ABSTRACT
Biologists have long been interested in understanding genetic constraints on the evolution of development. For example, noncoding changes in a gene might be favored over coding changes if they are less constrained by pleiotropic effects. Here, we evaluate the importance of coding‐sequence changes to the recent evolution of a novel anthocyanin pigmentation trait in the monkeyflower genus Mimulus. The magenta‐flowered Mimulus luteus var. variegatus recently gained petal lobe anthocyanin pigmentation via a single‐locus Mendelian difference from its sister taxon, the yellow‐flowered M. l. luteus. Previous work showed that the differentially expressed transcription factor gene MYB5a/NEGAN is the single causal gene. However, it was not clear whether MYB5a coding‐sequence evolution (in addition to the observed patterns of differential expression) might also have contributed to increased anthocyanin production in M. l. variegatus. Quantitative image analysis of tobacco leaves, transfected with MYB5a coding sequence from each taxon, revealed robust anthocyanin production driven by both alleles. Counter to expectations, significantly higher anthocyanin production was driven by the allele from the low‐anthocyanin M. l. luteus, a result that was confirmed through both a replication of the initial study and analysis by an alternative method of spectrophotometry on extracted leaf anthocyanins. Together with previously published expression studies, our findings support the hypothesis that petal pigment in M. l. variegatus was not gained by protein‐coding changes, but instead solely via noncoding cis‐regulatory evolution. Finally, while constructing the transgenes needed for this experiment, we unexpectedly discovered two sites in MYB5a that appear to be post‐transcriptionally edited—a phenomenon that has been rarely reported, and even less often explored, for nuclear‐encoded plant mRNAs.
Keywords: anthocyanin color patterning, cis‐regulatory evolution, digital image analysis, mRNA editing, transient transformation
The protein‐coding sequences of anthocyanin activator MYB5a/NEGAN, from both the high‐anthocyanin M. l. variegatus and the low‐anthocyanin M. l. luteus, are effective at stimulating anthocyanin production, further supporting the importance of previously documented cis‐regulatory divergence between the species.
1. Introduction
Phenotypic transitions in related taxa often share a common genetic basis, which suggests that constraints shape the process of evolution at the genetic level (Stern 2013). For example, noncoding mutations may contribute disproportionately to evolutionary change, due to being less pleiotropically constrained than coding mutations (Wray 2007).
In plants, transitions in anthocyanin pigmentation are well suited to investigating such patterns (Streisfeld and Rauther 2011; Sobel and Streisfeld 2013; Fairnie et al. 2022), particularly, in diverse yet tractable groups such as the monkeyflower genus Mimulus (synonym Erythranthe) (Yuan 2019). Repeated evolutionary changes in petal lobe anthocyanin (PLA) pigmentation, within and between species in the ancestrally yellow‐flowered luteus group of Mimulus, provide an opportunity to explore the relative importance of coding and noncoding changes (Figure 1). In Mimulus luteus var. variegatus, M. naiandinus, and M. cupreus, the recent gain of PLA is a derived single‐locus trait that maps to either of two genomic clusters of MYB transcription factors, located in QTLs pla1 and pla2 (Figure 1; Cooley et al. 2011). These tandemly duplicated gene arrays contain members of the anthocyanin‐activating Subgroup 6 of the MYB gene family (Stracke, Werber, and Weisshaar 2001). In M. l. variegatus, a combination of genetic mapping, stable transgenics, and transcriptomics shows that expression of mapped candidate gene MYB5a correlates with petal anthocyanin production and is both necessary and sufficient for the gain of PLA (Cooley et al. 2011; Zheng et al. 2021). However, this does not rule out that divergence in the protein‐coding sequence of MYB5a may also have contributed to the increased anthocyanin production seen in M. l. variegatus.
Figure 1.
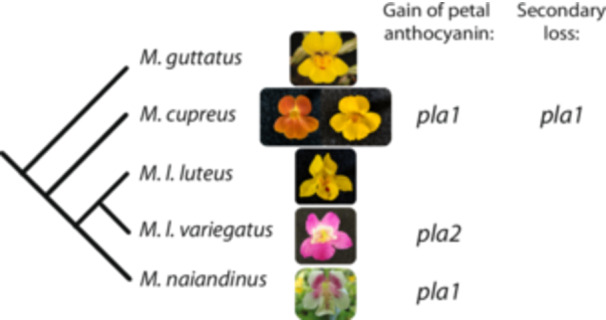
Petal lobe anthocyanin has been gained repeatedly in the luteus group of Mimulus. Closely related species outside the luteus group, like M. guttatus, are yellow‐flowered, with red anthocyanin pigmentation restricted to the nectar guide region of the corolla. Mimulus cupreus, M. l. luteus, and M. l. variegatus all produce a single type of anthocyanin pigment—cyanidin—which leads to floral colors ranging from orange to red to magenta depending on the relative intensities of cyanidin versus the yellow carotenoid pigments (Cooley, Carvallo, and Willis 2008). Mimulus cupreus and M. naiandinus have each gained petal lobe anthocyanin via a single locus change at genomic region pla1, which contains tandemly arrayed copies of candidate transcription factor genes MYB1, MYB2, and MYB3 (Cooley et al. 2011). The magenta‐petaled M. luteus var. variegatus gained petal lobe anthocyanin via a change at pla2, associated with an expansion in gene expression of the petal anthocyanin activator gene MYB5a (Zheng et al. 2021). A rare yellow‐flowered morph of M. cupreus, found in a single population in Chile, has lost petal lobe anthocyanin via a change at pla1 (Cooley and Willis 2009). Figure modified from Zheng et al. (2021). [Color figure can be viewed at wileyonlinelibrary.com]
MYB5a produces two splice variants, exon 1‐2‐3 and 1‐2‐4. In M. l. variegatus, only the 1‐2‐4 variant contains the Subgroup 6 anthocyanin‐activating motif (Stracke, Werber, and Weisshaar 2001), and altering the expression of this variant specifically was shown to alter PLA (Zheng et al. 2021). Consistent with a causal role in activating PLA in M. l. variegatus, exon 1‐2‐4 is the only splice variant that we have recovered from M. l. variegatus petal tissue (Zheng et al. 2021). In contrast, we have recovered both splice variants from M. l. luteus petal tissue, which lacks PLA. MYB5a coding sequence is quite similar between M. l. luteus and M. l. variegatus, with 98.9% identity across the exon 1‐2‐4 splice variants excluding indels (Supporting Information S1: Figure S1). Of the nine SNPs in this region, four (all in exon 4) are non‐synonymous, and there is also an 18‐bp indel in exon 4, thus leaving open the possibility of functional protein divergence between the luteus and variegatus alleles of MYB5a.
While sequencing the exon 1‐2‐4 splice variant of M. l. variegatus, we discovered a rarely occurring, unexpected new sequence. In two separate sequencing reactions, originating from two separate cDNA syntheses from a floral‐bud mRNA extraction, a “GG” sequence was found in which adenines at positions 582 and 686 (the “AA” sequence) were replaced with guanines (Figure 2; Supplemental Figure S1). We hypothesize that these “guanines” might, in fact, represent inosine, which is created by RNA editing of adenosine (Cattenoz et al. 2013), and test whether the “GG” variant is genomically encoded or whether it is unique to RNA.
Figure 2.

Putative A‐to‐I editing sites in the exon 1‐2‐4 splice variant of M. l. variegatus MYB5a. (A) Dark gray bars show the exon structure from start codon to stop codon. Light gray bars show the DNA‐binding R2 and R3 domains common to all members of the R2R3 MYB gene family (Stracke, Werber, and Weisshaar 2001). “Subgroup 6” is a sequence motif that is conserved across all R2R3 MYB genes that encode activators of anthocyanin biosynthesis (Stracke, Werber, and Weisshaar 2001). The two putative A‐to‐I editing sites are each marked as “edited site.” (B) Chromatograms from MYB5a Variant AA and Variant GG. The two polymorphic sites are both located in the fourth exon of MYB5, 584 and 686 nucleotides downstream of the translation start site. Nucleotide and amino acid differences are highlighted. Sequences were obtained using Sanger sequencing and were visualized using Geneious R10. [Color figure can be viewed at wileyonlinelibrary.com]
After determining that the “AA” variant is the only genomically encoded MYB5a sequence, we constructed “coding‐swap” transgenes and used transient transformation to compare the anthocyanin‐activation abilities of the genomically encoded exon 1‐2‐4 splice variants from M. l. variegatus versus M. l. luteus in leaves of the tobacco plant Nicotiana tabacum. Nicotiana is routinely used for such purposes, including tests of flower color genes from both Asterids and the more distantly related Rosids (Deluc et al. 2006, Montefiori et al. 2015). Ding and Yuan (2016) adapted methods from Nicotiana for use in Mimulus lewisii. In our hands, however, transient transformation caused substantial leaf tissue death in both M. lewisii and M. l. luteus. We therefore returned to Nicotiana, noting that Mimulus (an Asterid in the order Lamiales) is considerably more closely related to Nicotiana (order Solanales) than are many of the other species whose gene functions have been similarly tested in Nicotiana.
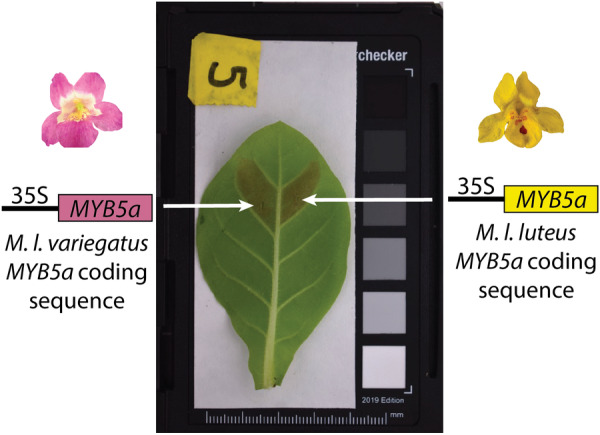
If the gain of PLA in the magenta‐flowered M. l. variegatus were caused solely by a cis‐regulatory‐driven spatial expansion of MYB5a function, then we predicted that coding sequences of MYB5a from M. l. variegatus and the yellow‐flowered M. l. luteus would be equally capable of stimulating anthocyanin production. We tested this hypothesis by transiently expressing each taxon's MYB5a coding sequence in leaves of N. tabacum, and digitally quantifying the resulting pigment. We then confirmed our findings by completely replicating the experiment, with the addition of spectrophotometry of extracted anthocyanins alongside digital image analysis.
Somewhat surprisingly, the luteus allele of MYB5a drove significantly stronger anthocyanin pigmentation than did the variegatus allele, across both datasets and both quantification techniques. This indicates that, in the context of Nicotiana at least, M. l. variegatus MYB5a has not evolved greater anthocyanin pigmentation ability via coding‐sequence changes, further pointing to cis‐regulatory evolution as the driver of the gain of PLA pigmentation in M. l. variegatus.
2. Methods
2.1. Plant Materials
Monkeyflower and tobacco seeds were planted and grown as in Zheng et al. (2021). For gene sequencing and transgene development we used 12 generations selfed M. luteus var. luteus EY7 and 11 generations selfed M. l. variegatus RC6, from seed collections described in Cooley, Carvallo, and Willis (2008). For tests of anthocyanin production, Data set 1 utilized N. tabacum cv. Petit Havana SR1 (Lehle Seeds). Data set 2 additionally used N. tabacum Havana, Havana 142, and Havana 608 (Victory seed company).
2.2. Determining the Genomically Encoded Sequence of MYB5a From M. l. variegatus
To test whether the less‐frequently‐cloned “GG” variant of M. l. variegatus MYB5a (Figure 2) might be encoded in the genome, either as residual heterozygosity or a gene duplicate, we cloned and sequenced a portion of genomic MYB5a from an F1 hybrid of M. l. luteus × M. l. variegatus using primers Myb5_64F and Myb5_57R (Supporting Information S1: Table S1 and Figure S1). We reasoned that, if the “GG” variant were from a paralogous gene, then we should be able to recover both variants from the F1 hybrid. If it instead represented residual heterozygosity, then any single F1 hybrid would contain only one of the two M. l. variegatus variants, AA or GG, along with the M. l. luteus allele of MYB5a. To further assess whether the “GG” variant represented residual heterozygosity, we cloned and sequenced genomic MYB5a from the M. l. variegatus inbred line.
A third hypothesis was that the “GG” variant was the result of posttranscriptional modification, in which case it should be absent from all genomic DNA, but present at some frequency among MYB5a mRNA and cDNA transcripts. To investigate this, we performed new mRNA extractions, and cloned and sequenced MYB5a cDNA from M. l. variegatus young buds, following Zheng et al. (2021).
2.3. Modeling the Effect of Two A‐to‐G Transitions on Protein Folding
Since both of the A‐to‐G transitions in MYB5a were nonsynonymous, AlphaFold (alphafoldserver.com) was used to predict folding of the polypeptides encoded by both the AA and the GG variants, in the presence versus absence of target DNA. The DNA sequence of the exon 1‐2‐4 splice variant from M. l. variegatus MYB5a was used to generate the predicted protein. Models were visualized with iCn3D (ncbi.nlm.nih.gov/Structure/icn3d).
2.4. Nucleic Acid Extraction, PCR, and Cloning
Genomic DNA was extracted from Mimulus leaf tissue using the Zyppy DNeasy Extraction kit (Zymo Research, CA, USA). RNA was extracted from young floral buds using the E.Z.N.A. Plant RNA Kit (Omega Bio‐Tek, GA, USA). cDNA was synthesized using the qScriptTM cDNA Synthesis Kit (Quanta BioSciences, Inc., MD, USA).
Regions of MYB5a spanning one or both adenine/guanine sites were PCR amplified (Supporting Information S1: Table S1) with G‐Biosciences Taq polymerase (St. Louis, MO, USA) and cloned into pGEM in Escherichia coli as in Zheng et al. (2021). Colonies were PCR‐screened using primers M13F(−20) and M13R(−24) and sequenced by Eton Biosciences (San Diego, CA, USA).
2.5. Plasmids Used to Test for Functional Equivalence of Two Coding Sequences
The “AA” allele of MYB5a exon 1‐2‐4 was amplified from M. l. variegatus petal cDNA, using primers cacc10F and Myb5_69R (Supporting Information S1: Table S1) and New England Biolabs Phusion High‐Fidelity DNA Polymerase, and directionally cloned using the pENTR‐D/TOPO Cloning Kits The protein‐coding region of M. l. luteus was synthesized by GENEWIZ (Plainfield, NJ, USA) based on published genomic sequence (Edger et al. 2017), and provided in a pUC57 vector. LR Clonase II (ThermoFisher Scientific, Waltham, MA, USA) was used to transfer inserts to the pEARLEYGATE101 destination vector (Earley et al. 2006). Colonies were PCR‐screened at each step, with a HinDIII diagnostic digest for additional verification. The A. tumefaciens‐compatible GFP expression plasmid pGFPGUSPlus was used as a negative control (Addgene plasmid #64401; Vickers et al. 2007).
2.6. Transient Transformation
Completed “coding‐swap” transgenes, and the negative control plasmid, were electroporated into A. tumefaciens GV3101. Putative transformants were PCR‐screened using primers pEG‐35S‐attB1_F and att‐R2 (Supporting Information S1: Table S1), and cultured and infiltrated into tobacco leaves following Ding and Yuan (2016).
In the first set of infiltrations, “Data set 1,” acetosyringone and Silwet L‐77 were not included in the resuspension solution as a possible solution for tissue damage previously observed in infiltrated M. lewisii (B. Ding, personal communication). Although eliminating acetosyringone and Silwet did not improve our results in M. lewisii, we maintained the protocol as we transitioned to Nicotiana. In a second set of infiltrations, “Data set 2,” acetosyringone was used at 150 µM after having qualitatively observed an increase in leaf damage at higher concentrations in initial trials and noting typically lower values used in other studies (i.e., Heidari‐Japelaghi et al. 2020). The addition of Silwet in any amount, from 0.1 to 1. mL/L, increased leaf damage and reduced anthocyanin production (Supporting Information S1: Figure S2), and thus continued to be excluded in Data set 2.
A 1‐mL disposable syringe with the needle removed was used to deliver A. tumefaciens cells to up to five leaves per plant of young (1–3 months) N. tabacum. The top of the leaf was held firmly while the leaf underside was injected until liquid had visibly spread past the site of injection (100–200 μL). The M. l. luteus and M. l. variegatus alleles were infiltrated in pairs, on either side of the leaf midvein. This paired design allows us to control for inevitable variation in conditions such as plant age, leaf age, temperature, cloud cover, pH, and days from infiltration to photography. pGFPGUSPlus negative controls were applied to other leaves. Photos are available on DataDryad (DOI: 10.5061/dryad.7d7wm3814).
2.7. Image Acquisition and Analysis
Whole leaves were imaged 3–12 days after infiltration in a dark room with a Nikon D3500 DSLR camera with 18–55 mm lens. For Data set 2, a 0.6‐cm diameter hole punch was then used to collect a disc of tissue from each infiltrated region, for photography followed by anthocyanin extraction.
The camera was fixed to a height of 50 cm above the sample. An overhead Sylvania Ceramic Metal Halide bulb was used for Data set 1. For Data set 2, samples were surrounded by three overhead bulbs: a 50‐W helical bulb and two 10‐W LED ring lights. Leaves were placed on a white paper background, and surrounded by a white paper cylinder to act as a light diffuser. VGG Image Annotator was used on.jpeg images of whole leaves to circumscribe the infiltrated region, which was generally visible as a slight color difference compared to uninfiltrated tissue even in the negative controls. If infiltrated tissue could not be distinguished from uninfiltrated tissue, or in the rare instance where pigment spread past a secondary vein into the leaf section above or below the infiltrated section, then the region circumscribed for analysis was bounded by the midvein, the leaf edge, and the secondary veins above and below the infiltration site. There was sometimes a small circle of pale or damaged tissue surrounding the injection site, and this was excluded from analysis. Leaf disks were algorithmically detectable in their images, and required no manual annotation.
A custom Python program imported RAW image files for processing, using a modified version of the MacDuff color chart detection algorithm (https://github.com/mathandy/python-macduff-colorchecker-detector) to automatically detect a reference color chart. Image pixel values were converted into normalized reflectance values based on a linear fit of the red, green, and blue signal strengths to the greyscale panels in the color chart. S green, the strength of the green channel relative to the total of all three color channels, was then averaged over all pixels of the annotated region of interest, minus a circular region surrounding the injection site if tissue damage from the injection syringe was observed. For leaf discs, S green was calculated for the entire disc.
S green is shown to correlate highly (R 2 ≥ 0.63, p < 0.001) with anthocyanin concentration across petals, stems, pedicels, and calyces of six plant species exhibiting a range of types of anthocyanin pigments (del Valle et al. 2018). For ease of interpretation, S green values were converted to “redness” values of 1 – S green.
2.8. Comparison of Digital Image Analysis to Spectrophotometric Analysis
Once photographed, leaf discs from Data set 2 were used for a spectrophotometry‐based anthocyanin quantification. Discs were placed in 2‐mL tubes containing a “hard tissue homogenizing mix” of 2.8 mm ceramic beads (VWR), frozen in liquid nitrogen, and ground to a powder using a Mini Bead Mill 4 (VWR). Anthocyanin was extracted by adding 1.2 mL of acidified methanol (70% MeOH:H2O with 1% HCl). Samples were stored at −20°C in the dark to avoid pigment degradation if not immediately used for analysis. Before analysis, centrifugation was used to spin down the tissue, and 1 mL supernatant was pipetted to cuvettes. Absorbance Units (AU) at 530 and 653 nm were measured, with a correction for chlorophyll absorbance following del Valle et al. (2018):
2.9. Data Visualization and Analysis
GraphPad Prism 10, Excel 16, and Adobe Illustrator 2024 were used to visualize results. Statistical analyses were performed in Rstudio 2024.04.2 and GraphPad Prism 10; R commands used are shown in the Supporting Information S1: Text. Redness was compared across all three treatments using one‐way ANOVAs followed by Tukey post hoc tests. After excluding the negative controls to enable paired analyses, luteus and variegatus infiltrations were compared using two‐tailed paired t tests and two‐tailed paired Wilcoxon signed rank tests. Correlation between image analysis and spectrophotometry data was tested using a linear model, and an adjusted R 2 value was calculated. Data set 2 was additionally visualized as a ratio of luteus:variegatus, and this ratio was tested for deviation from 1 using a one‐sample t test. The effect of Silwet on anthocyanin presence versus absence, and leaf damage presence versus absence, was assessed using a General Linear Model for binomial data.
3. Results
3.1. The “GG” Allele of MYB5a From M. l. variegatus Is Not Genomically Encoded, But Is Found in Flower‐Bud cDNA
Before this work, the “GG” variant of MYB5a had been found in two clones of petal‐bud cDNA, with the “AA” variant being more abundant, suggestive of the occasional editing of RNA transcripts.
Cloning MYB5a gDNA from a variegatus × luteus F1 hybrid yielded 29 colonies containing MYB5a (Supporting Information S1: Figure S3). Of these, 13 contained the “AA” variant and 16 contained the M. l. luteus allele. The “GG” variant was not discovered. Cloning MYB5a gDNA from a highly inbred line of M. l. variegatus yielded 37 colonies containing an MYB sequence (Supporting Information S1: Figure S3). All were the “AA” variant. Taken together, these results indicate that the unexpected GG variant is not genomically encoded.
However, we did recover the “GG” variant a third time from M. l. variegates flower‐bud cDNA, in one out of 30 colonies sequenced (Table 1), consistent with the GG variant arising from occasional RNA editing.
Table 1.
Sequencing MYB5a cDNA from M. l. variegatus developing flower bud tissue identifies a third case of A–“G” (putatively A‐to‐I) editing. “Inner petal” corresponds to the nectar‐guide‐spotted throat region of the flower and “outer petal” corresponds to the petal lobes. MYB5a activates anthocyanin production in both regions. N = 30 colonies sequenced from cDNA made from young flower bud tissue.
| MYB5a variant | Cloned sequences from inner petal | Cloned sequences from outer petal |
|---|---|---|
| Exons 1‐2‐4 with A | 10 | 19 |
| Exons 1‐2‐4 with “G” | – | 1 |
3.2. MYB5a From Both M. l. luteus and M. l. variegatus Drives Strong Anthocyanin Production
In Data set 1, consisting of whole‐leaf images from N. tabacum line SR1, both MYB5a transgenes resulted in significantly redder leaf tissue than did the negative control (Figure 3; Supporting Information S1: Table S2; F(2, 233) = 12.43; p = 7.44 × 10−6). Surprisingly, both a Tukey's post hoc test (p < 0.05 for each comparison) and a paired t test (t = 6.3656, df = 97, p = 6.46 × 10−9) show that the allele of MYB5a from the yellow‐flowered M. l. luteus drove significantly greater anthocyanin production than the corresponding allele from the magenta‐flowered M. l. variegatus. A paired, two‐sided Wilcoxon signed rank test showed the same (V = 4186, p = 4.47 × 10−10).
Figure 3.
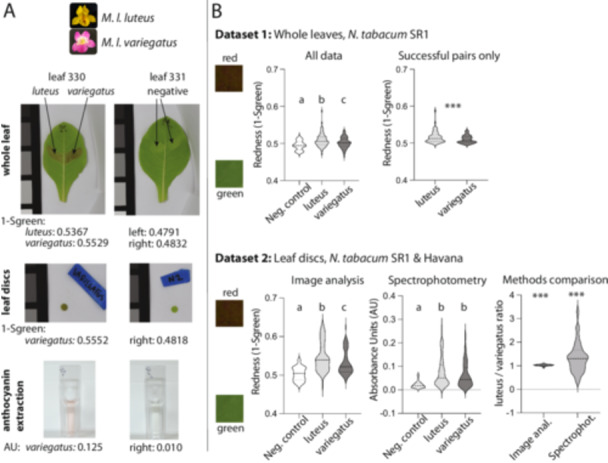
MYB5a coding sequence from yellow‐petaled M. l. luteus drives greater anthocyanin pigmentation than coding sequence from the magenta‐petaled M. l. variegatus. (A) Whole‐leaf, leaf disc, and anthocyanin extraction images for leaves 330 and 331 from Data set 2. Anthocyanin content is indicated underneath each photo. Anthocyanin quantification obtained by image analysis is reported as 1‐Sgreen; that obtained by anthocyanin extraction and spectrophotometry is indicated by Absorbance Units (AU) using the formula A 530‐(0.24 × A 653). In both cases, higher values indicate redder leaf tissue. Greyscale color squares, for scale, are 25 mm on each side. (B) Data set 1 results for (left) all samples with N = 40 (negative control), 98 (luteus coding sequence), 98 (variegatus coding sequence) (right) matched luteus‐variegatus pairs for which both members of the pair produced visible pigmentation, with N = 66 pairs. Data set 2 results are shown for whole leaves and leaf punches from lines SR1 and Havana, with image analysis results on the left and spectrophotometry results on the right. Data set 2 sample sizes are N = 38 (negative control), 80 (luteus coding sequence), and 80 (variegatus coding sequence). In the Methods Comparison panel, the ratios of paired luteus‐ versus variegatus‐infiltrated leaf discs are shown, for anthocyanin calculated by image analysis compared to spectrophotometry. While both ratios were significantly higher than one, variance was noticeably larger with the spectrophotometry approach. In each violin plot, the median is shown as a dashed line and quartiles as dotted lines. Statistics: Letters a, b, and c are significance groupings at p < 0.05 based on a one‐way ANOVA with Tukey's post hoc test; *** indicates p < 0.001 based on a two‐tailed paired t test (Data set 1) or a one‐sample t test compared to a value of 1 (Data set 2). [Color figure can be viewed at wileyonlinelibrary.com]
To exclude possible effects of varying infiltration success in Data set 1, we analyzed pigment production only from the pairs of infiltrations for which both members of the pair produced visible amounts of leaf anthocyanin pigment (Figure 3B, top right panel). In these pairs, a slightly smaller but still highly significant difference was found, again with the luteus coding sequences associated with higher redness (t = 4.4855, df = 65, p = 3.03 × 10−5). A paired, two‐sided Wilcoxon signed rank test produced a similar result (V = 1837, p = 3.02 × 10−6).
We used Data set 2 to compare our digital image values of redness to AUs obtained from spectrophotometry, for standardized discs of tissue punched out of the leaf near each infiltration site. The association between the two techniques was strongly positive and highly significant for the previously used SR1 line and the newly added Havana line, but less strong for Havana 142 (Supporting Information S1: Figure S4). Havana 608 yielded little to no anthocyanin pigmentation, and in fact a negative correlation was observed between the techniques (Supporting Information S1: Figure S4). We, therefore, confined the following analyses to lines SR1 and Havana.
Significant variation across treatments in Data set 2 was observed for both digital image analysis (F(2, 195) = 25.07, p = 2.04 × 10−10) and spectrophotometry (F(2, 195) = 13.71, p = 2.69 × 10−06; Figure 3; Supporting Information S1: Table S3). Both techniques confirmed our findings in Data set 1, of a slight but significant increase in redness associated with the luteus transgene compared to its paired variegatus transgene (image analysis: t = 6.659, df = 79, p = 3.33 × 10−09; spectrophotometry: t = 2.620, df = 79, p = 0.0105). Analysis of whole‐leaf images from Data set 2 showed the same pattern as the leaf discs (Supporting Information S1: Figure S5 and Table S4). Overall, image analysis showed greater discrimination amongst treatments and lower variance within treatments than did spectrophotometry (Figure 3B).
4. Discussion
What debt do species‐level novelties owe to coding mutations versus cis‐regulatory expression differences? We used transient transgenic assays to help investigate this question for the yellow‐flowered M. luteus var. luteus, which lacks PLA pigmentation, compared to the magenta‐flowered M. l. variegatus, which recently gained PLA via unknown mutation(s) within the MYB5a gene.
We report that there is a significant functional difference between the two protein‐coding regions when expressed in N. tabacum leaves, but in the opposite direction of what would be expected if coding sequence evolution were responsible for the increased pigmentation of M. l. variegatus. Together with a previous finding that MYB5a expression correlates with PLA, and that functional manipulations of MYB5a expression alter PLA (Zheng et al. 2021), this result—robustly replicated across two separate experiments—supports the hypothesis that cis‐regulatory evolution is solely responsible for the recent expansion of pigmentation in M. l. variegatus.
4.1. Improved Tools for Rapid Transgenic Assays in Mimulus
The success of the two Mimulus transgenes at activating anthocyanin production in N. tabacum is encouraging for future functional studies in Mimulus. Using N. tabacum for direct side‐by‐side comparison of Agrobacterium‐delivered transgenes takes advantage of leaf symmetry and the clearly delineated leaf sectors in N. tabacum to enable statistically powerful pairwise comparisons in an identical biological background. We believe this remains an underutilized strategy, given that Nicotiana is regularly used for heterologous tests of gene function (particularly, for anthocyanin‐related genes) from a wide range of species.
4.2. A‐to‐I Editing of mRNA
In the process of building MYB5a overexpression transgenes, we discovered what appears to be the first documented case of posttranscriptional editing in an anthocyanin‐related gene. Two sites within the M. l. variegatus allele are encoded as adenine in the genome, yet occasionally produce mRNA sequences that read as a guanine in Sanger sequencing. These “guanines” are presumably inosine bases, created through adenine deamination (Cattenoz et al. 2013). Since inosines are read by the cell as guanine, both sites are expected to result in an amino acid change: from asparagine to serine at nucleotide position 584, and from glutamic acid to glycine at position 686. Both sites are outside of the DNA‐binding R2 and R3 domains (Figure 1), but do alter folding at the C‐terminal end of the protein (Supporting Information S1: Figure S6). The functional effects of these changes are unknown, and may represent another layer in the molecular mechanisms of the evolution of PLA.
Although A‐to‐I editing of mRNA transcripts does not yet appear to have been described in plants, nuclear A‐to‐I posttranscriptional editing has been reported in plant tRNA (Delannoy et al. 2009), and the deaminase enzymes required for A‐to‐I editing have been discovered in Arabidopsis thaliana (Zhou, Karcher, and Bock 2014). mRNA A‐to‐I editing is widespread across the other domains of life, including fungi (Teichert 2018), animals (Knoop 2011), and bacteria (Bar‐Yaacov et al. 2017).
A variety of methods exist for confirming A‐to‐I editing, including chromatographic approaches (Wolf 2002) and “inosine chemical erasing” (ICE)‐Seq (Sakurai et al. 2010). However, A‐to‐I editing is commonly detected and quantified by the simple method used here, in which reverse transcription and sequencing of mRNA reveals unexpected “guanines” in some proportion of transcripts (Gu et al. 2012). Tests for mRNA A‐to‐I editing in other plant species would be intriguing and, if confirmed, could potentially open the door to new avenues of investigation.
4.3. Conclusions
M. luteus var. luteus and M. l. variegatus differ strikingly in floral phenotype, thanks to a derived loss of yellow carotenoid pigment and gain of magenta anthocyanin pigment in the latter. The expansion of anthocyanin to the petal lobes of M. l. variegatus has previously been tracked to MYB5a. Here we use transient transgenics coupled with quantitative digital image analysis to show that, in the context of Nicotiana, the protein‐coding region of MYB5a from the high‐anthocyanin M. l. variegatus fails to produce more pigment than the allele from the low‐anthocyanin M. l. luteus (in fact the reverse). This lack of evidence for pigment gain via protein evolution adds further support to the hypothesis that evolution in cis to MYB5a is the molecular mechanism for anthocyanin expansion in M. l. variegatus. We additionally report the discovery of what appears to be posttranscriptional mRNA editing in plants, leading to flexibility in the amino acid sequence and final structure of the M. l. variegatus MYB5a protein. Overall, our work highlights the utility of floral diversification for identifying the molecular mechanisms of evolution, as well as the scope for continued new discoveries in the realm of plant molecular genetics.
Conflicts of Interest
The authors declare no conflicts of interest.
Supporting information
Supporting information.
Acknowledgments
The authors thank N. Forsthoefel, B. Ding, and Y.‐W. Yuan for assistance with and advice on the transgenic techniques. We thank D. Vernon, B. Moss, J. Puzey, and their students for the discussion of some of the results presented here, and Oliver Baltzer for assistance with updates to the image analysis pipeline. A.M.C. was supported by NSF‐DEB‐1655311, NSF‐DEB‐1754075, and NSF‐IOS‐2031272. A.E.P. was additionally supported by a Whitman College Abshire Award for undergraduate research.
Data Availability Statement
Anthocyanin quantification data are available in Supporting Information. Leaf images are available on DataDryad (DOI: 10.5061/dryad.7d7wm3814). MYB5a sequences were previously published https://www.ncbi.nlm.nih.gov/nuccore/MT361119.1 (M. l. luteus/E. l. lutea) and https://www.ncbi.nlm.nih.gov/nuccore/2019733960 (M. l. variegatus/E. l. variegata). Image analysis code is at https://github.com/WhitmanOptiLab/PigmentSpotting.
References
- Bar‐Yaacov, D. , Mordret E., Towers R., et al. 2017. “RNA Editing in Bacteria Recodes Multiple Proteins and Regulates an Evolutionarily Conserved Toxin‐Antitoxin System.” Genome Research 27: 1696–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattenoz, P. B. , Taft R. J., Westhof E., and Mattick J. S.. 2013. “Transcriptome‐Wide Identification of A > I RNA Editing Sites by Inosine Specific Cleavage.” RNA 19: 257–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley, A. M. , Carvallo G., and Willis J. H.. 2008. “Is Floral Diversification Associated With Pollinator Divergence? Flower Shape, Flower Colour and Pollinator Preference in Chilean Mimulus.” Annals of Botany 101: 641–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley, A. M. , Modliszewski J. L., Rommel M. L., and Willis J. H.. 2011. “Gene Duplication in Mimulus Underlies Parallel Floral Evolution via Independent Trans‐Regulatory Changes.” Current Biology 21: 700–704. [DOI] [PubMed] [Google Scholar]
- Cooley, A. M. , and Willis J. H.. 2009. “Genetic Divergence Causes Parallel Evolution of Flower Color in Chilean Mimulus .” New Phytologist 183: 729–739. [DOI] [PubMed] [Google Scholar]
- del Valle, J. C. , Gallardo‐López A., Buide L., Whittall J. B., and Narbona E.. 2018. “Digital Photography Provides a Fast, Reliable, and Noninvasive Method to Estimate Anthocyanin Pigment Concentration in Reproductive and Vegetative Plant Tissues.” Ecology and Evolution 8: 3064–3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delannoy, E. , Le Ret M., Faivre‐Nitschke E., et al. 2009. “ Arabidopsis tRNA Adenosine Deaminase Arginine Edits the Wobble Nucleotide of Chloroplast tRNAArg(ACG) and Is Essential for Efficient Chloroplast Translation.” The Plant Cell 21: 2058–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluc, L. , Barrieu F., Marchive C., et al. 2006. “Characterization of a Grapevine R2R3‐MYB Transcription Factor That Regulates the Phenylpropanoid Pathway.” Plant Physiology 140: 499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, B. , and Yuan Y.‐W.. 2016. “Testing the Utility of Fluorescent Proteins in Mimulus lewisii by an Agrobacterium‐Mediated Transient Assay.” Plant Cell Reports 35: 771–777. [DOI] [PubMed] [Google Scholar]
- Earley, K. W. , Haag J. R., Pontes O., et al. 2006. “Gateway‐Compatible Vectors for Plant Functional Genomics and Proteomics.” Plant Journal 45: 616–629. [DOI] [PubMed] [Google Scholar]
- Edger, P. P. , Smith R., McKain M. R., et al. 2017. “Subgenome Dominance in an Interspecific Hybrid, Synthetic Allopolyploid, and a 140‐Year‐Old Naturally Established Neo‐Allopolyploid Monkeyflower.” Plant Cell 29: 2150–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairnie, A. L. M. , Yeo M. T. S., Gatti S., et al. 2022. “Eco‐Evo‐Devo of Petal Pigmentation Patterning.” Essays in Biochemistry 66: 753–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, T. , Buaas F. W., Simons A. K., Ackert‐Bicknell C. L., Braun R. E., and Hibbs M. A.. 2012. “Canonical A‐to‐I and C‐to‐U RNA Editing Is Enriched at 3′ UTRs and microRNA Target Sites in Multiple Mouse Tissues.” PLoS One 7: e33720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidari‐Japelaghi, R. , Valizadeh M., Haddad R., Dorani‐Uliaie E., and Jalali‐Javaran M.. 2020. “Production of Bioactive Human IFN‐γ Protein by Agroinfiltration in Tobacco.” Protein Expression and Purification 173: 105616. [DOI] [PubMed] [Google Scholar]
- Knoop, V. 2011. “When You Can't Trust the DNA: RNA Editing Changes Transcript Sequences.” Cellular and Molecular Life Sciences 68: 567–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montefiori, M. , Brendolise C., Dare A. P., et al. 2015. “In the Solanaceae, a Hierarchy of bHLHs Confer Distinct Target Specificity to the Anthocyanin Regulatory Complex.” Journal of Experimental Botany 66: 1427–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai, M. , Yano T., Kawabata H., Ueda H., and Suzuki T.. 2010. “Inosine Cyanoethylation Identifies A‐to‐I RNA Editing Sites in the Human Transcriptome.” Nature Chemical Biology 6: 733–740. [DOI] [PubMed] [Google Scholar]
- Sobel, J. M. , and Streisfeld M. A.. 2013. “Flower Color as a Model System for Studies of Plant Evo‐Devo.” Frontiers in Plant Science 4: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern, D. L. 2013. “The Genetic Causes of Convergent Evolution.” Nature Reviews Genetics 14: 751–764. 10.1038/nrg3483. [DOI] [PubMed] [Google Scholar]
- Stracke, R. , Werber M., and Weisshaar B.. 2001. “The R2R3‐MYB Gene Family in Arabidopsis thaliana .” Current Opinion in Plant Biology 4: 447–456. [DOI] [PubMed] [Google Scholar]
- Streisfeld, M. A. , and Rausher M. D.. 2011. “Population Genetics, Pleiotropy, and the Preferential Fixation of Mutations During Adaptive Evolution.” Evolution 65: 629–642. [DOI] [PubMed] [Google Scholar]
- Teichert, I. 2018. “Adenosine to Inosine mRNA Editing in Fungi and How It May Relate to Fungal Pathogenesis.” PLoS Pathogens 14: e1007231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers, C. E. , Schenk P. M., Li D., Mullineaux P. M., and Gresshoff P. M.. 2007. “pGFPGUS Plus, a New Binary Vector for Gene Expression Studies and Optimising Transformation Systems in Plants.” Biotechnology Letters 29: 1793–1796. [DOI] [PubMed] [Google Scholar]
- Wolf, J. 2002. “Tada, an Essential Trna‐Specific Adenosine Deaminase From Escherichia coli .” EMBO Journal 21: 3841–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray, G. A. 2007. “The Evolutionary Significance of Cis‐Regulatory Mutations.” Nature Reviews Genetics 8: 206–216. [DOI] [PubMed] [Google Scholar]
- Yuan, Y. W. 2019. “Monkeyflowers (Mimulus): New Model for Plant Developmental Genetics and Evo‐Devo.” New Phytologist 222: 694–700. [DOI] [PubMed] [Google Scholar]
- Zheng, X. , Om K., Stanton K. A., et al. 2021. “The Regulatory Network for Petal Anthocyanin Pigmentation Is Shaped by the MYB5a/NEGAN Transcription Factor in Mimulus .” Genetics 217: iyaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, W. , Karcher D., and Bock R.. 2014. “Identification of Enzymes for Adenosine‐to‐Inosine Editing and Discovery of Cytidine‐to‐Uridine Editing in Nucleus‐Encoded Transfer RNAs of Arabidopsis.” Plant Physiology 166: 1985–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Data Availability Statement
Anthocyanin quantification data are available in Supporting Information. Leaf images are available on DataDryad (DOI: 10.5061/dryad.7d7wm3814). MYB5a sequences were previously published https://www.ncbi.nlm.nih.gov/nuccore/MT361119.1 (M. l. luteus/E. l. lutea) and https://www.ncbi.nlm.nih.gov/nuccore/2019733960 (M. l. variegatus/E. l. variegata). Image analysis code is at https://github.com/WhitmanOptiLab/PigmentSpotting.


