperacillin (30 µg), piperacillin–tazobactam(30 –6 µg), cefalexin (30 µg), mecillinam (10 µg), cefoxitin (30 µg), cefixim(5 µg) cefotaxime (5 µg), ceftazidime (10 µg), cefepim (30 µg) aztreonam (30 µg), ertapenem (10 µg) imipenem (10 µg), gentamicin (10 µg), amikacin(30 µg), tobramicin(10 µg), netilimicin (10 µg), nalidixic acid (30 µg), norfloxacine (10 µg), ofloxacin (5 µg), ciprofloxacin(5 µg), chloramphenicol(30 µg), fosfomicin (200 µg), tigecycline (15 µg), nitrofurantoine (100 µg) and trimethoprime + sulfamethoxazole (1,25 − 23,75 µg). Minimum inhibitory concentrations (MICs) of FQs (ofloxacin, and ciprofloxacin) were determined for all E.coli isolates by agar dilution method The concentration ranges of antibiotics tested were 0,06 à 512 mg/L. The bacterial suspensions were deposited using a multipoint inoculator (Steers) and interpreted according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) criteria (http://www.eucast.org/clinical.breakpoints/2016-2018); the E-test (BioRad) method was used towards Qs (nalidixic acid) according to the manufacturer’s direction; and E. coli ATCC 25,922 was used as a quality control strain.
Characterization of FQs resistance mechanisms
Chromosomal mutations in the QRDRs of the gyrA and parC genes on E.coli isolates were assessed by polymerase chain reaction (PCR) amplification and sequencing20. The gyrA/parC PCR reaction (50 µL) consisted of 0.5 µmol/µL of each primer, 1,25 U/µL AmpliTaq Gold DNA polymerase, 5 µL AmpliTaq Gold buffer, 3 mM MgCl2 and 3 mM dNTP mix. The PCR program consisted of an initial denaturation at 95°C for 10 minutes, then 25 cycles of DNA denaturation at 95°C for 30 seconds, primer annealing at 56°C/55°C for 30 seconds, and extension at 72°C for 30 seconds. PCR products were purified enzymatically using the AmpliTaq DNA polymerase FS Dye Terminator Cycle Sequencing kit (Applied Biosystems, Courtaboeuf, France) The sequencing PCR program consisted an initial denaturation at 94°C for 5 minutes, then 30 cycles of DNA denaturation at 94°C for 30 seconds, primer annealing at 55°C for 30 seconds, and extension at 72°C for 30 seconds and sequenced with BigDye v.3.1 sequencing using ʺABI Prism 310ʺ (Applied Biosystems). Target gyrA (189 pb), primer sequence 5’3’ F-ACGTACATGGCAATGACTGG ; R-AGAAGTCGCCGTCGATAGAA ; Target parC (264 pb) Primer sequence 5’3’ F-TGTATGCGATGACTGAACTG ; R-CTCAATAGCAGCAGCTCGGATA. The gyrA and parC sequences were checked for similarity with the reference gene MG1655 (E. coli K-12) using NCBI’s BLAST.
PMQRs encoding genes, including qnrA, qnrB, qnrC, qnrD, qnrS, qepA, oqxAB, and the aac(6)-Ib-cr variant, were detected by PCR and subsequent sanger sequencing for allelic variation assessment10–21,22,23,24. Appropriate positive and negative controls were included in the assays for E. coli Jeg cr (+), E. coli qnrA (+), E. coli qnrB (+),E. cloacae qnrS (+), and K. pneumoniae 20,927 (oqx, qnrB, qnrS, and aac (6’)-Ib-cr)25.
Detection of E. coli phylogenetic groups and ST131 clone determination
Phylogenetic groups were determined by multiplex PCR targeting the following genetic markers, namely, chuA, yjaA, arpA TspE4.C2, ArpA, ArpAgpE, TrpAgpC TrpBA allowing the isolates to be classified into the main phylogenetic groups, A, B1, B2, C, D, E, F, G and cryptic clade I or II as described by Clermont et al.,26; membership of ST 131 was researched using an O25b-specific PCR method with pabB and trpA allele-specific primers27.
Pulsed-field gel electrophoresis
Clonal relatedness was investigated using pulsed-field gel electrophoresis (PFGE) with the restriction enzyme XbaI (Promega/Amersham Life Sciences, Uppsala, Sweden), as previously reported by26 using the PulseNet Europe protocol (www.pulsenetinternational.org) Clonal relationships and clusters were established following the criteria of Tenover28.
Results
Prevalence and identification of FQs-resistant isolates
Among the 131 patients undergoing the TRUS-BP, the detection of FQs-R isolates was revealed in 80 patients for whom colonies had grown from their rectal swabs on GDL culture medium supplemented with ciprofloxacin (0.05 µg/ml); i.e., a digestive carriage frequency of around 61,06% (80/131). The percentages of positive samples increased during the screening period from 40 to 67 to 72% during the years 2016, 2017, and 2018, respectively. Phenotypic and biochemical identification methods permit distinguishing that all the 80 strains isolated belong to E. coli species.
Antibiotic susceptibility
Susceptibility to Qs and FQs and determination of MIC values
All 80 E. coli strains were resistant to Qs (nalidixic acid), and 65 (81.25%) of them also presented a variable degree of resistance to FQs (ofloxacin and ciprofloxacin). As a result of the antibiogram interpretation and MICs values obtained, three resistance phenotypes of Qs and FQs resistance were distinguished and distributed as follows: (i) Profile I: 6 (7.5%) strains presented MIC50s to nalixdic acid of 24 mg/L (susceptible MIC breakpoint by EUCAST: 16 mg/L) and were all sensitive to ofloxacin and ciprofloxacin (susceptible MIC breakpoint by EUCAST: ≤0.25 mg/L); with a MIC50s level equal to 0.24 mg/L; (ii) Profile II: 9 (11.25%) strains, which are all highly resistant to nalidixic acid with MIC50s > 256 mg/L, showed intermediate sensitivity to ofloxacin with MIC50s of 0.5 mg/L and were all sensitive to ciprofloxacin with MIC50 of 0.24 mg/L. (iii) Profile III: 65 (81.25%) of Qs and FQs resistant strains exhibited MIC50s of nalidixic acid > 256 mg/L, and MIC50s of ofloxacin and ciprofloxacin were 32 mg/L (with MICs ranging from 1 to 128 mg/L) (Table 1).
Table 1.
Distribution of Qs-FQs resistance phenotype amongst the Ecoli isolates and impact of asssociated molecular mechanisms on MICs.
| Resistance phenotype | (N-%) | Qs MICs (mg/L) | FQs MICs (mg/L) | QRDRs mutations | PMQRs gene | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| NAL S ≤ 16 R > 16 |
OFX S ≤ 0,25 R > 0,5 |
CIP S ≤ 0,25 R > 0,5 |
Total number | gyrA | parC | ||||||
| Profile I | 6 (7,50%) | 24 | R | 0,24 | S | 0,24 | S | - | - | - | 1 |
| Profile II | 9 (11,25%) | 256< | R | 0,5 | I | 0,24 | S | 1 | 1 | - | - |
|
Profile III 65 (81,25%) |
9 (11,25%) | 256< | R | 2–4 | R | 1–4 | R | 2 | 1 | 1 | - |
| 10 (12,5%) | 256< | R | 1–64 | R | 1–32 | R | 1 | 1 | - | 1 | |
| 26 (32,5%) | 256< | R | 8–32 | R | 8–32 | R | 2 | 2 | - | - | |
| 16 (20%) | 256< | R | 8–64 | R | 8–64 | R | 3 | 2 | 1 | - | |
| 4 (5%) | 256< | R | 64–128 | R | 64–128 | R | 4 | 2 | 2 | - | |
PMQRs: plasmid resistance to fluoroquinolones, QRDRs: quinolone resistance determining regions MICs: Minimum inhibitory concentrations NA : Nalidixic acid; OFX, ofloxacine CIP, Ciprofloxacin,, S : Sensitive ; I : Intermediat ; R :Resistant.
Susceptibility to other classes of antibiotics
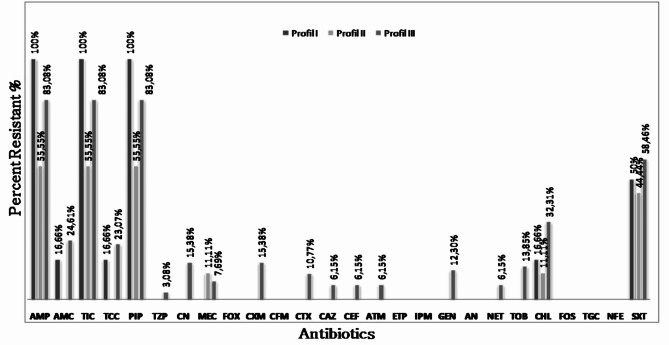
According to the different Qs- FQs resistance profiles (I, II, and III), the susceptibility rates of isolates studied to the other antibiotics tested are illustrated in (Fig. 1).
Fig. 1.
Others Antibiotic resistance rates in E. coli strains according to their Fluoroquinolones resistance profiles: Profile I: strains of resistance phenotype I :, Profile II : strains of resistance phenotype II, Profile III : strains of resistance phenotype III; AMP: Ampicillin; TIC: Ticarcillin; PIP: Piperacillin; AMC: Amoxicillin + Clavulanic Acid; TCC: Ticarcillin + Clavulanic Acid; TZP: Piperacillin + Tazobactam; MEC: Mecillinam; CN: Cefalexin; CXM: Cefuroxime; CTX: Cefotaxime; CAZ: Ceftazidime; CFM: Cefixime; CEF: Cefepime; FOX: Cefoxitin; ATM: Aztreonam; ETP: Ertapeneme; IPM: Imipeneme; GM: Gentamicin; TOB: Tobramycin; NET: Netilmicin; AN: Amikacin; CHL: Chloramphenicol; SXT: Trimethoprim + Sulfonamides; FOS: Fosfomycin; TGC: tigecycline; NFE: Furans.
Susceptibility rates for strains belonging to Profile I was as follows: mainly towards beta-lactams: ampicillin (100%), ticarcillin (100%), piperacillin (100%), amoxicillin + clavulanic acid (16.6%), ticarcillin + clavulanic acid (16.6%), mecillinam (16.6%) I.e. (16,66%) low-level penicillinase; In addition, (50%) of the strains were resistant to the combination trimethoprim and sulphonamide and (16.6%) to chloramphenicol. Susceptibility rates for strains belonging to Profile II were as follows: ampicillin (55.50%), ticarcillin (55.50%), piperacillin (55.50%), and mecillinam (11.10%), I.e. (22,22%) low-level penicillinase and high-level penicillinase (33,33%) resistance profile. In addition, (44.44%) of the strains were resistant to the combination trimethoprim + sulfonamide and (11.11%) to chloramphenicol.
Susceptibility rates for strains belonging to Profile III were as follows: mainly against ampicillin (83.10%), ticarcillin (83.10%), piperacillin (83.10%), amoxicillin + clavulanic acid (24.60%), Ticarcillin + .clavulanic acid (23.10%), Piperacillin + Tazobactam (3.10%), and mecillinam (7.70%).cefalexin (15.38%), cefuroxime (15.38%), cefotaxime (10.77%), ceftazidime (6.15%), and cefepime (6. 15%). For aztreonam, the resistance rate was (6.15%). Among these strains, one (1.53%) presented a resistance profile of hyperproduced cephalosporinase and one (1.53%) of penicillinase plus cephalosporinase. Seven (10.76%) had an extended-spectrum beta-lactamase (ESBL) profile. The rate of resistance to aminoglycosides was 12.30% for gentamicin, 13.85% for tobramycin, and 6.15% for netilmicin. In addition, 58.46% of the strains were resistant to the combination of trimethoprim and sulfonamides and 32.31% to chloramphenicol. While the three phenotype profiles were characterized by 0% resistance to carbapenemase,
Detection of chromosomal mutations in gyrA and parC genes
The alignment comparison obtained by sequencing the gyrA and parC genes led to determinate readable sequences for 73/80 (91.25%) strains. Six (8.21%) strains present a wild-type profile for the gyrA and parC genes. Sixty-seven 67/73 (91.78%) strains resulted in alterations in QRDRs genes. Of these, twenty-one strains (31.34%) showed one or more mutations in the gyrA gene only, and fifty-one strains (76.11%) presented co-carriage of one or more mutations in the gyrA and parC genes simultaneously. The amino acid variation observed in the QRDRs revealed, respectively, five different points of substitution in the gyrA gene on the subsequent codons Ser-83 and/or Asp-87 (S83L, S83T, S83Y, S83W, and D87N) and in the parC genes on codons Ser-80 and/or Glu-84 (S80I, S80L, E84V, E84G, and E84K). These amino acid changes, as well as their respective prevalence, were described in (Table 2). Codon 83 was the most frequent substitution detected on the gyr A gene, with a switch from Serine to Leucine (S83L) in 70.11% of strains, followed by changes from serine to tyrosine (S83Y) and Serine to Threonine (S83T), as well as a from Serine to Tryptophan (S83W) and from Asparatate to Asparagines in codon 87( D87N). Regarding parC mutation, 10% of the strains presented a substitution of Glutamate by Valine (E84V), followed by changes in Glutamate by Lysine (E84K), Serine by Isoleucine, Serine by Leucine (S80L), and Glutamic acid by Glycine (E84G).
Table 2.
Frequencies of QRDRs mutation amongst E.coli isolate highlighting co-occurrence of amino acid substitution within gyrase A (gyrA) and topoisomerase IV (parC).
| QRDRs mutations (N) | Isolates N (%) | Amino acid substitution | ||
|---|---|---|---|---|
| N (%) | GyrA | ParC | ||
| 1 | 11 (16,41%) |
7(10,44%) 1(1,49%) 1(1,49%) 2(1,49%) |
S83L S83T S83Y D87N |
- - - - |
| 2 | 11 (16,41%) |
5(7,46%) 4(5,97%) 1(1,49%) 1(1,49%) |
S83L + D87N S83L S83W D87N |
- S80I S80I S80I |
| 3 | 36 (53,73%) |
35(52,23%) 1(1,49%) |
S83L + D87N S83L + D87N |
S80I E84K |
| 4 | 9 (13,43%) |
7(10,44%) 1(1,49%) 1(1,49%) |
S83L + D87N S83L + D87N S83L + D87N |
S80I + E84V S80L + E84V S80I + E84G |
S: Serine; L: Leucine; I: Isoleucine; D: Aspartate; N: Asparagine; T: Threonine; Y: Tyrosine; W: Tryptophan; E: Glutamic acid; G: Glysine; V: Valine; K: Lysine.
Detection of plasmid-mediated quinolone and ESBLs resistance genes
Investigation into PMQRs harbored in the 80 E. coli strains revealed that twelve strains (15%) carry plasmid determinants, which are distributed as follows: Eleven qnr genes have been identified, including qnrB in four strains and qnrS in seven strains. Subsequently, the comparison of the nucleotide sequences of the qnr genes with those available in the databases allowed the identification of the allele qnrS1 in 4 strains and qnrS15 in 2 strains. The qnrB19 and qnrB81 type alleles were identified in one strain each. The aac(6’)-Ib-cr gene was discovered in one strain only. Furthermore, six (6.90%) of the strains harbouring PMQRs determinants shared simultaneously one or more QRDRs mutations. Tables 3, 4, 5 summarise the various chromosomal mutations, as well as those harbouring the PMQRs genes within the three resistant phenotype profiles of E. coli, isolates.
Table 3.
Characterization of E. coli isolates grouped on Profil I in terms of phenotypic resistance and its genotypic basis, phylogenetic groups, and clonal relatedness.
| Strains | Qs -FQs resistance phenotypes NAL NXN OFX CIP |
Qs-FQs MICs (mg/L) | ATB resistance profiles | QRDRs mutations | PMQRs gene | Phylogenetic group | ST131 clone | PFGE pulsotype/subtype |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NAL S ≤ 16 R > 16 |
OFX S ≤ 0.25 R > 0.5 |
C.I.P. S ≤ 0.25 R > 0.5 |
ß-lactams | Aminoglycosides | Other | gyrA | parC | ||||||
| E. coli (1) | RSSS | 24 | 0.24 | 0.24 | AMP, TIC, PIP | - | - | WP | WP | qnrS1 | B2 | + | Pulsotype I |
| E. coli (2) | RSSS | 24 | 0.24 | 0.24 | AMP, TIC, PIP | - | - | WP | WP | qnrS1 | B2 | + | Pulsotype I |
| E.coli (3) | RSSS | 24 | 0.24 | 0.24 | AMP, TIC, PIP | - | CHL, SXT | WP | WP | qnrB19 | Unknown | - | - |
| E. coli (4) | RSSS | 24 | 0.24 | 0.24 | AMP, TIC, PIP, AMC, TCC, MEC | - | - | WP | WP | qnrB81 | Clade I or II | - | - |
| E.coli (5) | RSSS | 24 | 0.24 | 0.24 | AMP, TIC, PIP | - | SXT | WP | WP | qnrB | B2 | + | Pulsotype VII |
| E.coli (6) | RSSS | 24 | 0.24 | 0.24 | AMP, TIC, PIP | - | SXT | WP | WP | qnrS15 | B2 | + | Subtype VIIIb |
MICs: minimum inhibitory concentration levels; S: sensitive ; R: resistant; NAL: Nalidixic acid, NXN: Norfloxacin, OFX: ofloxacin; CIP: ciprofloxacin; AMP: ampicillin; TIC: ticarcillin; PIP: piperacillin; TCC : Ticarcillin + Ac.clavulanic; MEC: mecillinam; CHL: chloramphenicol; SXT: co- trimoxazole; RPF: plasmid resistance to fluoroquinolones, QRDRs: quinolone resistance determining region, R: resistant; S: sensitive; WP: wild phenotype, +: positive; -: Negative.
Table 4.
Characterization of E. coli isolates grouped on profil II in terms of phenotypic resistance and its genotypic basis, phylogenetic groups, and clonal relatedness.
| Strains | Qs-FQs resistance phenotypes NAL NXN OFX CIP |
Qs -FQs MICs (mg/L) | ATB resistance profiles | QRDRs mutations | PMQRs gene | Phylogenetic group | ST131 clone | PFGE pulsotype/subtype |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NAL S≤16 R>16 |
OFX S≤0.25 R>0.5 |
C.I.P. S≤0.25 R>0.5 |
ß-lactams | Aminoglycosides | Other | gyrA | parC | ||||||
| E. coli (7) | RSSS | 256 | 0.5 | 0.24 | AMP, TIC, PIP | - | - | S83L | WP | - | B2 | - | - |
| E.coli (8) | RSSS | 256 | 0.5 | 0.24 | AMP, TIC, PIP | - | SXT | S83T | WP | - | B2 | - | - |
| E. coli (9) | RSSS | 256 | 0.5 | 0.24 | - | - | SXT | S83L | WP | - | Clade I or II | - | - |
| E.coli (10) | RSSS | 256 | 0.5 | 0.24 | AMP, TIC, PIP | - | - | S83L | WP | - | B2 | - | - |
| E. coli (11) | RSSS | 256 | 0.5 | 0.24 | AMP, TIC, PIP | - | CHL, SXT | S83L | WP | - | B2 | - | - |
| E.coli (12) | RSSS | 256 | 0.5 | 0.24 | - | - | - | D87N | WP | - | B2 | + | Pulsotype VI |
| E.coli (13) | RSSS | 256 | 0.5 | 0.24 | - | - | - | D87N | WP | - | Unknown | - | - |
| E.coli (14) | RSSS | 256 | 0.5 | 0.24 | MEC | - | - | S83L | WP | - | Clade I or II | - | - |
| E. coli (15) | RSSS | 256 | 0.5 | 0.24 | AMP, TIC, PIP | - | SXT | S83L | WP | - | Clade I or II | - | - |
MIC: minimum inhibitory concentration; S: sensitive ; R: resistant; NAL: Nalidixic acid, NXN: Norfloxacin, OFX: ofloxacin; CIP: ciprofloxacin AMP: ampicillin; TIC: ticarcillin; PIP: piperacillin; MEC: mecillinam; CHL: chloramphenicol; SXT: co- trimoxazole; PMQR: plasmid resistance to fluoroquinolones, QRDR: quinolone resistance determining region, R: resistant; S: sensitive; WP: wild phenotype; +: positive; -: Negative.
Table 5.
Characterization of E. coli isolates grouped on profil III in terms of phenotypic resistance and its genotypic basis, phylogenetic groups, and clonal relatedness.
| Strains | Qs-FQs resistance phenotypes NAL NXN OFX CIP |
Qs- FQs MICs (mg/L) | ATB resistance profiles | QRDRs mutations | PMQRs gene | Phylogenetic group | ST131 clone | PFGE pulsotype/subtype |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NAL S ≤ 16 R > 16 |
OFX S ≤ 0.25 R > 0.5 |
C.I.P. S ≤ 0.25 R > 0.5 |
ß-lactams | Aminoglycosides | Other | gyrA | parC | ||||||
| 1 QRDRs ( 1 gyrA mutation + PMQR ) | |||||||||||||
| E.coli (16) | R R RR | 256< | 1 | 1 | AMP, TIC, PIP | - | - | S83L | PS | qnrS1 | B2 | - | - |
| E.coli (17) | R RR R | 256< | 1 | 1 | - | - | - | S83Y | PS | qnrS15 | Clade I or II | - | - |
| 2 QRDRs ( 2 gyrA mutations) | |||||||||||||
| E.coli (18 ) | RRRR | 256 < | 32 | 32 | AMP, TIC, PIP | - | - | S83L + D87N | ND | - | B2 | + | Pulsotype III |
| E.coli (19) | RRRR | 256< | 8 | 8 | AMP, TIC, PIP | - | - | S83L + D87N | ND | - | B2 | - | Subtype IIId |
| 2QRDRs (1 gyrA mutation + 1 parC mutation) | |||||||||||||
| E.coli (20) | RRRR | 256< | 2 | 1 | AMP, TIC, PIP | - | SXT | S83L | S80I | - | B2 | + | - |
| E.coli (21) | RRRR | 256< | 1 | 1 | - | - | - | S83L | S80I | - | B2 | - | - |
| E.coli (22) | RRRR | 256< | 2 | 1 |
AMP, TIC , PIP |
- | SXT | S83W | S80I | - | B2 | + | - |
| E.coli (23) | RRRR | 256< | 4 | 4 | AMP, AMC, TIC, TCC, PIP | - | SXT | S83L | S80I | - | Clade I or II | - | Pulsotype VIII |
| E. coli (24) | RRRR | 256< | 4 | 4 | - | - | CHL, SXT | S83L | S80I | - | Unknown | - | - |
| E. coli (25) | RRRR | 256< | 4 | 4 | AMP, AMC, TIC, TCC, PIP, TZP, CN, MEC, CXM | - | CHL | D87N | S80I | - | Unknown | - | Pulsotype IX |
| 2 QRDRs (2 gyrA mutation ) + 1 PMQR gene | |||||||||||||
| E. coli (26) | RRRR | 256< | 64 | 32 | - | - | - | S83L + D87N | - | qnrB | B2 | + | Subtype IIIb |
| E.coli (27) | RRRR | 256< | 8 | 8 | AMP, TIC, PIP, AMC, CN, CXM, CTX, CAZ, CEF, ATM | TOB | SXT | S83L + D87N | - | aac(6)-Ib-cr | B2 | - | Subtype IIIc |
| E. coli (28) | RRRR | 256< | 64 | 32 | - | - | - | S83L + D87N | - | qnrS1 | B2 | + | - |
| 3QRDRs (2 gyrA mutations + 1 parC mutation) | |||||||||||||
| E.coli (29) | RRRR | 256< | 64 | 64 | AMP, TIC, PIP, CN, MEC, CXM, CTX, CEF, ATM | - | SXT | S83L + D87N | S80I | - | Clade I or II | - | - |
| E. coli (30) | RRRR | 256< | 32 | 16 | - | - | SXT | S83L + D87N | E84K | - | F | - | - |
| E.coli (31) | RRRR | 256< | 32 | 16 | AMP, TIC, PIP | - | CHL, SXT | S83L + D87N | S80I | - | Unknown | - | - |
| E. coli (32) | RRRR | 256< | 32 | 32 | AMP, TIC, PIP | - | SXT | S83L + D87N | S80I | - | Unknown | - | - |
| E. coli (33) | RRRR | 256< | 16 | 16 | AMP, TIC, PIP | - | SXT | S83L + D87N | S80I | - | Unknown | - | - |
| E.coli (34) | RRRR | 256< | 16 | 16 | AMP, AMC, TIC, TCC, PIP, TZP, MEC | - | SXT | S83L + D87N | S80I | - | B2 | + | - |
| Strains | Quinolone resistance phenotypes and FQs NAL NXN OFX CIP |
Qs - FQs MICS (mg/L) | ATB resistance profiles | QRDRs mutations | PMQR gene | Phylogenetic group | ST131 clone | PFGE pulsotype/subtype |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NAL S ≤ 16 R > 16 |
OFX S ≤ 0.25 R > 0.5 |
C.I.P. S ≤ 0.25 R > 0.5 |
ß-lactams | Aminoglycosides | Other | gyrA | parC | ||||||
| 3 QRDRs ( 2 gyrA mutations + 1 parC mutation ) | |||||||||||||
| E.coli (35) | RRRR | 256< | 16 | 16 | AMP, TIC, CBT, PIP | - | CHL, SXT | S83L + D87N | S80I | - | B2 | + | - |
| E. coli (36) | RRRR | 256< | 32 | 32 | AMP, TIC, PIP | - | SXT | S83L + D87N | S80I | - | Clade I or II | - | - |
| E.coli (37) | RRRR | 256< | 16 | 16 | AMP, AMC, TIC, TCC, PIP, MEC | - | SXT | S83L + D87N | S80I | - | B2 | + | - |
| E.coli (38) | RRRR | 256< | 64 | 64 | AMP, TIC, PIP, CN, CXM | - | SXT | S83L + D87N | S80I | - | Clade I or II | - | - |
| E.coli (39) | RRRR | 256< | 64 | 64 | AMP, TIC, PIP | GM, NET, TOB | CHL | S83L + D87N | S80I | - | B2 | + | - |
| E. coli (40) | RRRR | 256< | 64 | 64 | AMP, AMC, TIC, TCC, PIP, CN, CXM, CFM, CTX, CAZ, CEF, ATM | - | SXT | S83L + D87N | S80I | - | Clade I or II | - | Subtype VIIa |
| E.coli (41) | RRRR | 256< | 64 | 64 | AMP, AMC, TIC, TCC, PIP | GM, TOB | SXT | S83L + D87N | S80I | - | Clade I or II | - | Pulsotype VII |
| E.coli (42) | RRRR | 256< | 32 | 32 | AMP, TIC, PIP | - | SXT | S83L + D87N | S80I | - | B2 | + | - |
| E.coli (43) | RRRR | 256< | 16 | 16 | - | - | - | S83L + D87N | S80I | B2 | + | Subtype IIa | |
| E.coli (44) | RRRR | 256< | 32 | 16 | AMP, TIC, PIP | - | CHL, SXT | S83L + D87N | S80I | B2 | + | Subtype IIb | |
| E.coli (45) | RRRR | 256< | 32 | 32 | AMP, TIC, PIP | - | CHL, SXT | S83L + D87N | S80I | Clade I or II | - | Subtype IIc | |
| E.coli (46) | RRRR | 256< | 32 | 32 | AMP, TIC, PIP | - | CHL, SXT | S83L + D87N | S80I | Clade I or II | - | IId subtype | |
| E.coli (47) | RRRR | 256< | 32 | 16 | AMP, AMC, TIC, TCC, PIP | - | CHL, SXT | S83L + D87N | S80I | Clade I or II | - | Subtype VIIIa | |
| E.coli (48) | RRRR | 256< | 32 | 16 | AMP, TIC, PIP | GM, TOB | CHL, SXT | S83L + D87N | S80I | Unknown | - | - | |
| E.coli (49) | RRRR | 256< | 16 | 8 | AMP, AMC, TIC, TCC, PIP | GM, NET, TOB | CHL, SXT | S83L + D87N | S80I | B2 | + | - | |
| E.coli (50) | RRRR | 256< | 8 | 8 | AMP, ICT, PIP | - | CHL, SXT | S83L + D87N | S80I | B2 | + | - | |
| E. coli (51) | R RRR | 256< | 16 | 8 | AMP, AMC, TIC, TCC, PIP | - CHL | S83L + D87N | S80I | -B2 | + | |||
| E.coli (52) | RRRR | 256< | 32 | 32 | AMP, AMC, TIC, TCC, PIP | - | SXT | S83L + D87N | S80I | - | B2 | + | - |
| E.coli (53) | RRRR | 256< | 64 | 64 | AMP, AMC, TIC, TCC, PIP | GM, NET, TOB | CHL | S83L + D87N | S80I | - | B2 | + | - |
| E.coli (54) | RRRR | 256< | 64 | 32 | AMP, AMC, TIC, TCC, PIP | GM, NET, TOB | CHL | S83L + D87N | S80I | - | Clade I or II | - | - |
| E. coli (55) | RRRR | 256< | 32 | 32 | AMP, TIC, PIP | - | - | S83L + D87N | S80I | - | Clade I or II | - | - |
| E.coli (56) | RRRR | 256< | 32 | 32 | AMP, TIC, PIP, MEC | - | - | S83L + D87N | S80I | - | B2 | + | - |
| E. coli (57) | RRRR | 256< | 32 | 32 | AMP, ICT, PIP | GM, TOB | CHL, SXT | S83L + D87N | S80I | - | B2 | + | - |
| E. coli (58) | RRRR | 256< | 32 | 32 | AMP, AMC, TIC, PIP | - | - | S83L + D87N | S80I | - | Clade I or II | - | - |
| E.coli (59) | RRRR | 256< | 32 | 32 | - | - | - | S83L + D87N | S80I | - | Clade I or II | - | - |
| E.coli (60) | RRRR | 256< | 32 | 32 | - | - | - | S83L + D87N | S80I | - | B2 | + | - |
| E.coli (61) | RRRR | 256< | 32 | 16 | AMP, TIC, PIP | - | - | S83L + D87N | S80I | - | Clade I or II | - | - |
| E.coli (62) | RRRR | 256< | 32 | 32 | AMP, TIC, PIP, CN, MEC, CXM | - | - | S83L + D87N | S80I | - | Clade I or II | - | Subtype IXa |
| E.coli (63) | RRRR | 256< | 8 | 8 | AMP, TIC, PIP, MEC | - | SXT | S83L + D87N | S80I | - | B2 | + | Xa subtype |
| Strains | Qs-FQs resistance phenotype NAL NXN OFX CIP |
Qs-FQs MICS (mg/L) | ATB resistance profiles | QRDR mutations | PMQRs gene | Phylogenetic group | Clone ST131 |
PFGE pulsotype/subtype |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NAL S≤16 R>16 |
OFX S≤0.25 R>0.5 |
C.I.P. S≤0.25 R>0.5 |
ß-lactams | Aminoglycosides | Other | gyrA | parC | ||||||
| 3 QRDRs ( 2 mutations gyrA + 1 mutations parC) + 1 PMQR | |||||||||||||
| E. coli (64) | RRRR | >256 | 32 | 16 | AMP, TIC, PIP | SXT | S83L+D87N | S80I | qnrS | Unknown | |||
| 4 QRDRs ( 2 mutations gyrA + 2 mutations parC ) | |||||||||||||
| E. coli (65) | RRRR | 256< | 64 | 64 | AMP, TIC, PIP, CN, CXM, CTX | - | SXT | S83L+D87N | S80I+E84V | - | B2 | + | Subtype IIIa |
| E.coli (66) | RRRR | 256< | 64 | 64 | AMP, TIC, PIP | - | CHL | S83L+D87N | S80I+E84V | - | B2 | + | - |
| E. coli (67) | RRRR | 256< | 64 | 64 | - | - | - | S83L+D87N | S80I+E84V | - | B2 | + | - |
| E.coli (68) | RRRR | 256< | 128 | 128 | AMP, TIC, PIP | - | SXT | S83L+D87N | S80I+E84V | - | Unknown | - | - |
| E. coli (69) | RRRR | 256< | 128 | 128 | AMP, TIC, PIP | - | CHL, SXT | S83L+D87N | S80I+E84G | - | Clade I or II | - | Pulsotype IV |
| E.coli (70) | RRRR | 256< | 64 | 64 | AMP, TIC, PIP | - | CHL | S83L+D87N | S80I+E84V | - | B2 | - | Subtype Va |
| E. coli (71) | RRRR | 256< | 64 | 64 | AMP, TIC, PIP, CN, CXM, CTX | - | SXT | S83L+D87N | S80L+E84V | - | B2 | + | Pulsotype V |
| E. coli (72) | RRRR | 256< | 64 | 64 | AMP, TIC, PIP, CN, CXM, CTX, CAZ, CEF | - | SXT | S83L+D87N | S80I+E84V | - | B2 | + | - |
| E.coli (73) | RRRR | 256< | 64 | 64 | AMP, AMC, TIC, TCC, PIP, CN, CXM, CFM, CTX, CAZ, CEF, ATM | - | - | S83L+D87N | S80I+E84V | - | Clade I or II | - | - |
| Mutation not determined | |||||||||||||
| E. coli (74) | RRRR | 256< | 32 | 16 | AMP, TIC, PIP | - | CHL, SXT | ND | ND | - | B2 | + | - |
| E.coli (75) | RRRR | 256< | 128 | 128 | AMP, TIC, PIP | GM | SXT | ND | ND | - | Clade I or II | - | - |
| E. coli (76) | RRRR | 256< | 2 | 2 | AMP, TIC, PIP | - | - | ND | ND | - | Clade I or II | - | Subtype IVa |
| E.coli (77) | RRRR | 256< | 32 | 16 | - | - | - | ND | ND | - | Clade I or II | - | Subtype IVb |
| E. coli (78) | R RRR | 256< | 16 | 16 | AMP, TIC, PIP | - | SXT | ND | ND | - | Unknown | - | - |
| E. coli (79) | R RRR | 256< | 128 | 128 | AMP, TIC, PIP | GM, TOB | CHL | ND | ND | - | Clade I or II | - | - |
| E. coli (80) | R RRR | 256< | 32 | 32 | - | - | CHL | ND | ND | - | Unknown | - | - |
MIC: minimum inhibitory concentration; R: resistant; NAL: Nalidixic acid, NXN: Norfloxacin, OFX: ofloxacin; CIP: ciprofloxacin AMP: ampicillin; TIC: ticarcillin; PIP: piperacillin; TCC : Ticarcillin + Ac.clavulanic; TZP : Piperacillin + Tazobactam; MEK : Mecillinam; AMC: amoxicillin + clavulanic acid; CN: cephalexin; MEC: mecillinam; CXM: cefuroxime VI; CTX: cefotaxime; CAZ: ceftazidime; CEF: cefepime; ATM: aztreonam; GM: gentamicin; TOB: tobramycin; NET: Netilmicin; CHL: chloramphenicol; SXT: cotrimoxazole RPF: plasmid resistance to fluoroquinolones, QRDRs: quinolone resistance determining region, ND: not determined (illegible sequences) ; +: positive; -: Negative.
Impact of existing genetic mechanisms of resistance on MICS levels
According to the different MICs of Qs and FQs highlighted for E. coli isolates, they were categorized into three resistance profiles, suggesting the involvement of multiple resistance mechanisms. The results observed reveal that levels of resistance evolve proportionally to the type and frequency of genetic variation involved. Strains that exhibited resistance mechanisms of chromosomal origin and/or plasmid have shown a proportional variation between the number of chromosomal substitutions in the genes as well as the presence of (PMQRs) determinants and the resistance levels observed concerning the MICS of Qs and FQs. Thus, the phenotypic susceptibility profiles were linked to molecular characterization.
(i) Strains exhibiting the FQs resistance profile I (with MICs values of 24 mg/l for nalidixic acid and 0.24 mg/L for ofloxacin and ciprofloxacin) harbored one PMQRs determinant; but no QRDRs mutations were detected (Table 2). (ii) Those of resistance phenotype profile II showing MIC values > 256 mg/l for nalidixic acid and a MIC equal to 0.5 mg/L for ofloxacin as well as 0.24 mg/L for ciprofloxacin, were characterized by a single chromosomal mutation in the gyrA gene. (iii) Finally, the strain with resistance phenotype profile III, for which we determined nalidixic acid MICs values > 256 mg/L, presents the higher FQs MICS ranging from 1 to 128 mg/L that went through five steps of escalation related to the genetic mechanisms implicated. (i) MIC values concerning FQs varying between 1 and 4 mg/L were determined in the co-carriage of two chromosomal mutations on the gyrA gene and the absence of PMQRs. (ii) MIC values between 1 and 64 mg/L for ofloxacin and between 1 and 32 mg/L for ciprofloxacin were determined in association with one chromosomal mutation and one PMQRs, (iii) MIC values for ofloxacin and ciprofloxacin of 8 and 32 mg/L, respectively, were determined in the co-carriage of two chromosomal mutations in the gyrA gene, either one on gyrA + one on parC, (iv) MICs values between 8 and 64 mg/L concerning ofloxacin and ciprofloxacin, respectively, were determined in the co-carriage of three chromosomal mutations (2 in the gyrA gene and 1 in the parC gene). (V) MIC values varying between 64 and 128 mg/L concerning FQs were determined in the co-carriage of 4 chromosomal mutations (2 on the gyrA gene and 2 on the parC gene) (Table 2).
Phylogenetic groups and ST131 Clone determination
The phylogenetic groups determined for the 80 E. coli strains allowed assign 68(85%) of them to a distinctive phylogenetic group, resulting in the following distribution: There are 41 (51,25%) strains belonged to the B2 group, and only one (1,25%) strain in the F group. Then, 26 (32,50%) strains showed the cryptic Clade I or II, however, 12 (15%) showing none of the profiles studied were therefore not assigned to any of the phylogenetic groups studied in this work. Moreover, 28 (35%) among our strains were found to belong to the clone ST131 all assigned to phylogroup B2, thus we have noted that fifteen B2 group E. coli isolates did not belong to the ST131 clone. Tables 3, 4 and 5 summarize the strain’s characteristics based on their resistance profiles I, II, or III.
Clonal relatedness
The restriction profiles obtained by PFGE typing showed 53 different pulsotypes grouping one strain each, i.e., 66.25% of the strains proved to be sporadic without any clonality link. However, 27 (33.75%) of the strains are divided into 10 pulsotypes (analogous clone) designated from I to X, each grouping one or more subtypes with two or more related strains, among which we distinguish two pulsotypes with homogeneous restriction profiles: pulsotype I, which groups together (2) clonal strains, and pulsotype II, which groups together (4) clonal strains. The remaining eight pulsotypes are heterogeneous and divided into subtypes, each with one to four distinct bands from the original pulsotype. Pulsotype III has been subdivided into four subtypes (IIIa, IIIb, IIIc, and IIId), each grouping a single strain.; The pulsotypes IV and VIII have each been subdivided into two subtypes (IVa, IVb) and (VIIIa, VIIIb), each grouping together a single strain.; Pulsotypes V, VI, VII, IX, X, and XI have each been subdivided into a single subtype (Va, VIa, VIIa, IXa, Xa, and XIa), each of which regroups a single strain.E. coli strains of lineage ST131 showed a few similarities among them. Tables 3, 4 and 5 summarize the various Pulsotypes/subtypes groups based on the resistance profiles.
Discussion
Infectious complications caused by FQs resistance following (TRUS-BP) have increased globally, prompting the medical community in most countries to question whether FQs should continue to be the first drug of choice for such prophylaxis treatment. In fact, given the ever-growing emergence of organisms resistant to FQs, the traditional choice of these molecules as prophylactic antibiotics before (TRUS-BP) contributes to an increase in the rate of post-biopsy infectious complications. The prevalence of FQs-R E.coli from rectal carriage in our patients revealed an overall quite high percentage with an increasing evolution rate according to the years of the screening. These result are therefore in concordance with several similar studies, which reveal a carriage incidence of FQs-R strains that has been constantly increasing over the years30. We hypothesized that one of the main reservoirs of dissemination of antimicrobial resistance genes worldwide is non-pathogenic intestinal flora; thus, the detection of fecal carriage of FQs-Resistance in patients before (TRUS-BP) is a crucial element for the implementation of a preventive, targeted, and effective antibiotic therapy strategy to avoid infectious complications. The first similar study was carried out in 2010 in the UK31 and the FQs-R was found in only 10.6% of these patients. The following studies worldwide have described prevalence rates ranging from 13 to 92%32-35. To our knowledge, there have been no such previous studies in Tunisia. Our results showed an exclusivity of the species E. coli; following most of the published similar data which report the high preponderance of E. coli species36.
Chromosomal resistance
It is of crucial importance to understand the molecular mechanisms implicated in antibiotic resistance and their influence on levels of MICs to control acquired resistance emergence and dissemination. Considering the analysis of the gyrA and parC genes our results are consistent with previous findings6. Our study reported a high prevalence among isolates of one or several substitutions on the gyrA gene alone or combined with one or two substitutions on the parC gene. Conversely, no isolates had a parC mutation without a gyrA mutation. Similarly to other reports, this study found that substitutions at Ser-83 and Asp-87 in GyrA and Ser-80 and Glu-84 in ParC are the most common mutations observed among FQs-R E. coli strains6–35. Switching from Serine to Leucine (S83L) was the most frequent substitution37, followed by a change from Serine to Tyrosine (S83Y); then from Serine to Threonine (S83T), as well as from Serine to Tryptophan (S83W); likewise, it has been reported previously38. Another less frequent double mutation was found substituting Aspartate for Asparagine (D87N) on the gyrA gene, which was found mainly in Fqs-R E. coli isolates exhibiting higher levels of MICs. Therefore, the mutation at position 87 of gyrA appeared to contribute to high resistance to FQs, while the mutation at position 83 could contribute to both resistances to Qs and FQs which is consistent with other studies39. Concerning the parC the Serine by Isoleucine substitution (S80I) was the most frequent, which is in agreement with the literature40. To the best of our knowledge, the study presented here is the first report on the characterization of molecular mechanisms of FQs resistance in enteric isolates from men in Tunisia. Nevertheless, chromosomal mutations of the gyrA (Ser83Leu + Asp87Asn) and parC (Ser80Ile- Glu84Val- Glu84Gly) types were detected in the fecal carriage of FQs-R E. coli in healthy children as well as in various pathological samples; several simultaneous point mutations were also reported41.
Plasmid-mediated resistance
There is currently no phenotypic criterion for distinguishing between chromosomal and plasmid-mediated associated resistance (PMQRs) or different other mechanisms, their identification requires molecular biology techniques. It has been shown that (PMQRs) alone confer low levels of resistance (phenotypically sensitive or intermediate sensitivity) in Enterobacterales. The existence of these determinants is known to favor the selection of resistant mutants by additional chromosomal resistance mechanisms42. This has aroused interest in the detection of plasmid resistance mechanisms that generate low-level resistance to avoid the risk of selection of existing mutants that could favor the emergence of highly resistant strains. Our findings corroborate studies highlighting that alleles belonging to qnr are usually the most frequently detected in the Enterobacterales family;7 and that QnrS protein is the most prevalent Qnr protein in E. coli species28. Only Few Similar studies from other countries found the qnr gene plasmid determinant in 8.8% and 7.35% of Enterobacterales isolates, respectively43,44. In Tunisia, qnr genes were previously detected in 8.2% of clinical strains of E. coli isolated at Sahloul Hospital in Sousse, in 16.9% at a children’s hospital, and in 22% at Charles Nicolle Hospital in Tunis41,45,46. A worthy fact to notice in our finding is the earliest description of qnrB19 in a human-derived E. coli isolate, which had previously been detected in an animal isolate47. This aforementioned gene was initially described in 2008 from a clinical isolate of E. coli in Colombia, and it has been suggested that its acquisition should be linked to a mobile genetic element of the ISEcp1 type48; to the best of our knowledge this is also the first report of a qnrB81 determinant in an E.coli species. To date, the qnrB81 gene has been described only on a Citobacter freundii isolated from an urinary tract infection in a small private hospital in Sierra Leone49, and recently identified in Salmonella infantis in Europe50. Concerning the determinant qnrS, the prevalence reported in the literature is generally low7. In our country, it was already detected in clinical E. coli isolates45,46 sequencing of this aforementioned gene allowed us to identify the allele qnrS15, the detection of the latter has not yet been described in Tunisia; this allele was reported in 2018 in E.coli strain isolated from a pig slaughterhouse in Holland51. In our study, the AAC (6′)-Ib-cr determinant was detected only in one strain, while, in Tunisia, a high prevalence of aac(6’)-Ib-cr (77.5%) genes was found in qnr-positive Enterobacterales isolated from children, even those not overtreated with quinolones46, or from the intestinal microbiota of healthy children52; mostly amongst ESBL E.coli producers46 and hospital environments41. In a similar study, the prevalence of this gene was (89.5%) percent among fecal carriage isolates43. None of our strains harbored the qnrA type genes, nevertheless, previous Tunisian studies41 had detected a percentage of 0.3% of the qnrA6 gene; Also, two outbreaks of Providencia stuartii carrying qnrA6 in burn and intensive care units were reported in Tunisia54,55. On the other hand, we note the absence of qnrC, qnrD, qepA, and oqxA gene which corroborates the fact that they are rarely reported in the world7 and is also consistent with some previous clinical Tunisian studies45, 46. Conversely41, found genes encoding for the plasmid-mediated efflux pumps OqxAB with a frequency of (21.6%) among ESBL-producing Enterobacterales.
Impact of genetic basis involved on Qs-FQs MICs range amongst phenotypic profile
The strain belonging to profile I, identified as resistant to Qs and sensitive to FQs, was found to harbor PMQRs genes with no additional chromosomal mutation and showed a low level of resistance against Qs (MIC50s to nalidixic acid of 24 mg/L). The acquisition of a plasmid carrying qnr genes is not sufficient to transform a wild-type bacterium sensitive to FQs into a resistant strain according to the standard of sensitivity set by international scientific committees. Nonetheless, the PMQRs phenomenon plays an important role in increasing the MICs against FQs, thus offering bacteria an opportunity to generate a chromosomal mutation6. A single mutation in the gyrA gene only occurred in isolates with MICs of FQs ≤ 0.24 mg/L, on strains classified into profile II that conferred a high level of resistance to nalidixic acid only. These observations support the fact that detection of this first-level genetic variation is important from a preventive point of view; in fact, it facilitates molecularly the selection of a second mutation, which will lead to an increase in the degree of resistance56. These findings support recent suggestions that systematic reporting of nalidixic acid sensitivity could effectively identify numerous isolates already harboring early gyrA mutations, whether or not accompanied by parC mutations8. Indeed, these are precisely the isolates most likely to become fully resistant in the presence of selective antimicrobial pressure. In this perspective57, have shown for E. coli, that ciprofloxacin concentration prevention mutants (CPM) may influence susceptibility to select resistance, hence the importance of detecting and characterizing these low-level resistances to avoid the risk of increasing the selection of resistant mutants. The results found in this study don’t support the hypothesis that an alteration of a single amino acid in the GyrA QRDRs on codon 83 is sufficient to decrease susceptibility to ciprofloxacin, as previously noted44. This is in agreement with the fact stated that the combination of (PMQRs) genes and mutations in the QRDRs of GyrA and ParC contributes to high resistance to FQs. While a single stepwise accumulation of mutations in the gyrase and topoisomerase subunits or harboring only plasmid determinants can explain quinolone resistance. The presence of a double mutation in the gyrA and parC genes appears to be associated with rising levels of resistance to FQ. More specifically accumulation of amino acid changes in GyrA (codons 83 and 87) and simultaneous alterations of ParC (codons 80 and 84) seems to play a key role in the development of high-level resistance (MIC > 32 mg/L) to FQs. The strains listed in resistance profile III were highly resistant to Qs (≥ 256 mg/L) and had a stepwise increase in FQs-MICs (from 8 to 128 mg/L). They were characterized by the co-occurrence of multiple genetic mechanisms involving several (two to four) co-carriages of mutations in the gyrA and parC genes as well as the co-existence of one additional PMQRs gene. The co-carriage of 3 QRDRs mutations was the most frequently detected in isolates with rising levels of resistance to ciprofloxacin (CIP) (MICs 8–64 mg/mL), while co-carriage of 4 QRDRs mutation was detected in (5%) of highly resistant isolates (MICs 64–128 mg/mL). As a result, these findings support the notion that there is a close relationship between the addition of chromosomal mutations and the increase in steps of resistance. The acquisition of highly bacterial resistance to FQs is a phenomenon that occurs in stages, with the progressive accumulation of molecular resistance mechanisms “on a scale.” This addition of mutations within the gyrA and parC genes is therefore responsible for the achievement of levels of clinical resistance to FQs as previously reported by58, 59, 60. Thus, our observation emphasizes the fact that the rise of FQ-R E.coli with three QRDR mutations could present a significant clinical problem because the three-mutation strains are only one mutation away from becoming extremely highly resistant. To the best of our knowledge, our findings are the first full description in Tunisia of the impact on MICs of FQs-Resistance molecular mechanisms involved in human intestinal carriage isolates. Few such studies on men undergoing (TRUS-BP) in the literature provide information on the common presence of QRDRs and PMQRs among Enterobacterales isolates with a ciprofloxacin MIC ≥ 0.25 mg/L43. Our findings support the widely held belief that PMQR determinants may enhance mutations in the (QRDRs), contributing to rising FQ resistance in clinical settings and possibly playing a role in post-biopsy infection prevalence.
Associated resistances
Simultaneous resistance to other families of antibiotics was frequent in our strains. This suggests the possibility of acquiring plasmid-mediated antibiotic resistance factors in these strains. Indeed, resistance to several antibiotic families is frequently reported in FQs-R Enterobacterales60. These associated resistances can be explained by the frequent presence of acquired resistance genes in different families of antibiotics in the same population. Moreover, the multi-drug resistance of the strains harboring the PMQRs genes has been reported in several studies involving all classes of antibiotics at variable frequencies. They are increasingly isolated from human infections as well as from intestinal carriage42. Another phenomenon reported is inactivating enzymes that possibly target more than one antibiotic, such as the enzyme AAC-(6 ‘)-Ib-cr, the first enzyme capable of inactivating aminoglycosides and quinolones by acetylation. This was confirmed in our study by the case of E. coli number (27) harboring this gene. Thus, due to the multiresistant character of the strains, which harbor mechanisms of resistance to FQs, the therapeutic choice can be critical, causing an embarrassing situation for the prescriber. Moreover, these strains constitute an epidemiological risk since they ensure the co-selection of resistance determinants, thus promoting their dissemination. The appearance of such resistant strains in the intestinal carriage is another worrying phenomenon. The promotion of therapeutic alternatives as well as the rational use of FQs is essential to controlling the spread of resistance to other antibiotics and multiresistant Enterobacterales.
Phylogenetic groups, ST131 clone determination, and clonal link
The predominance of the B2 group amongst strains investigated in this study is worrying, since E.coli most pathogenic strains belong to group B2. The unidentified traits that B2 group possess yet enhance their survival in the human intestine, giving them superior capacity to persist in the intestinal microflora It appear to more frequently carry virulence-factor genes than do group A strains and group B1. On the other hand, it has been shown that strains that cause extraintestinal infections belong mostly to group B2 and, to a lesser extent, group D61. Of the cryptic clades, clade I is associated with disease in humans, while clades II to V are thought to potentially be of environmental origin and therefore more adapted to life outside a host62. The data on the distribution of phylogenetic groups of E. coli within the commensal flora are highly variable. Indeed, early studies have shown predominant phylogenetic groups are A and B163, with further work documenting a high proportion of B2 strains among commensal isolates from populations of industrialized countries64. A high prevalence of the notorious ST131 E. coli clone was noted in this study, our findings corroborate that the development of FQs-R was accompanied by a large surge in the ST131 population globally making this pandemic multi-drug resistant lineage remaining one of the most common FQs-R clonal groups65. Our study was conducted during 2016–2018, and no actual data concerning the evolution of the fecal carriage of this lineage among community settings in Tunisia are available. to the best of our knowledge, just one study report in a screening conducted from 2013 to detect two isolates of the pandemic clone ST131 (non-extended-spectrum-β-lactamase (ESBL)-producers) in culture swab from healthy children27. Therefore, this is the first report that emphasizes the dissemination of the ST131 clone among commensal E. coli strains recuperated from rectal carriage of men in our country. Thus reporting the emergence of such Extraintestinal pathogenic Escherichia coli (ExPEC) ST131 clone in the community is of great importance and remain alarming. The evolution of FQs-R therefore aptly demonstrates the influential role that small but significant foothold mutations play in MDR clone evolution. We therefore suggest that antimicrobial stewardship programmes should broaden their focus from just controlling infection by antibiotic adaptation in clinical settings. Greater focus should be on controlling the gut carriage of resistant bacteria, including screening for and decolonizing commensal carriage of antibiotic-resistant strains above all in the most vulnerable individuals. Furthermore, our findings support the fact that STs lineage strains carry mutations that give FQs resistance by targeting serine residues in DNA gyrase (GyrA-S83) and topoisomerase IV (ParC-S80). All E. Coli ST131 clones identified with high levels of ciprofloxacin resistance (MIC > 32 mg/L), carried double or triple mutations in gyrA in combination with single or double mutations in parC. These changes may confer a commensal fitness benefit in such clonal settings. Besides this increase in fitness may have helped these large clones proliferate widely in the healthcare context as well as colonize healthy people over time, allowing them to circulate in the population even in settings where FQs use is little or nonexistent19.
Genetic relatedness determined by PFGE indicated a high level of isolate heterogeneity permitting the distinction of a large amount of clonal variability. Only one abroad similar study conducted in Chicago indicates that the strains studied did not share a common origin66. In Tunisia, there have been a few studies that have looked at the distribution of phylogenetic groups among strains that have FQs-R determinants, and the results have been mixed41. Showed the predominance of group B2 followed by group A in clinical strains and the predominance of group A (51%) followed by group D (27.5%) in a collection of commensal strains. Indeed, resistance among Enterobacterales continues to rise, then generalizes with a typically non-clonal appearance, implying independent emergence67; nevertheless, a few of the strains could have a common origin. The intestinal microbiota of healthy subjects constitutes a potential reservoir for the emergence and diffusion of resistant strains. This dissemination could have a few possible explanations: the selection of different mutant strains, often attributed to overcrowding, basic hygiene problems, local antimicrobial prescribing and colonization capabilities success of multi-drug resistant lineage. It should also be noted that, even in the absence of selection pressure, due to recent antibiotic therapy, the food origin of resistant strains in the intestinal flora is strongly suggested. As long as strains showing chromosomal mutations and/or harboring PMQRs genes are frequently isolated from food products and many findings have suggested that the agricultural use of antimicrobial agents is implicated in human infections caused by drug-resistant bacteria68. Finally, given the diversity of commensal flora and heterogeneous phenotypic susceptibility, universal prophylaxis will no longer be able to provide optimal coverage for patients undergoing prostate biopsy. Indeed this hypothesis is well supported by our previous findings in which targeted antibiotic prophylaxis (TAP) adapted to rectal swab culture in patients undergoing (TRUS-BP) significantly reduced the rate of infectious complications69.
Conclusion
In conclusion, this study revealed the concerning incidence of FQs-R E.coli in digestive carriages of local population. there is a predominance of virulent phylogroup B2, as well as a worrying spread of colonization with the highly virulent multiresistant ST131 lineage in the community. This shows potential to cause post-infections using inappropriate prophylactic treatment. The possibility of cross-transmission as well as environmental dissemination, raises awareness among prescribing physicians about the dangers of antibiotic misuse. Thus, this justifies the contribution of rectal screening prior to TRUS-BP for adequate targeted antibiotic prophylaxis prescription to reduce the risk of infectious postoperative complications. Our investigation confirms, from a molecular point of view, that mutations in the drug target-encoding genes gyrA and parC are the major mechanisms involved in FQs-R in E. coli. It also confirms their impact, coupled with plasmid determinant acquisition, on resistance levels. MICs increase in a narrow escalation step proportionally to the co-occurrence of multiple molecular acquired mechanisms. These results elucidate the role of PMQRs determinants in promoting QRDRs mutations and the acquisition of high-level FQs-R in clinical settings, confirming the importance of detecting the low level of resistance to FQs in order to prevent the selection of highly resistant mutants. The occurrence of both plasmid-borne and chromosome-borne resistance towards FQs can drive movement of the resistance phenotype across bacterial species and vertical movement of resistance along the bacterial generations. It is worth noting that allelic variation in PMQRs genes allows us to report the earliest presence of qnrB81 in E. coli species isolate from human intestinal carriages. Our findings encourage efforts to mitigate challenges in order to adapt prophylactic therapy to control infection, as well as to monitor the emergence of multidrug-resistant strains. This is important to limit their spread and find solutions to improve the epidemiological situation. Thus, we underscore the role of infection control as one of the key interventions in fighting the threat of antibiotic resistance. There is a pressing need to strengthen the implementation of the “One Health” concept and monitor antibiotic prescriptions in both human and veterinary medicine on a global scale.
Acknowledgements
E. coli J96 reference strains was kindly given by Mobley H (USA), Moreno, E. (Spain), and Dobrindt, U. (Germany). E. coli qnrA(+), K. pneumoniae qnrB(+), E. cloacae qnrS(+) positive control strains were kindly given by Nordman, P. (France) and E. coliaac(6')-Ib-cr (+) by Berçot, B. (France).
Author contributions
Rehaiem Amel Was responsible for the design of this study and phenotypic analysis, including strains isolation and identification, antibiotic susceptibility testing and PCR experiments and data analysis, performed sequencing experiments ant writing of the original draft of manuscript; Bouzouita abderazzek Was responsible for the conceptualization of this study and choice of methods, data analysis, check and manuscript draft; Ferjani Sana Participation in investigation and analysing of gene sequencing; Saadi Ahmed Responsible for rectal swab, resources validation. Zrelli Mariem Investigation, formal analysis; Kanzari lamia and Ferjani asma Revised the draft paper critically for important intellectual content; Ben Slama Mohamed Riadh Supervising, Boutiba-Ben Boubaker IlhemWas the supervisor and main reviewer and editor of the final version of the manuscript. All authors have read and agreed to the published version of the manuscript. All authors gave final approval of the version to be published.
Funding
This work was supported by the Ministry of Scientific Research, Technology, and Competence Development of Tunisia.
Data availability
The data underlying this article will be shared on reasonable request to the corresponding author.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hooper, D. C.Mechanisms of action and resistance of older and newer fluoroquinolones. Clin. Infect. Dis.31(Suppl 2), S24–S28 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Correia, S., Poeta, P., Hébraud, M., Capelo, J. L. & Igrejas, G. Mechanisms of quinolone action and resistance: Where do we stand. J. Med. Microbiol.66(5), 551–559 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Jacoby, G.A., Strahilevitz, J., & Hooper, D.C. Plasmid-mediated quinolone resistance. Microbiol. Spectr.2(5), 3 (2014). [DOI] [PMC free article] [PubMed]
- 4.Strahilevitz, J., Jacoby, G. A., Hooper, D. C. & Robicsek, A. Plasmid-mediated quinolone resistance: A multifaceted threat. Clin. Microbiol. Rev.22(4), 664–689 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martínez-Martínez, L., Pascual, A. & Jacoby, G. A. Quinolone resistance from a transferable plasmid. Lancet. 351(9105), 797–799 (1998). [DOI] [PubMed] [Google Scholar]
- 6.Rodríguez-Martínez, J. M. et al. Plasmid-mediated quinolone resistance: Two decades on. Drug Resist. Updat. 29, 13–29 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Rodríguez-Martínez, J. M., Cano, M. E., Velasco, C., Martínez-Martínez, L. & Pascual, A. Plasmid-mediated quinolone resistance: An update. J. Infect. Chemother.17(2), 149–182 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Kotb, D. N., Mahdy, W. K., Mahmoud, M. S. & Khairy, R. M. M. Impact of co-existence of PMQR genes and QRDR mutations on fluoroquinolones resistance in Enterobacteriaceae strains isolated from community and hospital acquired UTIs. BMC Infect. Dis.19(1), 979 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robicsek, A., Jacoby, G. A. & Hooper, D. C. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect. Dis.6(10), 629–640 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Yamane, K. et al. New plasmid-mediated fluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate. Antimicrob. Agents Chemother.51(9), 3354–3360 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansen, L. H., Jensen, L. B., Sørensen, H. I. & Sørensen, S. J. Substrate specificity of the OqxAB multidrug resistance pump in Escherichia coli and selected enteric bacteria. J. Antimicrob. Chemother.60(1), 145–147 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Duboureau, H., Achkar, K., Stephan, R., Schmit, J. L. & Saint, F. Ecology and fluoroquinolon resistance profiles in febrile urinary tract infections (FUTI) after prostate needle biopsy: A retrospective study in 466 biopsies. Prog Urol.27(6), 345–350 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Tuppin, P. et al. Vers une évolution des pratiques de détection et de prise en charge du cancer de laprostate chez les hommes de 40 ans et plus en France (2009–2014)? Bulletin épidémiologique hebdomadaire. 156, 22 Mars 2016 BEH 9.
- 14.Briffaux, R. État de l’art: antibioprophylaxie pour les biopsies de prostate. Progrès en Urologie - FMC. 18(3), F15–F18 (2008). [Google Scholar]
- 15.Massot, M., Picard, B. & Denamur, E. Diversité des populations d’Escherichia coli et leurs variations au cours du temps au sein du microbiote intestinal. Revue Francophone des. Laboratoires. 486, 35–43 (2016). [Google Scholar]
- 16.Sieczkowski, M., Gibas, A., Bronk, M. & Matuszewski, M. Fluoroquinolone-based antimicrobial prophylaxis in patients undergoing transrectal ultrasound-guided prostate biopsy. Eur. J. Clin. Microbiol. Infect. Dis.34(9), 1815–1821 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adamczyk, P. et al. Fluoroquinolone-resistant Escherichia coli in intestinal flora of patients undergoing transrectal ultrasound-guided prostate biopsy - possible shift in biopsy prophylaxis. Cent. Eur. J. Urol.70(2), 192–196 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cummins, E. A., Snaith, A. E., McNally, A. & Hall, R. J. The role of potentiating mutations in the evolution of pandemic Escherichia coli clones. Eur J Clin Microbiol Infect Dis. (2021). 10.1007/s10096-021-04359-3. Epub ahead of print. PMID: 34787747. [DOI] [PubMed]
- 19.Fuzi, M. & Sokurenko, E. Commensal fitness advantage may contribute to the global dissemination of multidrug-resistant lineages of bacteria-the case of uropathogenic E. coli. Pathogens. 12(9), 1150. 10.3390/pathogens12091150 (2023). PMID: 37764958; PMCID: PMC10536240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Everett, M. J., Jin, Y. F., Ricci, V. & Piddock, L. J. Contributions of individual mechanisms to fluoroquinolone resistance in 36 Escherichia coli strains isolated from humans and animals. Antimicrob. Agents Chemother.40(10), 2380–2386 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cattoir, V., Poirel, L., Rotimi, V., Soussy, C. J. & Nordmann, P. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J. Antimicrob. Chemother.60(2), 394–397 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Cavaco, L. M., Hasman, H., Xia, S. & Aarestrup, F. M. qnrD, a novel gene conferring transferable quinolone resistance in Salmonella enterica serovar Kentucky and Bovismorbificans strains of human origin. Antimicrob. Agents Chemother.53(2), 603–608 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, H. B. et al. oqxAB encoding a multidrug efflux pump in human clinical isolates of Enterobacteriaceae. Antimicrob. Agents Chemother.53(8), 3582–3584 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang, M. et al. New plasmid-mediated quinolone resistance gene, qnrC, found in a clinical isolate of Proteus mirabilis. Antimicrob. Agents Chemother.53(5), 1892–1897 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sana, F. et al. Prevalence and characterization of uropathogenic Escherichia coli harboring plasmid-mediated quinolone resistance in a Tunisian university hospital. Diagn. Microbiol. Infect. Dis.79(2), 247–251 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Clermont, O. et al. The Clermont Escherichia coli phylo-typing method revisited: Improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep5(1), 58–65. [DOI] [PubMed]
- 27.Ferjani, S., Saidani, M., Maamar, E., Harbaoui, S., Hamzaoui, Z., Hosni, H., Amine, F.S. & Boubaker, I.B.B. Escherichia coli colonizing healthy children in Tunisia: High prevalence of extra-intestinal pathovar and occurrence of non-extended-spectrum-β-lactamase-producing ST131 clone. Int. J. Antimicrob. Agents. 52(6), 878–885.(2013). 10.1016/j.ijantimicag.2018.07.015. Epub 2018 PMID: 30036576. [DOI] [PubMed]
- 28.Kaufmann, M. E. Pulsed-field gel electrophoresis. Methods Mol. Med.15, 33–50 (1998). [DOI] [PubMed] [Google Scholar]
- 29.Tenover, F. C. et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: Criteria for bacterial strain typing. J. Clin. Microbiol.33(9), 2233–2239 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Besien, J., Uvin, P., Van den Abeele, A. M., Merckx, L. & Prevalence risk factors, and clinical relevance of fluoroquinolone-resistant organisms in rectal cultures: Should we target antibiotic prophylaxis prior to prostate biopsy. Adv. Urol.2016, 5392107 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Batura, D., Rao, G. G. & Nielsen, P. B. Prevalence of antimicrobial resistance in intestinal flora of patients undergoing prostatic biopsy: implications for prophylaxis and treatment of infections after biopsy. BJU Int.106(7), 1017–1020 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Minamida, S. et al. Prevalence of fluoroquinolone-resistant Escherichia coli before and incidence of acute bacterial prostatitis after prostate biopsy. Urology. 78(6), 1235–1239 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Siriboon, S., Tiengrim, S., Taweemongkongsup, T., Thamlikitkul, V. & Chayakulkeeree, M. Prevalence of antibiotic resistance in fecal flora of patients undergoing transrectal ultrasound-guided prostate biopsy in Thailand. Urol. Int.88(2), 187–193 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Lee, J. W. et al. Prevalence of antimicrobial resistance in normal rectal flora of patients undergoing transrectal ultrasonography-guided prostate biopsy in Korea. Int. J. Urol.21(8), 811–814 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Tsu, J. H. et al. Prevalence and predictive factors of harboring fluoroquinolone-resistant and extended-spectrum β-lactamase-producing rectal flora in Hong Kong Chinese men undergoing transrectal ultrasound-guided prostate biopsy. Urology. 85(1), 15–21 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Chamberland, R. R. Cutting to the core of the issue: Emerging strategies to reduce prostate biopsy-related infections. J. Clin. Microbiol.54(10), 2431–2435 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hooper, D. C. Mechanisms of fluoroquinolone resistance. Drug Resist. Updat. 2(1), 38–55 (1999). [DOI] [PubMed] [Google Scholar]
- 38.Varughese, L. R. et al. Analytical profiling of mutations in quinolone resistance determining region of gyrA gene among UPEC. PLoS One. 13(1), e0190729 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Araújo, B. F. et al. High frequency of the combined presence of QRDR mutations and PMQR determinants in multidrug-resistant Klebsiella pneumoniae and Escherichia coli isolates from nosocomial and community-acquired infections. J. Med. Microbiol.66(8), 1144–1150 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Qi, C. et al. Characterization of ciprofloxacin resistant Escherichia coli isolates among men undergoing evaluation for transrectal ultrasound guided prostate biopsy. J. Urol.190(6), 2026–2032 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Ferjani, S., Saidani, M., Amine, F. S. & Boutiba-Ben Boubaker, I. Prevalence and characterization of plasmid-mediated quinolone resistance genes in extended-spectrum β-lactamase-producing Enterobacteriaceae in a Tunisian hospital. Microb. Drug Resist.21(2), 158–166 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Nordmann, P. & Mammeri, H. Résistance plasmidique aux quinolones. Antibiotiques. 9 (4), 246–253 (2007). [Google Scholar]
- 43.Tigen, E. et al. Prevalence of CTX-M enzyme and qnrA, qnrB, qnrC, qnrS, aac-(6)-Ib genes among ESBL (Extended spectrum beta lactamase)-positive isolates in patients undergoing transrectal needle prostate biopsy in Turkey. Dis. Mol. Med.4(1), 1 (2016). [Google Scholar]
- 44.Piekarska, K. et al. The molecular mechanisms of fluoroquinolone resistance found in rectal swab isolates of Enterobacterales from men undergoing a transrectal prostate biopsy: The rationale for targeted prophylaxis. Ann. Clin. Microbiol. Antimicrob.20(1), 81 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dahmen, S., Poirel, L., Mansour, W., Bouallègue, O. & Nordmann, P. Prevalence of plasmid-mediated quinolone resistance determinants in Enterobacteriaceae from Tunisia. Clin. Microbiol. Infect.16(7), 1019–1023 (2010). [DOI] [PubMed] [Google Scholar]
- 46.Jlili, N. E. et al. Trend of plasmid-mediated quinolone resistance genes at the Children’s Hospital in Tunisia. J. Med. Microbiol.63(Pt 2), 195–202 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Sallem, R. B. et al. First detection of CTX-M-1, CMY-2, and QnrB19 resistance mechanisms in fecal Escherichia coli isolates from healthy pets in Tunisia. Vector Borne Zoonotic Dis.13(2), 98–102 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Cattoir, V., Nordmann, P., Silva-Sanchez, J., Espinal, P. & Poirel, L. ISEcp1-mediated transposition of qnrB-like gene in Escherichia coli. Antimicrob. Agents Chemother.52(8), 2929–2932 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leski, T. A. et al. Prevalence of quinolone resistance in enterobacteriaceae from sierra Leone and the detection of qnrB pseudogenes and modified lexA binding sites. Antimicrob. Agents Chemother.60(11), 6920–6923 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alba, P. et al. Molecular epidemiology of Salmonella Infantis in Europe: Insights into the success of the bacterial host and its parasitic pESI-like megaplasmid. Microb. Genom. 6(5), e000365 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cardenas, R. I. Y., Kant, A., Veldman, K. T. & Mevius, D. J. Novel plasmid mediated quinolone resistance QnrS gene from E. coli isolated from a slaughter pig in the Netherlands GenBank: MK303617. ; 1: 25–12. (2018).
- 52.Jouini, A. et al. Detection of unrelated Escherichia coli strains harboring genes of CTX-M-15, OXA-1, and AAC(6’)-Ib-cr enzymes in a Tunisian hospital and characterization of their integrons and virulence factors. J. Chemother.22(5), 318–323 (2010). [DOI] [PubMed] [Google Scholar]
- 53.Ferjani, S. et al. Community fecal carriage of broad-spectrum cephalosporin-resistant Escherichia coli in Tunisian children. Diagn. Microbiol. Infect. Dis.87(2), 188–192 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Arpin, C. et al. Evolution of an incompatibility group IncA/C plasmid harboring blaCMY-16 and qnrA6 genes and its transfer through three clones of Providencia stuartii during a two-year outbreak in a Tunisian burn unit. Antimicrob. Agents Chemother.56(3), 1342–1349 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mnif, B. et al. Nosocomial dissemination of Providencia stuartii isolates carrying bla OXA-48, bla PER-1, bla CMY-4 and qnrA6 in a Tunisian hospital. J. Antimicrob. Chemother.68(2), 329–332 (2013). [DOI] [PubMed] [Google Scholar]
- 56.Zhao, L. et al. Molecular epidemiology and genetic diversity of fluoroquinolone-resistant Escherichia coli isolates from patients with community-onset infections in 30 Chinese county hospitals. J. Clin. Microbiol.53(3), 766–770 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Olofsson, S. K., Marcusson, L. L., Komp Lindgren, P., Hughes, D. & Cars, O. Selection of ciprofloxacin resistance in Escherichia coli in an in vitro kinetic model: Relation between drug exposure and mutant prevention concentration. J. Antimicrob. Chemother.57(6), 1116–1121 (2006). [DOI] [PubMed] [Google Scholar]
- 58.Machuca, J. et al. Cellular response to ciprofloxacin in low-level quinolone-resistant Escherichia coli. Front. Microbiol.8, 1370 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Norouzi, A., Azizi, O., Hosseini, H., Shakibaie, S. & Shakibaie, M. R. Amino acid Substitution mutations analysis of gyrA and parC genes in clonal lineage of klebsiella pneumoniae conferring high-level quinolone resistance. JoMMID. 2(3), 109–117 (2014). [Google Scholar]
- 60.Salah, F. D. et al. Distribution of quinolone resistance gene (qnr) in ESBL-producing Escherichia coli and Klebsiella spp. in Lomé, Togo. Antimicrob. Resist. Infect. Control. 8, 104 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nowrouzian, F. L., Wold, A. E. & Adlerberth, I. Escherichia coli strains belonging to phylogenetic group B2 have superior capacity to persist in the intestinal microflora of infants. J. Infect. Dis.191(7), 1078–1083 (2005). [DOI] [PubMed] [Google Scholar]
- 62.Walk, S.T. The cryptic Escherichia. EcoSal Plus. 6, ecosalplusESP–0002 (2015). [DOI] [PMC free article] [PubMed]
- 63.Banerjee, R. & Johnson, J. R. A new clone sweeps clean: The enigmatic emergence of Escherichia coli sequence type 131. Antimicrob. Agents Chemother. [Internet]. 58(9), 4997–5004. (2014) http://aac.asm.org/lookup/doi/10.1128/AAC.02824-14 [DOI] [PMC free article] [PubMed]
- 64.Duriez, P. et al. Commensal Escherichia coli isolates are phylogenetically distributed among geographically distinct human populations. Microbiology (Reading). 147(Pt 6), 1671–1676 (2001). [DOI] [PubMed] [Google Scholar]
- 65.Moreno, E. et al. Structure and urovirulence characteristics of the fecal Escherichia coli population among healthy women. Microbes Infect.11(2), 274–280 (2009). [DOI] [PubMed] [Google Scholar]
- 66.Liss, M. A. et al. Prevalence and significance of fluoroquinolone resistant Escherichia coli in patients undergoing transrectal ultrasound guided prostate needle biopsy. J. Urol.185(4), 1283–1288 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Muylaert, A. & Mainil, J. G. Résistances aux fluoroquinolones: la situation actuelle. Ann. Méd. Vét., 157(1), 15–26 (2013).
- 68.O’Neel, S. et al. Icefield-to-ocean linkages across the northern pacific coastal temperate rainforest ecosystem. BioScience. 65(5), 499–512 (2015).
- 69.Bouzouita, A. et al. Antimicrobial prophylaxis protocol based on rectal swab culture before prostate biopsy to prevent infectious complications: A prospective randomized comparative study. Int. Urol. Nephrol.56(8), 2495–2502. 10.1007/s11255-024-03998-7 (2024). Epub 2024 Mar 7. PMID: 38448785. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author.