Abstract
Protective antibodies to the important childhood pathogen Haemophilus influenzae type b (Hib) are directed against the capsular polysaccharide (HibCP). Most of the antibody is encoded by a well-defined set of (“canonical”) immunoglobulin genes, including the Vκ A2 gene, and expresses an idiotypic marker (HibId-1). In comparison to noncanonical antibodies, the canonical antibody is generally of higher avidity, shows higher levels of in vitro bactericidal activity, and is more protective in infant rats. Using site-directed mutagenesis, we here characterize canonical HibCP antibodies expressed as antigen-binding fragments (Fabs) in Escherichia coli, define amino acids involved in antigen binding and idiotype expression, and propose a three-dimensional structure for the variable domains. We found that canonical Fabs, unlike a noncanonical Fab, bound effectively to HibCP in the absence of somatic mutations. Nevertheless, pronounced mutation-based affinity maturation was demonstrated in vivo. An almost perfect correlation was found between unmutated gene segments that mediated binding in vitro and those encoding canonical HibCP antibodies in vivo. Thus, the Vκ A2a gene could be replaced by the A2c gene but not by the highly homologous sister gene, A18b, corresponding to the demonstrated usage of A2c but not of A18b in vivo. Similarly, only Jκ1 and Jκ3, which predominate in the response in vivo, were able to facilitate binding in vitro. These findings suggest that the restricted immunoglobulin gene usage in HibCP antibodies reflects strict structural demands ensuring relatively high affinity prior to somatic mutations—requirements met by only a limited spectrum of immunoglobulin gene combinations.
Haemophilus influenzae type b (Hib) is a serious human pathogen that causes invasive diseases such as meningitis and septicemia among unvaccinated children. Protective antibody responses are directed against the capsular polysaccharide (HibCP), which consists of repeated units of 3-β-d-ribose-(1-1)-ribitol-5-phosphate (11). HibCP is a relatively rigid, unbranched, linear molecule, and most, if not all, HibCP antibodies recognize repeated linear epitopes comprising approximately three adjacent repeat units (20, 23, 38). Antibodies to the ends of the polysaccharide have not been described.
Antibodies to HibCP are predominated by molecules (mostly immunoglobulin G [IgG]) carrying a kappa light chain encoded by the variable (V) region VκII A2 gene (Immunogenetics database [IMGT] nomenclature, IGKV 2D-29) rearranged to one of the joining (J) genes, Jκ1, Jκ2, or Jκ3 (47). The VJ genes are only slightly mutated and have extended third complementarity-determining regions (CDR) (10 amino acids, codons 89 to 97) with a characteristic arginine in the place of VJ recombination (codon 95A; nomenclature according to Kabat and colleagues [27]) (1, 3, 6, 31, 46). Two highly homologous alleles at the A2 locus, A2a and A2c, have been used. The corresponding heavy chain is encoded by one of the highly homologous heavy chain V genes, either 3-23 or VH26, rearranged either directly to JH6b1 or through DN1 to JH4b1, resulting in an extremely short CDR3 region (six amino acids, codons 95 to 102) with a conserved glycine-tyrosine-glycine motif (codons 95 to 97) (4, 22, 39). Antibodies with these characteristics are called “canonical” with respect to the HibCP antibody response as proposed by Pinchuk et al. (39), using the terminology for Ig gene combinations dominating certain antibody responses in mice.
The canonical light chain expresses an idiotope (HibId-1) recognized by the monoclonal antibody LuC9 (31). Judged by expression of this idiotope, the canonical antibody has been detected in 85% of postvaccination sera constituting on average 60% of the HibCP-specific IgG (31). In comparison to noncanonical antibodies, the canonical antibody is generally of higher avidity, shows higher levels of in vitro bactericidal activity, and is more protective in infant rats (30, 36). A structural analysis may therefore improve our understanding of natural and vaccination-induced resistance to Hib disease. Furthermore, the antibody response to HibCP may be a model of more general relevance for human antibody responses to antigens with a limited number of epitopes.
MATERIALS AND METHODS
Sources of Ig sequences for antigen-binding fragment (Fab)-encoding constructs.
A set of canonical heavy (clone ToPG438) and light (clone ToP218) chains was selected among published plasmid clones of reverse-transcribed and PCR-amplified Ig mRNA (6, 22). The mRNA was derived from purified HibCP-specific antibody-secreting cells (AbSC) present in the circulation of a healthy adult male (22 years of age) 9 days after vaccination with a single dose of a HibCP-tetanus toxoid (TT) conjugate (ActHib; Pasteur Mérieux Serum et Vaccines, Lyon, France). The A18b germ line sequence was obtained from a published plasmid clone (A18b clone 002) derived from PCR-amplified genomic DNA (25). The IGVH 3-23 germ line sequence was obtained from a plasmid clone (To2317) from PCR-amplified DNA, and the JH6b1 germ line sequence was obtained from the clone ToPG335 (22).
PCRs for the construction of Fab-expressing vectors.
All PCRs were performed in a final volume of 50 μl containing 1× PFU reaction buffer, 0.2 mM deoxynucleoside triphosphate, 0.078 U of Pfu polymerase (Stratagene, La Jolla, Calif.), and 0.55 U of Taq polymerase (Life Technologies, Paisley, United Kingdom) mixed with 0.55 U of Taq-Start antibody (Clontech Laboratories, Palo Alto, Calif.) and 5 pmol of gene-specific primer pairs. After an initial denaturation for 4 min at 94°C, 20 to 30 PCR cycles, consisting of 30 s at 94°C, 1 min at 55°C, 1.5 min at 72°C, and a final 10-min step at 72°C, were performed.
Cloning of Fab-encoding constructs.
The cloning procedures used for Fab-encoding constructs, described below briefly, were previously described in detail (22).
(i) Cloning of the VH domain.
One hundred nanograms of the plasmid ToPG438 was used as a template for a 20-cycle PCR amplification of the VH domain sequence. Gene-specific primers were placed in framework region 1 (FR1) and FR4 and contained an NheI or ApaI site (primer 3-23Fab3′, 5′- CTCGCGAATTGGGCCCTTGGTGGAGGCTGAGGAGACGGTGACCGT-3′; primer 3-23Fab5′, GGATTGTTATTGCTAGCAGCACAGCCAGCAATGGCAGAGGTGCAGCTGTTGGAG-3′ (the restriction sites are underlined). The PCR product was size purified, digested with NheI and ApaI (New England Biolabs, Beverly, Mass.), and cloned into the phage display expression vector pFab73HHui (13) which was modified to express soluble Fabs as described elsewhere (22). The vector already contained the human Cκ and an IgG1 CH1 domain with a His6 tail appended at the carboxy terminus. The resulting pFab3-23/Hui phagemid was cloned and purified. The VH domain sequence was verified by sequencing. Two micrograms of plasmid DNA was digested with SfiI and AscI (New England Biolabs).
(ii) pFab3-23/A2a.
A canonical Fab was produced by incorporating the canonical light chain from ToP218 into the pFab3-23/Hui phagemid. The light chain was amplified in three parts and subsequently assembled by PCR. An RsrII site was introduced in codons 98 to 100 without changing the amino acid sequence of the light chain. The following primers were used: for codons 1 to 6, primer A2Fab5′ (5′-GATCCTCGCGAATTGGCCCAGCCGGCCATGGCAGATATTGTGATGACCCAG-3′); for codons 104 to 96, primer Jk3FabV (5′-TTTGGTCCCCGGTCCGAAAGTGAA-3′); for codons 96 to 104, primer Jk3FabC (5′-ACTTTCGGACCGGGGACCAAAGTG-3′); for codons 122 to 117, primer CK117rc (5′-CATCAGATGGCGGGAAGAT-3′); for codons 117 to 122, primer CK117 (5′-ATCTTCCCGCCATCTGATG-3′); and for codons 214 to 209, primer HCK.FORW (5′-GTCTCCTTCTCGAGGCGCGCCTCACTAACACTCTCCCCTGTTGAAGCT-3′) (SfiI, RsrII, RsrII, and AscI restriction sites are underlined). Each PCR was performed with Pfu and Taq polymerases with anti-Taq antibody for 20 cycles as described above. The resulting full-length kappa light chain PCR product was size purified, digested with SfiI and AscI, and cloned into pFab3-23/Hui.
(iii) pFab3-23/A2c.
The other functional allele of the A2 gene, A2c, differs from A2a only by a mutation in codon 43 of the FR2 coding for a single amino acid change. This change was introduced by site-directed PCR mutagenesis with primers VkA2cc43 (5′-AAGCCAGGCCAGTCTCCACAGCTC-3′) and VkA2cc43rc (5′-GAGCTGTGGAGACTGGCCTGGCTT-3′) (the site of mutation is shown in boldface) in combination with HCK.FORW and A2Fab5′, respectively, with pFab3-23/A2a as the template. The novel light chain construct was then cloned into the pFab3-23/A2a phagemid, replacing the A2a-derived light chain.
(iv) pFab3-23/A18b.
The sister gene of A2 is A18, of which four functional alleles are known, namely, A18b, A18c, A18d, and A18e (25), all encoding proteins with identical amino acid sequences but differing from the A2a gene product in four amino acid positions. We combined an A18b germ line sequence (A18b 002) with the rearranged sequence of the ToP218 clone by using primer A18Fab3′ (5′-TTTGGTCCCCGGTCCGAAAGTGAATCGAGGAAGGTGTATACCTTG-3′) (RsrII site underlined), for codons 103 to 90, in combination with A2Fab5′ for the PCR. The PCR product was cloned into pFab3-23/A2a, replacing the A2a-derived light chain sequence but leaving the site of rearrangement, the arginine in codon 95A, and the Jκ3 sequence in situ.
(v) pFab3-23gl/A18bJk3.
A 3-23 germ line sequence (To2317) was combined with the rearranged sequence of ToPG335 (using JH6b1 in germ line configuration [22]) by using two primer pairs complementary to codons 1 to 6 and 95 to 89 (3-23Fab5′ and 3-23c89 [5′-TCTTTCGCACAGTAATAT-3′], respectively) and to codons 89 to 96 and 113 to 108 (3-23glC [5′-GTATATTACTGTGCGAAAGGGTAC-3′] and 3-23Fab3′, respectively). The PCR product was cloned into pFab3-23/A18b, replacing the VH domain.
(vi) pFab3-23gl/A2aJk3.
pFab3-23gl/A2aJk3 was made exactly as pFab3-23gl/A18bJk3 was, by replacing the VH domain of pFab3-23/A2a.
(vii) pFab3-23gl/A2aJk1, pFab3-23gl/A2aJk2, pFab3-23gl/A2aJk4, pFab3-23gl/A2aJk5, pFab3-23gl/A18bJk1, pFab3-23gl/A18bJk2, pFab3-23gl/A18bJk4, and pFab3-23gl/A18bJk5.
These Fabs were constructed to detect the influence of the Jκ chain on the affinity for HibCP. The Vκ domain was amplified for 20 PCR cycles by using pFab3-23/A2a or pFab3-23/A18b as the template with the primer A2Fab5′ and one of the following Jκ primers (codons 103 to 90): Jk1Vrc, 5′-CTTGGTCCCTTGGCCGAACTGCCATCG(AG)GGAAG-3′; Jk2Vrc, 5′-CTTGGTCCCCTGGCCAAAATGGTATCG(AG)GGAAG-3′; Jk4Vrc, 5′-CTTGGTCCCTCCGCCGAAAGTGAGTCG(AG)GGAAG-3′; and Jk5Vrc, 5′-TCGTGTCCCTTGGCCGAAGGTGATTCG(AG)GGAAG-3′. The Cκ domain was amplified for 20 PCR cycles by using pFab3-23/A2a as the template with the primer HCK.FORW and one of the following Jκ primers (codons 99 to 110): Jk1C, 5′-GGCCAAGGGACCAAGGTGGAAATCAAACGAACTGTG-3′; Jk2C, 5′-GGCCAGGGGACCAAGCTGGAGATCAAACGAACTGTG-3′; Jk4C, 5′-GGCGGAGGGACCAAGGTGGAGATCAAACGAACTGTG-3′; and Jk5C, 5′-GGCCAAGGGACACGACTGGAGATTAAACGAACTGTG-3′. The PCR products were size purified on a 2% agarose gel and further purified with the Qiaex II gel extraction kit (Qiagen, Hilden, Germany). For each of the four Jκ genes, 1/20 of the corresponding Vκ and Cκ PCR products was mixed and used as the template in an assembly PCR with the primer set A2Fab5′-HCK.FORW. Each PCR was performed with Pfu and Taq polymerases with anti-Taq for 20 cycles. The resulting full-length kappa light chain PCR products were size purified and digested with SfiI and AscI and cloned into pFab3-23gl/A2a, replacing the light chain.
(viii) pFab3-23/A3.
A hybrid Fab was constructed by combining the heavy chain of one HibCP-specific Fab (Fab3-23/A2a) with the light chain of another HibCP-specific Fab (Fab3-73/A3) (22). The construction of the A3 light chain is described in detail elsewhere (22).
(ix) Recombinants of A2 and A18.
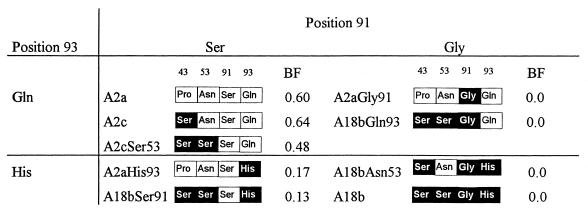
Six recombinants of A2 and A18 sequences were made by recombining the phagemid vectors pFab3-23/A2a, pFab3-23/A2c, and pFab3-23/A18b described above. The phagemids were digested with SacI, which cuts a single site upstream of the light chain leader, and by SphI or SnaI, which cuts the Vκ sequences in codons 88 and 92, respectively. After size purification, the V gene-containing small DNA fragments were exchanged between the clones and ligated with T4 DNA ligase (Boehringer Mannheim). For two of the recombinants, two sequential recombinatorial events were necessary. This procedure led to the construction of six phagemids: pFab3-23/A2cSer53, pFab3-23/A2aGly91, pFab3-23/A2aHis93, pFab3-23/A18bAsn53, pFab3-23/A18bSer91, and pFab3-23/A18bGln93.
DNA sequencing.
Plasmid DNA was purified by an alkaline lysis protocol (28a) and extracted with chloropane (Amresco, Solon, OH) before use as the template for sequencing. The dideoxy method of Sanger et al. (43) was used by means of the Ready Reaction kit (Perkin-Elmer Roche, Foster City, Calif.) and an ABI 373 automatic sequencer (Perkin-Elmer) as instructed by the manufacturer.
Production and purification of Fabs.
The production and purification of Fab fragments were done as described previously in detail (22). Briefly, the phagemid-infected TOP10/F′TetR cells were grown in 1 liter of LB medium (42) containing 50 mg of carbenicillin, 10 mg of tetracycline, and 20 mM MgCl2. Cultures were grown for 6 to 7 h at 37°C with shaking, induced with IPTG (isopropyl-β-d-thiogalactopyranoside; 1 mM) (Sigma, St. Louis, Mo.) and 2 mg of cyclic AMP (Sigma), and cultured overnight at 30°C with shaking. After harvesting, soluble Fabs were extracted from the periplasmic space and purified on a Ni-nitrilotriacetic acid superflow resin (Qiagen) in a Poly-Prep column (Bio-Rad, Hercules, Calif.). The column was washed with 20 ml of column washing buffer (300 mM NaCl, 50 mM sodium phosphate, 10% glycerol [pH 7.8]) containing 20 mM imidazole and then with 4 ml of washing buffer containing 50 mM imidazole. After washing, the Fabs were eluted with column washing buffer containing 250 mM imidazole. The buffer was changed to phosphate-buffered saline (PBS), and the Fabs were concentrated in a Centricon-30 centrifugal concentrator (Amicon, Beverly, Mass.). The Fab preparations were analyzed by unreduced sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by silver staining to ensure proper molecular weight and degree of purity. Concentrations were determined by an enzyme-linked immunosorbent assay (ELISA) with a highly purified Fab preparation as a reference (22).
ELISA. (i) Determination of Fab concentrations.
Each well of the ELISA plates (Costar, Cambridge, Mass.) was coated overnight at 4°C with 100 μl of a 10-μg/ml concentration of goat antibodies to F(ab)2 fragments of human IgG (Pierce, Rockford, Ill.). After four washings in PBS containing 0.05% Tween 20, the plates were blocked for 1 h at 37°C with 3% bovine serum albumin (BSA) in PBS. Then, 50-μl volumes of purified Fab at 20 ng/ml and twofold dilutions in PBS with 1% BSA were incubated in triplicate at 37°C for 1 h. As a concentration standard, a highly purified Fab preparation, described before (22), was used (20 ng/ml and twofold dilutions). After four washings, goat anti-human kappa L chain antibodies conjugated with alkaline phosphatase (AP) (Sigma), diluted 1/500 in PBS with 1% BSA, were added (50 μl/well). After 1 h at 37°C, the wells were washed and p-nitrophenyl phosphate in AP substrate buffer (MgCl2, 2.03 g/liter; Na2CO3, 8.4 g/liter; sodium azide, 1.0 g/liter [pH 9.8]) was added (50 μl/well). The optical density at 410 nm (OD410) was measured after ∼60 min at room temperature.
(ii) Evaluation of HibCP binding.
ELISA plates (Immulon 2; Dynatech, Chantilly, Va.) were coated overnight at room temperature with HibCP oligomer (100 μg/ml, 20 repeat units) coupled to human serum albumin (HibCP-HSA) (HbO-HA lot no. 15 D; Lederle-Praxis Biochemicals). After four washings in PBS containing 0.05% Tween 20, the plates were blocked with 3% BSA in PBS for 1 h at 37°C. Then, 50 μl of purified Fabs (20 μg/ml in PBS with 1% BSA and twofold dilutions of this concentration) were incubated at 37°C for 2 h (all in duplicate). The remaining ELISA procedures were performed as described above. In some experiments, binding was inhibited by an initial 1-h incubation of the Fabs (10 μg/ml) with different concentrations of soluble HibCP polymers (Connaught Laboratories Inc.) or with Escherichia coli K100CP (1 mg/ml; kindly supplied by Uffe Skov Sørensen, Statens Seruminstitut, Copenhagen, Denmark) at 37°C to demonstrate specificity.
(iii) Cross-reactivity with other polysaccharides.
ELISA plates (catalog no. 269620; Nunc, Roskilde, Denmark) were coated (100 μl/well) with one of six phenylated pneumococcal capsular polysaccharides (4-μg/ml concentrations of types PP1, PP4, PP6B, PP7F, PP14, or PP18C; all kindly supplied by Uffe Skov Sørensen) in PBS overnight at room temperature. After the plates were washed and blocked, 50 μl of purified Fabs (5 μg/ml) was incubated at 37°C for 2 h. The ELISA plates were then developed as described above. As a positive control, 1:100 and 1:1,000 dilutions of a serum pool (HSP1) made from 10 donors vaccinated with pneumococcal capsular polysaccharides or HibCP-conjugated with TT or diphtheria toxoid were used.
(iv) HibId-1 expression and cross-reactivity with TT.
ELISA plates (Maxisorp; Nunc) were coated overnight at 4°C with a murine monoclonal antibody defining the HibId-1 idiotype (LuC9; 100 μl/well, 10 μg/ml in PBS) or with TT (1 μg/ml in PBS). After the plates were washed and blocked, 50 μl of purified Fabs (10-μg/ml concentration and twofold dilutions of this) was incubated at 37°C for 1 h. After washings, a 1/500 dilution of biotinylated mouse anti-human kappa L chain antibodies (Zymed Laboratories, South San Francisco, Calif.) were added at 50 μl/well in PBS with 1% BSA and incubated for 1 h at 37°C. After further washings, a 1/500 dilution of streptavidin conjugated with AP (Kirkegaard & Perry Laboratories, Gaithersburg, Md.) was added in PBS with 1% BSA (50 μl/well). As a positive control, a preparation of HibId-positive HibCP antibody (0.7 ng/ml purified from a serum pool kindly supplied by Alexander Lucas) was used. Inhibition by soluble HibCP was performed by incubation of 5 μg of Fab per ml with 1 mg of HibCP per ml for 1 h at 37°C before testing in the HibId-1 ELISA.
(v) Measurement of relative Kd values.
ELISA plates (Immulon 2) were coated overnight at room temperature with 100 μl of HibCP-HSA per well (2 μg of HibCP oligosaccharide/ml). After washing and blocking, 100 μl of twofold dilutions of purified Fabs (initial concentrations, 2.5 μg of Fab3-23/A2a per ml, 10 μg of Fab3-23gl/A2aJk1 per ml, and 80 μg of Fab 3-23gl/A2aJk3 per ml, all in PBS with 1% BSA) were incubated in duplicate at 37°C for 24 h to measure binding at equilibrium. Other triplicate sets of wells were incubated with 100 μl of each Fab (2.5, 10, or 80 μg/ml, respectively), but after 21.5 h of incubation, the concentrations of free Fab in these wells were determined by transferring the 100 μl to new wells. At the same time, new dilution series were made starting with 2.5, 10 or 80 μg of Fab per ml, respectively, to serve as a reference. After the remaining 2.5 h of incubation, the plate was washed four times and incubated for 1 h at 37°C with 100 μl of goat anti-human kappa light chain antibodies conjugated with AP diluted 1/200 in PBS with 1% BSA and developed by using p-nitrophenyl phosphate in AP substrate buffer at 100 μl/well. The dissociation constant Kd is defined in the Law of Mass Action by the equation:
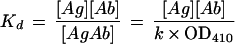 |
1 |
where [Ag] and [Ab] are the concentrations of unbound (free) antigen and antibody (in this situation Fab) at equilibrium, respectively. [Ab] was measured from the OD values after 2.5 h of incubation of the transferred supernatants in comparison with the standard curve obtained after 2.5 h of incubation. [AgAb] is the concentration of antigen-antibody complexes. Because care was taken to ensure that neither the amount of secondary antibody nor of the substrate were limiting factors in the ELISA, [AgAb] was proportional with OD410 (equation 1). This OD value was measured after 24 h of incubation with Fab but represented [AgAb] at the time of supernatant transfer (21.5 h) due to the state of equilibrium reached at this time point as confirmed by independent experiments (data not shown). The constant, k, was unknown, but equal for all Fabs binding to the same epitope on the HibCP oligosaccharide.
The ratio between the Kd values of two Fabs could then be determined by the following equation:
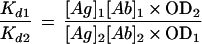 |
2 |
RESULTS
Construction of canonical and noncanonical HibCP-specific Fabs from a vaccinated individual.
Cloned cDNAs from HibCP-specific AbSC participating in the vaccine response of a 22-year-old healthy male volunteer were used for the construction of Fabs. Circulating AbSC were recovered 9 days after vaccination with a single dose of HibCP-TT. The purification of HibCP-specific cells (21), reverse transcription-PCR, cloning, and analysis of the utilized VL (kappa) and VH (IgA and IgG) genes have been described in detail elsewhere (6, 22). A total of 42 representative kappa light chain sequences (6) and 58 heavy chain sequences (41 IgA and 17 IgG) were analyzed (22).
The AbSC response was dominated by the clonal progeny of a single cell which used a noncanonical set of V genes (6, 22). This clone used a slightly mutated light chain encoded by VκII A3/A19 rearranged to Jκ3 and a somewhat more mutated heavy chain (IgA1 and IgA2) encoded by VHIII 3-73 rearranged to D3-22 (DXP3) and JH4b1. A representative set of light and heavy chains was expressed as a Fab which was named Fab3-73/A3 after the utilized heavy and light chain germ line V genes. This noncanonical Fab is used for comparison in the present work. Its construction and ability to bind to HibCP and to cross-react with E. coli K100CP have been described elsewhere (22).
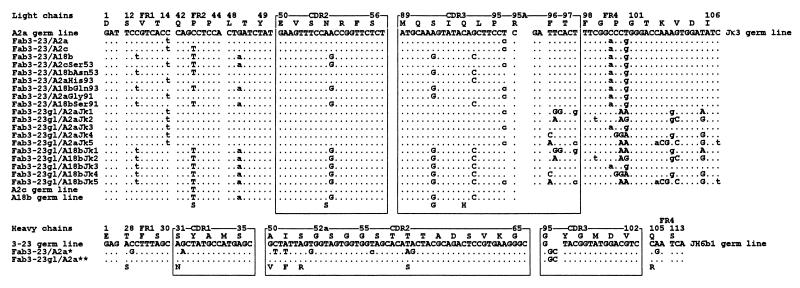
A minor part of the AbSC response of the volunteer involved the use of canonical genes. Thus, 3 of 42 kappa sequences used VκII A2 rearranged to Jκ3 with the characteristic arginine in position 95A. The clone ToP218 (6) was selected for the construction of a canonical Fab (Fab3-23/A2a) because it contained no amino acid-replacing mutations (Fig. 1). Twelve of the 58 heavy chain sequences (including 10 of 17 IgG sequences) used canonical rearrangements comprising VHIII 3-23 rearranged directly with JH6b1 (9 sequences) or through DN1 to JH4b1 (3 sequences), in both cases under the formation of the characteristic six-amino-acid CDR3 with a conserved non-germ-line-encoded glycine in position 95. The least mutated of the nine 3-23/JH6b1 sequences, that of ToPG438, was selected for Fab construction (Fab3-23/A2a) (Fig. 1).
FIG. 1.
Light and heavy chain V domain sequences of 19 Fabs analyzed in this paper. The canonical germ line genes VκII A2a, A2c, A18b, Jκ3, VHIII 3-23, and JH6b1 (accession no. M31952, U41644, U41645, J00242, M99660, and X86355) are shown for comparison. For codons not listed, all were identical with the germ line sequences through the entire VL and VH domains. Dots indicate nucleotide identity. Uppercase and lowercase letters indicate amino acid replacements and silent substitutions, respectively. Mutant amino acids are given below the sequences (except Jκ3). The canonical HibCP-specific fragment, Fab3-23/A2a, was encoded by light and heavy chain sequences derived from affinity-purified, circulating B cells obtained 9 days after immunization of a healthy adult male with a HibCP-TT conjugate. ∗, All the fragments with the prefix Fab3-23/ shared the same mutated heavy chain which characteristically lacked a D segment. The two last nucleotides of heavy chain codon 95 were probably N additions. This heavy chain sequence is available from EMBL/GenBank/DDBJ under accession no. Z98723. ∗∗, The Fabs with the prefix Fab3-23gl used the heavy chain gene in germ line configuration.
Binding of HibCP by the canonical Fab.
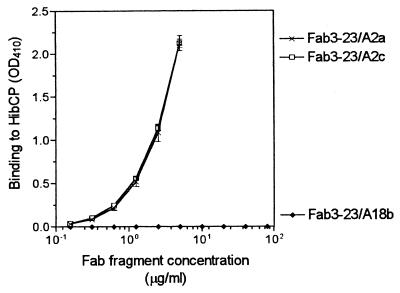
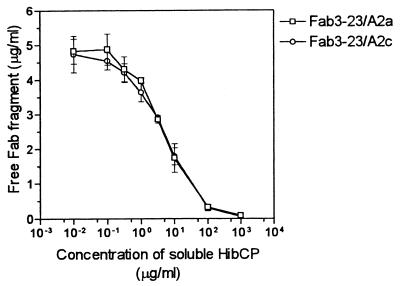
As illustrated in Fig. 2, the canonical Fab, Fab3-23/A2a, bound immobilized HibCP oligomer in a dose-dependent manner. The binding could be completely inhibited by preincubation with high-molecular-weight HibCP in solution showing reactivity with the native molecule (Fig. 3). The Fab was not polyreactive as no cross-reactivity to any of six tested pneumococcal polysaccharides or to TT was detected (Table 1). The fine specificity was tested by replacing HibCP with soluble capsular polysaccharide from the E. coli strain K100 (K100CP) in an inhibition experiment. This isomeric polysaccharide could not inhibit the binding to HibCP oligomer even at a concentration of 1 mg/ml (Table 1). In contrast, this concentration of K100CP almost totally blocked the binding of the noncanonical Fab, Fab3-73/A3, to HibCP.
FIG. 2.
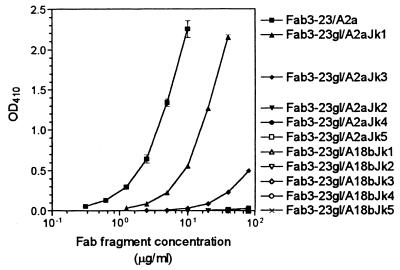
Effect of highly homologous light chain V gene replacements on the binding of canonically rearranged Fabs to HibCP oligosaccharides immobilized on a solid phase (ELISA technique) at 37°C. The Fabs had identical heavy chains but different light chains as indicated in Fig. 1. Data are given as mean (± standard deviation) of three independent measurements (two for nonbinders). Nonbinders were tested up to 80 μg/ml. Net OD410s are given.
FIG. 3.
Inhibition of binding of Fabs to solid-phase immobilized HibCP oligosaccharides by various concentrations of soluble high-molecular-weight HibCP. A fixed concentration of Fabs (5 μg/ml) was used. After preincubation of Fabs with soluble HibCP for 1 h at 37°C, the amount of free Fab was determined by ELISA using an uninhibited sample of the same Fab as reference. Data are given as described in the legend to Fig. 2.
TABLE 1.
Specificity of purified Fabs determined by ELISA
| Fab | Net OD410 after binding to different solid-phase antigensa
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| HibCP | HibCP plus free K100CPc | PP1 | PP4 | P6B | PP7F | PP14 | PP18C | TT | |
| Fab3-23/A2a | ++++ | ++++ | − | − | − | − | − | − | − |
| Fab3-23/A2c | ++++ | ++++ | − | − | − | − | − | − | − |
| Fab3-23/A2cSer53 | ++++ | ++++ | − | − | − | − | − | − | − |
| Fab3-23/A2aGly91 | − | − | − | − | − | − | − | − | |
| Fab3-23/A2aHis93 | ++ | ||||||||
| Fab3-23/A18b | − | − | − | − | − | − | − | − | |
| Fab3-23/A18bAsn53 | − | − | − | − | − | − | − | − | |
| Fab3-23/A18bSer91 | + | − | − | − | − | − | − | − | |
| Fab3-23/A18bGln93 | − | ||||||||
| Fab3-23gl/A2aJk1 | + | − | − | − | − | − | − | − | |
| Fab3-23gl/A2aJk2 | − | − | − | − | − | − | − | − | |
| Fab3-23gl/A2aJk3 | −d | − | − | − | − | − | − | − | |
| Fab3-23gl/A2aJk4 | − | − | − | − | − | − | − | − | |
| Fab3-23gl/A2aJk5 | − | − | − | − | − | − | − | − | |
| Fab3-23gl/A18bJk1 | − | − | − | − | − | − | − | − | |
| Fab3-23gl/A18bJk2 | − | − | − | − | − | − | − | − | |
| Fab3-23gl/A18bJk3 | − | − | − | − | − | − | − | − | |
| Fab3-23gl/A18bJk4 | − | − | − | − | − | − | − | − | |
| Fab3-23gl/A18bJk5 | − | − | − | − | − | − | − | − | |
| Fab3-23/A3 | − | ||||||||
| Fab3-73/A3 | ++++ | − | − | − | − | − | − | − | − |
| HSP1 (dilution 1:1,000)b | ++++ | +++ | ++++ | ++++ | ++++ | ++++ | ++++ | +++ | |
Results are given as net OD410 after 1 h using a Fab concentration of 5 μg/ml. ++++, OD > 2.0; +++, OD = 1.0 to 2.0; ++, OD = 0.5 to 1.0; +, OD = 0.1 to 0.5; −, OD < 0.1. The mean background OD value in the HibCP-specific ELISA was 0.036 (range, 0.025 to 0.053).
HSP1 is a pool of immune sera from individuals immunized with HibCP, pneumococcal polysaccharides, and TT.
OD value in HibCP ELISA after preincubation of the Fabs with 1 mg of soluble E. coli K100CP per ml.
Binding was demonstrated with this Fab at higher concentrations as shown in Fig. 6.
The A2c allele may replace A2a in the sequences coding for the canonical Fab.
The product of the other functional allele of the A2 germ line gene, A2c, differs from that of A2a only in one amino acid position (serine instead of proline at position 43). To test the possible consequences of this allotypic variation for canonical HibCP antibodies, a Fab that deviated from Fab3-23/A2a by only a serine at position 43 of the light chain (Fab3-23/A2c) was made by site-directed mutagenesis (Fig. 1). As shown in Fig. 2 and 3, Fab3-23/A2c showed binding to HibCP oligomers and inhibition by native HibCP that were indistinguishable from those of Fab3-23/A2a. Again, polyreactivity and cross-reactivity with K100CP were absent (Table 1).
The functional A18 alleles cannot replace A2 in the sequences coding for the canonical Fab.
To detect whether the highly homologous germ line gene, A18b, could replace A2a in canonical HibCP-specific antibodies, the A2a gene-encoded part of Fab3-23/A2a (codons 1 to 95) was replaced with the A18b germ line gene product, resulting in Fab3-23/A18b (Fig. 1). The characteristic arginine encoded by codon 95A and the Jκ3 gene were conserved from the canonical Fab. As illustrated in Fig. 2, Fab3-23/A18b did not show any binding to the HibCP oligomer, not even when tested at a concentration of 80 μg/ml, which was at least 100 times higher than the concentration needed for detectable binding of Fab3-23/A2a and Fab3-23/A2c (Fig. 2). Neither was any binding to pneumococcal polysaccharides or TT detected (Table 1).
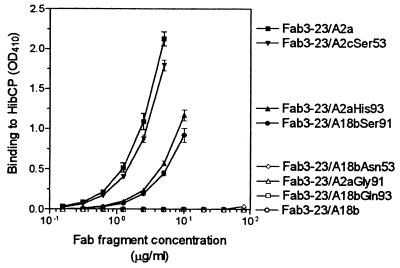
Analysis of the effect of individual amino acid differences on HibCP binding.
Because the serine at position 43 of the A18b gene-encoded light chain was also present in the HibCP binding Fab3-23/A2c, this amino acid could not be responsible for the lack of HibCP binding by Fab3-23/A18b. Thus, one or more of the three remaining amino acid differences (serine 53, glycine 91, and histidine 93) had to be responsible for the lack of HibCP binding. To identify which of these were involved, six Fabs expressing hybrid A2/A18b light chains were constructed (Fig. 1) and tested for binding to HibCP. As illustrated in Fig. 4 and 5, the A18b gene-encoded serine 53 (CDR2) had only a slightly negative effect on HibCP binding, while both CDR3 positions 91 and 93 turned out to be more important. Thus, the introduction of histidine at position 93 (Fab3-23/A2aHis93) (Fig. 4) reduced binding to a level where threefold more Fab was necessary to obtain the same level of binding to HibCP as that of the canonical Fab (Fab3-23/A2a). Substitution at the same position in the A18b gene-encoded Fab, Fab3-23/A18b, with the A2a gene-encoded glutamine could not, however, confer the ability to bind HibCP (Fab3-23/A18bGln93) (Fig. 4). Therefore, a crucial importance of residue 91 was evident, and indeed substitution of the A2a gene-encoded glycine in that position by the A18-encoded serine completely abrogated binding, while the opposite substitution restored binding in all Fabs (Fig. 4 and 5). The failure of A18b to induce HibCP binding was therefore largely due to the presence of serine rather than glycine at position 91.
FIG. 4.
Effect of specific amino acid substitutions on the binding of Fabs to HibCP oligosaccharides immobilized on a solid phase (ELISA technique) at 37°C. All Fabs carried the same mutated heavy chain but differed at specific amino acid positions of the light chain (Fig. 1). Data are given as described in the legend to Fig. 2.
FIG. 5.
HibCP binding efficacies of Fabs using the same heavy chain but light chains with different recombinations of A2a and A18b gene-encoded amino acids arranged after the most important residues are shown (data from Fig. 2 and 4). A2a gene-encoded amino acid residues are indicated by open boxes, while A18b gene-encoded residues are indicated by solid boxes. The figure illustrates that HibCP binding is largely determined by the CDR3 positions 91 and 93. Fabs with histidine at position 93 demonstrate reduced binding to HibCP, while fragments with a glycine at position 91 are totally unable to bind HibCP irrespective of the nature of residue 93. BF, binding factor. Binding factor is defined as the reciprocal of the concentration of Fab (micrograms per milliliter) resulting in an OD410 signal of 0.75 after 1 h.
Affinity maturation by mutation of the canonical heavy chain.
To determine whether somatic hypermutations influenced the affinity for HibCP, a Fab expressing the deduced amino acid sequence of the involved immunoglobulin genes prior to mutation was produced. Because the light chain did not contain any replacement mutations compared with the published germ line gene products, only the heavy chain needed to be modified. The published germ line sequences for VHIII 3-23 and JH6b1 (33, 48) were used, and the conserved glycine residue at position 95 was preserved at the VH-JH junction.
As evident in Fig. 6, the Fab carrying the unmutated canonical heavy chain together with the unmutated canonical light chain (Fab3-23gl/A2aJk3) indeed bound to HibCP, but with a lower affinity than that of the Fab with a mutated heavy chain (Fab3-23/A2a). The ratio between the Kd values of Fab3-23gl/A2aJk3 (Kd1) and Fab3-23/A2a (Kd2), which differ by only seven amino acids in the heavy chain (Fig. 1), could be measured by using the Law of Mass Action (see Materials and Methods, equation 2):
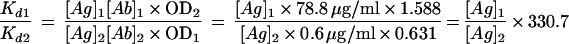 |
Because binding to the solid-phase oligosaccharides was not saturable at the available Fab concentrations, the exact values for free antigen, [Ag]1 and [Ag]2, were not determined. However, a rather narrow interval for the ratio [Ag]1/[Ag]2 could be estimated because doubling the total concentration of Fab3-23/A2a led to approximately a doubling of the OD value, showing that less than half of the antigenic epitopes were occupied at a free Fab concentration of 0.6 μg/ml (data not shown). Therefore, the ratio [Ag]1/[Ag]2 had to lie between 1 (corresponding to virtually all antigen unbound) and [1 − 0.5 × (0.631/1.588)]/[1 − 0.5] = 1.60 (half saturation at [Ab]2). An estimate for the ratio of dissociation constants of the two Fabs was therefore 331 < Kd1/Kd2 ≤ 530. It could therefore be concluded that whereas the unmutated Fab (representing the putative virgin B cell that had given rise to the mutated progeny) clearly showed detectable binding to HibCP, a considerable increase in affinity had occurred in vivo in the canonical HibCP antibody.
FIG. 6.
Effect of different Jκ chains on the binding of Fabs to HibCP oligosaccharides immobilized on a solid phase (ELISA technique) at 37°C. All Fabs except for Fab3-23/A2a (used as a reference) carried the same heavy chain in germ line configuration but differed at specific amino acid positions of the Jκ chain (Fig. 1). Data are given as described in the legend to Fig. 2.
Influence of the Jκ gene on HibCP binding of unmutated Fabs.
Using the unmutated heavy chain construct fixed, the unmutated light chain was modified by site-directed mutagenesis to encode the amino acids of the known Jκ germ line genes (33). The A2 gene-encoded part of the light chain and the extra arginine in position 95A were left unchanged (Fig. 1). Figure 6 shows that the choice of the Jκ chain had considerable impact on the binding of the unmutated antibody to HibCP. Thus, binding was detectable only when Jκ3 was replaced by Jκ1, not by Jκ2, Jκ4, or Jκ5. Interestingly, Jκ1 was considerably more effective than Jκ3 in mediating binding. By using measurements of free and bound Fab concentrations at equilibrium, the ratio between the Kd values of Fab3-23gl/A2aJk3 (Kd1) and Fab3-23gl/A2aJk1 (Kd2) was estimated as described above. It was found that 40.4 < Kd1/Kd2 ≤ 61.0. This shows that a virgin B cell using the Jκ1 gene in combination with the other canonical gene segments has approximately 50-times-higher affinity of the antigen receptor for HibCP than one using the Jκ3 gene. Other Jκ genes yield much lower affinities, if any. It is notable that the Fab using Jκ1 had an affinity only eight times lower than that of the highly affinity-maturated antibody represented by Fab3-23/A2a.
The failure of A18b was not due to mutations of the heavy chain.
While the mutated heavy chain in vivo was selected together with an A2 gene-encoded light chain, it was possible that some of the seven heavy chain mutations could be responsible for the failure of A18b to replace A2 in the canonical Fab. To exclude this, we constructed Fab3-23gl/A18bJk3 by combining the germ line version of the canonical heavy chain with the A18b-substituted canonical light chain. Also the possibility that A18b might be able to participate in the formation of HibCP antibodies in combination with other Jκ genes was studied. Figure 6 shows that the A18b gene-encoded Fabs did not bind to HibCP in concentrations as high as 80 μg/ml irrespective of which Jκ gene was used. Because the recently described c, d, and e alleles of the A18 gene all translate into the same amino acid sequence as that of A18b (25), none of the known functional A18 alleles are likely to replace A2a or A2c in the canonical HibCP antibodies in vivo due to very low (if any) affinity of the unmutated B-cell receptor.
Mapping of the HibId-1 idiotope.
The HibId-1 expression of the Fabs was evaluated by ELISA (Fig. 7). The canonical Fab3-23/A2a bound to the solid-phase immobilized anti-idiotypic antibody, LuC9, in a dose-dependent manner, whereas no binding was found for the noncanonical Fab, Fab3-73/A3 (Fig. 7a). Lucas et al. (31) have shown that LuC9 does not react with the canonical heavy chain in Western blots. However, to exclude binding to a native version of the heavy chain, we tested the ability of a Fab combining the canonical heavy chain with a noncanonical light chain, Fab3-23/A3, to bind to LuC9. Figure 7b shows that this Fab did not bind LuC9, indicating that the A2 gene-encoded light chain alone contains most if not all of the HibId-1 idiotope. In agreement with that, no effect on the affinity for LuC9 was seen when the heavy chain was changed into the mutated (seven amino acid positions) version (Fig. 7a, compare Fab3-23/A2a and Fab3-23gl/A2aJk3 results).
FIG. 7.
Binding of Fabs to solid-phase bound LuC9 monoclonal antibody determined in an ELISA. LuC9 defines the HibId-1 idiotope expressed on canonical HibCP antibodies. (a) All Fabs except for Fab3-23/A2a and Fab3-73/A3 (used as references) carried the same canonical heavy chain in germ line configuration but differed at specific amino acid positions of the Jκ chain (Fig. 1). (b) All Fabs carried the same mutated heavy chain but differed at specific amino acid positions of the light chain (Fig. 1). Data are given as described in the legend to Fig. 2.
A certain level of binding to LuC9 was detected for all Fabs carrying an A2a or A18b gene-encoded light chain in all Jκ combinations (Fig. 7a). Gross differences in binding efficacy were, however, evident. In general, Fabs employing A18b-derived light chains were relatively poor binders. The best binding was to Fab3-23gl/A2aJk1, which also was the best HibCP binder among the unmutated Fabs. The two HibCP nonbinders, Fab3-23gl/A2aJk4 and Fab3-23gl/A2aJk5, also bound relatively poorly to LuC9. There was, however, no absolute correlation between the ability to bind HibCP and the expression of the HibId-1 epitope. This was evident from the fact that Fab3-23gl/A2aJk2 bound effectively to LuC9 but not to HibCP.
The different structural requirements of the HibCP antibody paratope and the HibId-1 idiotope were even more evident when the effects of single amino acid residues were studied. To this end, Fab3-23/A2c and the six A2/A18b hybrid Fabs were analyzed for LuC9 binding (Fig. 7b). The Fab with the A2c gene-encoded light chain, Fab3-23/A2c, showed binding identical to that of Fab3-23/A2a, demonstrating that the change from proline to serine in FR2 position 43 did not influence the HibId-1 expression. In contrast, changing asparagine 53 in CDR2 to serine reduced the ability to bind LuC9 (Fig. 7b, Fab3-23/A2cSer53), and the introduction of asparagine in the A18-derived Fab (Fab3-23/A18bAsn53) improved binding of that Fab considerably. Also, the light chain position 91 was important for binding to LuC9, though not as crucial as it was for HibCP binding. Thus, exchange of the A2a gene-encoded serine for the A18b gene-encoded glycine reduced the ability to bind LuC9 significantly but did not abrogate binding as was the case for HibCP binding.
Finally, position 93 turned out to be involved in LuC9 binding, too, but somewhat surprisingly, introduction of the A18b gene-encoded histidine residue in the otherwise A2a gene-encoded Fab increased LuC9 binding (Fig. 7b, Fab3-23/A2aHis93), while the same substitution reduced the binding affinity for HibCP (Fig. 4). In agreement with the positive effect of histidine 93 on LuC9 binding, the replacement of that residue by glutamine in Fab3-23/A18b reduced binding further to a level almost undetectable in the ELISA (Fig. 7b, Fab3-23/A18bGln93). It may be concluded that the HibId-1 idiotope overlaps considerably with the light chain part of the antibody paratope comprising at least some parts of the CDR2- and CDR3-encoded areas. In contrast, no major contribution of the heavy chain is likely.
Modeling of the unmutated, major canonical HibCP antibody.
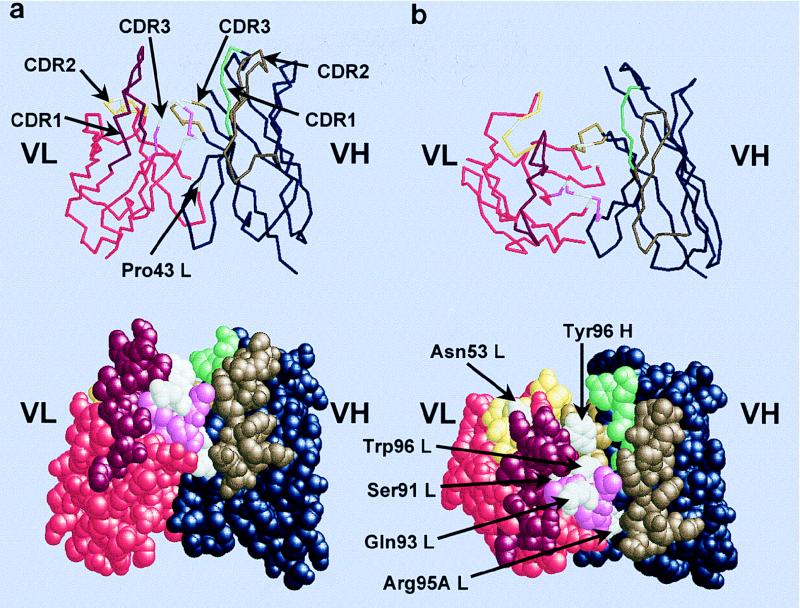
A model for the VH and VL domains of Fab3-23gl/A2aJk1 is shown in Fig. 8. As demonstrated above, this Fab has a relatively high affinity for HibCP in the absence of mutations and strongly expresses HibId-1. Furthermore, Jκ1 is the J gene used most often by canonical HibCP antibodies in vivo. This Fab may therefore be considered the prototypic canonical HibCP antibody. The three-dimensional structure of Fab3-23gl/A2aJk1 was predicted by using the ABGEN software package (32). ABGEN finds the optimal candidate scaffolding structures based on residue numbers and sequence homology from a database of known crystallized antibody structures. The final model is generated by using molecular mechanics algorithms of energy minimization. Figure 8 shows the resulting model. It appears from Figure 8a that the short heavy chain CDR3 together with the light chain CDR3 forms the floor of a groove flanked by the CDR1 and CDR2 of both chains on each side. This groove is likely to accommodate the linear polysaccharide epitope because it contains the light chain amino acids found to be most important for binding (Gln 93 and Gly 91) while the less important residue Asn53 is located in the VL CDR2 loop flanking the groove (Fig. 8b). Furthermore, the groove contains the conserved light chain residue Arg 95A. Centrally, the tryptophan 96 of the light chain is evident. It is the only Jκ gene-encoded amino acid of the light chain CDR3 which differs between the five Jκ genes and that position could be important for the different efficacies of the five Jκ genes. In close proximity to the Trp 96 of the light chain, the Tyr 96 of the heavy chain is indicated. It constitutes the apex of the heavy chain CDR3 loop and resides in the middle of the conserved Gly-Tyr-Gly motif. The light chain amino acid position 43, which differs between A2a and A2c gene-encoded antibodies and was found to be without any detectable influence on HibCP affinity or HibId-1 expression in this study, is exposed on the surface of the Fv fragment but far from the putative paratope and HibId-1 idiotope.
FIG. 8.
Model of the prototypic unmutated canonical HibCP antibody (Fv fragment). (a) Side view of the Fv fragment as a backbone and spacefill model. The CDRs form a groove in the antigen binding site because of the very short VH CDR3 and the long Vκ CDR1. The long linear HibCP molecule is expected to be placed in the groove. (b) Top view of the Fv fragment displaying the putative paratope for HibCP as well as the HibId-1 idiotope. The three-dimensional structure was predicted by using the ABGEN algorithm on the VH and VL amino acid sequences of Fab3-23gl/A2aJk1 (Fig. 1). Amino acids discussed in the text are indicated in white. The remaining residues in the VL sequence are indicated by the following colors: red, FRs; bordeaux, CDR1; yellow, CDR2; pink, CDR3. The remaining residues in the VH sequence are indicated by the following colors: blue, FRs; green, CDR1; brown, CDR2; curry, CDR3. Atomic coordinates are available from the Protein Data Bank (the Research Collaboratory for Structure Bioinformatics [RCSB]) under the entry 1HOU.
DISCUSSION
In this study, we constructed a Fab, Fab3-23/A2a, showing characteristics of the dominating HibCP-specific antibodies in humans, i.e., it is HibId-1 positive and it binds to HibCP but not to K100CP. The only difference between HibCP and K100CP is a 1—2 glycoside bond (K100CP) instead of a 1—1 glycoside bond (HibCP) between the ribose and the ribitol phosphate units of the polysaccharides. Cross-reactivity with K100CP is the rule for noncanonical HibCP antibodies but has never been detected for canonical HibCP antibodies. The heavy and light chains were derived from antigen-purified B cells participating in the antibody response of an HibCP-vaccinated adult and were probably closely related to or identical to the configuration in an original HibCP-specific B cell from that individual. The VL region carried a canonical gene combination (A2a, Jκ3) without replacement mutations and possessing the characteristic 10-amino-acid CDR3 with the conserved, non-germ-line-encoded arginine at position 95A (1, 3, 31, 46, 47). No or only very few amino acid-replacing mutations have been seen in HibCP-specific hybridomas using this light chain (1, 3, 39). The VH region was highly homologous to the VH regions of four of five published HibCP-specific heterohybridomas known to use the canonical light chain (ANN2, D3F3, and Gar6E8) (4, 39) or to be HibId-1 positive (ED8.4) (4). All of these hybridomas use the 3-23 germ line gene or the VH26 gene (probably an allele of the 3-23 germ line gene which only differs by a single G-to-A change in CDR1 codon 35 encoding a serine-to-asparagine mutation), and all have a heavy chain CDR3 region (codons 95 to 102) of only six amino acids which cover the site of V-(D)-J rearrangement. Similar to three of the four hybridomas, the VH gene of Fab3-23/A2a had rearranged directly to JH6 under the formation of a glycine-encoding codon 95. This glycine is probably essential for HibCP binding because all canonical heavy chains sequenced to date have glycine in that position despite the fact that only the first nucleotide of this codon (G) is germ line encoded (from the VH gene) while the last two are likely to have arisen by N addition. In agreement with selection for glycine, the second nucleotide is always G while all four nucleotides have been detected in the third (wobble) position (22, 28).
Like all canonical VH regions sequenced to date, the VH domain of Fab3-23/A2a contained a number of somatic mutations. Of nine mutations, seven involved amino acid replacements and all but two were placed in the CDRs (Fig. 1). Analysis of the VH regions of seven HibCP-specific heterohybridomas employing the canonical VH gene combinations revealed an average of 17 replacement mutations (range, 7 to 28) per VH region (4, 39). A similar number of mutations was seen among the sequences from individual To, but the least mutated sequence was chosen for Fab construction. No single substitution was conserved among all sequences, suggesting that no particular mutation was especially important for HibCP binding. It is therefore concluded that the Fab3-23/A2a is representative of the major canonical HibCP antibody involving the JH6b1 germ line gene.
It has been suggested that HibCP antibodies are encoded by Ig genes which are close to optimal for binding in the germ line configuration and that somatic mutations therefore contribute little to affinity (1, 2, 24). This is known from the murine antiphosphocholine antibody response where germ line-encoded T15 antibodies predominate in the primary antibody response and protect the animals, while mutations of these antibodies in secondary responses result in decreased binding (10). This notion has been supported by sequence analyses of the light chain of purified HibCP-specific antibodies and hybridomas which have shown a relatively low number of somatic mutations and lower ratios of replacement to silent mutations in the CDR relative to those of average memory B cells in peripheral blood (1, 8, 24). However, all heavy chains have been mutated, and in this report we clearly demonstrate that these mutations may increase the affinity of a canonical HibCP antibody dramatically (>300-fold). We have recently found similar mutation-based affinity maturation in a noncanonical HibCP antibody (22), suggesting that it is a general phenomenon in the HibCP response. This does, however, not exclude that the canonical A2 gene-encoded light chain could be close to optimal for binding already in the germ line version and therefore an inefficient target for affinity-increasing mutations.
Recently, it has become clear that the human kappa locus is rather polymorphic (44). Thus, an apparently functional allele (A2c) of the A2 gene and a minor defect allele, A2b, have recently been described in Native American Navajos. This opens up the possibility that the ability to form canonical HibCP antibodies may differ between individuals due to genetic makeup. The apparently functional allele, A2c, differs from the wild-type allele, A2a, only by a mutation in codon 43 of the FR2 resulting in a single amino acid replacement (46). In Navajos, the A2c gene frequency is approximately 27%. The prevalence of this gene in other populations is unknown. The A2c allele has been detected in canonical HibCP antibodies on two occasions only (3, 41). The demonstration in this report that canonical Fabs involving the A2a and A2c gene products have very similar HibCP and LuC9 binding abilities strongly suggests that they have almost identical paratopes and are equally effective as light chain in the canonical HibCP antibody. This is in agreement with the fact that amino acid residue 43 in most crystallized antibodies is located far from the paratope (27). In the predicted model for the prototypic canonical HibCP antibody shown in Fig. 8, position 43 is located on the Fv surface opposite the paratope (Fig. 8a). Because the regulatory elements of transcription and rearrangement are identical between A2a and A2c to the extent they have been sequenced (5), we predict that the relative utilization of A2a and A2c in canonical HibCP antibodies in the population simply reflects the gene frequencies of these alleles.
The canonical light chain gene, A2, is localized in the kappa locus on chromosome 2. Besides one Cκ gene and five Jκ genes, the 1,800-kb, large locus usually contains 76 V genes, of which 40 are placed in a Jκ-proximal region and 36 are placed in a Jκ-distal region separated from the proximal by approximately 800 kb. The distal group of V genes has arisen by gene duplication after the speciation of humans approximately 5 million years ago (14), and most of the distal genes therefore have a highly homologous “sister” gene located in the proximal group. The A2 gene is located in the distal group, and its sister gene in the proximal group is A18. For unknown reasons, the genes of the distal kappa gene group are rarely used in human antibody responses. Thus, of 55 sequenced kappa mRNAs from peripheral blood mononuclear cells, only a single sequence was derived from a gene of the distal group (26). With this in mind, it is remarkable that the distal A2 gene is able to dominate the antibody response to HibCP. The ability to invoke the A2 gene is more or less restricted to HibCP since the international sequence databases have registered only one antibody of known specificity other than HibCP which involves the A2 gene (50). This suggests that the structure of the A2 gene-encoded light chain satisfies unique requirements of the HibCP antigen which are not easily met by other V genes. In fact, the highly homologous sister V gene in the proximal group, A18, has never been found in canonical HibCP antibodies, although the products of the functional alleles of this deviate by only four amino acids from the A2a gene product and by three amino acids from the A2c gene product. This may, however, be due to the fact that most individuals studied have been Caucasians—a population in which most individuals are homozygous for the nonfunctional A18a (IMGT IGKV 2-29) allele carrying a stop codon in position 88 (25). Very recently, however, several potentially functional A18 alleles (A18b, -c, -d, and -e; all encoding the same amino acid sequence) have been described and found to be common in other populations. Thus, the functional A18b allele is present in 15% of Caucasians, 77% of Eskimos (25), and 54% of Native American Navajos (5), while 61% of black Africans (Mozambicans) carry the A18b, -c, -d, or -e alleles (25).
If functional A18 alleles can replace the A2 gene in the sequence coding for canonical antibody to HibCP, their presence might affect the natural immunity and vaccination responses to Hib in populations carrying the functional alleles—not least in populations with high frequencies of the deficient A2b allele like Native American Navajos, a population with relatively poor antibody responses to HibCP vaccines and a high prevalence of invasive Hib diseases (5, 15). Thus, the demonstration in this report that A18 could not replace the A2 gene in the sequence coding for canonical antibody has several implications. One is that individuals lacking a functional A2 gene most likely are unable to form canonical HibCP antibodies even if they carry functional A18 alleles in the proximal V gene group. This goes for individuals homozygous for the haplotype 11 lacking the entire distal group (44) and probably also for individuals homozygous for the A2b allele which is inefficient due to defects of the recombination signal sequences (35). The consequences that this may have for the susceptibility of these individuals to Hib infection and for the quality of the antibody they make upon Hib vaccination are presently unknown, but increased susceptibility and qualitatively poor antibody responses are indeed possible. It should be noted, however, that no history of Hib meningitis has been reported for the few individuals known to be homozygous for haplotype 11 (45). This does not of course exclude an effect on the levels of populations.
Another implication of the inability of A18b to replace A2 in the canonical HibCP antibody is that it indicates specific structural requirements of the canonical light chain. To aid the interpretation of the effects of individual amino acids on the function of the antibody, a model for the VH and VL domains of a prototypical canonical HibCP antibody was produced. The model (Fig. 8) predicts that the short CDR3 of the heavy chain together with the extended light chain CDR3 forms the floor of a groove flanked by the CDR1s and CDR2s of the two chains. CDR1 of the Vκ A2 gene product is four to five amino acids longer than CDR1 in most Vκ gene products. The model structure is homologous to the so-called groove-type dextran antibodies described by Padlan and Kabat and by Wang et al. (37, 49).
Only three amino acid positions separate the fully efficient A2c allele product from the completely inefficient A18b allele product. In fact, all three positions turned out to be important for the function of the canonical light chain, and in all three positions the amino acid residue encoded by A2 was superior to that encoded by A18b. Asparagine 53 was slightly better in facilitating binding than the serine encoded by the A18b allele, suggesting contact between HibCP and the light chain CDR2, where residue 53 is usually surface exposed in crystallized antibodies (9). In the model, Asn53 is indeed surface exposed but placed somewhat laterally with respect to the groove axis. A much more important role was found, however, for the CDR3 residues at positions 91 and 93, which are also surface exposed but located centrally in the presumed antigen-binding groove (Fig. 8). Changing the glutamine at position 93 to the A18b allele-encoded histidine reduced binding considerably, suggesting direct interaction between HibCP and this amino acid residue. Most pronounced was, however, the effect of changing serine 91 into the A18b allele-encoded glycine, which completely abrogated antigen binding. In fact, the reversal of this amino acid change was sufficient to change the nonbinder Fab3-23/A18b into a binder despite the negative influences of serine 53 and histidine 93. Thus, it is clear that the light chain residue at position 91 plays a pivotal role in the canonical HibCP antibody. There are several possible explanations for this. One is that a serine in that position is crucial due to direct engagement in antigen binding. The alternative is that a glycine residue in the light chain position 91 is deleterious due to some structural changes affecting other residues important for antigen binding. Our data do not allow us to discriminate between these possibilities. It is noteworthy, however, that the hydroxyl group of serine 91 is indeed accessible on the surface centrally in the paratope and could engage in hydrogen bond formation to the polysaccharide (Fig. 8). Model considerations, however, indicate that changing serine 91 to glycine induces a dislocation of the aromatic residue of light chain position 96 (Phe in Fab3-23/A2a and Trp in Fab3-23gl/Jk1) (Fig. 1) and changes the orientation of Tyr 96 of the heavy chain (Fig. 8). Because both of these residues are likely to be directly engaged in antigen binding (see below), a glycine in light chain position 91 might eliminate binding indirectly through these effects. The third possibility, that Gly 91 is incompatible with the V domain structure, is not likely for two reasons. First, a glycine is quite common in position 91 in crystallized antibodies, suggesting that it is compatible with normal loop structure (34). Second, the finding in this report that LuC9 binding was not abrogated by the introduction of this amino acid points to conservation of the overall structure of the paratope.
The close spatial relation between the crucial serine at position 91 and the aromatic residue at position 96 of the canonical light chain is interesting. The light chain residue at position 96 is the only Jκ gene-encoded amino acid which differs between all five Jκ genes, and this amino acid is often engaged in antigen binding. Only the residues at positions 96 and 97 are surface exposed in the antigen-binding area, and the latter is threonine irrespective of the Jκ gene. Therefore, the residue at position 96 may determine which Jκ genes are suitable for canonical HibCP antibodies and which are not. Indeed, the two Jκ gene products that have never been seen in canonical HibCP antibodies, Jκ4 and Jκ5, have aliphatic residues in that position (Leu and Ile, respectively), while the products of three Jκ genes found among canonical HibCP antibodies all have large aromatic residues (Trp, Tyr, and Phe for Jκ1, Jκ2, and Jκ3, respectively). In this report, we found excellent binding when Jκ1 was used to encode the FR4 of the canonical light chain and reasonable binding when Jκ3 was used, while Jκ2 did not yield detectable binding in the unmutated Fabs. The canonical light chains sequenced to date reveal accordingly that Jκ1 is used most often (12 of 24) (6, 24, 31, 46, 47), followed by Jκ3 (7 of 24). Jκ2 was used in 5 of 24 sequenced antibodies only. A closer look into these Jκ2 sequences showed that a Tyr 96 residue was present in only one of them (a mutated light chain with unknown heavy chain (could be noncanonical)) (47). In the second one, the residue at position 96 could not be ascertained by amino acid sequencing which, by the technique used, suggested that it was a Trp rather than a Tyr (46). In the third and fourth sequences, the codon at position 96 was, in fact, encoding Trp while the remaining part of the Jκ gene-encoded segments was Jκ2-like (24). The fifth sequence encoded a Cys at position 96 as the only difference from the Jκ2 gene-encoded sequence (24), but the ability of this PCR-derived sequence to code for an HibCP-binding antibody remains to be demonstrated. Together these data strongly suggest that only Jκ1, followed by Jκ3 is effective in canonical HibCP antibodies in the germ line versions because of the important role for a large hydrophobic aromatic amino acid (Trp or Phe) in position 96 of the light chain. The use of Jκ2 apparently requires introduction of a tryptophan at position 96 either in the process of rearrangement or by somatic mutation. The latter possibility would require some affinity by the germ line-encoded antibody with a tyrosine in position 96 in order to account for the selection of the virgin B cell.
The murine LuC9 monoclonal antibody detecting HibId-1 expression has been widely used to detect canonical HibCP antibodies (17, 18, 29, 31). Because LuC9 inhibits antigen binding, it has even been possible to quantitate the canonical antibody by inhibition in Farr assays (18, 29, 31). Early, it was shown that the HibId-1 idiotope is present on the isolated canonical light chain (31). Reason and Lucas (40) recently showed that the HibId-1 idiotope is also expressed on isolated rearranged gene products of A18b and irrespective of the presence of the arginine at position 95A characteristic for the canonical light chain CDR3. They concluded that HibId-1 is not confined to HibCP-specific antibodies but can be expressed by antibodies using either A2 or A18b irrespective of antigen specificity (40). The finding in this report of the inability of A18b to participate in HibCP antibodies using the canonical heavy chain indicates that inhibition by LuC9 in anti-HibCP Farr assays is still a reliable way of detecting HibCP antibodies employing the A2 gene product. Our finding of identical binding curves for A2a- and A2c-encoded light chains indicates that these two versions of the canonical antibody are likely to be detected equally.
Concerning the location of HibId-1 in the canonical antibody, this report shows involvement of the light chain CDR2 amino acid position 53 as well as CDR3 positions 91 and 93. No contribution from the heavy chain was found. Because these three light chain positions were also involved in HibCP binding, a rather precise overlapping of the idiotope and the light chain part of the paratope was evident. As expected, though, the specific requirements of the various amino acid residues differed between HibCP and LuC9. In fact, a glutamine-to-histidine change at position 93 improved binding to LuC9 but reduced binding to HibCP.
The recurrent involvement of certain combinations of V, (D), and J genes (i.e., canonical genes) in the antibody response to certain antigens is well known from murine studies of immune responses to haptens (12, 19) and polysaccharides (16). The HibCP response probably constitutes the best-characterized human antibody response showing similar genetic restriction. Some differences from the murine homologs should, however, be noted. Whereas the canonical antibodies in the murine systems usually carry characteristic mutations and tend to be replaced by antibodies employing other genes in secondary and tertiary antibody responses (7), the human canonical HibCP antibodies persist in recall antibody responses (17) and tend to use the VκII A2 gene-encoded light chain in an unmutated or only slightly mutated form (1, 3, 6, 24, 31, 39, 46, 47).
The mechanism behind this restriction is unknown. The antigen systems in which it has been demonstrated (haptens and polysaccharides) are characterized by a limited number of possible epitopes, and it is therefore possible that the restriction is a general feature of the antibody response to a single epitope. Several mechanisms could operate. Canonical rearrangements could be very common in the primary repertoire due to preferences of the recombination machinery or to selective forces acting on the primary repertoire prior to exposure to external antigens. This is, however, not the case for the canonical HibCP antibodies because they utilize the Vκ A2 gene product (which is relatively rarely utilized in the general repertoire) rearranged under the formation of a CDR3 of extended length due to incorporation of an extra arginine (a rare event in light chain rearrangements).
The most straightforward explanation for the recurrent use of canonical rearrangements is that these, prior to mutations, encode an antigen receptor with higher affinity for that specific epitope than other rearrangements available in the repertoire and that B cells with that receptor are much more effectively expanded early in the B-cell response than are B cells with other receptors. The finding in this report of effective binding of Fabs of the unmutated canonical B-cell receptor to HibCP in vitro supports this hypothesis, especially because we recently have failed to demonstrate any detectable binding of a Fab representing the predominant noncanonical HibCP antibody of the same individual (To) when the Fab was back-mutated to germ line configuration (22). However, these findings should be extended to include more noncanonical antibodies in order to be conclusive on their own. The conclusion is, however, also supported by the demonstration in this study of a correlation between the gene segments facilitating binding in vitro and those utilized in vivo. Especially, the demonstration that the Jκ chain usage in vivo correlated with the affinity obtained in vitro when different Jκ genes were introduced in the canonical light chain fits well with the affinity hypothesis. We therefore propose that the recurrent usage of canonical gene segments in the antibody response to HibCP is due to these segments forming an antigen receptor on the virgin B cell which has a relatively high affinity for HibCP prior to somatic mutation. Strict structural requirements of the paratope limit this to a few gene segments and excludes even highly homologous ones. It follows logically from this proposal that there exists only one (repeated) epitope on the HibCP molecule suitable for canonical antibody formation.
ACKNOWLEDGMENTS
This study was supported by the Gerda & Aage Haensch Foundation, the Novo Nordisk Foundation, the Lundbeck Foundation, and the Danish Medical Research Council (grants 9503060, 9700609, and 9601791).
We thank Marianne Petersen and Ingrid Alsing for excellent technical assistance, Uffe Skov Sørensen, Morten Dziegiel, Klaus Rieneck, and Alexander Lucas for the generous supply of reagents, D. Scott Linthicum for Fv modeling, and Carsten Heilmann for critically reviewing the manuscript.
REFERENCES
- 1.Adderson E E, Shackelford P G, Insel R A, Quinn A, Wilson P M, Carroll W L. Immunoglobulin light chain variable region gene sequences for human antibodies to Haemophilus influenzae type b capsular polysaccharide are dominated by a limited number of Vκ and Vλ segments and VJ combinations. J Clin Investig. 1992;89:729–738. doi: 10.1172/JCI115649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adderson E E, Shackelford P G, Quinn A, Carroll W L. Restricted Ig H chain V gene usage in the human antibody response to Haemophilus influenzae type b capsular polysaccharide. J Immunol. 1991;147:1667–1674. [PubMed] [Google Scholar]
- 3.Adderson E E, Shackelford P G, Quinn A, Wilson P M, Carroll W L. Diversity of immunoglobulin light chain usage in the human immune response to Haemophilus influenzae type b capsular polysaccharide. Pediatr Res. 1993;33:307–311. doi: 10.1203/00006450-199303000-00022. [DOI] [PubMed] [Google Scholar]
- 4.Adderson E E, Shackelford P G, Quinn A, Wilson P M, Cunningham M W, Insel R A, Carroll W L. Restricted immunoglobulin VH usage and VDJ combinations in the human response to Haemophilus influenzae type b capsular polysaccharide. Nucleotide sequences of monospecific anti-Haemophilus antibodies and polyspecific antibodies cross-reacting with self antigens. J Clin Investig. 1993;91:2734–2743. doi: 10.1172/JCI116514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atkinson M J, Cowan M J, Feeney A J. New alleles of IGKV genes A2 and A18 suggest significant human IGKV locus polymorphism. Immunogenetics. 1996;44:115–120. [PubMed] [Google Scholar]
- 6.Barington T, Hougs L, Juul L, Madsen H O, Ryder L P, Heilmann C, Svejgaard A. The progeny of a single virgin B cell predominates the human recall B cell response to the capsular polysaccharide of Haemophilus influenzae type b. J Immunol. 1996;157:4016–4027. [PubMed] [Google Scholar]
- 7.Berek C, Milstein C. Mutation drift and repertoire shift in the maturation of the immune response. Immunol Rev. 1987;96:23–41. doi: 10.1111/j.1600-065x.1987.tb00507.x. [DOI] [PubMed] [Google Scholar]
- 8.Brezinschek H P, Brezinschek R I, Lipsky P E. Analysis of the heavy chain repertoire of human peripheral b cells using single-cell polymerase chain reaction. J Immunol. 1995;155:190–202. [PubMed] [Google Scholar]
- 9.Chothia C, Lesk A M, Gherardi E, Tomlinson I M, Walter G, Marks J D, Llewelyn M B, Winter G. Structural repertoire of the human VH segments. J Mol Biol. 1992;227:799–817. doi: 10.1016/0022-2836(92)90224-8. [DOI] [PubMed] [Google Scholar]
- 10.Claflin J L, Berry J. Genetics of the phosphocholine-specific antibody response to Streptococcus pneumoniae. Germ-line but not mutated T15 antibodies are dominantly selected. J Immunol. 1988;141:4012–4019. [PubMed] [Google Scholar]
- 11.Crisel R M, Baker R S, Dorman D E. Capsular polymer of Haemophilus influenzae, type b. I. Structural characterization of the capsular polymer of strain Eagan. J Biol Chem. 1975;250:4926–4930. [PubMed] [Google Scholar]
- 12.Cumano A, Rajewsky K. Clonal recruitment and somatic mutation in the generation of immunological memory to the hapten NP. EMBO J. 1986;5:2459–2468. doi: 10.1002/j.1460-2075.1986.tb04522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engberg J, Andersen P S, Nielsen L K, Dziegiel M, Johansen L K, Albrechtsen B. Phage-display libraries of murine and human antibody Fab fragments. Methods Mol Biol. 1995;51:355–376. doi: 10.1385/0-89603-275-2:355. [DOI] [PubMed] [Google Scholar]
- 14.Ermert K, Mitlöhner H, Schempp W, Zachau H G. The immunoglobulin kappa locus of primates. Genomics. 1995;25:623–629. doi: 10.1016/0888-7543(95)80003-5. [DOI] [PubMed] [Google Scholar]
- 15.Feeney A J, Atkinson M J, Cowan M J, Escuro G, Lugo G. A defective Vκ A2 allele in Navajos which may play a role in increased susceptibility to Haemophilus influenzae type b disease. J Clin Investig. 1996;97:2277–2282. doi: 10.1172/JCI118669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandez C. Identical VHD and DJH junctions in monoclonal antibodies derived in response to dextran B512 could be the result of developmental selection. Scand J Immunol. 1994;40:581–590. doi: 10.1111/j.1365-3083.1994.tb03509.x. [DOI] [PubMed] [Google Scholar]
- 17.Granoff D M, Holmes S J, Osterholm M T, McHugh J E, Lucas A H, Anderson E L, Belshe R B, Jacobs J L, Medley F, Murphy T V. Induction of immunologic memory in infants primed with Haemophilus influenzae type b conjugate vaccines. J Infect Dis. 1993;168:663–671. doi: 10.1093/infdis/168.3.663. [DOI] [PubMed] [Google Scholar]
- 18.Granoff D M, Shackelford P G, Holmes S J, Lucas A H The Collaborative Vaccine Study Group. Variable region expression in the antibody responses of infants vaccinated with Haemophilus influenzae type b polysaccharide-protein conjugates. Description of a new lambda light chain-associated idiotype and the relation between idiotype expression, avidity, and vaccine formulation. J Clin Investig. 1993;91:788–796. doi: 10.1172/JCI116298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffiths G M, Berek C, Kaartinen M, Milstein C. Somatic mutation and the maturation of immune response to 2-phenyl oxazolone. Nature. 1984;312:271–275. doi: 10.1038/312271a0. [DOI] [PubMed] [Google Scholar]
- 20.Hennessey J P, Jr, Bednar B, Manam V. Molecular size analysis of Haemophilus influenzae type b capsular polysaccharide. J Liquid Chromatogr. 1993;16:1715–1729. [Google Scholar]
- 21.Hougs L, Barington T, Madsen H O, Ryder L P, Svejgaard A. Rapid analysis of rearranged kappa light chain genes of circulating polysaccharide-specific B lymphocytes by means of immunomagnetic beads and the polymerase chain reaction. Exp Clin Immunogenet. 1993;10:141–151. [PubMed] [Google Scholar]
- 22.Hougs L, Juul L, Ditzel H J, Heilmann C, Svejgaard A, Barington T. The first dose of a Haemophilus influenzae type b conjugate vaccine reactivates memory B cells. Evidence for extensive clonal selection, intraclonal affinity maturation, and multiple isotype switches to IgA2. J Immunol. 1999;162:224–237. [PubMed] [Google Scholar]
- 23.Insel R A, Anderson P W J. Cross-reactivity with Escherichia coli K100 in the human serum anticapsular antibody response to Haemophilus influenzae type b. J Immunol. 1982;128:1267–1270. [PubMed] [Google Scholar]
- 24.Insel R A, Varade W S, Chu Y W, Marin E, Fuleihan R, Geha R S. Somatic mutation of human immunoglobulin V genes: bias, rate, and regulation. Ann N Y Acad Sci. 1995;764:158–169. doi: 10.1111/j.1749-6632.1995.tb55820.x. [DOI] [PubMed] [Google Scholar]
- 25.Juul L, Hougs L, Andersen V, Garred P, Ryder L P, Svejgaard A, Høgh B, Lamm L, Graugaard B, Barington T. Population studies of the human Vκ A18 gene polymorphism in Caucasians, Blacks, and Eskimos. New functional alleles and evidence for evolutionary selection for a more restricted antibody repertoire. Tissue Antigens. 1997;49:595–604. doi: 10.1111/j.1399-0039.1997.tb02807.x. [DOI] [PubMed] [Google Scholar]
- 26.Juul L, Hougs L, Andersen V, Svejgaard A, Barington T. The normally expressed kappa immunoglobulin light chain gene repertoire and somatic mutations studied by single-sided specific polymerase chain reaction. Frequent occurrence of features often assigned to autoimmunity. Clin Exp Immunol. 1997;109:194–203. doi: 10.1046/j.1365-2249.1997.4341332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kabat E A, Wu T T, Perry H M, Gottesman K S, Foeller C. Sequences of proteins of immunological interest. 5th ed. Bethesda, Md: U.S. Department of Health and Human Services; 1991. [Google Scholar]
- 28.Lausen, B. 1998. Unpublished data.
- 28a.Jones D C S, Schofield J P. A rapid method for isolating high quality plasmid DNA suitable for DNA sequencing. Nucleic Acids Res. 1990;18:7463–7464. doi: 10.1093/nar/18.24.7463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lucas A H, Azmi F H, Mink C M, Granoff D M. Age-dependent V region expression in the human antibody response to the Haemophilus influenzae type b polysaccharide. J Immunol. 1993;150:2056–2061. [PubMed] [Google Scholar]
- 30.Lucas A H, Granoff D M. Functional differences in idiotypically defined IgG1 anti-polysaccharide antibodies elicited by vaccination with Haemophilus influenzae type B polysaccharide-protein conjugates. J Immunol. 1995;154:4195–4202. [PubMed] [Google Scholar]
- 31.Lucas A H, Langley R J, Granoff D M, Nahm M H, Kitamura M Y, Scott M G. An idiotypic marker associated with a germ-line encoded κ light chain variable region that predominates the vaccine-induced human antibody response to the Haemophilus influenzae b polysaccharide. J Clin Investig. 1991;88:1811–1818. doi: 10.1172/JCI115502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mandal C, Kingery B D, Anchin J M, Subramaniam S, Linthicum D S. ABGEN: a knowledge-based automated approach for antibody structure modeling. Nat Biotechnol. 1996;14:323–328. doi: 10.1038/nbt0396-323. [DOI] [PubMed] [Google Scholar]
- 33.Mattila P S, Schugk J, Wu H, Mäkelä O. Extensive allelic sequence variation in the J region of the human immunoglobulin heavy chain gene locus. Eur J Immunol. 1995;25:2578–2582. doi: 10.1002/eji.1830250926. [DOI] [PubMed] [Google Scholar]
- 34.Mian I S, Bradwell A R, Olson A J. Structure, function and properties of antibody binding sites. J Mol Biol. 1991;217:133–151. doi: 10.1016/0022-2836(91)90617-f. [DOI] [PubMed] [Google Scholar]
- 35.Nadel B, Tang A, Lugo G, Love V, Escuro G, Feeney A J. Decreased frequency of rearrangement due to the synergistic effect of nucleotide changes in the heptamer and nonamer of the recombination signal sequence of the V kappa gene A2b, which is associated with increased susceptibility of Navajos to Haemophilus influenzae type b disease. J Immunol. 1998;161:6068–6073. [PubMed] [Google Scholar]
- 36.Nahm M H, Kim K H, Anderson P, Hetherington S V, Park M K. Functional capacities of clonal antibodies to Haemophilus influenzae type b polysaccharide. Infect Immun. 1995;63:2989–2994. doi: 10.1128/iai.63.8.2989-2994.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Padlan E A, Kabat E A. Model-building study of the combining sites of two antibodies to alpha (1→6)dextran. Proc Natl Acad Sci USA. 1988;85:6885–6889. doi: 10.1073/pnas.85.18.6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pillai S, Ciciriello S, Koster M, Eby R. Distinct pattern of antibody reactivity with oligomeric or polymeric forms of the capsular polysaccharide of Haemophilus influenzae type b. Infect Immun. 1991;59:4371–4376. doi: 10.1128/iai.59.12.4371-4376.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pinchuk G V, Nottenburg C, Milner E C B. Predominant V-region gene configurations in the human antibody response to Haemophilus influenzae capsule polysaccharide. Scand J Immunol. 1995;41:324–330. doi: 10.1111/j.1365-3083.1995.tb03574.x. [DOI] [PubMed] [Google Scholar]
- 40.Reason D C, Lucas A H. Functional expression of the IGKV A18b gene and its idiotypic crossreactivity with the A2 variable region. Immunogenetics. 1997;45:343–344. doi: 10.1007/s002510050214. [DOI] [PubMed] [Google Scholar]
- 41.Reason D C, Wagner T C, Lucas A H. Human Fab fragments specific for the Haemophilus influenzae b polysaccharide isolated from a bacteriophage combinatorial library use variable region gene combinations and express an idiotype that mirrors in vivo expression. Infect Immun. 1997;65:261–266. doi: 10.1128/iai.65.1.261-266.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 43.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5468. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schäble G, Rappold G A, Pargent W, Zachau H G. The immunoglobulin kappa locus: polymorphism and haplotypes of Caucasoid and non-Caucasoid individuals. Hum Genet. 1993;91:261–267. doi: 10.1007/BF00218268. [DOI] [PubMed] [Google Scholar]
- 45.Scott M G, Crimmins D L, McCourt D W, Chung G, Schäble K F, Thiebe R, Quenzel E M, Zachau H G, Nahm M H. Clonal characterization of the human IgG antibody repertoire to Haemophilus influenzae type b polysaccharide. IV. The less frequently expressed VL are heterogeneous. J Immunol. 1991;147:4007–4013. [PubMed] [Google Scholar]
- 46.Scott M G, Crimmins D L, McCourt D W, Zocher I, Thiebe R, Zachau H G, Nahm M H. Clonal characterization of the human IgG antibody repertoire to Haemophilus influenzae type b polysaccharide. III. A single VKII gene and one of several JK genes are joined by an invariant arginine to form the most common L chain V region. J Immunol. 1989;143:4110–4116. [PubMed] [Google Scholar]
- 47.Scott M G, Zachau H G, Nahm M H. The human antibody V region repertoire to the type b capsular polysaccharide of Haemophilus influenzae. Int Rev Immunol. 1992;9:45–55. doi: 10.3109/08830189209061782. [DOI] [PubMed] [Google Scholar]
- 48.Tomlinson I M, Walter G, Marks J D, Llewelyn M B, Winter G. The repertoire of human germline VH sequences reveals about fifty groups of VH segments with different hypervariable loops. J Mol Biol. 1992;227:776–798. doi: 10.1016/0022-2836(92)90223-7. [DOI] [PubMed] [Google Scholar]
- 49.Wang D, Hubbard J M, Kabat E A. Modeling study of antibody combining sites to (alpha 1-6)dextrans. Predictions of the conformational contribution of VL-CDR3 and J kappa segments to groove-type combining sites. J Biol Chem. 1993;268:20584–20589. [PubMed] [Google Scholar]
- 50.Youngblood K, Fruchter L, Ding G, Lopez J, Bonagura V, Davidson A. Rheumatoid factors from the peripheral blood of two patients with rheumatoid arthritis are genetically heterogeneous and somatically mutated. J Clin Investig. 1994;93:852–861. doi: 10.1172/JCI117040. [DOI] [PMC free article] [PubMed] [Google Scholar]