Abstract
Objectives
Due to the variety of modern diet and lifestyle changes, China has become the world’s largest number of people with T2DM. How to prevent and cure T2DM has become one of the urgent public health events in China. Numerous studies have demonstrated vitamin D (VitD) was independently correlated with insulin sensitivity and β cells function. VitD deficiency occurs in about 70% to 80% of patients with T2DM. However, the reason of T2DM patients suffering from VitD deficiency is not very clear. The aim of this project is to identify biomarkers to explore potential mechanism of VitD deficiency in patients with T2DM.
Methods
We used the iTRAQ-coupled LC-MS/MS technique to screen differential expression proteins between VitD deficiency group and VitD sufficiency group in T2DM patients. Then we carried out hierarchical clustering analysis, Gene Ontology classification and enrichment analysis, KEGG pathway enrichment analysis, protein-protein interaction network (PPI) analysis and ELISA validation.
Results
We identified 63 differentially expressed proteins, 17 proteins were up-regulated and 46 proteins were down-regulated (VitD sufficiency vs. VitD deficiency). We ultimately selected four proteins, Podocalyxin (PODXL), ICAM3, MMP9, ApoF for further verification. As a result, the level of MMP9 and ICAM3 was higher in VitD sufficiency group than VitD deficiency group.
Conclusions
Our study provided a solid theoretical foundation for the study of biomarkers and their mechanisms in most patients with T2DM who suffer from vitamin D deficiency. In addition, MMP9 and ICAM3 may play critical roles in the process of VitD deficiency in T2DM.
Supplementary Information
The online version contains supplementary material available at 10.1007/s40200-024-01456-w.
Keywords: iTRAQ, Biomarkers, Vitamin D deficiency, T2DM
Introduction
Type 2 diabetes mellitus (T2DM) is a chronic metabolic disease, and the incidence of T2DM is increasing worldwide. According to The International Diabetes Federation (IDF), international number of patients with diabetes is estimated to rise to 783.2 million by 2045 [1]. Studies have shown that T2DM patients generally suffer from vitamin D deficiency [2]. Vitamin D deficiency may participate in the pathogenesis of T2DM disease [3], and vitamin D supplementation may improve pancreatic islet function and even delay the disease process of T2DM [4, 5].
Vitamin D is a steroid hormone known to regulate calcium and phosphorus absorption in bone and is often used to prevent or treat bone metabolic diseases such as rickets and osteoporosis [6]. However, in the past decade, the extra-skeletal action of vitamin D has become a research hotspot. Vitamin D receptors(VDR) exist in various organs and tissues throughout the body. By binding with VDR, vitamin D can regulate cell proliferation, differentiation, apoptosis, inflammatory response, vascular activity, immune function, etc [7, 8]. It has been confirmed that there are nuclear receptors and membrane receptors of vitamin D in islet beta cells [9]. There are more and more studies on the correlation between VitD and insulin metabolism and T2DM. The specific relationship and potential mechanism between VitD deficiency and insulin resistance still need further research.
Since the study of rickets in the early 20th century, VitD has been found to be closely related to calcium and phosphorus metabolism and bone health. The extraskeletal effects of VitD include effects on cardiovascular system, metabolism, immunity, tumorigenesis, fetal development, etc [10].Vitamin D is converted into a non-biologically functional 25 hydroxyvitamin D3 [25 (OH) D3] by the action of liver 25 hydroxylase, and then enters the kidneys or other tissues. After secondary hydroxylation, it becomes a biologically active 1,25 (OH) 2D3, which binds to its receptor and functions [11]. 25(OH) D3 is the main storage form of vitamin D in the body, and most clinical studies have used 25(OH) D3 to evaluate vitamin D levels in the body [12].
In recent years, studies have confirmed that the early decline in insulin secretion function in T2DM patients is related to the low level of 25(OH)D3 [13, 14]. Serum 25(OH)D3 was positively correlated with basal insulin secretion, early and late phase insulin secretion [15, 16]. However, the specific research data on the mechanism of 25(OH)D3 affecting insulin secretion is still limited, and more studies are needed to further confirm it. Vitamin D has become a hot spot in clinical and basic research.
Numerous studies have been reported that inflammatory factors are one of the pathogenesis of T2DM, and 1,25(OH)2D3 can limit the expression of chemokines, reduce the density of major histocompatibility complex (MHC-I) like proteins, and reduce the effect of T cells infiltration’ on islets β cells, improvement the function of β cells [17]. Vitamin D can also inactivate inflammatory factors related to insulin resistance, thereby increasing insulin sensitivity [18]. In addition, studies have reported that active vitamin D supplementation can improve insulin resistance and better control blood glucose in newly diagnosed T2DM patients [19, 20]. Finally, the islet function was improved [4]. However, at present, the specific molecular mechanism between VitD and the development of T2DM has not fully elucidated. Accordingly, the research, we wanted to screen some biomarkers by plasma proteomics to explore the potential mechanism of low 25 (OH) D, that is, vitamin D deficiency, which is more common in T2DM patients.
Materials and methods
Participants and clinical data
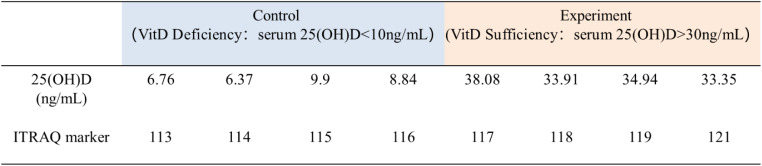
First, we collected peripheral blood from T2DM patients, four each with VitD deficiency(< 10 µg/L) and VitD sufficiency(> 30 µg/L), and separated plasma for proteomic analysis. The enrolled patients were from the Center for Endocrine Metabolism and Immune Diseases of our hospital. All participants were enrolled in a program at the National Center for Metabolic Management(MMC) [21]. All T2DM participants visited endocrine centers for consultation between 2018 and 2019, had no history of bone metabolic diseases, skin diseases, or other autoimmune related diseases. None of the participants were supplemented with active vitamin D or its analogues (Table 1). All participants provided written informed consent. This study was approved by the Ethics Committee of our hospital, and obeyed the ethical guidelines of the Declaration of Helsinki. Basic clinical and laboratory data of participants, including age, sex, serum intact PTH, serum 25 (OH) D, glucose (GLU), alanine aminotransferase (ALT), aspartate transaminase (AST), total cholesterol (TC), high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), triglyceride levels (TG), white blood cell (WBC) count and so on, recorded by experienced Medical Technologists.
Table 1.
Basic information of samples for proteomics
Sample preparation, protein digestion and iTRAQ labeling
A total of 8 human plasma liquid samples were collected, including 4 cases in the VitD-deficient group and 4 cases in the VitD-sufficient group. In order to better conduct the experiment, the sample numbers were renumbered, as detailed in Table 1. Serum albumin and immunoglobulin G were removed from blood samples using the Pierce Top 12 Abundant Protein Depletion Spin Columns (Fisher Scientific). Using a 10 kDa molecular weight cut-off filter for ultrafiltration, the final volume of each sample is 100 µL. Using BCA protein assay to determine protein concentration in supernatant, and then 100 µg of protein for each condition was transferred to a new tube and adjusted with 100mM TEAB (triethyl ammonium bicarbonate) to a final volume of 100µL. The protein samples then underwent sequence-grade modified trypsin (Promega, Madison, WI) digestion, and the resultant peptide mixture were collected. Isobaric tags for a relative and absolute quantification (iTRAQ) reagent (ITRAQ 8 standard kit, AB Sciex) were added to each tube and desalted using C18 SPE column (Sep-Pak C18, 1 cc(100 mg), Waters Corporation, `Milford, MA) and dried in vacuo.
High pH reverse phase separation and LC-MS/MS analysis
Fractionation of peptide mixtures using the Aquity UPLC system (Waters Corporation, Milford, MA) connected to a reverse column (BEH C18 column, Waters Corporation) for high pH separation. Maintain column flow rate at 250 µL/min and column temperature at 45℃. A total of 12 fractions were collected, each of which was dried in a vacuum concentrator. These fractions were then separated by nanoLC and analyzed by online electrospray tandem mass spectrometry. The experiments were carried out on a Nano Aquity UPLC system (Waters Corporation), which was connected to a quadrupole-Orbitrap mass spectrometer (Q-Exactive, Thermo Fisher Scientific, Germany) equipped with an online nano-electrospray ion source. Q-Exactive mass spectrometer operated in data related mode to automatically switch between MS and MS/MS acquisition. After inputting the mass spectrum data into Proteome Discoverer software (PD) (version 1.4.0.288, Thermo Fisher Scientific), the software first screens the mass spectrum. The PD extracted spectrogram is searched with the Mascot (version 2.3.2, Matrix Science, London, UK). After the search, the PD software performs quantitative analysis based on the Mascot search results and the spectrogram filtered in the first step. Analyze the results using the Unipro-Swiss Prot database (Taxonomy: Homo sapiens, 20,411 entries).
Data Management and Bioinformatics analysis
The original data contains 8 experimental samples, from which 773 Proteins are extracted. To better analyze the data, we have carried out a series of management and normalization on the raw data. It mainly includes the screening of missing value and the screening of the number of unique peptide segments of the protein: unique peptide ≥ 1. After pretreatment, 751 detection proteins were retained. We used statistical methods to screen for differentially expressed proteins. The screening criteria were p-value < 0.05 and fold-change < 0.83 or fold-change > 1.2. The functions of differentially expressed proteins were analysed, including Hierarchical Clustering Analysis, KEGG Pathway Analysis (KEGG database), GO Annotation Enrichment Analysis (http://www.geneontology.org/), Protein-Protein Interaction Network Analysis (STRING database. v10, string-db.org).
Verification of differentially expressed proteins
Populations and clinical data
T2DM patients with high vitamin D levels (21 cases) and low vitamin D level (24 cases) were selected from the Center for Endocrine Metabolism and Immune Diseases of our hospital. The rest is the same as part of 1.1.
Plasma sample preparation and ELISA detection
The remaining peripheral venous blood (about 3 ml, EDTA anticoagulant collection vessel) was collected from the enrolled population for HbA1c detection, and the upper plasma was extracted after centrifugation and stored at -80℃. The levels of ApoF, PODXL, MMP9 and ICAM3 in fasting plasma were measured by ELISA(Enzyme-Linked Immunosorbent Assay) following the manuals (ApoF: SEB879Hu, Cloud-Clone Corp; PODXL: SEA768Hu, Cloud-Clone Corp; MMP9: Lot#:0412190173, RayBiotech; ICAM3: Lot#:0412192032, RayBiotech).
Detection of total protein
Utilizing Pierce ® BCA Protein Assay Kit (Thermo scientific, Germany) for the determination of total plasma protein levels. According to the instruction adding 25µL of A-G bovine serum albumin standard or protein sample to be tested into wells of a 96-well plate in triplicate. Next adding 200 µL aliquot of BCA working solution to each well, which was then covered. After incubation at 37℃ for 30 min, measure the absorbance at 560 nm using a multifunctional ELISA reader(PerkinElmer, EnSpire).
Statistical analyses
Statistical analyses were performed using the GraphPad Prism 9 software (San Diego, CA, USA) and SPSS 17.0 (SPSS Inc, Chicago, IL, USA). Student’s t-test were used to assess differences in clinical data between VitD sufficiency group and VitD deficiency group. Data are excluded if it deviates from mean with > 3SDs, p-value < 0.05 was considered as statistically significant.
Results
Screening of differentially expressed proteins
In comparison with the control group (VitD deficiency: serum 25(OH)D < 10ng/mL), the experiment group (VitD sufficiency: serum 25(OH)D > 30ng/mL), we used statistical methods to screen out 63 differentially expressed proteins. Among them, 17 proteins were up-regulated and 46 proteins were down-regulated. The analysis results of screening differentially expressed proteins were visualized in the form of Volcano Plot (Fig. 1). The selection criteria are p-value < 0.05 and fold change < 0.83 or > 1.2.
Fig. 1.
Volcano map showing differentially expressed proteins. Each dot in the figure represents a test protein, and the abscissa represents the fold change of differentially expressed proteins (taken as the base 2 logarithm), proteins with significant differences are distributed at both ends. The ordinate represents the student's T-test P-VALUE (taken the negative of the logarithm base 10), the larger the multiple of differences, the more significant the T-test for proteins. The greater the absolute value of the abscissa, the greater the difference between the two groups. The larger the ordinate value, the more significant the difference in protein expression between groups, and the more reliable the results. The scatter color represents the final screening result, the significantly up-regulated differentially expressed proteins are shown in red, the significantly down-regulated differentially expressed proteins are shown in blue, and the non-significantly differentiated proteins are shown in gray
Hierarchical clustering analysis of differentially expressed proteins
In order to further study the function of screened proteins, proteins with similar functions are grouped together by clustering. Hierarchical clustering analysis and visualization by heat map of differentially expressed proteins can help us to discover the variation pattern of differentially expressed proteins among experimental groups. The basic process of differential expression protein classification cluster analysis was as follows: Firstly, the expression values of differentially expressed proteins obtained from each group were corrected and converted into normal distribution, which as an input to the hierarchical clustering algorithm. Then, in the process of calculating the distance matrix, Euclidean distance was used for distance and Complete Linkage Method was used for linkage. Finally, the heat map of hierarchical clustering analysis is obtained (Fig. 2)
Fig. 2.

Hierarchical Cluster Analysis Thermodynamic Map of VitD sufficiency vs. VitD deficiency Group. The abscissa represents different groups, the experimental group was VD sufficiency group, and the control group was VD deficiency group. The ordinate represents differential expression proteins, the color blocks at different positions represent relative expression of proteins at corresponding positions, the red represents high expression, and the blue represents low expression. It can be seen that the pattern of differentially expressed proteins grouping appeared in the figure
Annotation enrichment analysis of differentially expressed proteins
GO database, the full name of which is Gene Ontology, divides the functions of genes into three aspects: biological processes involved in genes, cell locations and molecular functions played by genes. It organizes the functional concepts of different concepts into DAG (directed acyclic graph). Using the GO database, we can get our target genes at the three levels of Biological Process (BP), Cellular Component (CC), Molecular Function (MF), which are mainly related to. It unifies t the classification system of gene function of each species in the form of a directed acyclic graph. Thus, the functional information of genes is summarized comprehensively, and the confusion of dimensions which is common in traditional functional classification system is corrected. In the analysis of gene expression profiles, GO is often used to provide the background knowledge of gene function classification tags and gene function studies.
Using the knowledge system and structural features of the GO database, the aim is to discover the combination of single characteristic gene functional class or multiple characteristic functional class associated with differential gene expression. In this study, we mapped genes to each node of the GO database and perform functional enrichment analysis with GO (http://www.geneontology.org/). Then we classified and displayed the differentially expressed proteins in each group according to three independent ways: BP, CC, MF(Fig. 3).
Fig. 3.
Histogram(A) and bubble chart(B) of GO enrichment analysis and classification of differentially expressed proteins(experiment vs. control). (A) The abscissa is GO Term, the ordinate is the number of differential expression proteins mapped. The red, green andblue represent annotation information for Biological Process (BP), Cellular Component (CC), Molecular Function (MF), respectively, and the transparency is p-value size. The darker the color, the smaller the p-value. (B)In the figure, the abscissa is the rich factor value and the ordinate is GO Term. Among them, the size of the circle represents the number of differentially expressed proteins mapped, and the larger the circle, the more the quantity; the color of the circle represents the size of the corrected p-value
KEGG pathway analysis of differentially expressed proteins
In addition to the annotation of the function of genes themselves, we also know that genes participate in various pathways in the human body, and the database formed based on human pathways is the database related to the pathway. KEGG(the Kyoto Encyclopedia Genes and Genomes Pathway database, www.kegg.jp/kegg/pathway.html) is a kind of pathway related database, stores the function of gene and genome information. It is guided by biological processes such as gene assembly, transcription, regulation, modification, translation and cellular environmental variation, and constructs a set of visual and interactive models of gene and non-gene coding activities such as various metabolic pathways. By analyzing the metabolic pathways of significant enrichment of differentially expressed proteins, we can find out which pathways undergo significant and systemic changes under different experimental conditions. Enrichment analysis results of pathways in experiment group compared with contrast group in the form of bubble chart and histogram (Fig. 4). We found that the most differential expression proteins are mapped to the focal adhesion pathway.
Fig. 4.
Bubble(A) and Histogram(B) Analysis of KEGG Metabolic Pathway Enrichment Analysis of Differentially Expressed Proteins(experiment vs. control) (A) The abscissa is rich factor value of enrichment degree, the larger the abscissa, the more reliable the enrichment significance of gene products in the pathway. The ordinate is KEGG Pathway information. Among them, the size of the circle represents the number of differential expression proteins in the mapping pathway, and the larger the circle, the more the quantity. The color of the circle represents the size of the corrected p-value, the redder the color, the smaller the p-value. (B) In the figure, the abscissa is - log10 (p-value), and the ordinate is the name of the pathway. Among them, the length of the column indicates the size of p-value. The longer the column, the smaller the p-value, the more red the color, and more differentially expressed proteins were mapped to the pathway
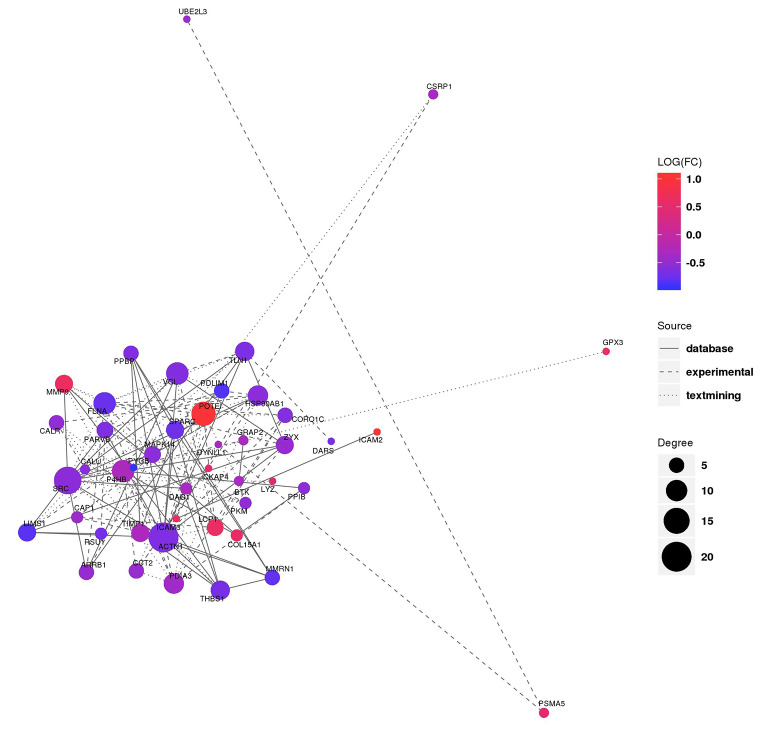
Analysis of PPI network construction of differentially expressed proteins
The analysis of Protein-Protein Interaction (PPI) Network is important for understanding protein function and relationship, and is also one of the important research contents of proteomics. The PPI network is formed to maintain the coordination of time and space when proteins perform biological functions. By constructing the interaction network of differentially expressed proteins, we can identify the change trend of differential expression proteins at the proteome level, and further help us identify the key nodes of differential expression proteins. In this project, protein interaction information for the PPI network was constructed using STRING online database (v10, string-db.org). The protein interaction relationship in STRING database was queried for differentially expressed proteins, and the network interaction map of differential expression proteins was constructed (Fig. 5). PPI Network Analysis showed that the maximum number of genes/proteins interacting with ACTN1 was 20 (p = 0.0258). However, our literature review found no reports of ACTN1 associated with diabetes, so there is no further study at present. Based on all the above analysis results, and by consulting the relevant references, we identified four proteins that may be involved in the pathogenesis of low plasma 25(OH)D levels in T2DM patients. They are PODXL(Podocalyxin), ICAM3(Intercellular adhesion molecule 3), MMP9(Matrix metalloproteinase 9) and ApoF (Apolipoprotein F).
Fig. 5.
Protein-Protein interaction network map of experiment group compared with control group. In the figure, the color indicates the level of differential expression protein, the red indicates a significant increase in the difference, and the blue indicates a significant decrease in the difference. The size of the circle indicates the degree of connectivity of differential expression proteins (The number of connections between the node and the outside world is called degree). The higher the degree of connectivity, the greater the circle. The type of connection represents the source of the interaction relationship, and the solid line ndicates that the interaction relationship comes from the database. The dashed and dotted line represent the interaction relationship from experiments and text mining, respectively
Verification of differentially expressed proteins
Participant demographic and clinical characteristics
Table 2 showed all patients and controls’ clinical information. And analysis also suggested that the variables of ALT, AST, TC, HDL-C, LDL-C, TG, OSTEOC and WBC count had no statistical difference, while variables of 25(OH)D, age, GLU and iPTH were significantly different between experimental group and control group (p < 0.05).
Table 2.
Demographic and clinical data of participants in verification tests
After enlarging the sample size, ELISA was used to verify that 4 differentially expressed proteins, MMP9 and ICAM3 had changed
The level of each protein in the serum is expressed by the concentration of the protein divided by the concentration of the total protein. Compared with the VitD deficiency group, the expression of MMP9 and ICAM3 protein in the VitD sufficiency group tended to increase, but the p value was not significant(p > 0.05). However, there was no statistically significant difference in the levels of ApoF and PODXL proteins between the two groups(Fig. 6).
Fig. 6.
ELISA assay to examine those 4 differential proteins between VitD sufficiency group and VitD deficiency group. (A-D) The levels of ApoF, PODXL, MMP9, and ICAM3 in plasma. Proteins expression was normalized to total protein
Discussion
Adipose tissue is the main storage site of vitamin D [22, 23], and adipose cells are the expression site of vitamin D receptor (VDR). Vitamin D is active in all levels of adipose cells and can regulate the expression of adipogenic genes. Therefore, vitamin D plays an important role in lipid metabolism. Vitamin D deficiency can cause significant lipid metabolism disorders, and experiments have confirmed that vitamin D deficiency is associated with decreased high density lipoprotein(HDL) concentration and increased low density lipoprotein (LDL) concentration [24, 25]. 1,25 (OH)2D3 can modify adipocytes through VDR, inhibit activated B cell (NF-κB) signal transduction and reduce the expression of pro-inflammatory cytokines in adipose tissue [26]. At the same time, vitamin D is associated with cholesterol transport and lipid deposition [27]. In addition, vitamin D can directly or indirectly regulate the expression of a variety of proteins related to the artery wall, inhibit angiogenesis, smooth muscle cell proliferation, and play an immunomodulatory role through VDR mediating immune cells to reduce the occurrence of atherosclerosis [28]. Studies have shown that 1,25 (OH)2D3 can regulate inflammation and immune response, and has a significant inhibitory effect on immunity, regulating the adhesion and migration of macrophages [29], inhibiting the proliferation and differentiation of T-assisted (Th) cells, and regulating the production of cytokines [30].
The proteins in the peripheral plasma of T2DM patients were screened by iTRAQ technology combined with LC-MS/MS(liquid chromatography-tandem mass spectrometry). The levels of the above four proteins were different between the high VitD group and the low VitD control group. However, the results were not the same when these proteins were examined in a larger number of samples and only the change trend of MMP9 and ICAM3 proteins between the two groups is the same as that of proteomics. So, we analyzed the possible reasons: (1) Selection bias– the selected samples were not exactly the same between screening tests and verification tests. (2) Different detection methods–iTRAQ combined with LC-MS/MS technology had higher sensitivity than ELISA technology. (3) Individual differences in patients–the larger the sample number, the higher the accuracy. Therefore, it will be necessary to increase sample sizes when undertaking further analyses of these proteins in the future.
In summary, 63 proteins were differentially screened by iTRAQ combined with LC-MS/MS technology. MMP9 and ICAM3 were confirmed as a differentially expressed protein between VitD deficiency and VitD sufficiency groups in a larger sample using ELISA technology. The two proteins may play a vital role in the pathogenesis of patients with T2DM have lower serum 25 (OH) D level but further studies to elucidate the regulatory mechanism of MMP9 and ICAM3 protein expression in peripheral blood are required. It was previously reported that ICAM3 is associated with a lower risk of T2DM [31]. One study indicated that VitD treatment attenuated particles-induced lung injury and promoted tissue repair by inhibiting of TGFβ1 signal pathway and upregulating of MMP9 expression [32]. Compared with healthy controls, MMP9 and hsa-miR-21-5p levels were down-regulated and up-regulated in T2DM and diabetic nephropathy(DN) patients, respectively, results from another study. [33]. And it may also help us to find out novel treatments of T2DM. Further study of the relationship between VitD and diabetes can not only improve the theoretical mechanism of VitD participation in the pathogenesis of diabetes mellitus, but also may be used to intervene in high-risk groups of diabetes mellitus and as an auxiliary means of clinical treatment of diabetes, which may have a positive impact on the prevention of diabetes mellitus and the improvement of living standards of patients.
We have previously published studies on the relationship between VitD levels and macrophage typing in T2DM. The objective of this study was to screen the differentially expressed proteins in the serum of T2DM patients with different VitD levels by proteomic methods, in order to explore the potential mechanism of vitamin D deficiency and the regulation of VitD on glucose in T2DM patients, and to explore the possible mechanism of VitD regulating the polarization typing of macrophages and thus affecting the progression of T2DM. The research in this paper is the continuation and deepening of the previous research.
Conclusion
With the increase of VitD level, MMP9 and ICAM3 have an increasing trend, they have the potential to play a significant role in causing VitD deficiency in patients with T2DM.The specific role of the two proteins in the process of VitD involved in T2DM also needs further exploration. We’re doing it, too. This study can help us to find some clues in the mechanism of VitD participating in the pathogenesis of diabetes.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We acknowledge Beijing Luhe Hospital, Capital Medical University for providing financial and technical support for this study. The authors also thank the patients involved in this study for their cooperation.
Author contributions
Lijie Zhang and Zongwei Wang contributed equally as first authors. Lijie Zhang involved in the conception and design, Zongwei Wang and Xiaobo Wang analysis and interpretation of the data; Lingling Wei and Baoyu Zhang were responsible for drafting of the paper, Longyan Yang made critical revision for intellectual content and the final approval of the version to be published. The population and clinical data were also collected by Longyan Yang. All authors agree to be accountable for all aspects of the work.
Data availability
Data to support the findings of this study are available upon request from the corresponding authors.
Declarations
Ethics approval and consent to participate
This project has been approved by the Institutional Ethics Committee, Beijing Luhe Hospital, China (Registration no: 2019LH-WZ-007). All procedures carried out in research involving human participants comply with the ethical standards of institutional and/or national research councils, as well as the 1964 Helsinki Declaration and subsequent amendments or similar ethical standards. Informed written consent was obtained from all individual participants involved in the study.
Conflict of interest
The authors declare that they have no conflicts of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Lijie Zhang and Zongwei Wang shared first authorship.
Contributor Information
Baoyu Zhang, Email: Zhangby@ccmu.edu.cn.
Longyan Yang, Email: lyyang15@ccmu.edu.cn.
References
- 1.Sun H, Saeedi P, Karuranga S, et al. IDF Diabetes Atlas: Global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045 [J]. Diabetes Res Clin Pract. 2022;183:109119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Md Isa Z, Amsah N, Ahmad N. The impact of vitamin D Deficiency and Insufficiency on the outcome of type 2 diabetes Mellitus patients: a systematic review [J]. Nutrients, 2023, 15(10). [DOI] [PMC free article] [PubMed]
- 3.Ehrampoush E, Mirzay Razzaz J, Arjmand H, et al. The association of vitamin D levels and insulin resistance [J]. Clin Nutr ESPEN. 2021;42:325–32. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y, Xue Y, Zhang D et al. Effect of vitamin D supplementation on Glycemic Control in Prediabetes: a Meta-analysis [J]. Nutrients, 2021, 13(12). [DOI] [PMC free article] [PubMed]
- 5.Niroomand M, Fotouhi A, Irannejad N, et al. Does high-dose vitamin D supplementation impact insulin resistance and risk of development of diabetes in patients with pre-diabetes? A double-blind randomized clinical trial [J]. Diabetes Res Clin Pract. 2019;148:1–9. [DOI] [PubMed] [Google Scholar]
- 6.LebofF MS, Bischoff-ferrari HA. The effects of Vitamin D Supplementation on Musculoskeletal Health: the VITAL and DO-Health trials [J]. J Gerontol Biol Sci Med Sci. 2023;78(Supplement1):73–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouillon R, Marcocci C, Carmeliet G, et al. Skeletal and extraskeletal actions of vitamin D: current evidence and outstanding questions [J]. Endocr Rev. 2019;40(4):1109–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sintov AC, Yarmolinsky L, Dahan A, et al. Pharmacological effects of vitamin D and its analogs: recent developments [J]. Drug Discov Today. 2014;19(11):1769–74. [DOI] [PubMed] [Google Scholar]
- 9.Dunlop TW, Vaisanen S, Frank C, et al. The human peroxisome proliferator-activated receptor delta gene is a primary target of 1alpha,25-dihydroxyvitamin D3 and its nuclear receptor [J]. J Mol Biol. 2005;349(2):248–60. [DOI] [PubMed] [Google Scholar]
- 10.Eisman JA, Bouillon R, Vitamin D. Direct effects of vitamin D metabolites on bone: lessons from genetically modified mice [J]. Bonekey Rep. 2014;3:499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christakos S, Dhawan P, Verstuyf A, et al. Vitamin D: metabolism, molecular mechanism of Action, and Pleiotropic effects [J]. Physiol Rev. 2016;96(1):365–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ganmaa D, Bromage S, Khudyakov P, et al. Influence of vitamin D supplementation on growth, body composition, and Pubertal Development among School-aged children in an Area with a high prevalence of vitamin D Deficiency: a randomized clinical trial [J]. JAMA Pediatr. 2023;177(1):32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szymczak-pajor I, Sliwinska A. Analysis of Association between Vitamin D Deficiency and insulin resistance [J]. Nutrients, 2019, 11(4). [DOI] [PMC free article] [PubMed]
- 14.Zhang Q, Chen W, Yun C, et al. The application value of serum 25(OH)D3, uric acid, triglyceride, and homeostasis model assessment of insulin resistance in male patients with hyperuricemia combined with hypogonadism [J]. BMC Endocr Disord. 2021;21(1):102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo J, Xiao Z, Xue X, et al. 25-Hydroxyvitamin D is closely related with the function of the pancreatic islet beta cells [J]. Pak J Med Sci. 2013;29(3):809–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Y, Zhang X, Bao M, et al. Effect of serum 25-hydroxyvitamin D3 on insulin resistance and beta-cell function in newly diagnosed type 2 diabetes patients [J]. J Diabetes Investig. 2016;7(2):226–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gysemans CA, Cardozo AK, Callewaert H, et al. 1,25-Dihydroxyvitamin D3 modulates expression of chemokines and cytokines in pancreatic islets: implications for prevention of diabetes in nonobese diabetic mice [J]. Endocrinology. 2005;146(4):1956–64. [DOI] [PubMed] [Google Scholar]
- 18.Cheng Q, Li YC, Boucher BJ, et al. A novel role for vitamin D: modulation of expression and function of the local renin-angiotensin system in mouse pancreatic islets [J]. Diabetologia. 2011;54(8):2077–81. [DOI] [PubMed] [Google Scholar]
- 19.Pittas AG, Jorde R, Kawahara T, et al. Vitamin D supplementation for Prevention of type 2 diabetes Mellitus: to D or not to D? [J]. J Clin Endocrinol Metab. 2020;105(12):3721–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rasouli N, Brodsky IG, Chatterjee R, et al. Effects of vitamin D supplementation on insulin sensitivity and secretion in prediabetes [J]. J Clin Endocrinol Metab. 2022;107(1):230–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Wang W, Ning G. Metabolic Management Center: an innovation project for the management of metabolic diseases and complications in China [J]. J Diabetes. 2019;11(1):11–3. [DOI] [PubMed] [Google Scholar]
- 22.Abbas MA. Physiological functions of Vitamin D in adipose tissue [J]. J Steroid Biochem Mol Biol, 2017, 165(Pt B): 369– 81. [DOI] [PubMed]
- 23.Bennour I, Haroun N, Sicard F, et al. Recent insights into vitamin D, adipocyte, and adipose tissue biology [J]. Obes Rev. 2022;23(8):e13453. [DOI] [PubMed] [Google Scholar]
- 24.Dziedzic EA, Smyk W, Sowinska I et al. Serum level of vitamin D is Associated with Severity of Coronary atherosclerosis in Postmenopausal women [J]. Biology (Basel), 2021, 10(11). [DOI] [PMC free article] [PubMed]
- 25.Faridi KF, Zhao D, Martin SS, et al. Serum vitamin D and change in lipid levels over 5 y: the atherosclerosis risk in communities study [J]. Nutrition. 2017;38:85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moreno-Santos I, Castellano-Castillo D, Lara MF et al. IGFBP-3 interacts with the Vitamin D Receptor in Insulin Signaling Associated with obesity in visceral adipose tissue [J]. Int J Mol Sci, 2017, 18(11). [DOI] [PMC free article] [PubMed]
- 27.Hamouda HA, Mansour SM, Elyamany MF. Vitamin D combined with Pioglitazone mitigates Type-2 diabetes-induced hepatic Injury through targeting inflammation, apoptosis, and oxidative stress [J]. Inflammation. 2022;45(1):156–71. [DOI] [PubMed] [Google Scholar]
- 28.Shen L, Ma C, Shuai B, et al. Effects of 1,25-dihydroxyvitamin D(3) on the local bone renin-angiotensin system in a murine model of glucocorticoid-induced osteoporosis [J]. Exp Ther Med. 2017;13(6):3297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nieuwland AJ, Kokje VB, Koning OH, et al. Activation of the vitamin D receptor selectively interferes with calcineurin-mediated inflammation: a clinical evaluation in the abdominal aortic aneurysm [J]. Lab Invest. 2016;96(7):784–90. [DOI] [PubMed] [Google Scholar]
- 30.Wang F, Zhou L, Zhu D, et al. A retrospective analysis of the Relationship between 25-OH-Vitamin D and Diabetic Foot Ulcer [J]. Diabetes Metab Syndr Obes. 2022;15:1347–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pletsch-Borba L, Watzinger C, Turzanski Fortner R et al. Biomarkers of Vascular Injury and Type 2 diabetes: a prospective study, systematic review and Meta-analysis [J]. J Clin Med, 2019, 8(12). [DOI] [PMC free article] [PubMed]
- 32.Tao S, Zhang H, Xue L, et al. Vitamin D protects against particles-caused lung injury through induction of autophagy in an Nrf2-dependent manner [J]. Environ Toxicol. 2019;34(5):594–609. [DOI] [PubMed] [Google Scholar]
- 33.Khokhar M, Roy D, Bajpai NK et al. Metformin mediates MicroRNA-21 regulated circulating matrix metalloproteinase-9 in diabetic nephropathy: an in-silico and clinical study [J]. Arch Physiol Biochem, 2021: 1–11. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data to support the findings of this study are available upon request from the corresponding authors.