Abstract
Secreted aspartyl proteinases are putative virulence factors in Candida infections. Candida albicans possesses at least nine members of a SAP gene family, all of which have been sequenced. Although the expression of the SAP genes has been extensively characterized under laboratory growth conditions, no studies have analyzed in detail the in vivo expression of these proteinases in human oral colonization and infection. We have developed a reliable and sensitive procedure to detect C. albicans mRNA from whole saliva of patients with oral C. albicans infection and those with asymptomatic Candida carriage. The reverse transcription-PCR protocol was used to determine which of the SAP1 to SAP7 genes are expressed by C. albicans during colonization and infection of the oral cavity. SAP2 and the SAP4 to SAP6 subfamily were the predominant proteinase genes expressed in the oral cavities of both Candida carriers and patients with oral candidiasis; SAP4, SAP5, or SAP6 mRNA was detected in all subjects. SAP1 and SAP3 transcripts were observed only in patients with oral candidiasis. SAP7 mRNA expression, which has never been demonstrated under laboratory conditions, was detected in several of the patient samples. All seven SAP genes were simultaneously expressed in some patients with oral candidiasis. This is the first detailed study showing that the SAP gene family is expressed by C. albicans during colonization and infection in humans and that C. albicans infection is associated with the differential expression of individual SAP genes which may be involved in the pathogenesis of oral candidiasis.
Candida albicans is a commensal fungus commonly colonizing human mucosal surfaces. Under conditions of immune dysfunction, C. albicans can become an opportunistic pathogen causing recurrent chronic oral and vaginal candidiasis and life-threatening disseminated infections (38). Putative attributes which contribute to C. albicans virulence include adhesion, formation of hyphae, phenotypic switching, and proteinase secretion (1, 15, 39, 48). Secreted aspartyl proteinases (Saps) exhibit broad substrate specificity since they are able to degrade many human proteins found at lesion sites, such as albumin, hemoglobin, keratin, collagen, mucin, and secretory immunoglobulin A (sIgA) (13, 22, 41).
To date, nine different SAP genes have been identified in C. albicans. Each SAP gene in C. albicans is regulated at the transcriptional level and processed by a signal peptidase and a Kex2-like proteinase (37, 50, 53). Recently, the KEX2 gene of C. albicans has been disrupted, resulting in impaired Sap secretion and hyphal growth (37). The presence of nine SAP genes points to their importance in Candida pathogenesis and cell biology. Many studies have explored the regulation of SAP expression under laboratory conditions, primarily through the analysis of mRNA and protein synthesis under various nutritional and ionic conditions and in different strains. SAP2 is the most frequently expressed SAP gene; it is expressed in nearly all strains of C. albicans during log-phase growth in proteinase-inducing media (23, 53), and most studies which predate the recognition of the complexity of this gene family have most likely described the properties and characteristics of this proteinase. SAP1 and SAP3 are regulated during phenotypic switching (35, 36, 52), but SAP3 has also been detected in some strains when SAP2 is expressed (23, 46, 52). In vitro studies suggest that SAP1, SAP2, and SAP3 are expressed by yeast cells only. SAP4, SAP5, and SAP6 expression has been detected in C. albicans undergoing a transition from yeast to hyphae at neutral pH (23, 52), although S1 nuclease assays indicated that SAP6 is the predominant transcript (52). SAP7 expression has never been detected under any laboratory growth conditions (23). While SAP1, SAP2, and SAP3 comprise a subfamily distinct from that of SAP4, SAP5, and SAP6 by sequence homology, SAP7 appears to be the most divergent, showing only 20.4% similarity to SAP1 and about 26% homology to SAP4, SAP5, and SAP6 (34). SAP8 transcripts have been detected in yeast cells grown at 25°C in a defined medium with bovine serum albumin as the sole source of protein, and SAP9 is preferentially expressed in later growth phases, when the expression of other SAP genes has decreased (33).
The importance of the secreted proteinases as a virulence factor has been investigated in model systems of candidiasis. In the rat vaginitis model, only proteinase-producing Candida species were shown to be pathogenic (2), and in C. albicans infection, SAP1 and SAP2 expression has been demonstrated by Northern analysis in vivo (17). Sap2 was shown to be actively secreted during experimental vaginitis in the rat (47), and anti-Sap2 polyclonal antibodies appeared to confer partial protection against infection (8). In mouse and guinea pig models of disseminated candidiasis, mutant strains of C. albicans in which individual SAP genes had been disrupted exhibited attenuated virulence and reduced accumulation in host organs (24, 44). More recently, SAP expression was investigated in vitro by using reconstituted human epithelium as a model for human oral candidiasis (45). SAP1 and SAP3 were the first proteinases to be expressed, followed by SAP6, SAP8, and finally, SAP2. However, it is difficult to know whether this expression profile is representative of the initial stages of human oral C. albicans infection in vivo.
The expression pattern of members of the SAP gene family, as established under laboratory growth conditions, is under complex regulatory control. Due to the range of environmental conditions which regulate SAP gene expression in vitro, the collective and individual roles of Saps during infection are incompletely understood. In the present paper, we describe methods that allow direct assay of the expression of individual SAP genes in the oral cavity. The aims of this study were to develop a reliable and sensitive method for the detection of C. albicans SAP1 to SAP7 mRNA from whole saliva by using reverse transcription (RT)-PCR. Using this protocol, we compared the in vivo expression of genes SAP1 to SAP7 in oral candidiasis patients with that in asymptomatic Candida carriers.
MATERIALS AND METHODS
Strains, media, and culture conditions.
The Candida reference strains used in this study were C. albicans NCPF 3156, C. stellatoidea ATCC 11006 (type I), C. dubliniensis NCPF 3949, C. tropicalis ATCC 750, C. parapsilosis ATCC 22019, and C. guilliermondii NCPF 3099. Strains were maintained on Sabouraud (SAB) dextrose agar plates (Oxoid Ltd., Basingstoke, England). C. albicans NCPF 3156 was used for all optimization experiments and as a positive control for SAP2 expression throughout the study. For total RNA isolation from C. albicans NCPF 3156, cultures were grown in YCB-BSA medium (1.17% [wt/vol] yeast carbon base, 1% bovine serum albumin) for 24 h at 27°C. For DNA isolation, yeast cultures were grown in SAB broth (Oxoid) for 2 days at 27°C. For all cultures, 10 ml of test medium was inoculated with 10 μl of a 2-day culture of Candida grown in SAB broth at 27°C and shaken on an orbital incubator.
Study populations and sample collection.
The following three study populations were used: (i) a Candida-negative group, consisting of seven subjects whose saliva samples were consistently Candida negative by culture six times over a period of 3 months (four males and three females; mean age ± standard deviation, 31.3 ± 3.9 years); (ii) an asymptomatic Candida carrier group, consisting of eight subjects without any clinical signs or symptoms of candidiasis but harboring 50 to 800 C. albicans CFU/ml of saliva on six occasions over a period of 3 months (seven males and one female; mean age, 37.1 ± 10.9 years); and (iii) patients with oral Candida infection, consisting of 10 patients with clinical signs and symptoms of candidiasis and salivary counts of >2 × 103 C. albicans CFU/ml (14) (six males and four females; mean age, 45.5 ± 12.5 years). All the Candida carriers were volunteers that were not tested for human immunodeficiency virus (HIV) infection. Six of the oral candidiasis patients were HIV seropositive, and four were HIV seronegative. No individual in our patient group had been treated with anti-HIV proteinase drugs prior to or at the time of sample collection. Although a number of these patients had previously been treated with antifungal drugs, none were being treated with either antibiotic drugs or antimycotic drugs at the time of sampling.
Two unstimulated saliva samples were collected from each subject; one was immediately frozen on dry ice at −70°C to preserve the integrity of RNA for SAP analysis, and the other was used to determine the number of Candida CFU per milliliter of saliva. Yeast isolates were identified as C. albicans by using the API 32C AUX system (bioMerieux, Lyon, France), CHROMagar Candida (CHROMagar, Paris, France), germ tube formation, and growth at 42°C.
Germ tube formation and growth at 42°C.
One milliliter of 10% horse serum in brain heart infusion broth was inoculated with each strain and incubated at 35°C for 3 h. Production of germ tubes was observed microscopically under magnification at ×40. Each strain was observed for growth on SAB plates after incubation at 42°C for 48 h.
RNA and DNA isolation.
The use of glass beads and the length of vortexing were critical for optimal RNA isolation from Candida cells. All experiments were performed in 1.5-ml microcentrifuge tubes. Glass beads of different sizes (400 to 600 μm or 700 to 1,100 μm) and volumes (0.1 to 0.5 ml) were added to 0.5- or 1-ml C. albicans suspensions containing 104 cells/ml. Lysis of cells was measured at intervals during 30 min of vortexing by plating 100-μl samples onto SAB agar to evaluate percent survival. Optimal RNA recovery was obtained by vortexing a 1-ml volume of Candida cell culture with 0.5 ml of glass beads (size, 400 to 600 μm) for 30 min. This resulted in 100% killing of Candida cells.
Total RNA was prepared by a modified version of the procedure described by Chomczynski and Sacchi (11). C. albicans cells grown in culture or whole saliva samples were centrifuged, and 1 ml of Tri-reagent (Sigma, Poole, United Kingdom) and 0.5 ml of glass beads (size, 400 to 600 μM; Sigma) were added to the pellet prior to vortexing for 30 min. After the addition of 0.2 ml of chloroform, samples were centrifuged at 12,000 × g for 15 min at 4°C. Total RNA was precipitated from the aqueous fraction with isopropanol and centrifuged at 12,000 × g for 30 min at 4°C. Total RNA was treated with DNase (40 mM Tris-Cl [pH 7.9], 1 mM NaCl, 6 mM MgCl2, 10 mM CaCl2, 0.5 U of RNasin [Promega, Southampton, United Kingdom], 20 to 50 U of RQ-1 DNase [Promega]), reextracted twice with phenol-chloroform-isoamyl alcohol (25:24:1, vol/vol/vol) at pH 4.3 and twice with chloroform-isoamyl alcohol (24:1, vol/vol), and finally precipitated with 2.5 volumes of ethanol. Total RNA concentrations were determined by using a GeneQuant II spectrophotometer (Amersham Pharmacia Biotech, Little Chalfont, Buckinghamshire, United Kingdom). Purified RNA from each sample was confirmed to be DNA free by the absence of an amplified product after PCR performed by using fungus-specific primers complementary to the 5.8S rRNA gene (rDNA) plus intergenic sequence regions (54).
For DNA isolation, yeasts grown in SAB broth were harvested after 2 days, washed in sterile water, and resuspended in 0.4 ml of lysis buffer (0.2 M NaCl, 0.4% sodium dodecyl sulfate, 0.1 M Tris-Cl [pH 7.5], 5 mM EDTA [pH 8]). Equal volumes of phenol (pH 8) and glass beads were added, and the mixture was vortexed for 30 min. DNA was extracted twice with phenol-chloroform-isoamyl alcohol (25:24:1, vol/vol/vol) at pH 8 and twice with chloroform-isoamyl alcohol (24:1, vol/vol) and precipitated with 2.5 volumes of ethanol. DNA concentrations were determined on a GeneQuant II spectrophotometer.
Selection of SAP-, actin-, and fungus-specific primers.
One primer pair each for SAP1, SAP2, SAP3, and SAP7 and one primer pair for SAP4, SAP5, and SAP6, which have near-identical sequence homology, were chosen (Table 1). Two primer pairs were also selected to detect the C. albicans actin gene (ACT1); the pairs consisted of two primers that recognize separate sequences on the plus strand and one common primer that recognizes the minus strand (Table 1). Primers detecting the 5.8S rDNA plus intergenic spacer regions have been described previously (54) (Table 1). None of the primer sets used in this study amplified regions containing introns. The SAP1 to SAP7 and ACT1 primers were tested against C. albicans DNA to check for accurate amplification of the correct genes and against a panel of genomic DNAs isolated from different Candida species to test for cross-reactivity. The two actin primer sets reacted with all of the Candida species tested, yielding PCR products of the same size as for C. albicans, and thus served as a positive control for Candida colonization. Two sets of actin primers were used as a rigorous positive control; our criteria required that the results obtained with both actin primer pairs concurred to confirm the presence or absence of Candida. This study did not analyze the expression of SAP8 and SAP9 because these sequences became available only toward the completion of the work.
TABLE 1.
Oligonucleotide primer sets detecting ACT1 and SAP1 to SAP7 gene mRNAs and 5.8S rRNA
| Gene (reference) | Accession no. | Primer | Sequence | RT-PCR product size (bp) | Primer positions |
|---|---|---|---|---|---|
| SAP1 (25) | X56867 | Upstream | 5′-TCAATCAATTTACTCTTCCATTTCTAACA-3′ | 96–124 | |
| Downstream | 5′-CCAGTAGCATTAACAGGAGTTTTAATGACA-3′ | 161 | 237–256 | ||
| SAP2 (55) | M83663 | Upstream | 5′-AACAACAACCCACTAGACATCACC-3′ | 94–117 | |
| Downstream | 5′-TGACCATTAGTAACTGGGAATGCTTTAGGA-3′ | 178 | 242–271 | ||
| SAP3 (53) | L22358 | Upstream | 5′-CCTTCTCTAAAATTATGGATTGGAAC-3′ | 151–176 | |
| Downstream | 5′-TTGATTTCACCTTGGGGACCAGTAACATTT-3′ | 231 | 352–381 | ||
| SAP4 (32) | L22388 | Upstream | 5′-CATTCATTCCTTTAATACCGACTATC-3′ | 69–94 | |
| Downstream | 5′-GGTAACAAACCCTGTAGATCTTTTAAC-3′ | 156 | 198–224 | ||
| SAP5 (34) | 30191 | Upstream | Same as SAP4 | 149–174, 174–199, 199–224 | |
| Downstream | Same as SAP4 | 156, 181, 206 | 328–354 | ||
| SAP6 (34) | 30192 | Upstream | Same as SAP4 | 105–130, 130–155, 155–180 | |
| Downstream | Same as SAP4 | 156, 181, 206 | 284–310 | ||
| SAP7 (34) | 30193 | Upstream | 5′-GAAATGCAAAGAGTATTAGAGTTATTAC-3′ | 280–307 | |
| Downstream | 5′-GAATGATTTGGTTTACATCATCTTCAACTG-3′ | 196 | 446–475 | ||
| ACT1 (29) | XI6377 | Upstream A | 5′-GGCTGGTAGAGACTTGACCAACCATTTG-3′ | 304 | 2224–2251 |
| Upstream B | 5′-GATTTTGTCTGAACGTGGTAACAG-3′ | 271 | 2257–2280 | ||
| Downstream | 5′-GGAGTTGAAAGTGGTTTGGTCAATAC-3′ | 2502–2527 | |||
| 5.8S rRNA (30) | L07796 | Upstream | 5′-TCCGTAGGTGAACCTGCGG-3′ | 196–214 | |
| Downstream | 5′-TCCTCCGCTTATTGATATGC-3′ | 534 | 710–729 |
RT-PCR SAP1 to SAP7 mRNA expression in clinical whole saliva samples.
Ten RT-PCRs were performed for each sample of RNA isolated from each patient saliva specimen. These included the five experimental reactions for SAP detection (SAP1, SAP2, SAP3, SAP4 to SAP6, and SAP7) and five different control reactions, as follows: two actin control reactions to demonstrate the presence or absence of Candida species, a negative (water) control reaction, a positive control reaction carried out by using total RNA isolated from SAP2 mRNA-expressing C. albicans NCPF 3156 cells (SAP2 RNA), and a salivary inhibitor control reaction whereby SAP2 RNA was added to the saliva RNA preparation to check if the latter was inhibitory to the RT-PCR.
RT-PCR experiments with the actin and SAP primers were performed at an annealing temperature of 62°C by using Access RT-PCR reagents (Promega). RT-PCR conditions were optimized by modified Taguchi methods (12) and included a touchdown protocol. Template RNA was added to an RT-PCR mixture containing 1× avian myeloblastosis virus-Tfl buffer (Promega), 1 mM MgSO4, 0.1 mM deoxynucleoside triphosphates, 0.6 μM primers, 3.75 U of avian myeloblastosis virus reverse transcriptase, and 1 μCi of 32P-labelled dCTP (ICN, Thame, United Kingdom). Radioactive labelling was used to maximize sensitivity. After RT (48°C for 45 min), the cDNA-RNA hybrid was denatured at 94°C for 3 min and 2.5 U of Tfl DNA polymerase was added to the reaction mixture. Annealing temperatures used for touchdown cycling were as follows: 66°C for 2 cycles, 65°C for 2 cycles, 64°C for 2 cycles, and 63°C for 2 cycles, followed by 62°C for 35 cycles. Cycling times were as follows: denaturation at 94°C, annealing at 62°C, and extension at 72°C, each for 30 s. A final extension step of 72°C for 10 min followed the cycling.
For control experiments detecting the 5.8S rDNA plus intergenic spacer regions, RT-PCRs and PCRs were carried out at an annealing temperature of 55°C. For PCRs testing the cross-reactivity of the actin and SAP primers against genomic DNAs isolated from different Candida species, an annealing temperature of 62°C was used. PCR was performed by using Taq polymerase (Boehringer Mannheim, Lewes, United Kingdom). PCR conditions were optimized by modified Taguchi methods (12). Template DNA was added to a PCR mixture containing 2.5 mM MgCl2, 0.1 mM deoxynucleoside triphosphates, 0.6 μM primers, 1 U of Taq DNA polymerase, and 1 μCi of 32P-labelled dCTP. DNA was denatured at 94°C for 3 min and amplified for 35 cycles at the following cycling times: denaturation at 94°C, annealing at 62 or 55°C, depending on the primers used, and extension at 72°C, each for 30 s. A final extension step of 72°C for 10 min followed the cycling.
All radiolabelled RT-PCR and PCR products were mixed with formamide loading buffer (80% formamide, 50 mM Tris-borate buffer [pH 8.3], 1 mM EDTA, 0.1% [wt/vol] xylene cyanol, 0.1% [wt/vol] bromophenol blue), incubated at 95°C for 5 min, and cooled on ice. The products were electrophoresed through a denaturing 7% polyacrylamide gel (7 M urea), exposed to autoradiography film at −70°C, and developed.
Sensitivity and specificity of RT-PCR.
SAP2-expressing C. albicans cells grown as described above were added to Candida culture-negative saliva at concentrations of 100 to 106 cells/ml. Total RNA was isolated from the spiked saliva, and SAP2 mRNA expression was assayed by RT-PCR. In a separate experiment, the sensitivity of the RT-PCR method was assessed by adding decreasing amounts (from 105 to 100 pg) of SAP2 RNA from cells grown as described above.
Selected PCR products were sequenced to assess the specificity of the reaction. After electrophoresis through a 2% agarose gel, PCR products were purified by the QIAquick gel extraction protocol as described by the manufacturer (Qiagen, Crawley, United Kingdom). Each PCR product was sequenced by using the automated ABI Prism 377 sequencing system at the Advanced Biotechnology Centre, London, United Kingdom.
RESULTS
SAP gene expression is extremely sensitive and responsive to changes in environmental growth conditions, pH, and metal ion concentrations. Candida is also a commensal organism and is present at relatively low concentrations in asymptomatic Candida carriers. Therefore, we needed to differentiate between individuals who were carriers and those individuals who were truly Candida negative. Thus, assessments of the sensitivity, as well as the specificity, of SAP gene expression in vivo from the oral cavity were necessary.
Sensitivity of the RNA isolation and RT-PCR protocols.
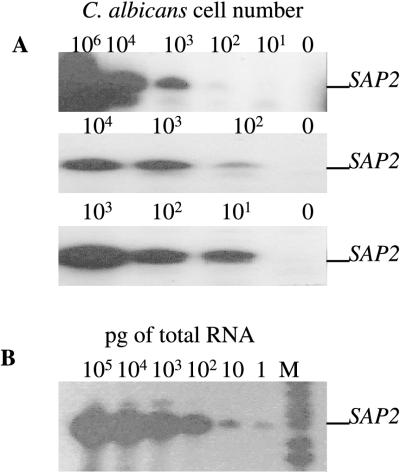
The sensitivity of the SAP mRNA detection protocol was tested in spiking experiments whereby saliva (Candida negative by culture) was spiked with 101 to 106 SAP2-expressing C. albicans cells/ml of saliva; this concentration range is representative of the range of C. albicans cells present in asymptomatic Candida carriers and candidiasis patients. SAP2 mRNA from samples containing between 10 and 100 SAP2 mRNA-expressing C. albicans cells was consistently amplified by RT-PCR (Fig. 1A) by using the cell lysis and RNA isolation protocols described above. The RT-PCR method, optimized by modified Taguchi methods (12), could detect as little as 1 pg of SAP2 RNA (Fig. 1B), although it is difficult to relate this to Candida cell numbers in vivo.
FIG. 1.
Sensitivity of the RNA isolation protocol and RT-PCR system. (A) SAP2 mRNA amplification from total RNA isolated from Candida culture-negative saliva spiked with different amounts of SAP2-expressing C. albicans NCPF 3156 cells (101 to 106). The autoradiographs from triplicate experiments are shown. (B) SAP2 mRNA amplification from RT-PCR mixtures containing between 100 and 105 pg of total RNA isolated from SAP2-expressing C. albicans NCPF 3156 cells. M denotes radiolabelled pBR322 MspI-digested molecular size markers (from the bottom, 147, 160, 180, 190, 201, 217, and 240 bp).
Specificity of the SAP1 to SAP7 primers.
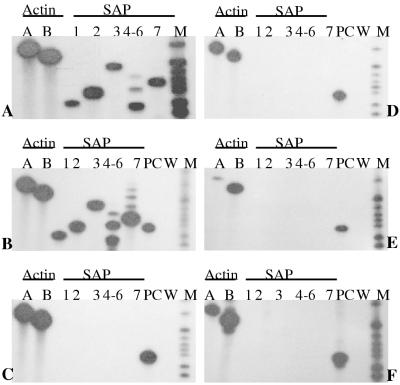
PCR amplification primers were chosen from upstream transcribed but untranslated regions of each of the C. albicans SAP1 to SAP7 gene sequences and resulted in products ranging in size from 156 to 231 bp (Table 1). SAP4, SAP5, and SAP6 are nearly identical in the sequences of both translated and untranslated regions of their respective mRNAs. Therefore, a single set of PCR primers which produces a 156-bp product derived from any one member or all three members of the SAP4, SAP5, and SAP6 gene subfamily was chosen. While SAP4 contains only a single target DNA sequence complementary to the 5′ PCR primer, upstream of both SAP5 and SAP6, this sequence is repeated twice in tandem, resulting in two additional PCR products of 181 and 206 bp. Thus, the absence of these two bands in the presence of the common 156-bp signal indicates that SAP4 is uniquely expressed. Two primer sets designed to detect the Candida actin gene (ACT1) served as a positive control for the presence of Candida species in the absence of SAP gene expression. We used two sets of actin primers which produced PCR products of 304 and 271 bp. Using RT-PCR, both actin primer sets were consistently positive or negative, confirming the presence or absence of Candida in the saliva samples.
Each of the five different SAP primer sets was specific for each of the SAP genes (Fig. 2A), excluding the possibility that any one SAP primer set could elicit a false-positive result. Cross-reactivity with other SAP genes was assayed by PCR by using the specific SAP1 to SAP7 primer sets and C. albicans genomic DNA. The specificity and identity of the SAP1, SAP2, SAP3, and SAP7 PCR products were confirmed by DNA sequence analysis (data not shown). Inefficient separation of the SAP4, SAP5, and SAP6 gene products complicated sequence analysis, but the derived sizes of the PCR products were consistent with the amplification of the correct gene fragments. In occasional saliva samples obtained from Candida carriers and Candida-infected individuals, the SAP2 primer set amplified a 165-bp fragment in conjunction with the expected 178-bp SAP2 mRNA product. The 165-bp fragment was sequenced and found to correspond to mRNA from human cytochrome b, a common, highly expressed mitochondrial protein (data not shown).
FIG. 2.
Reactivity of SAP1 to SAP7 primer sets with genomic DNAs isolated from different Candida species. (A) C. albicans NCPF 3153; (B) C. stellatoidea ATCC 11006 (type I); (C) C. dubliniensis NCPF 3949; (D) C. tropicalis ATCC 750; (E) C. parapsilosis ATCC 22019; (F) C. guilliermondii NCPF 3099. In autoradiographs B to F, SAP2 detection from C. albicans NCPF 3156 DNA was included as a positive control (PC) and water was used as a negative control (W). M denotes radiolabelled pBR322 MspI-digested molecular size markers (from the bottom, 147, 160, 180, 190, 201, 217, 240, and 307 bp). The expected SAP1 to SAP7 gene products are amplified from DNAs isolated from C. albicans (A) and C. stellatoidea (B). The autoradiographs for the other four species (C to F) indicate a lack of any SAP gene products, suggesting that the SAP1 to SAP7 primer pairs are specific for C. albicans species.
In addition to the ability to recognize the appropriate C. albicans SAP genes, each of the SAP1 to SAP7 primer sets was also checked for specificity by attempting to amplify each of the SAP genes from genomic DNAs isolated from the following five other pathogenic Candida species: C. stellatoidea, C. dubliniensis (a new Candida species closely related to C. albicans), C. tropicalis, C. parapsilosis, and C. guilliermondii. The SAP1 to SAP7 primer pairs did not react with any of these species except C. stellatoidea (Fig. 2). All of the C. albicans SAP1 to SAP7 primer sets produced the appropriate amplification products from C. stellatoidea (Fig. 2B), although this was expected since it is generally accepted that C. stellatoidea is a variant of C. albicans (27, 38).
Sample collection.
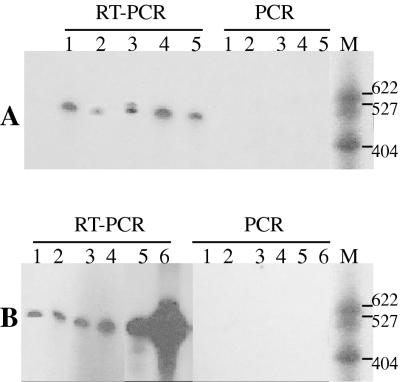
For assay of SAP mRNA expression in vivo, whole saliva was chosen, as it is a quickly and conveniently collected sample and provided reliable and consistent SAP mRNA detection. The sample volumes and their concentrations of Candida CFU for saliva collected from all subjects who participated in this study are shown in Table 2. Between 0.7 and 8 ml of whole saliva was collected for total RNA isolation; salivary Candida counts varied from 70 to >104 CFU/ml (excluding the Candida culture-negative group). Consequently, the minimal number of yeast cells present for total RNA isolation was 420, indicating that all experimental samples were above the detection limit determined for this assay (Fig. 1A). The absence of fungal DNA in the isolated total RNA preparations was confirmed by PCR by using generic fungus-specific primers which failed to detect the 5.8S rDNA plus intergenic sequence regions (54) (Fig. 3).
TABLE 2.
Salivary volumes and Candida counts in the three study populations
| Group | Saliva vol (ml) | Candida count (CFU/ml) | Total Candida count in extraction (CFU) |
|---|---|---|---|
| Candida culture-negative controls | |||
| A | 2.5 | 0 | 0 |
| B | 5 | 0 | 0 |
| C | 5 | 0 | 0 |
| D | 2.5 | 0 | 0 |
| E | 5 | 0 | 0 |
| F | 2.5 | 0 | 0 |
| G | 2.5 | 0 | 0 |
| Candida carriersa | |||
| A | 6 | 300 | 1,800 |
| B | 6.5 | 800 | 5,200 |
| C | 8 | 300 | 2,400 |
| D | 4.5 | 200 | 900 |
| E | 5.5 | 200 | 1,100 |
| F | 7 | 780 | 5,460 |
| G | 7 | 680 | 4,760 |
| H | 6 | 70 | 420 |
| Oral candidiasis patientsa | |||
| A | 1.5 | (1,720)b | (2,580)b |
| B | 2 | 4,800 | 9,600 |
| C | 1 | 5,360 | 5,360 |
| D | 4 | >10,000 | >40,000 |
| E | 1 | (1,620)b | (1,620)b |
| F | 2 | 2,880 | 5,760 |
| G | 2.3 | >10,000 | >23,000 |
| H | 1 | 4,000 | 4,000 |
| I | 0.7 | >10,000 | >7,000 |
| J | 2.5 | >10,000 | >25,000 |
Subjects harbored only C. albicans.
Counts were not performed at time the saliva sample was taken. However, post-thawed counts (in parentheses) indicated high initial C. albicans counts.
FIG. 3.
Detection of DNA and RNA in DNase-treated total RNA preparations in five Candida carrier subjects (A) and six patients with oral candidiasis (B). An absence of PCR-generated products confirmed that the preparations were DNA free, while RT-PCR generated the expected products from RNA. RT-PCR and PCR used primers specific for the 5.8S rDNA plus intergenic spacer regions. All of the total RNA preparations were DNA free. Differences in mobility were due to a gel artifact. M denotes radiolabelled pBR322 MspI-digested molecular size markers. Molecular sizes are indicated on the right in base pairs.
In vivo expression of C. albicans SAP1 to SAP7 genes in saliva.
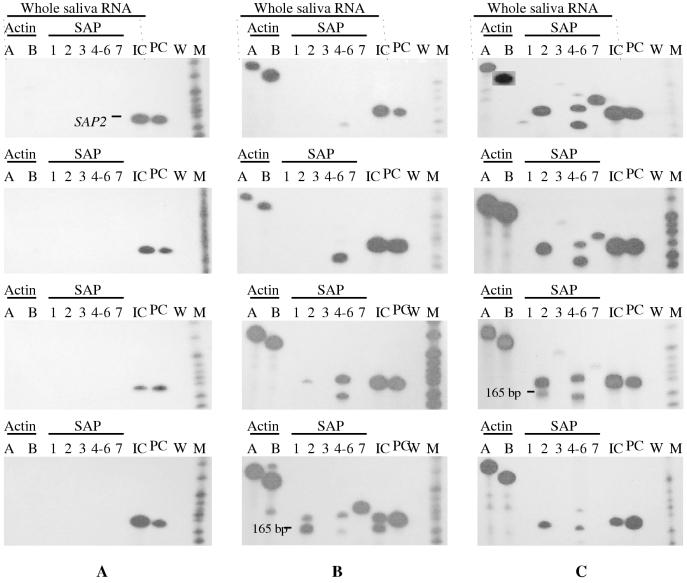
All seven Candida culture-negative subjects were negative for actin mRNA and all seven SAP genes, confirming that Candida species were absent from the saliva samples (Fig. 4A and Table 3). The absence of either specific or nonspecific RT-PCR amplification products also demonstrated that the primer sets were not cross-reactive with other unknown RNA sequences present in the saliva samples. We also observed that occasional saliva RNA preparations were inhibitory to the RT-PCR. To control for this eventuality, SAP2 RNA was added to each saliva RNA sample; synthesis of a SAP2 amplification product indicated that these saliva RNA preparations did not inhibit the RT-PCR.
FIG. 4.
Detection of C. albicans ACT1 and SAP1 to SAP7 mRNAs from clinical saliva samples. Four representative autoradiographs are presented for each of the following study groups: Candida culture-negative controls (A), asymptomatic Candida carriers (B), and oral candidiasis patients (C). Lanes A and B show the two primer pairs used for the detection of actin. The presence or absence of actin signals correlated with the presence (B and C) or absence (A) of C. albicans in the saliva samples. Lanes 3 to 7 show the five primer pairs used to detect SAP1 to SAP7 transcripts (SAP1, SAP2, SAP3, SAP4 to SAP6, and SAP7). Expression of SAP1 to SAP7 mRNA was absent in group A and variable in groups B and C. The following three controls detecting SAP2 mRNA were included in each assay: an inhibition control (lane 8) whereby SAP2 RNA was added to the saliva RNA to determine if the latter was inhibitory to the RT-PCR (lane IC), a positive control (lane 9) with SAP2 RNA alone (lane PC), and a negative water control (lane W). M denotes radiolabelled pBR322 MspI-digested molecular size markers (from the bottom, 147, 160, 180, 190, 201, 217, 240, 307, and 404 bp). The 165-bp fragment (B, bottom panel) was sequenced and found to correspond to mRNA from human cytochrome b, a common, highly expressed mitochondrial protein.
TABLE 3.
Summary of in vivo expression of C. albicans ACT1 and SAP1 to SAP7 mRNAs in Candida culture-negative control, Candida carrier, and oral candidiasis study populations
| Study population | HIV statusa | Actinb |
SAP gene expressionb
|
||||
|---|---|---|---|---|---|---|---|
| SAP1 | SAP2 | SAP3 | SAP4– SAP6 | SAP7 | |||
| Total no. of controls positivec | NDd | 0 | 0 | 0 | 0 | 0 | 0 |
| Candida carriers | |||||||
| A | ND | + | − | − | − | + | − |
| B | ND | + | − | − | − | + | − |
| C | ND | + | − | + | − | + | + |
| D | ND | + | − | + | − | + | − |
| E | ND | + | − | + | − | + | − |
| F | ND | + | − | + | − | + | − |
| G | ND | + | − | + | − | + | − |
| H | ND | + | − | + | − | + | + |
| Total no. positive | ND | 8 | 0 | 6 | 0 | 8 | 2 |
| Oral candidiasis patients | |||||||
| A | + | + | − | + | + | + | + |
| B | − | + | − | + | − | + | − |
| C | + | + | − | + | − | + | − |
| D | + | + | + | + | + | + | + |
| E | + | + | − | + | − | + | − |
| F | + | + | − | + | + | + | − |
| G | − | + | − | + | + | + | + |
| H | − | + | − | + | − | + | + |
| I | + | + | + | + | + | + | + |
| J | − | + | − | + | + | + | + |
| Total no. positive | 6 | 10 | 2 | 10 | 6 | 10 | 6 |
+, HIV-seropositive patient; −, HIV-seronegative patient.
+, presence of mRNA detected by RT-PCR; −, absence of mRNA determined by RT-PCR.
Seven Candida culture-negative controls were studied.
ND, not determined.
The eight specimens obtained from asymptomatic Candida carriers were each positive for both actin primer sets, confirming the presence of Candida mRNA in the saliva RNA samples. The yeast species was previously identified as C. albicans. The presence of RT-PCR products from Candida carriers with low CFU counts (Table 2) confirmed the sensitivity of the protocol, which was capable of detecting fewer than 100 C. albicans cells in saliva (Fig. 1A). Two of the eight asymptomatic carriers were positive for SAP4, SAP5, or SAP6 mRNA only; four were positive for SAP2 and SAP4, SAP5, or SAP6 transcripts; two were positive for SAP2, SAP4, SAP5, or SAP6, and SAP7 mRNA. All Candida carriers were positive for SAP4, SAP5, or SAP6 transcripts, but none were positive for SAP1 or SAP3 mRNA (Fig. 4B and Table 3).
The primer sets designed to detect SAP4, SAP5, or SAP6 mRNA produced three RT-PCR fragments of 156, 181, and 206 bp. However, it was noted that samples from some Candida carriers gave rise only to the 156-bp fragment (Fig. 4B). If SAP5 and SAP6 transcripts were present, the primers might be expected to amplify the 181- and 206-bp fragments in addition to the 156-bp fragment. Their absence suggests that only SAP4 mRNA is expressed in these Candida carriers. Whether SAP4 mRNA alone or a combination of SAP4, SAP5, and SAP6 transcripts is present in these RNA preparations, the results clearly demonstrate that the SAP4 to SAP6 gene subfamily was expressed in all of the saliva samples containing C. albicans. As SAP4 mRNA has never been detected under laboratory conditions (23, 52), the expression of this proteinase alone, or in conjunction with SAP5 and SAP6, may be different in vivo.
All 10 patients with oral C. albicans infection were positive for actin, confirming the presence of Candida mRNA in the saliva RNA sample. Two patients expressed the full repertoire of all seven SAP genes, and three patients were positive for SAP2, SAP3, SAP4, -5, or -6, and SAP7 mRNA but not SAP1 mRNA. One patient was positive for SAP2, SAP3, and SAP4, SAP5, or SAP6 transcripts, and one other patient expressed SAP4, SAP5, or SAP6 and SAP2 and SAP7 transcripts. Three patients expressed SAP2 and SAP4, SAP5, or SAP6 transcripts only. All 10 patients were positive for SAP2 and SAP4, SAP5, or SAP6 mRNA transcripts (Fig. 4C and Table 3).
DISCUSSION
This study has established a convenient, reliable, and highly sensitive method for the detection of C. albicans SAP mRNA expression directly in patient samples. Specific primers constructed for seven SAP genes (SAP1 to SAP7) showed that all seven are expressed in vivo and provided preliminary data indicating which of the seven proteinases are commonly expressed in individuals with C. albicans oral colonization and infection. SAP mRNA expression patterns observed in vivo appear to be different from those previously demonstrated under laboratory culture conditions.
The SAP1 to SAP7 primers were shown to be C. albicans specific against a panel of genomic DNAs isolated from various Candida species. The SAP primers reacted only with DNAs isolated from C. albicans and C. stellatoidea (Fig. 2A and B, respectively), the latter of which is considered to be a variant of C. albicans (27, 38). The SAP primers did not react with DNA isolated from C. dubliniensis, C. tropicalis, C. parapsilosis, or C. guilliermondii (Fig. 2C, D, E, and F, respectively). C. dubliniensis has recently been shown to have its own SAP family comprised of at least seven genes (20), and the lack of cross-reactivity of the C. dubliniensis DNA with the C. albicans SAP1 to SAP7 primers supports the species distinction between C. dubliniensis and C. albicans.
Analysis of the differential expression of genes SAP1 to SAP7 in vivo indicated that SAP2 and SAP4, SAP5, and SAP6 appear to be the predominant genes expressed in the oral cavities of both asymptomatic Candida carriers and patients with oral candidiasis, with SAP4, SAP5, or SAP6 mRNA being found in all of the subjects examined so far (Table 3). This study also demonstrated the expression of SAP1 and SAP3 transcripts in oral candidiasis patients but not in Candida carriers (Table 3). Expression of SAP1 and SAP3 has been associated with phenotypic switching in vitro, and this association with oral infection may implicate phenotypic switching in the virulence of C. albicans. A further observation was the expression of SAP7 mRNA, which has never before been demonstrated (Table 3). This study suggests that progression from colonization to symptomatic infection with C. albicans is reflected by the expression pattern of certain proteinases, most likely due to different environmental conditions in the oral cavities of carriers and those of diseased individuals.
Data acquired in vitro suggest that Sap2 is the main proteinase secreted in protein-containing media (22, 52, 55). Our results are consistent with the premise that Sap2 is a predominant proteinase of C. albicans, as SAP2 expression was observed in all patients with oral candidiasis and all but two Candida carrier individuals (Table 3). Under laboratory conditions, Sap2 is capable of degrading numerous substrates, including extracellular matrix proteins, mucin, and sIgA (13, 22, 41), all of which constitute host proteins in the oral cavity. Digestion of these (and other) nutrients in vivo may help C. albicans to acquire essential nitrogen for growth and/or to attach to and penetrate oral mucosa. In addition, digestion of sIgA (which is normally resistant to proteolysis) may assist C. albicans to evade the immune response.
The samples from the two Candida carriers that did not express SAP2 were positive for SAP4, SAP5, or SAP6 transcripts, suggesting that this subfamily of proteinases may have functional properties similar to those of, and may be able to substitute for, Sap2 in the oral cavity. Studies of a rat model have demonstrated SAP1 and SAP2 expression in C. albicans experimental vaginitis (17) and indicated that Sap2, but not Sap4, Sap5, or Sap6, may be important in vaginal candidiasis in rats (16). In our study, we observed SAP4, SAP5, or SAP6 expression in samples collected from the oral cavities of all C. albicans-positive individuals, indicating that differential expression of this proteinase family in the human oral cavity may be very different from that in the rat vagina. This is supported by a recent study suggesting that pH-dependent genes are differentially regulated in systemic and vaginal infections (19).
The expression of SAP4, SAP5, or SAP6 transcripts in all Candida carriers and patients with oral candidiasis suggests that this proteinase subfamily may have an important role in oral C. albicans colonization and infection. This is supported by a study of animal models of disseminated candidiasis in which a sap4 sap5 sap6 triple null mutant showed marked attenuation of virulence (44). The Sap4, Sap5, and Sap6 subfamily may also play a role in immune evasion, as their production appears to partially protect C. albicans from phagocytic killing by murine macrophages (6). Sanglard et al. (44) also speculated that SAP4, SAP5, or SAP6 expression may be required in the process of SAP2 induction. In patient samples, we did not observe SAP2 mRNA without the expression of SAP4, SAP5, or SAP6, suggesting that this regulation may possibly occur in vivo.
There is evidence acquired in vitro that Sap antigens are expressed on the surface of C. albicans following attachment to epithelial cells (5) and on the surface of hyphal forms (42), which are known to be more adherent than yeast forms (43). White and Agabian (52) and Hube et al. (23) also showed that only SAP4, SAP5, and SAP6 transcripts were expressed during the transition from yeast to hyphae at neutral pH in vitro. This suggests that the secretion of Sap4, Sap5, and Sap6 in the oral cavity may facilitate attachment of C. albicans to the oral mucosa. However, a recent study showed that a sap4 sap5 sap6 null mutant exhibited increased adherence to buccal epithelial cells in vitro (51). The presence of hyphae, and hence the expression of SAP4, SAP5, or SAP6 mRNA, may be expected in oral candidiasis patients. However, the association of hyphae with the carrier state has not been thoroughly investigated; therefore, the expression of these three proteinases in all of the Candida carriers is suggestive of the presence of hyphae. Alternatively, in vivo, yeast cells may express SAP4, SAP5, and SAP6. Although there is no in vitro evidence that SAP4, SAP5, and SAP6 genes are expressed by yeast cells, the expression of the three proteinases they encode in Candida carriers and the first demonstration of SAP7 expression in vivo (vide infra) confirm that the oral milieu is quite different from laboratory culture conditions used to study SAP mRNA expression.
Similar patterns of specific gene expression have been observed in studies of other organisms. For example, during infection with the spirochete Borrelia burgdorferi, several genes have been shown to be selectively induced in vivo, and different Borrelia gene products have been detected in different sites of the body (3, 10). The extended repertoire of SAP genes in C. albicans and their differential expression in vivo indicate that different SAP genes may be selectively expressed at different stages and in different clinical forms of candidiasis, such as pseudomembranous, erythematous, atrophic, and hyperplastic forms (4, 28). Further studies using the techniques developed in this study will address this issue.
This is the first study to detect expression of the SAP7 gene. It also indicates that the proteinase encoded by this gene, if translated, may be associated with C. albicans infection, as SAP7 transcripts were detected in 60% of oral candidiasis patients (6 of 10 patients), as opposed to 25% of Candida carriers (2 of 8 carriers) (Table 3). The function of Sap7 is unknown, but its apparent differential expression in oral candidiasis patients is noteworthy and clearly warrants further investigation.
Interestingly, SAP1 and SAP3 mRNA transcripts were detected only in patients with oral C. albicans infections and not in Candida carriers (Table 3). Under laboratory conditions, the expression of SAP1 and SAP3 is coordinately regulated during phenotypic switching (23, 35, 36, 52). Therefore, the detection of SAP1 and SAP3 transcripts in oral candidiasis suggests that phenotypic switching occurs in vivo, and furthermore, that it may be associated with the development of infection. This is supported by the observation that C. albicans isolated from infected patients exhibits, on average, higher rates of phenotypic switching than commensal strains from the oral cavity (21). Recently, Schaller et al. (45) investigated the temporal expression of SAP genes in an in vitro model of oral candidiasis using reconstituted human epithelium. They showed that SAP1 and SAP3 were the first SAP genes to be expressed by C. albicans. In addition, they noted that the expression of these two genes coincided with the development of lesions and suggested that their model reflects the infectious state. This is supported by our findings, since we did not detect SAP1 or SAP3 mRNA expression in any of the Candida carriers tested, only in the oral candidiasis group.
In this study, we have found differences in the expression profiles of SAP1 through SAP7 mRNA between patients with oral C. albicans infections and asymptomatic Candida carrier subjects. We do not believe these differences are due to the greater numbers of C. albicans cells present in the samples obtained from patients with oral candidiasis, because in some cases, the total numbers of C. albicans cells present in the RNA extraction were similar in the two study groups (Table 2). In addition, two Candida carriers with lower total C. albicans cell numbers expressed more SAP genes than three other Candida carriers who had greater total C. albicans cell counts (compare Table 2 with Table 3). Furthermore, the detection of C. albicans actin mRNA in all of the Candida carrier saliva samples indicated that the methods employed were sufficiently sensitive to detect the presence of SAP mRNA. In terms of SAP mRNA expression, we realize that the assay used does not quantify exact levels of individual mRNAs and that comparative levels of different SAP mRNAs may vary.
Two additional elements that may possibly affect SAP gene expression in C. albicans in vivo are HIV infection and antifungal drug therapy, which may select for more pathogenic C. albicans strains (48). Selection of C. albicans strains is thought to occur early in HIV infection and may be associated with increased Sap production (18, 40), increased adherence to oral epithelial cells (49), and genotypic alterations (9). Antifungal drug therapy, which has been correlated with colonial morphology changes (31) and phenotypic differences (7, 26), may also be a major cause of Candida strain selection. A number of the patients had previously been treated with antifungal drugs, although none were being treated with either antibiotic or antimycotic drugs at the time of sampling; 6 of 10 patients with oral candidiasis were HIV seropositive. However, no correlation between HIV infection or antimicrobial treatment history and SAP expression was evident.
In summary, this is the first study to show that genes SAP1 to SAP7 are expressed in vivo, providing evidence of a role for this proteinase family in human oral candidiasis. It further indicates that the pathogenesis of oral C. albicans infections may be associated with the differential expression of individual SAP genes. In addition, SAP mRNA expression patterns observed in vivo appear to be different from those first demonstrated under laboratory culture conditions, indicating that the involvement of secreted aspartyl proteinases in C. albicans pathogenesis in vivo may not be necessarily inferred from conventional laboratory growth and analysis.
ACKNOWLEDGMENTS
We thank Sue Howell for kindly supplying the Candida reference strains used in this study.
This work was supported by the Dunhill Medical Trust and NIH grants AI33317 and POI-DE-07946.
REFERENCES
- 1.Agabian N, Odds F C, Poulain D, Soll D R, White T C. Pathogenesis of invasive candidiasis. J Med Vet Mycol. 1994;32:229–237. doi: 10.1080/02681219480000861. [DOI] [PubMed] [Google Scholar]
- 2.Agatensi L, Franchi F, Mondello F, Bevilacqua R L, Ceddia T, De Bernardis F, Cassone A, Ruchel R, Zimmermann F, Boning-Stutzer B, Helmchen U. Vaginopathic and proteolytic Candida species in outpatients attending a gynaecology clinic. J Clin Pathol. 1991;419:199–202. doi: 10.1136/jcp.44.10.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akins D R, Porcella S F, Popova T G, Shevchenko D, Baker S I, Li M, Norgard M V, Radolf J D. Evidence for in vivo but not in vitro expression of a Borrelia burgdorferi outer surface protein F (OspF) homologue. Mol Microbiol. 1995;18:507–520. doi: 10.1111/j.1365-2958.1995.mmi_18030507.x. [DOI] [PubMed] [Google Scholar]
- 4.Axell T, Baert A, Brocheriou C, Challacombe S J, Greenspan D, Ten Kate W, Laskaris G, Mano Azul A, Pindborg J J, Reichart P, Schulten E A J M, Scully C, Syrjanen S, Van der Wall I, Williams D. An update of the classification and diagnostic criteria of oral lesions in HIV infection. J Oral Pathol Med. 1991;20:97–100. [PubMed] [Google Scholar]
- 5.Borg M, Watters D, Reich B, Rüchel R. Production and characterization of monoclonal antibodies against secretory proteinase of Candida albicans CBS 2730. Zentbl Bakteriol Mikrobiol Hyg Ser A. 1988;268:62–73. doi: 10.1016/s0176-6724(88)80116-0. [DOI] [PubMed] [Google Scholar]
- 6.Borg-von Zepelin M, Beggah S, Boggian K, Sanglard D, Monod M. The expression of the secreted aspartyl proteinases Sap4 to Sap6 from Candida albicans in murine macrophages. Mol Microbiol. 1998;28:543–554. doi: 10.1046/j.1365-2958.1998.00815.x. [DOI] [PubMed] [Google Scholar]
- 7.Bruatto M, Vidotto V, Marinuzzi G, Raiteri R, Sinicco A. Candida albicans biotypes in human immunodeficiency virus type 1-infected patients with oral candidiasis before and after antifungal therapy. J Clin Microbiol. 1991;29:726–730. doi: 10.1128/jcm.29.4.726-730.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cassone A, Boccanera M, Adriani D, Santoni G, De Bernardis F. Rats clearing a vaginal infection by Candida albicans acquire specific, antibody-mediated resistance to vaginal reinfection. Infect Immun. 1995;63:2619–2624. doi: 10.1128/iai.63.7.2619-2624.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Challacombe S J, Muir J, Howell S A, Sweet S P. Genetic variability of Candida albicans in HIV infection. Microb Ecol Health Dis. 1995;8:63–70. [Google Scholar]
- 10.Champion C I, Blanco D R, Skare J T, Haake D A, Giladi M, Foley D, Miller J N, Lovett J A. A 9.0-kilobase-pair circular plasmid of Borrelia burgdorferi encodes an exported protein: evidence for expression only during infection. Infect Immun. 1994;62:2653–2661. doi: 10.1128/iai.62.7.2653-2661.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 12.Cobb B D, Clarkson J M. A simple procedure for optimising the polymerase chain reaction (PCR) using modified Taguchi methods. Nucleic Acids Res. 1994;22:3801–3805. doi: 10.1093/nar/22.18.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colina A R, Aumont F, Deslauriers N, Belhumeur P, de Repentigny L. Evidence for degradation of gastrointestinal mucin by Candida albicans secretory aspartyl proteinase. Infect Immun. 1996;64:4514–4519. doi: 10.1128/iai.64.11.4514-4519.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coogan M M, Sweet S P, Challacombe S J. Immunoglobulin A (IgA), IgA1, and IgA2 antibodies to Candida albicans in whole and parotid saliva in human immunodeficiency virus infection and AIDS. Infect Immun. 1994;62:892–896. doi: 10.1128/iai.62.3.892-896.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cutler J E. Putative virulence factors of Candida albicans. Annu Rev Microbiol. 1991;45:187–218. doi: 10.1146/annurev.mi.45.100191.001155. [DOI] [PubMed] [Google Scholar]
- 16.De Bernardis F, Arancia S, Morelli L, Hube B, Sanglard D, Schäfer W, Cassone A. Evidence that members of the secretory aspartyl proteinases gene family (SAP), in particular SAP2, are virulence factors for Candida vaginitis. J Infect Dis. 1999;179:201–208. doi: 10.1086/314546. [DOI] [PubMed] [Google Scholar]
- 17.De Bernardis F, Cassone A, Sturtevant J, Calderone R. Expression of Candida albicans SAP1 and SAP2 in experimental vaginitis. Infect Immun. 1995;63:1887–1892. doi: 10.1128/iai.63.5.1887-1892.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Bernardis F, Chiani P, Ciccozzi M, Pellegrini G, Ceddia T, D’Offizzi G, Quinti I, Sullivan P A, Cassone A. Elevated aspartic proteinase secretion and experimental pathogenicity of Candida albicans isolates from oral cavities of subjects infected with human immunodeficiency virus. Infect Immun. 1996;64:466–471. doi: 10.1128/iai.64.2.466-471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Bernardis F, Muhlschlegel F A, Cassone A, Fonzi W A. The pH of the host niche controls gene expression in and virulence of Candida albicans. Infect Immun. 1998;66:3317–3325. doi: 10.1128/iai.66.7.3317-3325.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilfillan G D, Sullivan D J, Haynes K, Parkinson T, Coleman D C, Gow N A R. Candida dubliniensis: phylogeny and putative virulence factors. Microbiology. 1998;144:829–838. doi: 10.1099/00221287-144-4-829. [DOI] [PubMed] [Google Scholar]
- 21.Hellstein J, Vawter-Hugart H, Fotos P, Schmid J, Soll D R. Genetic similarity and phenotypic diversity of commensal and pathogenic strains of Candida albicans isolated from the oral cavity. J Clin Microbiol. 1993;31:3190–3199. doi: 10.1128/jcm.31.12.3190-3199.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hube B. Candida albicans secreted aspartyl proteinases. Curr Top Med Mycol. 1996;7:55–69. [PubMed] [Google Scholar]
- 23.Hube B, Monod M, Schofield D A, Brown A J P, Gow N A R. Expression of seven members of the gene family encoding secretory aspartyl proteinases in Candida albicans. Mol Microbiol. 1994;14:87–99. doi: 10.1111/j.1365-2958.1994.tb01269.x. [DOI] [PubMed] [Google Scholar]
- 24.Hube B, Sanglard D, Odds F C, Hess D, Monod M, Schafer W, Brown A J, Gow N A. Disruption of each of the secreted aspartyl proteinase genes SAP1, SAP2, and SAP3 of Candida albicans attenuates virulence. Infect Immun. 1997;65:3529–3538. doi: 10.1128/iai.65.9.3529-3538.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hube B, Turver C J, Odds F C, Eiffert H, Boulnois G J, Kochel H, Ruchel R. Sequence of the Candida albicans gene encoding the secretory aspartate proteinase. J Med Vet Mycol. 1991;29:129–132. [PubMed] [Google Scholar]
- 26.Imbert-Bernard C, Valentin A, Mallie M. Involvement of Candida albicans cell wall proteins in the adherence of blastospores to human buccal epithelial cells. Exp Mycol. 1995;19:247–253. doi: 10.1006/emyc.1995.1031. [DOI] [PubMed] [Google Scholar]
- 27.Kwon-Chung K J, Riggsby W S, Uphoff R A, Hicks J B, Whelan W L, Reiss E, Magee B B, Wickes B L. Genetic differences between type I and type II Candida stellatoidea. Infect Immun. 1989;57:527–532. doi: 10.1128/iai.57.2.527-532.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lehner T. Oral candidosis. Dent Pract. 1967;17:209–216. [PubMed] [Google Scholar]
- 29.Losberger C, Ernst J F. Sequence of the Candida albicans gene encoding actin. Nucleic Acids Res. 1989;17:9488. doi: 10.1093/nar/17.22.9488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lott T J, Kuykendall R J, Reiss E. Nucleotide sequence analysis of the 5.8S rDNA and adjacent ITS2 region of Candida albicans and related species. Yeast. 1993;9:1199–1206. doi: 10.1002/yea.320091106. [DOI] [PubMed] [Google Scholar]
- 31.McCullough M J, Ross B C, Dwyer B D, Reade P C. Genotype and phenotype of oral Candida albicans from patients infected with the human immunodeficiency virus. Microbiology. 1994;140:1195–1202. doi: 10.1099/13500872-140-5-1195. [DOI] [PubMed] [Google Scholar]
- 32.Miyasaki S H, White T C, Agabian N. A fourth secreted aspartyl proteinase gene (SAP4) and a CARE2 repetitive element are located upstream of the SAP1 gene in Candida albicans. J Bacteriol. 1994;176:1702–1710. doi: 10.1128/jb.176.6.1702-1710.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monod M, Hube B, Hess D, Sanglard D. Differential regulation of SAP8 and SAP9, which encode two new members of the secreted aspartic proteinase family in Candida albicans. Microbiology. 1998;144:2731–2737. doi: 10.1099/00221287-144-10-2731. [DOI] [PubMed] [Google Scholar]
- 34.Monod M, Togni G, Hube B, Sanglard D. Multiplicity of genes encoding secreted aspartic proteinases in Candida species. Mol Microbiol. 1994;13:357–368. doi: 10.1111/j.1365-2958.1994.tb00429.x. [DOI] [PubMed] [Google Scholar]
- 35.Morrow B, Ramsey H, Soll D R. Regulation of phase-specific genes in the more general switching system of Candida albicans strain 3153A. J Med Vet Mycol. 1994;32:287–294. doi: 10.1080/02681219480000361. [DOI] [PubMed] [Google Scholar]
- 36.Morrow B, Srikantha T, Soll D R. Transcription of the gene for a pepsinogen, PEP1, is regulated by white-opaque switching in Candida albicans. Mol Cell Biol. 1992;12:2997–3005. doi: 10.1128/mcb.12.7.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newport G, Agabian N. KEX2 influences Candida albicans proteinase secretion and hyphal formation. J Biol Chem. 1997;272:28954–28961. doi: 10.1074/jbc.272.46.28954. [DOI] [PubMed] [Google Scholar]
- 38.Odds F C. Candida and candidosis. Philadelphia, Pa: Bailliere Tindall; 1988. [Google Scholar]
- 39.Odds F C. Candida species and virulence. ASM News. 1994;60:313–318. [Google Scholar]
- 40.Ollert M W, Wende C, Gorlich M, McMullan-Vogel C G, Borg-von Zepelin M, Vogel C W, Korting H C. Increased expression of Candida albicans secretory proteinase, a putative virulence factor, in isolates from human immunodeficiency virus-positive patients. J Clin Microbiol. 1995;33:2543–2549. doi: 10.1128/jcm.33.10.2543-2549.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rüchel R. A variety of Candida proteinases and their possible targets of proteolytic attack in the host. Zentbl Bakteriol Hyg A. 1984;257:266–274. [PubMed] [Google Scholar]
- 42.Rüchel R, Zimmermann F, Boning-Stutzer B, Helmchen U. Candidiasis visualised by proteinase-directed immunofluorescence. Virchow’s Arch Abt A Pathol Anat. 1991;419:199–202. doi: 10.1007/BF01626348. [DOI] [PubMed] [Google Scholar]
- 43.Sandin R L, Rogers A L, Patterson R J, Beneke E S. Evidence for mannose-mediated adherence of Candida albicans to human buccal cells in vitro. Infect Immun. 1982;35:79–85. doi: 10.1128/iai.35.1.79-85.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanglard D, Hube B, Monod M, Odds F C, Gow N A. A triple deletion of the secreted aspartyl proteinase genes SAP4, SAP5, and SAP6 of Candida albicans causes attenuated virulence. Infect Immun. 1997;65:3539–3546. doi: 10.1128/iai.65.9.3539-3546.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schaller M, Schäfer W, Korting H C, Hube B. Differential expression of secreted aspartyl proteinases in a model of human oral candidosis and in patient samples from the oral cavity. Mol Microbiol. 1998;29:605–615. doi: 10.1046/j.1365-2958.1998.00957.x. [DOI] [PubMed] [Google Scholar]
- 46.Smolenski G, Sullivan P A, Cutfield S M, Cutfield J F. Analysis of secreted aspartic proteinases from Candida albicans: purification and characterization of individual Sap1, Sap2 and Sap3 isoenzymes. Microbiology. 1997;143:349–356. doi: 10.1099/00221287-143-2-349. [DOI] [PubMed] [Google Scholar]
- 47.Stringaro A, Crateri P, Pellegrini G, Arancia G, Cassone A, De Bernardis F. Ultrastructural localization of the secretory aspartyl proteinase in Candida albicans cell wall in vitro and in experimentally infected rat vagina. Mycopathologia. 1997;137:95–105. doi: 10.1023/a:1006897208863. [DOI] [PubMed] [Google Scholar]
- 48.Sweet S P. Selection and pathogenicity of Candida albicans in HIV infection. Oral Dis. 1997;3:S88–S95. doi: 10.1111/j.1601-0825.1997.tb00383.x. [DOI] [PubMed] [Google Scholar]
- 49.Sweet S P, Cookson S, Challacombe S J. Candida albicans isolates from HIV-infected and AIDS patients exhibit enhanced adherence to epithelial cells. J Med Microbiol. 1995;43:452–457. doi: 10.1099/00222615-43-6-452. [DOI] [PubMed] [Google Scholar]
- 50.Togni G, Sanglard D, Quadroni M, Foundling S I, Monod M. Acid proteinase secreted by Candida tropicalis: functional analysis of preproregion cleavages in C. tropicalis and Saccharomyces cerevisiae. Microbiology. 1996;142:493–503. doi: 10.1099/13500872-142-3-493. [DOI] [PubMed] [Google Scholar]
- 51.Watts H J, Cheah F S, Hube B, Sanglard D, Gow N A R. Altered adherence in strains of Candida albicans harbouring null mutations in secreted aspartic proteinase genes. FEMS Microbiol Lett. 1998;159:129–135. doi: 10.1111/j.1574-6968.1998.tb12851.x. [DOI] [PubMed] [Google Scholar]
- 52.White T C, Agabian N. Candida albicans secreted aspartyl proteinases: isoenzyme pattern is determined by cell type, and levels are determined by environmental factors. J Bacteriol. 1995;177:5215–5221. doi: 10.1128/jb.177.18.5215-5221.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.White T C, Miyasaki S H, Agabian N. Three distinct secreted aspartyl proteinases in Candida albicans. J Bacteriol. 1993;175:2997–3005. doi: 10.1128/jb.175.19.6126-6133.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.White T J, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. New York, N.Y: Academic Press, Inc.; 1990. pp. 315–322. [Google Scholar]
- 55.Wright R J, Carne A, Hieber A D, Lamont I L, Emerson G W, Sullivan P A. A second gene for a secreted aspartate proteinase in Candida albicans. J Bacteriol. 1992;174:7848–7853. doi: 10.1128/jb.174.23.7848-7853.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]