Abstract
Colorectal cancer (CRC) patients with mismatch repair (MMR)-deficient (dMMR) but not MMR-proficient (pMMR) tend to benefit from immune checkpoint blockade (ICB) therapy. To profile the tumor microenvironments (TME) underlying these varied therapeutic responses, we integrate spatial enhanced resolution omics-sequencing (Stereo-seq), single-cell RNA sequencing, and multiplexed imaging analysis to create high-definition spatial maps of tumors from treatment-naïve and ICB-treated CRC patients. Our results identify the spatial organization and immune status of the tumor-stroma boundary as a distinctive feature of dMMR and pMMR CRCs, which associates with ICB response. The physical interactions and abundance of LAMP3+DCs and CXCL13+T cells may shape the ICB-responsive tumor-stroma boundary, whereas CXCL14+cancer-associated fibroblasts tend to remodel extracellular matrix to form a structural barrier in non-responders. Our work therefore points out the importance of the molecular and cellular spatial structures of tumors in ICB response, raising the possibility of reprogramming tumor-stroma boundary for sensitizing immunotherapies in the majority of CRCs.
Subject terms: Cancer immunotherapy, Immunotherapy, Colorectal cancer
Colorectal cancer patients with mismatch repair-deficient, but not MMR-proficient tend to benefit from immune checkpoint blockade therapy. Here, the authors identify interactions among tumor and immune cells that are associated with the responsiveness to immunotherapy in colorectal cancer.
Introduction
Immune checkpoint blockade (ICB) therapy has redefined the management of patients with colorectal cancer (CRC). In general, CRC patients with DNA mismatch repair (MMR)-deficient/microsatellite instability-high (dMMR/MSI-hi) tumors have a high mutational burden and are tend to be sensitive to ICB therapy, whereas MMR-proficient (pMMR) ones often have a low mutational burden and are mostly unresponsive1–4. We recently reported a 93.75% (15/16) overall response rate (12 were complete response) to anti-program death 1 (PD1) monoclonal antibody (mAb, sintilimab) in the dMMR cohorts, which potentially spare these patients from radical surgeries5. In contrast, ICB has exhibited limited efficacy in pMMR cases, with a 0% response rate to anti-PD1 monotherapy and up to 27% of stable disease (SD) or partial response (PR) reported in early-stage patients to combinatory approaches4,6. The abundance of cytotoxic T cells, numbers and functional status of myeloid cells as well as expression levels of immune checkpoint molecules including PD1 and its ligand 1 (PD-L1) are commonly recognized as main contributors to govern the ICB responsive difference between dMMR and pMMR CRCs7–9. Based on the advancement of single cell RNA-sequencing (scRNA-seq), Pelka et al. reported shared and distinct features of dMMR and pMMR CRCs by profiling 371,223 cells from tumors of treatment naïve CRC patients10. Their data revealed that the existence of a dMMR-enriched immune hub with activated T cells and C-X-C Motif Chemokine Receptor 3 (CXCR3) ligand-expressing malignant and myeloid cells may associate with ICB response10. In addition, a recent study further showed that the crosstalk between FAP+fibroblasts and SPP1+macrophages may stimulate the formation of immune-excluded desmoplastic structure and limit T cell infiltration in CRC tumors, which correlate with low ICB response11. Although these substantial progresses have uncovered numerous key cell subsets in the tumor microenvironment (TME) that contribute to variable immune responses of CRC, how these cells are spatially organized and interacted within dMMR or pMMR tumors and how they response towards ICB treatment remain poorly understood.
While conventional histology and multiplex-immunofluorescence (mIF) can provide positional information of cells, these methods do not supply the detailed molecular information needed to identify and phenotype cells precisely. The dissociated single cell methods of scRNA-seq or high-dimensional flow cytometry that widely used in studying the molecular and cellular heterogeneity, however, lack information of cell-cell interaction and spatial architectures of tissues. Several high throughput spatial transcriptomic technologies are therefore developed to dissect the detailed information of molecular and cellular features in tissues in situ, including commercialized Visium by 10X Genomics, CosMx SMI and DBiT-seq12, as well as the spatial enhanced resolution omics-sequencing (Stereo-seq) developed by us13,14. The stereo-seq provides customized resolution by binning neighboring nanoballs as a minimal spot for further analysis, e.g., at anatomical level with square bins or at single cell level with cell bins13,14. However, the resolution is often compromised by the tissue nature in practice, because smaller spots contain fewer detected transcripts. Considering the smaller size of immune cells and their physical overlay on the stromal cells in the tumors, here we applied an integrative analysis of scRNA-seq and Stereo-seq at 50 µm resolution to in-depth dissect the gene regulatory programs and cell-cell interactions underlying ICB response in CRC patients.
In this work, through analysis in 25 tumor specimens from CRC patients of treatment naïve dMMR, pMMR and anti-PD1-treated dMMR patients that included responders (complete response (CR)/PR) and non-responders (stable disease, SD), we generated a CRC spatial transcriptomic atlas and uncovered a 300 micrometer (μm) boundary region (0 ± 150 μm) that regulated immune cell influx to the tumor center region (>150 μm). In this microscopic structure of tumor-stroma boundary, the active interplays among tumor cells, fibroblasts, macrophage/dendritic cells (DCs) and T cell subsets associated with the distinctive immune status of dMMR and pMMR CRCs patients, which may associate with their diverse ICB responses. Overall, our study uncovered a mix of transcriptomic and spatial programs underlying specific TME formation, which highlighted the importance of considering the interplay of innate, adaptive and structural cells in TME for therapeutic approach development.
Results
Spatial transcriptome atlas of human CRCs by Stereo-seq in CRC patients under ICB treatment
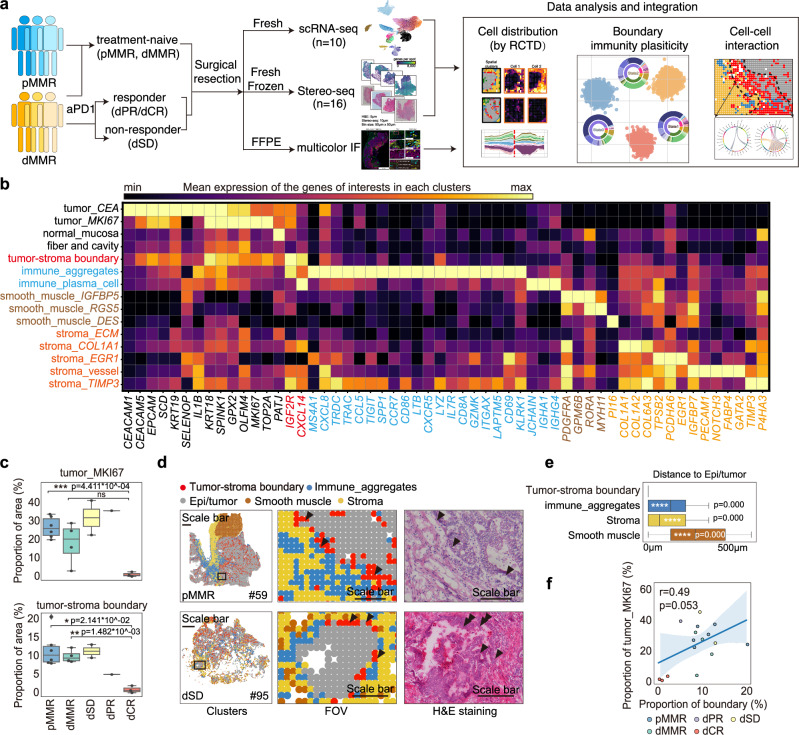
To study the spatial organization and heterogeneity of cellular components and their responses toward ICB treatment in CRC patients, we performed scRNA-seq and Stereo-seq on surgical tissues from 23 CRC patients (Supplementary Data 1). In detail, 16 samples from 15 patients were sequenced by Stereo-seq, and 10 samples from 10 patients (2 were previously released12) were applied for scRNA-seq (Fig. 1a). 10 pMMR and 4 dMMR patients with no systemic treatment before surgery, i.e. treatment naïve CRC patients, and 11 dMMR patients received neoadjuvant anti-PD1 antibody treatment (Supplementary Data 1) that experienced stable disease (dSD, n = 5), partial response (dPR, n = 2) or complete response (dCR, n = 4) were included (Supplementary Fig. 1a). After pre-processing on the raw data generated by Stereo-seq (see Methods), the spatial transcriptome map was lassoed out and matched to the tissue edge (https://www.stomics.tech/sap/home.html). As the single-cell resolution was insufficient to generate an adequate number of genes per bin for cell type annotation and follow-up analysis in tumor tissues (Supplementary Fig. 1b), we adjusted the bin size to bin100, which allowed us to obtain a sufficient gene count for transcriptomic analysis at a resolution of 50μm (Supplementary Fig. 1b–d). After quality control and batch effect correction, 205,362 spatial bins with a median gene count of 1849 (Supplementary Fig. 1c) and 27,154 single cells with a median gene count of 1,115 (Supplementary Fig. 1d) were integrated into a joint map, clustered and subjected to region annotation (Fig. 1b, Supplementary Fig 1e-f). Using Leiden algorithm13, we next performed an integration of unsupervised clustering analysis of the spatial transcriptomics data with H&E staining image. This approach led to the generation of signature score and enrichment score, enabling the deconvolution of the cellular composition of spatially defined bins. As a result, we identified 15 spatial clusters, which captured a wide spectrum of regions of the 6 major anatomical structures: normal epithelia/tumor (epi/tumor), smooth muscle, tumor-stroma boundary, stroma, immune aggregates and low mRNA-enriched (fiber and cavity) regions in the tissues from 15 patients by Stereo-seq analysis (Fig. 1b and Supplementary Fig. 2a). Further assessment of copy number variation (CNV) score12,14 (https://github.com/broadinstitute/infercnv/) clearly showed that the tumor region exhibited higher CNV scores compared to other regions (Supplementary Fig. 2b, c). In addition, we observed a similar gene expression pattern in the gene set used to define the tumor-stroma boundary in breast cancer15 within our spatial cluster (Supplementary Fig. 2d), supporting the robustness of our spatial cluster definition. Next, we analyzed the spatial clusters in indicated patient groups (Supplementary Fig. 2e). Using the expressions of differentially expressed genes (DEGs) in each cluster and taking into account previous reports on cell type annotation from scRNAseq and image data10,11,16, we named the 15 spatial clusters based on their phenotype and predominant DEGs (Fig. 1b). For example, tumor_CEA referred to tumor cell cluster with predominant expressions of CEA cell adhesion molecules CEACAM1 and CEACAM5 (Fig. 1b). As expected, specimens from the three dCR patients contained limited proportions of tumor_MKI67 compared to the treatment-naïve and dSD patients (Fig. 1c and Supplementary Fig. 2e). Although the proportions of tumor-stroma boundary were similar between treatment naïve dMMR and pMMR patients, dCR patients displayed a significant lower proportion compared to the treatment-naïve and the dSD patients (Fig. 1c and Supplementary Fig. 2e). In addition, the representative field of view (FOV) data showed that pMMR displayed a well-organized barrier-like structure in tumor-stroma boundary region that clearly separated the tumor region from the immune aggregates and stromal regions, whereas the structure of spatial clusters in dMMR was unorganized, which was further supported by Moran index analysis (Fig. 1d and Supplementary Fig. 2f). Since ICB treatment is now a first line therapy for dMMR CRCs, we next compared the spatial transcriptomics data of dMMR patients after anti-PD1 treatment. Interestingly, dMMR patients who experienced lower clinical benefit towards anti-PD1 therapy, i.e. dSD displayed a well-organized tumor-stroma boundary structure similar to pMMR, commonly recognized as ICB insensitive group (Fig. 1d). In line with the lower proportions of proliferative tumor clusters (tumor_MIK67), the proportion of tumor-stroma boundary was also lower in dCR patients (Fig. 1c). The proximity (Fig. 1e) and the correlation (Fig. 1f) between the boundary and epi/tumor clusters further indicated that the presence and spatial organization of tumor-stroma boundary may associate with ICB response in CRC patients.
Fig. 1. Spatial transcriptome atlas of human CRCs by Stereo-seq reveals ICB responsiveness.
a Schematic of single cell and spatial transcriptomic analysis in CRC patients with or without neoadjuvant anti-PD1 treatment. b The top differential express gene (DEG) expressions in each spatial cluster are shown in the matrix plot. c The box and whisker plot shows the proportions of spatial clusters tumor_MKI67 and tumor-stroma boundary in indicated patient groups. The smooth muscle is not included to remove sampling bias. Data are represented as mean±IQR and analyzed by unpaired 2-tailed Student-t test with Bonferroni correction. Ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ***, p < 0.0001. N number: pMMR=6; dMMR=4; dSD=2; dPR=1; dCR=3. d Representative images of FOV and H&E staining in treatment naïve pMMR (patient #59), and anti-PD1 treated dMMR with stable disease (SD) (patient #95). Low mRNA-enriched region (fiber and cavity) is not displayed. Scale bars were set at 1200 μm for the global spatial plot (left) and 300μm for the FOVs (middle and right). e The box and whisker plot shows the average distance from epi/tumor cluster to the clusters of tumor-stroma boundary, immune aggregates, stroma and smooth muscle. Data are represented as mean ± IQR and analyzed by unpaired 2-tailed Student t test with Bonferroni correction. Ns not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ***, p < 0.0001. N number of the spots: tumor-stroma boundary=17,639; immune aggregates=16,304; stroma=28,012; smooth muscle=24,172. f Pearson correlation between spatial clusters tumor_MKI67 and tumor-stroma boundary. The dot plot color represents indicated patient group. The line and the band present the linear regression model and confidence interval (95%) respectively. N number of the patients: pMMR=6; dMMR=4; dSD=2; dPR=1; dCR=3.
dMMR and pMMR tumor-stroma boundaries display distinguished cellular components and spatial distributions
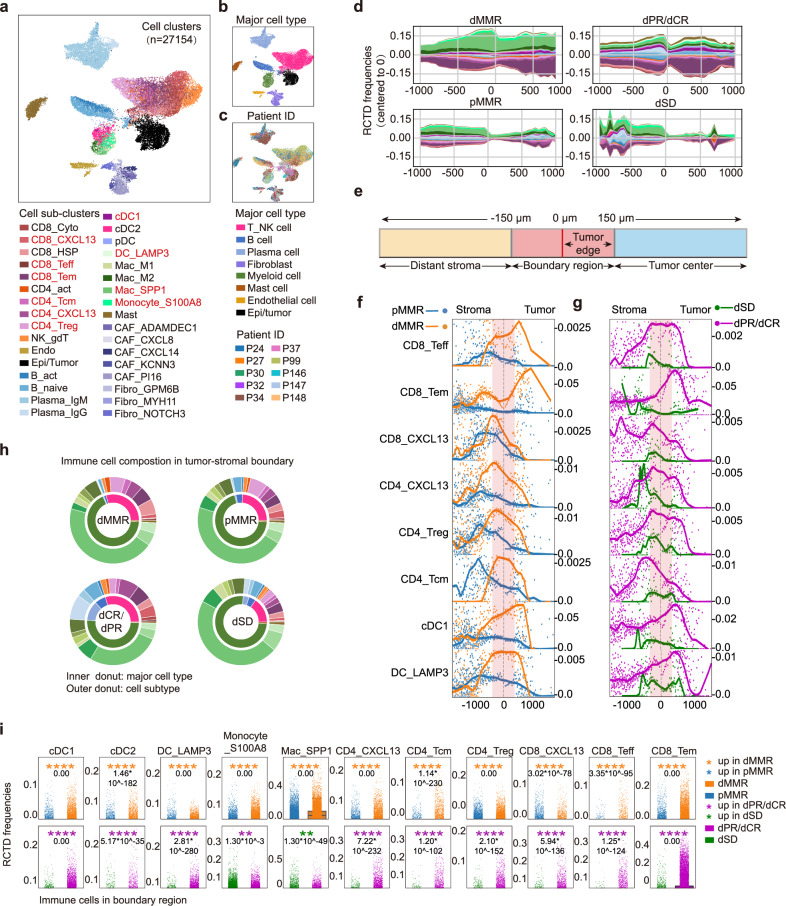
To in-depth analyse the cellular composition and their spatial organization in TME, we leveraged the scRNA-seq data of 27,154 cells from 5 treatment-naïve pMMR and 5 anti-PD1-treated dMMR CRC patients (Supplementary Fig. 1a). We integrated the cells by Batch Balanced K Nearest Neighbors (BBKNN) algorithm to correct the batch effect, performed cluster analysis, and annotated cell clusters manually based on markers of interests17. In total, we identified 8 major cell clusters (including 33 sub-clusters), covering 23 immune cell and 10 stromal cell subtypes (Fig. 2a–c, Supplementary Data 2–3). The cell counts were comparable in the patients except for patient #24 and #34 (Supplementary Fig. 3a). The stromal cells contained 8 cell clusters of fibroblasts (COL1A1), epi/tumor cells (KRT18, EPCAM), and endothelial cells (PECAM1) (Supplementary Fig. 3b–d). Among immune cells, we categorized T cells into 10 subclusters, including CD4_CXCL13 (CXCL13+CD4+T cells), CD4_Tcm (central memory CD4+T cell), CD4_Treg (regulatory CD4+T cell), CD8_Tcyto (cytolytic CD8+T cell), CD8_Teff (effector CD8+T cell), CD8_Tem (effector memory CD8+T cell), CD8_CXCL13 (CXCL13+CD8+T cells), and myeloid cells into 8 clusters covering cDC1 (conventional dendritic cell 1), cDC2, plasmacytoid DC (pDC), DC_LAMP3 (LAMP3+DC), Mac_M1 and Mac_M2 (M1 and M2-like macrophage), Mac_SPP1 (SPP1+macrophage), monocyte_S100A8 (S100A8+monocyte) and mast cell (Supplementary Fig. 4a–e), which were commonly reported by scRNA-seq analysis in CRC patients10. Of note, CD8_CXCL13 and CD4_CXCL13 clusters exhibited a typical T cell exhaustion signature represented by high expressions of TIGIT, LAG3, HAVCR2 (encodes TIM3) and PDCD1 (encodes PD1) (Supplementary Fig. 4a, b).To further explore the spatial localization and proportions of these cell subsets in TME, we next used Robust Cell Type Decomposition (RCTD)18 to deconvolute the spatial transcriptome with gene signatures generated from the scRNA-seq dataset, and interpreted their patterns into the spatial map. As expected, the majority of cell types accumulated in the tumor-stroma boundary were epithelial/tumor cells, followed by fibroblasts in either dMMR or pMMR, except for patient #25 and #61 who showed complete response to the anti-PD1 therapy (Supplementary Fig. 4f). This finding is consistent with previous report that describes the tumor-stroma boundary as a niche composed of malignant cells in the outermost circle of solid tumor and non-malignant cells that are closely adjacent in spatial architecture, bridging these distinct spatial regions19. Since immune cells are the direct targets of anti-PD1 therapy, we next reanalysed the immune cell components and spatial distributions as the first step. By continuous mapping the mean RCTD frequencies of each immune cell in our spatial transcriptomic data, we generated stacked stream plots that covered ±1000μm to tumor-stroma boundary (0μm) to study spatial distribution patterns of the immune cells (Fig. 2d, Supplementary Fig. 4g). Interestingly, we observed continued accumulation and increased abundance of immune cell clusters in the spatial proximity within ±500 μm to the tumor-stroma boundary in dMMR, with majority of them were myeloid cells and T_NK cells (Fig. 2d, Supplementary Fig. 4g). In contrast, immune cells showed a discontinuous distribution in the tumor-stroma boundary of pMMR, with a trend to accumulate in the stromal region (<−500 μm) but not the tumor region (>500μm) (Fig. 2d, Supplementary Fig. 4g). Of note, after anti-PD1 treatment, a clearer discontinuous curve that reflected limited immune cell infiltration was noticed in dSD patients (Fig. 2d, Supplementary Fig. 4g). These data suggested that: (1) the abundance of immune cell clusters in the tumor-stroma boundary may contribute to a better anti-PD1 response; (2) the limited infiltration of immune cells in the tumor region may be associated with a restricted anti-PD1 response in pMMR and dSD, potentially attributable to the spatial features of the tumor-stroma boundary.
Fig. 2. dMMR and pMMR tumor-stroma boundaries display distinguished cellular components and spatial distributions.
a Uniform manifold approximation and projection (UMAP) of the transcriptome of 27,154 single cells from 10 CRC patients (5 pMMR; 2 dPR/CR; 3 dSD). Cells are colored by single cell subclusters, b major cell types or (c) patient ID. The names of the cell of interests are highlighted in red. d The stacked stream plots of immune cell distribution patterns from distal stroma (–1000 μm, left) to tumor center (1000 μm, right) in indicated patient groups are shown. The mean RCTD frequencies of each immune cell in each 1 mm interval was smoothed using slinger model and colored by cell sub-clusters in accordance with (a). e Schematic diagram illustrates the definitions of the distant stroma (<-150 μm; light blue), the tumor-stroma boundary (0 ± 150 μm, light red), including the tumor edge (0 μm to 150 μm), and tumor center (> 150 μm; light yellow). f Scatter-curve plot of the distributive patterns of indicated cell clusters in treatment naïve dMMR (orange) and pMMR (blue), or (g) dCR/dPR(light pink) and dSD (green). The tumor-stroma boundary is highlighted in light red. The RCTD frequencies at the same distance from the boundary were averaged and represented as a single dot, which was smoothed using a loess model. h Donut charts show the proportions of immune cells at the central of the tumor-stroma boundary (0 μm). The inner donut represents major immune cell types, while the outer donut represents immune cell subclusters. The colors are in accordance with (a). i The RCTD frequencies of indicated immune cell subclusters in the tumor-stroma boundary of the four groups of patients. Data are analyzed by unpaired 2-tailed Student t test. P values are labeled within each plot.
Since a distinct discontinuous of immune cells was observed in the spatial maps of pMMR and dSD that covering ±150 μm from the center of boundary (0 μm) (Fig. 2d, Supplementary Fig. 4g), we therefore defined the spatial regions to distant stroma (<−150μm), tumor-stroma boundary (0 ± 150 μm) that cover the tumor edge region (0 μm–150 μm), and tumor center (>+150 μm, Fig. 2e) for further analysis. By dissecting the spatial distribution pattern of each immune cell subsets in details, we observed that immune cell subsets of CD8_Teff, CD8_Tem, CD8_CXCL13, CD4_CXCL13, CD4_Treg, CD4_Tcm, cDC1 and DC_LAMP3, but not other clusters showed significant enrichment peaks within the tumor-stroma boundary in treatment naïve dMMR compared to pMMR (Fig. 2f), which were also significantly higher in dPR/dCR compared to dSD (Fig. 2g). Interestingly, when we compared the RCTD frequencies of these immune cell subsets in the center of boundary (0 μm), we observed that the majority of immune cells were Mac_SPP1 in both treatment naïve dMMR and pMMR (Fig. 2h). In addition, the proportion of Mac_SPP1 was higher in treatment naïve dMMR compared to pMMR, while lower in dPR/CR compared to dSD (Fig. 2h, i). Previous reports on the immune cell frequencies in CRC tumors have demonstrated that SPP1-expressing macrophages (SPP1, APOE, APOC1) (Supplementary Fig. 4c) were commonly detected and their abundance are reported to correlate with less therapeutic benefit from anti-PD1 therapy10,11. This observation further supported the consistency and importance of immune cells in the tumor-stroma boundary. Of note, decreased RCTD frequencies of myeloid clusters (inner donut, dark green) and increased T cell clusters (inner donut, light pink) were observed in patients with dPR/dCR compared to dSD (Fig. 2h). The strip plot graphs comparing RCTD frequencies further validated that treatment naïve dMMR displayed significantly higher proportions of cDC1 and cDC2, DC_LAMP3, Monocyte_S100A8, CD4_CXCL13, CD4_Tcm, CD4_Treg, CD8_Teff, CD8_Tem and CD8_CXCL13 in boundary region, compared to pMMR patients (Fig. 2i). As the RCTD frequencies of these cell clusters were also higher in dPR/dCR patients compared to dSD (Fig. 2i), the result indicated their possible associations towards ICB response. Indeed, accumulating data from scRNA-seq of CRCs have demonstrated that the increased cDC1 (XCR1, CLEC9A, BATF3) (Supplementary Fig. 4c) correlates with better ICB response, whereas CD8_Teff (IFNG, CD69, CCL5, GZMA) and CD8_Tem cells (IL7R, TCF7) (Supplementary Fig. 4a) are reported to be the key intratumoral CD8+T cell subsets responding to ICB treatment20,21. Moreover, DC_LAMP3, which highly expressed signature genes including LAMP3, FSCN1, CCR7, FSCN1, CD274 (Supplementary Fig. 4c), was reported to represent a common matured DC subset in tumors that exhibited potential to regulate multiple subtypes of lymphocytes22–24. Although CD4_Treg is normally considered to be immunosuppressive, CXCL13-producing CD4+ T cells are demonstrated to drive the formation of tertiary lymphoid structure25, a well-recognized biomarker for a better ICB response10,26. Unlike the previously accepted poor prognostic value of CD8_CXCL13 in other cancers, the accumulation of CD8_CXCL13 was reported to favor the clinical outcome in ICB treatment of CRC patients, when accompanied with CXCL13-producing CD4+T cells27. Indeed, the representative FOV data suggested increased accumulations and abundances of CD8_Teff, CD8_Tem, CD8_CXCL13, CD4_CXCL13, CD4_Treg, CD4_Tcm, cDC1 and DC_LAMP3 in the tumor center (>+150μm) of dMMR (Supplementary Fig. 5). Anti-PD1 treatment was associated with the increased abundance of these subsets (excluded CD4_Tcm) into the tumor center in dPR/dCR (Supplementary Fig. 5), suggesting their potential contributions to a better ICB response in CRCs. Taken together, our scRNA-seq and spatial transcriptomic data analysis uncovered the cellular heterogeneity and spatial organizations of tumor ecosystem in CRCs, which allowed us to further investigate the gene expression profile reminiscent of cell subsets and dissect cell-cell interplays in ICB response in details.
LAMP3+DC and T cell subsets tend to accumulate in the tumor-stroma boundary of ICB responders
Since we have pointed out the potential importance of cDC1, DC_LAMP3, CD4_CXCL13, CD4_Treg, CD8_Teff, CD8_Tem and CD8_CXCL13 in ICB response, we next explored the potential mechanism of why these cell subsets were more abundant in the tumor-stroma boundary of dMMR but not pMMR. By comparing the gene expressions in the tumor-stroma boundary, we detected a significant enrichment of positive regulation of positive chemotaxis (GO:0050927) in treatment naïve dMMR compared to pMMR by gene set enrichment analysis (GSEA) (FDR = 0.009, p = 0.015; Supplementary Fig. 6a). Specifically, the expressions of CCL2/5/13/15/21 and CXCL9/10/11/12/13 were significantly higher in the tumor-stroma boundary of treatment naïve dMMR compared to pMMR (Supplementary Fig. 6b). In parallel, consistent higher mRNA levels of CCL5, CXCL9, CXCL10 and CXCL13 were detected in MSI-hi (MSI score≥10) compared to MSI-lo (MSI score≤4) CRC from colorectal adenocarcinoma (COAD) TCGA dataset (Supplementary Fig. 6c). Moreover, the expressions of CCL2/5/21 and CXCL9/10/12/13 were significantly higher in dPR/dCR compared to dSD (Supplementary Fig. 6b). CCL2/5 and CXCL9/10 are demonstrated to direct T cell migration, whereas CCL21 and CXCL12 are key chemokines that mediate DCs/macrophages trafficking28. Recent advancements on single cell analysis have also pinpointed the importance of CXCL13 in recruiting CXCR5+T cells for a better ICB responsiveness in CRC29. Therefore, we hypothesized that the T cell and DC subsets may be trapped by chemotaxis pathway to tumor-stroma boundary in ICB responders. Next, we detected the expressions of corresponding receptors toward CCL2/5/21 and CXCL9/10/13 on the immune cells that displayed higher abundance in treatment naïve dMMR or dPR/dCR using scRNA-seq dataset. The dot plot maps showed that CCR5, the corresponding receptor of CCL5, was highly expressed by CD8_CXCL13 and other T cell subsets, whereas CCL21 receptor CCR7 was predominantly expressed on DC_LAMP3, followed by cDC1 (Supplementary Fig. 6d, e). Consistent with previous report, the CXCL9/10 receptor CXCR3 was expressed by T cell subsets but not cDC1 and DC_LAMP330 (Supplementary Fig. 6d, e). Interestingly, the CXCL13 receptor CXCR5 was also expressed by a small proportion (~10%) of CD4_CXCL13, CD4_Treg, CD8_Teff, CD8_Tem and CD8_CXCL13 (Supplementary Fig. 6d, e). Taken together, our results indicated that these DCs and T cell subsets may be recruited to tumor-stroma boundary of ICB responders via chemotaxis pathways.
As chemotaxis is one of the common pathways that mediates cell-cell interaction, we further explored the potential cell-cell interactions by investigating the cellular resources of CCL2/5/21 and CXCL9/10/12/13 in the tumor-stroma boundary. Of note, CCL2, CCL5, CCL21, and CXCL13-expressing cells but no other chemokine-expressing cells showed significant higher abundances in treatment naïve dMMR compared to pMMR, which were consistently higher in dPR/dCR compared to dSD (Supplementary Fig. 6f), indicating that the CCL21-CCR7, CXCL13-CXCR5 and CCL5-CCR5 may be the key chemotaxis pathways in controlling the recruitment of DC and T cell subsets in the tumor-stroma boundary of ICB responders. Interestingly, epithelial/tumor cell cluster showed limited expressions of these chemokines (Supplementary Fig. 6g). CCL21 was predominantly expressed by endothelial cells (Supplementary Fig. 6g), suggesting that the accumulation of cDC1 and DC_LAMP3 in the tumor-stroma boundary of potential ICB responders may be largely dependent on its crosstalk with endothelial cells. In comparison, CCL5 and CXCL13 were predominantly expressed by NK_gdT or CD4_CXCL13 and CD8_CXCL13, respectively (Supplementary Fig. 6g), reflecting the possible existence of auto-reinforce chemotaxis pathways for T cells recruitment in ICB responders, but not non-responders. Therefore, the crosstalk between DCs and endothelial cells, as well as among different T cell subsets through chemotaxis pathways may govern DC and T cell subset accumulation in tumor-stroma boundary, thereby contribute to a better ICB response.
The juxtaposition and interactions between LAMP3+DCs and CXCL13-expressing T cells may correlate with a better ICB response
Next, we explored the potential crosstalk among cDC1, DC_LAMP3, CD4_CXCL13, CD4_Treg, CD8_Teff, CD8_Tem and CD8_CXCL13 cells that presented at higher levels in the tumor-stroma boundary of treatment naïve dMMR by calculating their Pearson’s correlations and spatial distances. Interestingly, we observed strong positive associations between DC_LAMP3 but not cDC1 towards CD4_CXCL13, CD4_Treg, CD8_Teff, CD8_Tem and CD8_CXCL13 in the tumor-stroma boundary using Stereo-seq dataset (Supplementary Fig. 7a). As the sample size of our in-house datasets was limited, we also validated these associations using the bulk-RNAseq datasets of 572 CRC patients with complete clinical information from COAD TCGA. Consistently, the DC_LAMP3 gene signature (top10 DEGs in RNA-seq datasets, Supplementary Data 2) was also positively associated with CD4_CXCL13, CD4_Treg, CD8_Tem and CD8_CXCL13 signatures (Supplementary Fig. 7b). In addition, the average distance between DC_LAMP3 towards these T cell subsets was less than 200 μm (Supplementary Fig. 7c), which was the maximal distance for juxtracrine signals31. These data suggested that DC_LAMP3 may interact with multiple T cell subsets and regulate their functional status in the tumor-stroma boundary of dMMR.
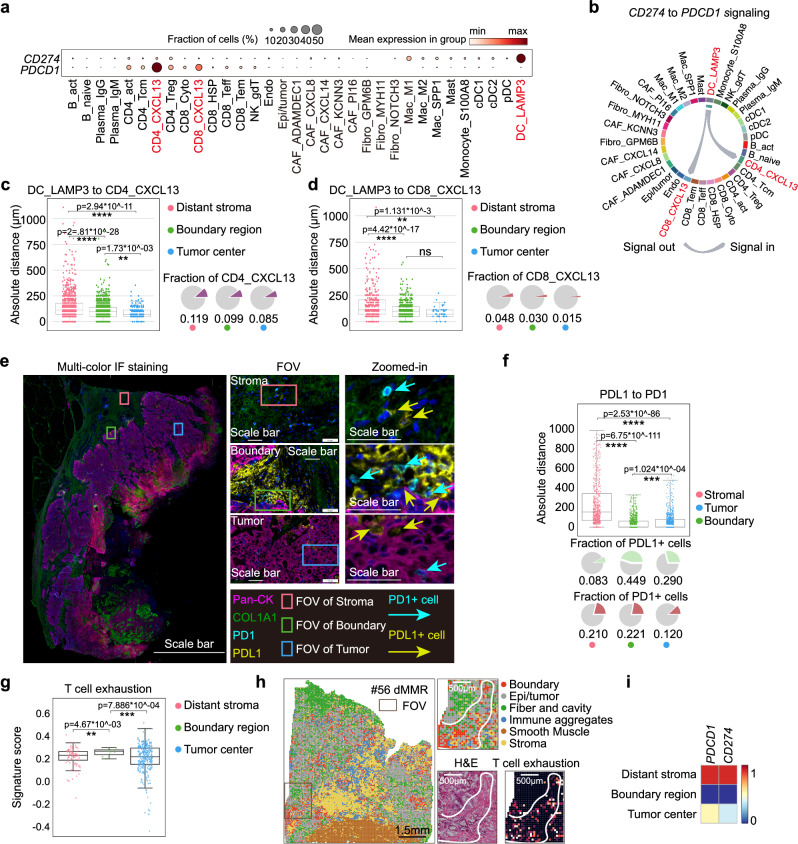
Although LAMP3+DCs are well characterized in hepatocellular carcinoma (HCC) by scRNAseq analysis22, its potential regulatory roles in CRC immunotherapy remain to be underestimated. Interestingly, the expression profiling of PD1-PD-L1 axis, the key factor in determining the therapeutic efficacy of anti-PD1 treatment32, confirmed that CD274 was predominantly expressed on DC_LAMP3, while CD4_CXCL13 and CD8_CXCL13 subsets showed higher PDCD1 expression (Fig. 3a). Of note, the ligand-receptor pair analysis of PD1-PD-L1 axis indicated the potential interactions of DC_LAMP3 towards CD4_CXCL13 and CD8_CXCL13, but not other cell types (Fig. 3b). In parallel, DC_LAMP3 may also interact with CD4_CXCL13 via CD80-CD28 axis in the tumor-stroma boundary of treatment naïve dMMR and dPR/dCR patients (Supplementary Fig 7d, e). Moreover, by calculating the absolute distance in the spatial maps, we found that the CD274-expressing DC_LAMP3 resided closely to the PDCD1-expressing CD4_CXCL13 and CD8_CXCL13 cells in the tumor-stroma boundary as well as tumor center regions (with a proximity less than 100 µm) (Figs. 3c, d). mIF staining data of specimens from treatment naïve dMMR CRC further verified the potential physical juxtaposition (mean distance <100 µm) of PD1- and PD-L1-expressing cells in the tumor-stroma boundary (Fig. 3e, f). Furthermore, when we scored the Stereo-seq data using the exhaustion-related gene signatures generated from a pan-cancer T cell study33,34 in our spatial transcriptomic dataset, we found that T cell exhaustion score was significantly higher in the tumor-stroma boundary when compared to distant stroma or tumor center (Fig. 3g). When further applied these exhausted signatures and PD1-PD-L1 axis into our spatial transcriptomics FOV analysis, our data confirmed an elevation of PDCD1/CD274 and T cell exhaustion signature in the tumor-stroma boundary of treatment naïve dMMR patients (Fig. 3h, i). Interestingly, we noticed that DC_LAMP3 also highly expressed other co-inhibitory molecules, like LGALS9, and co-stimulatory molecules, including CD80, CD86, ICOSLG, CD70, PVR (Supplementary Fig. 7f–h). Since the GSEA analysis showed significant enrichment of regulation of lymphocyte activation in the tumor-stroma boundary of treatment naïve dMMR compared to pMMR (FDR > 3.246 × 10–3; p = 0) (Supplementary Fig. 7i), our data suggested that LAMP3+DCs may co-opt surrounding environment to recruit and regulate multiple aspects of T cells, thereby leading to increased responding T cell numbers for anti-PD1 targeting in dMMR patients.
Fig. 3. The juxtaposition and interactions between LAMP3+DCs and CXCL13-expressing T cells contribute to better ICB response.
a Bubble plots represent the expression profiling of PDCD1 and CD274 in single cell clusters from scRNA-seq dataset. The plots are sized by the fraction of cells with positive gene expression, while the color represents the gene expression level. b The interactions among indicated cell clusters through PD1-PD-L1 axis from scRNA-seq dataset. c The box and whisker plot shows show the nearest distance from DC_LAMP3 to CD4_CXCL13 (N number of the counted cell-cell distances: distant stroma =918, boundary region=1867; tumor center=251) or (d) to CD8_CXCL13 (N number of the counted cell-cell distances: distant stroma =368, boundary region=567; tumor center=45) in distance stroma, tumor-stroma boundary and tumor center from treatment naïve dMMR patients. Data are represented as mean±IQR and analyzed by unpaired 2-tailed Student t test. ns, not significant; *, p < 0.05; **, p < 0.01. Pie charts represent the fractions of CD4_CXCL13 or CD8_CXCL13 in the indicated regions stratified by colors in accordance with (c) d. Data are analyzed by unpaired Student-t test. ns, not significant; ****, p < 0.0001. e Representative mIF images of panCK, COL1A1, PD-L1 and PD1 in treatment naïve dMMR patients (5 samples were analyzed). DAPI was used as a positive control for cell nuclei staining. Scale bars were set as 5 mm for global image (left) and 50μm for FOVs (middle and right). f The box and whisker plot shows the nearest distances from PD1+cells to PDL1+cells in distance stroma, tumor-stroma boundary and tumor center from 7 treatment naïve dMMR patients. Each dot represents a PD1-expressing cell. N number of the counted cell-cell distances: stroma = 750; boundary = 935; tumor = 892. Data are represented as mean ± IQR and analyzed by unpaired 2-tailed Student t test with Bonferroni correction. Ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ***, p < 0.0001. g The box and whisker plot shows the T cell exhaustion scores in the distant stroma, tumor-stroma boundary and tumor center from treatment naïve dMMR patients. Data are represented as mean±IQR and analyzed by unpaired 2-tailed Student t test. Ns not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ***, p < 0.0001. N number of the spots: distant stroma =86, boundary region=12; tumor center=287. h Representative images of H&E staining, FOV, and spatial map of T cell exhaustion scores in the tumor-stroma boundary of the treatment naïve dMMR patient #56. Scale bars were set as 1.5 mm for global image (left) and 500 μm for FOVs71. i Heatmap of PDCD1 and CD274 expressions in the distant stroma, tumor-stroma boundary and tumor center of treatment naïve dMMR patients.
The plasticity of CAF correlates with the immune status of tumor-stroma boundary in ICB non-responders
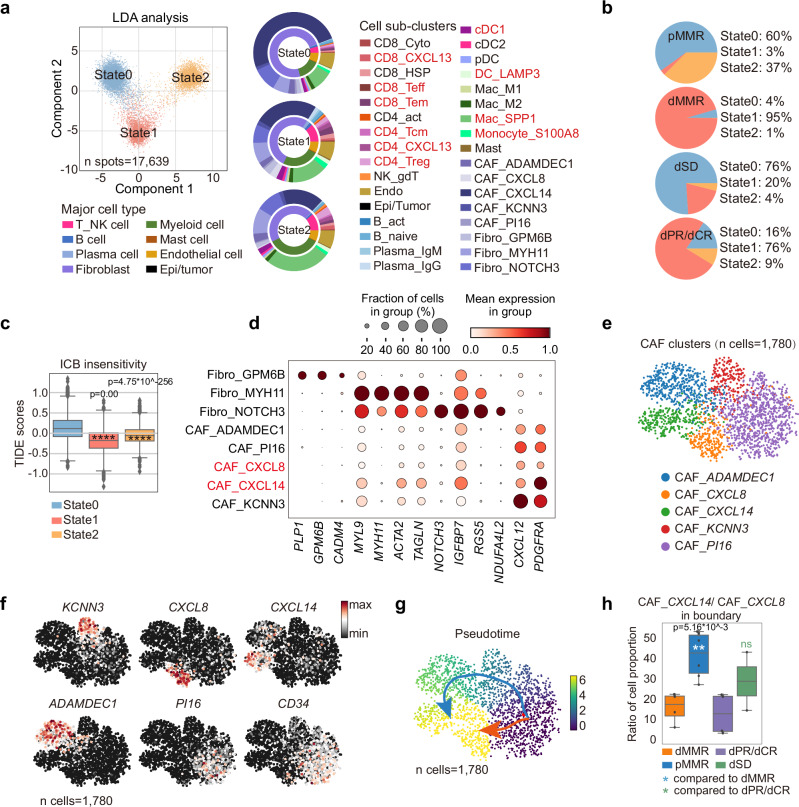
Compared to dMMR, pMMR CRC patients experience limited benefit upon ICB treatment6. As we have pinpointed the potential of tumor-stroma boundary in contributing to ICB respone, we next focused on analysing the spatial transcriptomic features of treatment-naïve pMMR and dSD. We extracted the uncured boundary spatial bins of the four patient groups and applied DDRTree algorithm to study the immune status35–37. Interestingly, three distinctive immune states were identified and projected into 2D axis using linear discriminant analysis (LDA) (Fig. 4a). When applied these immune states into patient groups, we found that the proportion of state 0 was higher in treatment naïve pMMR, whereas dMMR displayed the enrichment towards 95% of state 1 (Fig. 4a, b). In addition, anti-PD1 treatment was associated with the reduction of state 1 and expansion of state 2 in dPR/dCR, but an elevation of state 0 in dSD (Figs. 4a, b). The result suggested that the polarization towards state 0 may correlate with ICB insensitivity of CRC. Indeed, the Tumor Immune Dysfunction and Exclusion (TIDE) analysis, which integrates the expression signatures of T cell dysfunction and T cell exclusion to predict ICB response38,39, verified that state 0 showed a higher TIDE score indicating an immune dysfunction and exclusion status (Fig. 4c). Since we have shown that the frequencies of T cell subsets were significantly lower in the tumor-stroma boundary of pMMR and dSD (Fig. 2 and Supplementary Fig. 5), together, these results suggested that T cell exclusion in the tumor-stroma boundary may contribute to ICB low response in pMMR and dSD.
Fig. 4. The plasticity of CAF determines the immune status of tumor-stroma boundary in ICB non-responders.
a Linear discriminant analysis (LDA) of the three immune states in the tumor-stroma boundary (n spots=17,639). Nested donut charts show the proportions of cell clusters in the three states identified from scRNAseq data. The inner donut represents major immune cell types, while the outer donut represents immune cell subclusters. b Pie charts show the constitutions of immune states in the tumor-stroma boundary of the four patient groups. c The box and whisker plot shows the TIDE scores in the three identified cell states by TIDE algorithm are shown. Data are represented as mean±IQR and analyzed by unpaired 2-tailed Student t test. Ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ***, p < 0.0001.. N number of the boundary spots: state 0 = 8835; state2 = 4450; state 3 = 4354. d Bubble plots of marker gene expressions in fibroblast subsets from the scRNA-seq dataset are shown. The plots are sized by the fraction of cells with positive gene expression, while the color represents the gene expression level. The name of the two CAFs are highlighted in red. e UMAP of CAF subclusters (n cells =1780; n samples: 5 pMMR, 2 dPR/CR, 3 dSD) are shown and (f) the representative marker genes are highlighted in each cluster from scRNAseq dataset. g Pseudo-time inference analysis of the 5 CAF clusters (n cells =1,780; n samples: 5 pMMR, 2 dPR/CR, 3 dSD) by Slingshots. Cells on the trajectories are aligned to the possible differentiation routes by the colored arrows. h The box and whisker plot shows the ratios of CAF_CXCL14/CXCL8 in the tumor-stroma boundary of indicated patient groups. Data are represented as mean±IQR and analyzed by unpaired Student t test. Ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001; ***p < 0.0001. N number of the patients: dMMR = 4; pMMR = 6; dPR/dCR = 4; dSD = 2.
To investigate which cell component(s) contribute to the formation of T cell exclusive tumor-stroma boundary of treatment naïve pMMR and dSD, we exported the RCTD frequencies of cell clusters in each branch from spatial transcriptomics dataset. Consistently, higher proportions of T cell and myeloid cell subsets were observed in state 1 and 2 compared to state 0 (Fig. 4a, nested donut chart). Interestingly, we found that over 70% of cells in state 0 were fibroblasts. In addition, by plotting the top 10 genes of each state, we observed that the top expressed genes in state 0, CXCL14, SELENOP and PYGB were mainly expressed by fibroblast subsets, while MUC12, SELENOP, CEACAM6, PYGB, CEACAM5, CD24 were expressed by epithelial/tumor cells (Supplementary Fig. 8a, b), indicating the potential regulatory role of fibroblasts in the tumor-stroma boundary of treatment naïve pMMR and dSD. We thus identified 5 cancer-associated fibroblasts (CAFs) subsets from other fibroblast clusters by their distinguished expressions of CXCL12 and PDGFRA, which represented the mesenchymal-derived CAF phenotype (Fig. 4d)25,40–42. We then re-embedded these cells in a new UMAP for trajectory inference (Fig. 4e). These 5 CAF subsets were marked by ADAMDEC1, CXCL8, CXCL14, KCNN3 and PI16, respectively (Fig. 4e, f). CAF_PI16 that highly expressed CD34 is reported to be a stem-like fibroblast subset and preserved potent differentiation potentials43. Consistently, our trajectory analysis using Slingshot algorithm44 reflected two distinctive differentiation paths of CAF_PI16 into either CAF_CXCL8 or CAF_CXCL14, with sequential gene expression changes (Fig. 4g and Supplementary Fig. 8c). Transition state genes were identified as incrementally upregulated or downregulated by pseudo-time analysis, in particular CXCL8 and CXCL14 were remarkably upregulated in terminal differentiated CAF_CXCL8 or CAF_CXCL14, respectively (Fig. 4f and Supplementary Fig. 8c). Of note, we found that the ratio of CXCL14/CXCL8-expressed fibroblasts was significantly higher in treatment naïve pMMR compared to dMMR, or dSD compared to dPR/CR (Fig. 4h), thus highlighting the potential role of fibroblast plasticity in ICB response of CRCs. Together, these results prompted us to in depth analyse the phenotype and functions of fibroblast subsets in the tumor-stroma boundary.
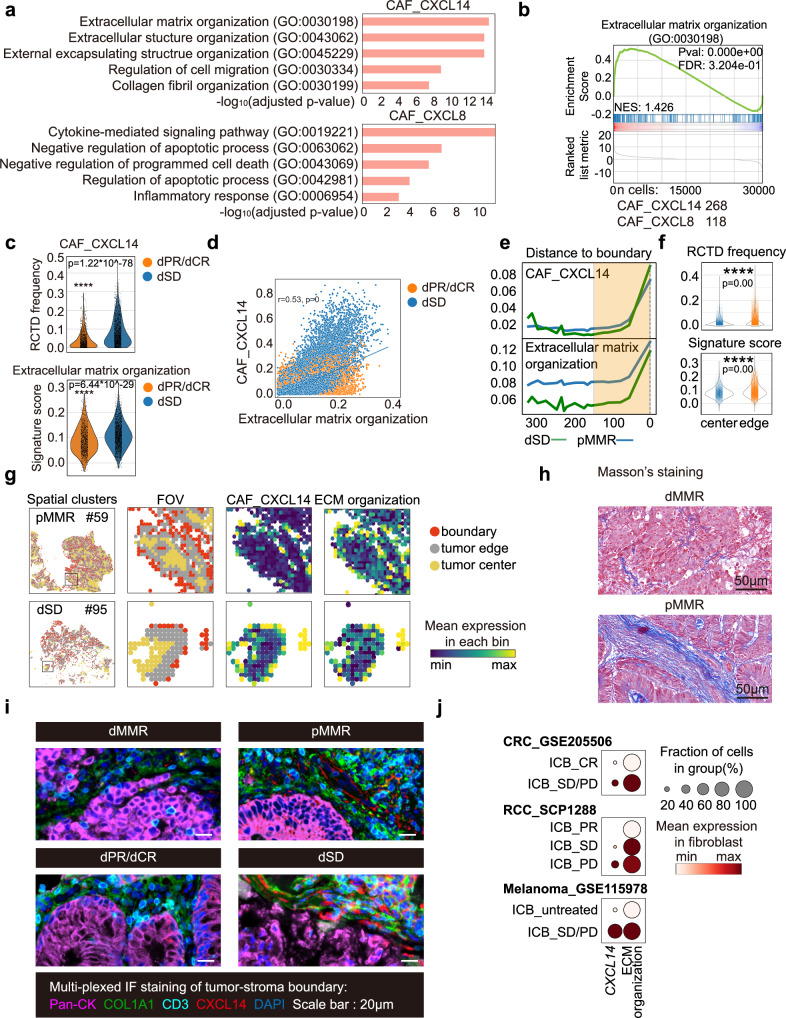
CXCL14+CAFs may contribute to the well-organized matrix structure and T cell exclusion in TME of ICB non-responders
To investigate the potential functions of CAF_CXCL8 or CAF_CXCL14 in the tumor-stroma boundary, gene ontology (GO) term analysis was performed using the top 200 DEGs (ranked by fold change) in CAF_CXCL14 or CAF_CXCL8, respectively. Of note, extracellular matrix (ECM) and structure organization were the top 2 most significantly enriched pathways in CAF_CXCL14 cluster, whereas CAF_CXCL8 showed an enrichment of cytokine-mediated signaling pathway (Fig. 5a). GSEA analysis further confirmed the significant enrichment of ECM organization pathway, which was positively correlated with increased CAF_CXCL14 RCTD frequency in the tumor stroma boundary of dSD, compared to dPR/dCR (r = 0.53, p = 0; Fig. 5b–d). Moreover, the spatial distribution of CAF_CXCL14 RCTD frequency and ECM organization signature displayed similar trend that gradually increased when approaching to the boundary (0-150μm) of treatment naïve pMMR and dSD (Figs. 5e, f). Furthermore, the representative FOV data of treatment naïve pMMR patient #59 and dSD patient #95 suggested the existence of well-organized barrier structure in the tumor-stroma boundary, which was constituted by CAF_CXCL14 and ECM organization (Fig. 5g). To further evaluate the distribution of collagen fiber and CAF_CXCL14, Masson’s trichrome staining and mIF staining were performed. As expected, a well-organized matrix structure and higher level of COL1A1+CXCL14+cells were clearly observed in the tumor-stroma boundary of treatment naïve pMMR but not dMMR (Fig. 5h, i). In addition, higher frequency of COL1A1+CXCL14+cells was also detected in the tumor-stroma boundary of dSD compared to dPR/dCR (Fig. 5i).
Fig. 5. CXCL14+CAFs may contribute to the well-organized matrix structure and T cell exclusion in TME of ICB non-responders.
a The top 5 GO terms of DEGs (p value < 0.001; fold change >2) identified from CAF_CXCL14 and CAF_CXCL8 clusters are shown. Comparison is made by two-tailed t test. b GSEA plot of the upregulated genes related to extracellular matrix organization in CAF_CXCL14 (n cells=268) compared to CAF_CXCL8 (n cells=118) is shown. Comparison is made by two-tailed t test. c RCTD frequencies of CAF_CXCL14 (up) and ECM organization signature scores (below) in the tumor-stroma boundary of dPR/dCR (n spots=1436) and dSD (n spots=2244) patients are shown as violim plots. Data are analyzed by unpaired 2-tailed Student t test. Ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ***, p < 0.0001. d Pearson correlation of RCTD frequencies of CAF_CXCL14 and ECM organization signature scores in dPR/dCR and dSD patients. e Loess smoothed curves and (f) violin plots of RCTD frequencies of CAF_CXCL14 (up) and ECM organization signature scores (below) in tumor ( + 300 μm, n spots = 9313) to the tumor-stroma boundary (0 μm, n spots = 56,357) from treatment naïve pMMR and dSD patients are shown. The tumor edge region ( + 150 μm to 0 μm) is highlighted in the yellow frame. g Representative images of FOV, CAF_CXCL14 and ECM organization scores in the tumor-stroma boundary of treatment naïve pMMR patient #59 and dSD patient #95 are shown. h Representative images of Masson’s trichrome staining from treatment naïve dMMR and pMMR patients (3 samples were analyzed in each group). Scale bar = 50 μm. i Representative mIF images of panCK, COL1A1, CD3 and CXCL14 in indicated patient groups (3 samples were analyzed in each group). DAPI was used as a positive control for cell nuclei staining. Scale bars, 50 μm. j CXCL14 expression level and ECM organization scores in fibroblasts from the three public scRNAseq datasets16,70,71.
A recent study by Pelka et al. reported that CXCL14 is mainly expressed by CAFs from pMMR, but not dMMR patients that may contribute to lower T cell activity10. Consistently, a well-organized extracellular matrix structure was also demonstrated to prevent T cells infiltration45. Therefore, our data suggested that CXCL14+CAF may contribute to the formation of structural barrier by the reprogramming ECM organization and structure, thereby leading to a T cell-exclusive TME of ICB non-responders. Using independent ICB-treated CRC patient cohort (GSE205506) of which transcriptomic and clinical data were available, we further observed a significantly higher CXCL14 expression and ECM organization signature score in CAFs from ICB non-responders compared to responders (Fig. 5j). Consistent higher expressions of CXCL14 were also found in ICB non-responders of the patients with renal cell carcinoma (RCC, SCP1288) and melanoma (GSE115978) (Fig. 5j). These results implicate an important role of CXCL14 in ICB resistance, supporting that targeting CXCL14+CAFs to destroy the structural barrier in the tumor-stroma boundary may sensitize ICB therapeutic efficacy in the majority of pMMR CRCs.
Tumor cells may promote CXCL14+CAFs via IHH/PTCH1 axis to constrain ICB efficacy in pMMR CRC patients
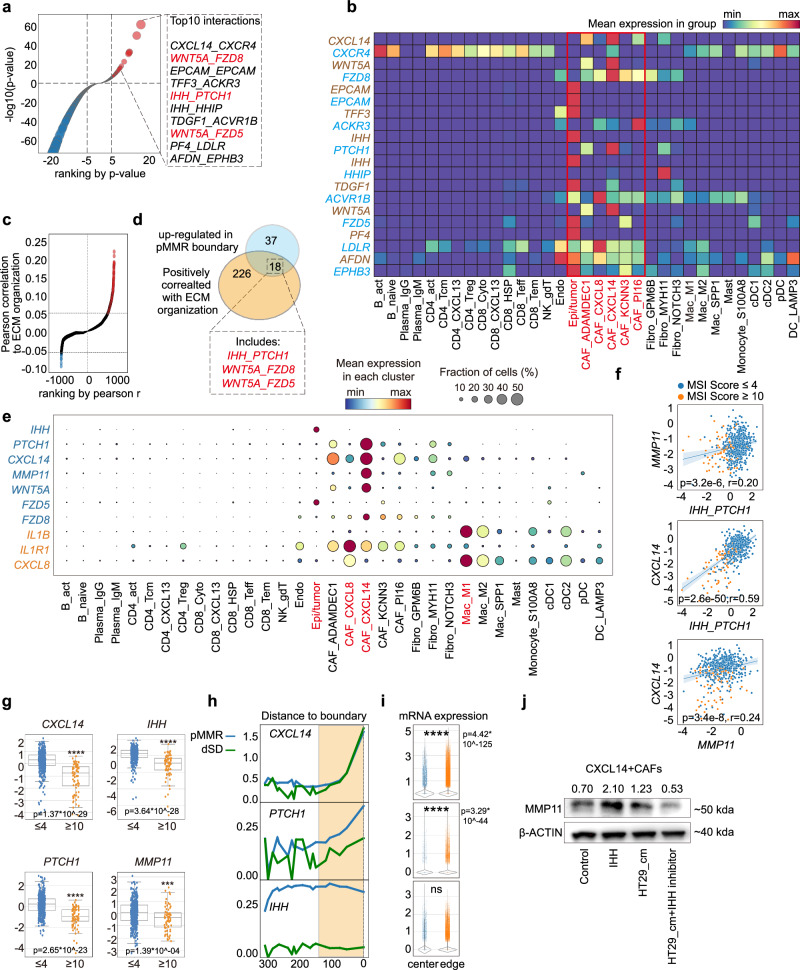
Finally, we explored the potential mechanism that may drive the CAF differentiation into CXCL14+ ones in the tumor-stroma boundary of ICB non-responsive CRCs. Since cell-cell communication in a close proximity is one of the key factors in driving cell differentiation, we next applied our Stereo-seq data into the stLearn algorithm, which co-considers the contributions of cell-cell distance and ligand-receptor expressions in the evaluation46. By comparing the tumor-stroma boundary of treatment naïve dMMR and pMMR, we identified 55 ligand-receptor pairs that were upregulated in pMMR (p < 0.001, fold change > =2, Fig. 6a and Supplementary Data 4). In accordance with the CAF abundance in treatment naïve pMMR, the top10 ranked ligand-receptors exhibited remarkably enriched expressions on epithelia/tumor or CAF_CXCL14, but not immune cells in the tumor-stroma boundary (Fig. 6b). Taken the functional significance of CXCL14+CAFs on ECM organization, we screened 18 ligand-receptor pairs that were up-regulated in the tumor-stroma boundary of treatment naïve pMMR and positively associated with ECM organization scores (Pearson’s correlation r > 0.05; Fig. 6c, d). Of note, we found that IHH_PTCH1 and WNT5A_FZD5/8 were primarily expressed by tumor cells or CAF_CXCL14 (Fig. 6e). Interestingly, the expression of Matrix metallopeptidase 11 (MMP11), a downstream target of IHH/PTCH1/GLI pathway47,48 was predominantly expressed by CAF_CXCL14 (Fig. 6e). Moreover, the expressions of MMP11, IHH, PTCH1 and CXCL14 were significantly higher in MSI-lo tumors (MSI sensor score≤4) compared to MSI-hi ones (MSI sensor score≥10) from the COAD TCGA dataset (n = 572), which were also positively correlated (Fig. 6f, g). Furthermore, by plotting the expression of IHH/PTCH1/CXCL14 from tumor center to boundary ( + 300 μm ~ 0 μm), we found the significant increased expression pattern of CXCL14, PTCH1 and IHH when approaching to the boundary region ( + 150 μm ~ 0μm) in both treatment naïve pMMR and dSD (Fig. 6h, i), which was in accordance with the distributions of CAF_CXCL14 and ECM organization (Fig. 5e, f). As expected, the tumor cell-derived IHH exhibited no difference between tumor center and tumor edge (Fig. 5e, f). Interestingly, we observed higher expression of IHH in pMMR CRC cell lines HT29 and SW620 compared to dMMR CRC cell lines, including HCT116, HCT8 and HCT1549 (Supplementary Fig. 9a). Treatment of CXCL14+CAFs (Guangzhou Saliai Stem cell Science and Technology Co., Ltd) with IHH recombinant protein (Glpbio, 5 μg/mL) resulted in a notable increase in MMP11 release (Supplementary Fig. 9b–d). Furthermore, exposure of CXCL14+CAFs to conditional medium (CM) from pMMR CRC HT29 cells also let to a similar upregulation of MMP11. Importantly, this upregulation could be suppressed by IHH inbitior Vismodegib (Selleckchem, 25 µM) (Fig. 6j). Taken together, these results indicated the potential interaction between pMMR tumor cells and CAF_CXCL14 through IHH_PTCH1 axis.
Fig. 6. Tumor cells promote CXCL14+CAFs via IHH/PTCH1 axis to constrain ICB efficacy in pMMR CRC patients.
a Ligand-receptor pair enrichment analysis (ranked by p value) in tumor-stroma boundary of Stereo-seq dataset. The top 10 enriched ligand-receptor pairs in pMMR (-log10 p value ≥ 5) are highlighted. Comparison is made by two-tailed t test. b The heatmap represents the expressions of the top 10 enriched ligand-receptor pairs in single cell clusters from scRNA-seq dataset. Ligands are labeled with brown, while receptors are labeled with light blue. Epithelia cells and CAFs are highlighted in red. c Pearson correlation of ligand-receptor pairs towards ECM organization scores in tumor-stroma boundary of pMMR patients. The positive correlations (r > 0.05) are labeled in red, and the negative correlations (r < 0.05) are labeled in blue. d Venn diagram of ligand-receptor pairs upregulated in the tumor-stroma boundary of treatment naïve pMMR patients and positively correlated with ECM organization identified 18 overlapped ligand-receptor pairs. e Bubble plots of expression profiling of identified ligand-receptor pairs and IL1B, IL1R1, CXCL8 in single cell clusters from scRNA-seq dataset are shown. The plot are sized by the fraction of cells with positive gene expression, while the color represents the gene expression level. f Pearson correlations and (g) expressions of IHH_PTCH1, MMP11 and CXCL14 from COAD TCGA dataset are shown. Patients are stratified to MSI-hi (MSI sensor score≥10, patient number = 78) and MSI-lo (MSI sensor score≤4, patient number = 494) accordingly. IHH_PTCH1 scores are calculated as sum of IHH and PTCH1 Z-scores. Data are represented as mean±IQR and analyzed by unpaired 2-tailed Student t test. Ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ***, p < 0.0001. The line and the band present the linear regression model and confidence interval (95%) respectively. h Loess smoothed curve and (i) violin plots represent the distribution and expressions of PTCH1, CXCL14, and MMP11 from tumor center ( + 300 μm) to the tumor-stroma boundary (0 μm) in dSD and pMMR patients. Data are represented as mean ± IQR and analyzed by unpaired 2-tailed Student t test with Bonferroni correction. Ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ***, p < 0.0001. N number of the spots: center=9313; edge=56,357. j Western blot analysis of MMP11 in indicated group is shown. β-actin serves as loading control. Number indicates the relative expression towards β-actin. This result represents 3 independent experimental repetitions.
Discussion
Although ICB therapies have revolutionized the treatment paradigm of dMMR CRC patients, the majority of CRC patients with pMMR are barely benefit. A number of recent studies have therefore explored the TME features of dMMR and pMMR tumors from CRC patients at single-cell resolution, revealing the heterogenicity and gene expression programs of fibroblasts and immune cells, and their functional consequences10,11. However, what are the spatial features of TME in dMMR and pMMR patients that may contribute their distinctive response rate to ICB treatment are still unclear. Herein, we charted the cellular and molecular landscape of ICB responsive and non-responsive TME, combining scRNA-seq, spatial transcriptomics, mIF and integrative bioinformatics of 23 treatment naïve or ICB-treated CRC patients. We identified spatially organized cell-cell interactions that contribute to a coordinated multi-cellular tumor-stroma boundary (0 ± 150 µm) in dMMR and pMMR CRC patients, which may contribute to the ICB response. Our data suggested the importance of spatial distributions and regulatory landscape of LAMP3+DCs and CXCL13+T cells in shaping the tumor-stroma boundary of ICB responders. In addition, we pinpointed that the tumor cell-CXCL14+CAF interactions may cause the formation of structural barrier in the tumor-stroma boundary, leading to an immune-exclusive and ICB insensitive TME in pMMR CRC patients. These results clarify the effects of TME spatial architecture, in particular the tumor-stroma boundary on ICB efficacy and highlight the potential of sensitizing ICB response by targeting CXCL14+CAFs in pMMR CRCs.
It has long been shown that TME is a well-organized complex ecosystem, functioning in tumor development, recurrence, metastasis and therapeutic responsiveness. However, studies that underestimate the potential importance of the TME spatial architecture are unable to completely elucidate its complexity. Using high-flux and high-dimensional scRNA-seq and Stereo-seq, we mapped the spatial landscapes of TME in dMMR and pMMR CRC patients, with or without ICB treatment. Our results pinpointed that the immune cells spatial distribution, instead of abundance may differentiate ICB sensitive and insensitive TME in CRC patients. By employing the bifurcation analysis, we revealed that distinctive immune states in the tumor-stroma boundary, instead of the tumor center or stroma region, associate with the clinical outcomes towards ICB treatment. In general, the tumor-stroma boundary of treatment naïve dMMR patients displayed an immune activation state (state 1 & 2) compared to an immune exclusive one (state 0) in pMMR patients. This distinction was also observed in ICB-treated patients, where dPR/dCR displayed an immune activation state (state 1 & 2), whereas dSD demonstrated an immune exclusive state (state 0). Consistently, it was reported that the pre-existence of CD8+T cells, which distinctly located at the invasive tumor margin, were associated with the expression of the PD-1/PD-L1 immune inhibitory axis and may predict response to ICB therapy in melanoma50. Nirmal et al. further highlighted the concept of tumor-stroma boundary in melanoma, where invasion and immunoediting may coexist to regulate tumor progression and metastasis51. A most recent study in HCC patients also identified a tumor immune barrier structure that consisted by SPP1+macrophage and CAFs in ICB non-responders52. Although it is still unclear exactly which spatial features are important in dictating ICB response of individual cancer types as TME spatial characterizations are just at the beginning, taken together with our analysis in CRC patients, it strongly suggests that the distributions and aggregative structures of stroma and immune cells are important in ICB clinical implications across cancer types. Our integrative scRNA-seq and Stereo-seq analysis also enabled the visualization of coordinated cellular neighborhood into a prominent accuracy over other spatial and single transcriptomics integration tools in a benchmark test53. Through continuous mapping of mean RCTD frequencies in our spatial transcriptomic data, we delivered a high-fidelity readout of immune cell distribution curves that covered ±1000μm to tumor-stroma boundary (0μm) to investigate spatial patterns of dMMR and pMMR tumors, as well as their possible alterations under anti-PD1 treatment. Hence, we witnessed a strong interaction between CXCL13+T cells and LAMP3+DCs in dMMR and dPR/CR mediated by the PD1-PD-L1 axis, which was proximity dependent and took place within a distance less than 250μm in tumor-stroma boundary and tumor center, but not in distant stroma. Thus, considering distance between different cells would be essential for juxtracrine signal and cell-cell interaction analysis.
CXCL13 is widely reported to be crucial for the formation of TLS that correlates with better outcomes of ICB treatment in various type of cancers, including breast cancer, melanoma, non-small cell lung cancer (NSCLC) and HCC32. Mechanistically, CXCL13 is mainly secreted by CD4+T cells to track CXCR5+T cells and help facilitate B cell response54. In addition to CD4+ T cells, studies also identified the existence of CXCL13-expressing PD1hiCD8+T cells in TLS with predictive potential in late-stage NSCLC patients under anti-PD1 treatment55. In this study, we additionally observed obvious expressions of CXCL13 in both CD4+ and CD8+ T cell subsets from treatment naïve dMMR compared to pMMR, which we defined as CD4_CXCL13 and CD8_CXCL13. In parallel, anti-PD1-treated dPR/dCR also displayed higher proportions of these CD4_CXCL13 and CD8_CXCL13 subsets, compared to anti-PD1-treated dSD patients. Interestingly, these T cell subsets also expressed high level of PDCD1, granzyme A (GZMA), TIGIT1, LAG3 and HAVCR2, suggesting their possible plasticity in the tumor-stroma boundary. How to drive these T cell subsets into potential effectors instead of exhausted ones for a better ICB efficacy is still unclear. In addition, we found that these T cells were closely located and interacted with LAMP3+DCs via PD1-PD-L1 axis in the tumor-stroma boundary of treatment naïve dMMR and anti-PD1-treated dPR/dCR. In consistent with our findings, previous study in HCC also showed that LAMP3+DCs highly expressed CD274 (PD-L1) and exhibited the physical juxtaposition towards PDCD1-expresing T cell subsets22. Our additional analysis of COAD TCGA data indicated strong associations between LAMP3+DC signature and T cell exhaustion as well as T cell activation signature. Furthermore, the scRNAseq and Stereo-seq data showed high expressions of both immune checkpoint ligands (e.g., CD274) and maturation markers (e.g., CD80, CD86, CD40) in LAMP3+DCs from ICB responders but not non-responders, implying the plasticity of DC functions, independent of their maturation features.
Another important question is to explore the possible mechanisms underlying ICB insensitivity of pMMR CRCs. Our integrative analysis demonstrated that the existence of the structural barrier, induced by ECM organization dysregulation, contributed to the discontinuous of immune influx and the formation of immune exclusive TME. As dMMR patients who showed insensitive to anti-PD1 therapy (dSD) displayed similar structural barrier, we spectulated that this spatial structure in the tumor-stromal boundary may contribute to ICB low response in pMMR CRCs. Our observation was supported by a recent publication in triple negative breast cancer (TNBC), showing that the collagen fiber alignment in the tumor margin determines immune cell tumor infiltration and the formation of ‘cold’ TME56. Importantly, our data further revealed that the abundance of CXCL14+CAFs in the tumor-stroma boundary may contribute to the dysregulation of ECM organization to restrict immune cell tumor infiltration in pMMR. Furthermore, the CXCL14+CAF differentiation may mainly be driven by IHH/PTCH1 pathway through interaction with pMMR tumors. Interestingly, CXCL14+CAFs from pMMR also showed high expression of WNT5A, whereas its receptors FZD5 and FZD8 were expressed by epi/tumor cells or fibroblasts, respectively, suggesting the potential of WNT/β-catenin signaling in modulating CXCL14+CAFs in CRCs. Taken the importance of hedgehog and WNT/β-catenin signals in tumors48,57,58, inhibitors that targeting these pathways may show great potential to sensitize ICB treatment in pMMR by co-opting both tumor cells and CXCL14+CAFs. In summary, our analyses established an integrated molecular and spatial landscape of CRC patients under ICB treatment and provided a rich dataset for therapeutic targeting of the tumor-stroma boundary underlying ICB insensitive TME formation.
While this study provides a thorough characterization of the in vivo architecture of human CRCs, it is subject to certain limitations. Firstly, due to the limited dMMR patient number that showed partial response to the current ICB neoadjuvant therapy, we only have 2 dPRs. Therefore, we are unable to further clarify the differences between partial and complete responders who would display survival or relapse difference. For dMMR patients, ICB has exhibited limited efficacy, with a 0% response rate to anti-PD1 monotherapy and up to 27% PR/SD rate reported in early-stage patients to combinatory approaches4,59. Instead, CRC patients with pMMR showed improved disease-free survival with neoadjuvant chemoradiotherapy, especially for stage III disease60. Therefore, neoadjuvant anti-PD1 therapy is infrequently employed in the clinical management of CRC patients with pMMR. As a consequence, we were also unable to collect tissues from anti-PD1-treated pMMR patients in our current study. In parallel, we were only able to collect patient specimens at a single time point, either before or after ICB treatment, thereby lacking dynamic analysis in the spatial map of TME. New technology like live-seq61, which could provide a time course transcriptome profile at a single-cell resolution, may help Stereo-seq to co-generate an authentic spatiotemporal profiling of TME. Integrative analysis by including new technologies will be essential for a better understanding of how ICB treatment modulates TME and drives the responsiveness in patients. Secondly, the Stereo-seq technique has not reached the single-cell resolution when analyzing heterogenous and complex tumor specimens in the current study. The nanoscale resolution (capture spot diameter: 220 nm; center-to-center distance: 500 nm) of Stereo-seq merely supported an estimate of 1-10 cells in each bin in CRC tumor tissues compared to previous analysis in mouse embryos and axolotl brain62–64. One probable explanation may be the difference on the average cell size and the complexity among different tissues. This observation indicated the limitation of our Stereo-seq platform, which may not be universally applicable, particularly in tissues enriched with small-sized immune cells, such as tumors, spleen and thymus. A more precise spatial map, if we can develop in the future, will be essential for better understanding of how to therapeutically target the spatiotemporal heterogeneity of the complex TME of CRCs, as well as other cancers. For example, enrichment of the variable regions of T and B cell receptor mRNA accompanied with the current Stereo-seq platform would present a compelling strategy for mapping the immune cells, at least T cell and B cells at single cell resolution in tumors in situ. Thirdly, it remains unclear how to therapeutically remodel the tumor-stroma boundary by our identified targets, including CXCL14+CAF, IHH/PTCH1 to sensitize the ICB antibodies in non-responders. Pre-clinical orthotopic mouse models that we previously developed28 may serve as proof of concept for evaluating the therapeutic efficacy of our identified combinatory treatment strategies in the future. Clearly, additional work in preclinical and clinical trials will be needed to better assess the future potential for sensitizing ICB therapy by remodeling the spatial architecture of TME in ICB non-responders.
Methods
Patients
Tumor tissues were collected from 23 patients with CRC who underwent colon resection with or without neoadjuvant ICB treatment at Sun Yat-Sen University Cancer Center. Tumors were staged with the 8th edition of the American Joint Committee of Cancer (AJCC) tumor node-metastasis (TNM) staging classification for CRC. Enhanced CT scans of the chest and abdomen, and MRI scans of the rectum, were used to ascertain the TNM stage. Transrectal ultrasonography or endoscopic ultrasound was used to ascertain the tumor and nodal stage for patients unsuitable for MRI tests because of metal implants or other reasons. Mismatch repair was determined by immunohistochemistry (IHC) for mismatch repair proteins and microsatellite instability status was determined by PCR for microsatellite instability markers. Patient response to neoadjuvant ICB treatment was determined by image-based evaluation such as endoscopy or MRI using Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1 as well as postoperative pathological evaluation according to the criteria from National Comprehensive Cancer Network (NCCN) Tumor Regression Grade (TRG) system. The clinical information including age, gender, MMR/MSI status, TNM stage, anti-PD1 monoclonal antibodies used, course of treatment and tumor response grade, as well as the stratification of the patients in the current study were listed in Supplementary Data 1. This study was done in accordance with the Declaration of Helsinki (B2023-178-01). The protocol was reviewed and approved by The Institutional Review Board of BGI Ethical (BGI-IRB21083-T1). All participants provided written informed consent and the clinical information was collected at Sun Yat-Sen University cancer center.
Sample preparation and processing for Stereo-seq and scRNAseq
For Stereo-seq, tumor specimens were embedded in Tissue-Tec OCT (Sakura, 4583) within 30 min after surgical resection or biopsy collection. Tissue sectioning (5 μm) was performed using Leica Cryostat (CM1950) at –20 °C. The RNA integrity was evaluated and tissue sections with RIN value over 6.0 were proceeded for Stereo-seq analysis. The sections were stained with Hematoxylin and Eosin staining (H&E) (Beyotime, C0105S) and the image were generated using Motic EasyScan System. The tissues sectioned onto the Stereo-seq chips (10 μm) were dried at 37 °C for 4 min, fixed with methanol at –20 °C for 30 min, and then stained using Qubit™ ssDNA Assay Kit (ThermoFisher, Q10212). The tissues were washed using 0.1x SSC solution and permeabilized as described previously62. After permeabilization, in situ reverse transcription was performed at 42 °C for 1.5 h, followed by tissue removal at 55 °C for 1 h. The cDNAs, which included coordinate identifier and unique molecular identifier (MID), were released from Stereo-seq chip and sequenced using BGISEQ-T10.
For scRNA-seq, single cell isolateion was performed using Tumor Dissociation Kit according to the manufacturer’s protocols (Eppendorf ThermoMixer® C). Cell viability was determined using Acridine Orange/Propidium Iodide staining kit (NEXCELOM, CS2-0106). Specimens with a cell viability over 85% were proceeded to scRNA-seq using droplet-based DNAelab-C4 High-throughput Single-cell RNA Library Preparation Set V2.0 (MGI Tech Co., Ltd, Cat. No.: 940-000519-00). The cDNA library was sequenced using BGISEQ-T1.
Sequencing and Raw data processing
The cDNA was sequenced by paired ends. For Stereo-seq, barcodes (CID:1–25 bp, MID: 26–35 bp) and cDNA inserts (100 bp) were located in the forward reads (read1) and the reverse reads (read2), respectively. In scRNA-seq, barcodes (cell barcode 1, 1–10 bp; cell barcode 2, 11–20 bp; and MID: 21–30 bp) and cDNA inserts (100 bp) were in read1 and read2, respectively. The sequencing parameters remained consistent with those previously described62. The raw data processing was carried out using the spatial_RNA_visualization tool (version 5) on the STOMICS online platform (https://cloud.stomics.tech/home.html). In details, the read1 was split into CID sequences (1–25 bp) and UMI sequences (26–35 bp) based on the read length. The CID sequences were then mapped to the white CID list (the BarcodeToMap.h5 file). A single base mismatch was allowed to account for errors in sequencing and amplification (PCR). MID sequences containing either N bases or more than 2 bases with a quality score lower than 10 were filtered out. The remaining valid reads were then aligned to the reference genome (hg38) using STAR65. Aligned reads with a mapping quality (MAPQ) score greater than 10 were counted and annotated as described previously62. Reads sharing the same UMI and CID were considered PCR replicates and collapsed into a single read. The output of spatial_RNA_visualization (version 5) included two gem files: one for the entire slide and one for the tissue cut. These gem files consisted of four columns, containing the gene symbol, x and y coordinates, and UMI counts. After online processing, the tissue cut gem file was downloaded and further analyzed.
Data quality control, integration, clustering and annotation
For stereo-seq, the spot size was set to bin100 (50 μm × 50 μm). The spots with gene numbers lower than 500 were filtered out. The gene expression matrix was normalized using scanpy.pp.normalize_total and logarithmized using scanpy.pp.log1p. The log-transformed matrix was then subjected to batch effect correction using Seurat (V4) built-in canonical correlation analysis, which was achieved by setting the nfeatures parameter to 10,000 and the batch to specimens66. The aligned expression matrix was further processed using SCANPY built-in functions, which included principal components analysis (PCA), neighborhood graph computing (n_pcs=20) and unsupervised clustering using Leiden algorithm (resolution = 3.0). The cluster annotation was determined by the top DEGs in each cluster and the image data.
For scRNA-seq, cells were filtered based on gene number and mitochondria-related gene percentage. Cells with a gene count lower than 300 or higher than 5000, or with over 50% mitochondria-related genes, were excluded from analysis. Genes detected in fewer than 3 cells were filtered out for analysis. After normalization and log transformation, transcriptome annatation was performed using two rounds of batch effect correction with batch balanced k nearest neighbors (BBKNN)17 and unsupervised clustering by Leiden13. In the first round, the BBKNN-aligned single cell transcriptome (set_op_mix_ratio=0.5, batch_key=sample ID) was subjected to unsupervised clustering (resolution=1) and annotated using gene of interests. The 8 major cell clusters including epi/tumor cells (KRT18, EPCAM), T_NK cells (CCL5, TRBC2, CD3E, NGLY), myeloid cells (LYZ, CD14), fibroblasts (COL1A1, PDGFRA), mast cells (GATA2, TPSAB1), endothelial cells (PECAM1), B cells (CD19, MS4A1) and plasma cells (JCHAIN, IGHA1, IGHG4) were annotated. For each cell type, a gene expression matrix (with raw counts) was constructed and subjected to another independent run of batch effect correction (BBKNN), unsupervised clustering, and annotation of cell sub-clusters as indicated in Supplementary Fig. 3b.
Calculation of boundary distance and construction of immune influx stream
To determine the distances from the tumor-stroma boundary, we first set the boundary spots as 0. For other spots, we calculated their center-to-center distances to the nearest boundary spot, which served as their distances to the boundary. The distances to the boundary were used to define different regions: stroma, immune aggregates, smooth muscle, fiber, and cavity regions were considered outside the tumor (indicated by negative numbers), while the boundary distances of the epi/tumor were represented as positive numbers (Fig. 2d). To analyze the distribution of immune cell types in the spatial map, we averaged the RCTD frequencies for immune cells at the same distances. These averaged frequencies were then aligned and stacked, starting from the furthest stroma to the tumor center. To visualize the influx stream, we centered the stacked Y-axis around zero by using the ‘wiggle’ baseline in the matplotlib.stackplot function. For data presentation, we uniformed the Y-axis range to (-0.15, 0.15).
Cell cluster deconvolution
Deconvolution of cell clusters in the spatial map was calculated using RCTD algorithm in R package spacexr-2.0.018 in combined with scRNAseq dataset. The raw counts of scRNA-seq (with subtype annotation) and the stereo-seq were input to construct the RCTD object. For stereo-seq data, we set the minimal UMI count to 0 to avoid filing out any spots. Since the cell diameters typically ranged from 5 to 12 μm, the 50 μm-binned spatial spots may contain multiple cell types that might not be consistent across the spot. Therefore, we ran the RCTD in full mode, without restrictions on the number of cell types. In this study, the normalized probabilities of cell types were referred to as “RCTD frequencies”, which were exported and integrated into SCANPY h5ad objects for further visualization and analysis.
Immune state analysis
The immune state of the tumor-stroma boundary spots was identified using Monocl35–37. The raw counts of the tumor-stroma boundary spots were input to construct the CellDataSet object by setting expressionFamily as negbinomial.size. Genes expressed in fewer than 10 spots and spots with fewer than 100 detected genes were filtered out. Next, the spots were subjected to stratification using AddCellType function, with CXCL14 and LAPTM5 serving as markers to define the “cold” spots (CXCL14 counts ≥1) and “hot” (CXCL14 counts <1 & LAPTM5 counts >1) spots. To construct the state transition model, we reduced the dimensionality of the boundary spots using DDRTree and ordered the spots on the tree-like structures using orderCells function. The resulting tree-like structure consisted of three branches. Spots assigned to the same branch were annotated as the same state. These annotated boundary spots were then used as input for linear discriminant analysis, employing the scikit-learn machine learning tool67.
Analysis of CAF trajectories and pseudo-temporal gene expressions
The CAF trajectory inference was performed using the python implementation of the Slingshot pseudotime algorithm44. To account for batch effects, the raw counts of the 5 CAF subpopulations were extracted for a new round of batch effect correction (by BBKNN) and UMAP embedding. The resulting UMAP was then used as input for the Slingshot analysis, with the starting node set as PI16_CAF. To examine the pseudo-temporal gene expression pattern, the Monocle software was employed, specifically utilizing the built-in function for branched expression analysis modeling (BEAM_res <- BEAM(rds, branch_point = 1, cores = 1)). Subsequently, the top 100 genes ranked by log2 fold change were visualized using the plot_pseudotime_heatmap function.
Cell lines and culture
The human cancer-associated fibroblasts68 were obtained from Guangzhou Saliai Stem Cell Science and Technology (Cat# CatiCel1-0030a)68. The human cancer-associated fibroblasts68 were obtained from Guangzhou Saliai Stem Cell Science and Technology (Cat# CatiCel1-0030a), and the human colorectal cancer cell lines HCT116, HCT8, HCT15, SW480, SW620, and HT29 were obtained from the American Type Culture Collection (ATCC). These cell lines were subjected to short tandem repeat (STR) testing at the time of purchase. The STR testing results were provided by the vendor and third-party testing agencies, ensuring the authenticity of the cell lines.
A commercial mycoplasma detection kit (Vazyme, Cat# D101-01) was used for monthly contamination testing of the cell lines. The procedure involved adding the cell culture supernatant to the reaction system, incubating at 60 °C for 1 h, and then determining the contamination status of the cell lines by comparing the results with negative and positive controls. Only cell lines with a negative mycoplasma status were used in this study.
All cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, New York, USA) or RPMI-1640 (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS, HyClone, Massachusetts, USA) and 1% penicillin/streptomycin (Gibco, USA) in a 37 °C humidified chamber with 5% CO2.
Real-time quantitative PCR analysis (qRT‒PCR)
Total RNA from cells was extracted using TRIzol reagent (Thermo Fisher, Cat# 15596018). The quality and quantity of total RNA were determined by measuring absorbance at 260 nm/280 nm using NanoDrop Spectrophotometer ND-2000 (NanoDrop Biotechnologies). 1 μg RNA was reverse transcribed to cDNA using HiScript III RT SuperMix( + gDNA wiper). Quantitative PCR (qPCR) was performed in triplicates by qPCR SYBR Green Master Mix (Yeasen, Cat# 11201ES08) on a LightCycler 480 System (Roche). Relative gene expression was determined using the 2-ΔΔCT method and the primer sequences using in this study were listed as follows:
MMP11 forward: 5ʹ-CCGCAACCGACAGAAGAGG-3ʹ
MMP11 reverse: 5ʹ-ATCGCTCCATACCTTTAGGGC-3ʹ
ACTB forward: 5ʹ-CATGTACGTTGCTATCCAGGC-3ʹ
ACTB reverse: 5ʹ-CTCCTTAATGTCACGCACGAT-3ʹ
Western blotting
Protein lysates from cells were prepared using RIPA lysis buffer (Merck Millipore, Cat# 20-188), containing protease and phosphatase inhibitor cocktail (Beyotime, Cat# P1046). Protein concentration was measured by DC Protein Assay (Beyotime, Cat# P0010). 20–80 μg of protein lysates were separated by sodium dodecyl sulfate polycrylamide gel electrophoresis (SDS-PAGE) (EpiZyme) and transferred to 0.45 μm PVDF membrane (Merck Millipore). After blocking with 5% non-fat milk in 1 × TBST, membranes were probed with primary antibodies β-actin (Proteintech, Cat# 20536-1-AP, 1:1000) and MMP11 (Abcam, Cat# ab53143, 1:1000) overnight at 4 °C, followed by secondary antibodies conjugated with horseradish peroxidase (HRP) (Proteintech, Cat# SA00001-2, 1:5000) for 2 h at room temperature. Antibody-antigen complexes were detected with Enhanced Chemiluminescence (GE Healthcare Life Sciences) and ChemDoc Imaging System (Bio-Rad).
Enzyme-linked immunosorbent assay (ELISA)
Secretion of MMP11 in the culture supernatant was masured using MMP11 ELISA kit (ELK Biotechnology, Cat# ELK2113) following the manufacturer’s instructions.
Masson’s trichrome staining
The paraffin sections were dewaxed and rehydrated, and then stained with Weigert iron hematoxylin for 10 min. Differentiation was performed using 1% hydrochloric acid alcohol after water wash. The sections were washed with water until the blue color was restored (approximately 15 min). Next, the sections were stained with Ponceau Acid Magenta for 10 min, followed by a water wash and treatment with phosphomolybdic acid for 5 min. Toluidine Blue staining was applied for 5 min, and the sections were then treated with 1% glacial acetic acid for 1 min. After dehydration and rendering them transparent, the sections were sealed with neutral gum. The image was taken by microscope (Olympus, Germany).
Multiplex immunofluorescent (mIF) staining
Human tissues were collected and fixed in 4% paraformaldehyde (Sigma-Aldrich) for 24 h, washed in 70% ethanol and embedded in paraffin. Five-millimeter sections from paraffin embedded tumor tissues were deparaffinized, rehydrated, and rinsed in distilled water. Antigen retrieval was done by using a pressure cooker with 1 mM EDTA buffer, pH 8.0, for 10 min. The endogenous peroxidase activity was then blocked by incubating the slides in 3% hydrogen peroxide in methanol for 30 min. The sections were then stained with Hematoxylin and Eosin staining (H&E). The mIF staining was carried out using PDOne 6-plex TSA-Rab IF staining kit (Cat No. 10236100100; Paovue, Beijing, China) for several cycles following the manufacturer’s instructions. For each cycle, primary antibodies were stained at room temperature for 1 h, followed by HRP conjugated secondary antibodies at room temperature for 10 min. The fluorescence dye was then applied to the slides at room temperature for 10 min. The signal was examined by VS200 (Olympus, Germany), in conjunction with UPLXAPO 20x objective lens.
Calculation of cell proportions and cell-to-cell distance
The calculation of cell proportions and cell to cell distance was applied to mIF images and spatial map. The cell-to-cell distance was calculated using Euclidean distance. The field of view (FOV) encompassing the tumor, tumor-stroma boundary, and stroma was manually selected based on the expressions of COL1A1 and panCK. The selected FOV was then analyzed using QuPath software for precise cell identification and counting. The cell proportion was calculated using the formula:
1. .
Statistics and reproducibility
All box and whisker plots are presented as the mean ± interquatile range (IQR: 25–75%) with bars indicating upper and lower extremes. Statistical significance was determined using a two-tailed unpaired Student’s t test (with Bonferroni correction for multiple comparison). Exact P values were labeled in the figures. Difference was considered to be significant if P < 0.05, (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). For the reproducibility of the results, all the experiments were repeated independently for over 3 times with similar results.
Materials availability
The antibodies for mIF staining are listed as follow:
| Reagent and resource | |||
| antibodies | Dilution | resource | identifier |
| Anti-PD1 rabbit mAb | 1:200 | Cell Signaling | CST86163 |
| Anti-COL1A1 rabbit mAb | 1:500 | Cell Signaling | CST72026S |
| Anti-IL-8 rabbit mAb | 1:200 | Cell Signaling | CST94407 |
| Anti-PDL1 antibody | 1:200 | Abcam | Ab213524 |
| Anti-pan cytokeratin antibody | 1:200 | Abcam | Ab7753 |
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Information
Source data
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (Grant No. 82002628 and 82473398), Guangdong Basic and Applied Basic Research Foundation (Grant No. 2021A1515010096 and 2023A1515010243), Chinese Society of Clinical Oncology Foundation (Grant Nos. Y-HR2018-319, Y-L2017-002, and Y-JS2019-009), Sun Yat-sen University Basic Research Fund (Grant No. 19ykpy180), and the open research funds from the Sixth Affiliated Hospital of Guangzhou Medical University, Qingyuan People’s Hospital (202011-103, 202301-314). This work was also supported by State Key Laboratory of Oncology in South China and Guangdong Provincial Key Laboratory of Human Disease Genomics. We thank China National GeneBank, BGI Research, Shenzhen for data storage, open use, and management.
Author contributions
Study design: Y.F., W.M. and R.Z., X.D.; Patient recruitment and sample preparation: R.Z. Library construction and data generation: Y.F., W.M., R.Z., Y.G., X.Z., M.L., M.W., and A.c.; Pathological annotation and tumor regression evaluation: Z.P. Data analysis and coding: Y.F., Y.Z., Y.L., Y.L.; Experimental validation: Y.Z.; Manuscript drafting: Y.F., J.Z.; Review & editing: D.K., Y.Z., L.L., J.Z. and R.Z.; Conceptualization and supervision: G.C., L.L., J.Z. and R.Z..
Peer review
Peer review information
Nature Communications thanks Noel (F) De Miranda, Helmut Dolznig, and Elisabeth Letellier for their contribution to the peer review of this work. A peer review file is available.
Data availability
The raw sequencing FASTQ files of the scRNA-seq and stereo-seq data generated in this study could be accessed on Genome Sequence Archive (accession number: PRJCA020107, https://ngdc.cncb.ac.cn/gsa-human/browse/HRA005647). The processed matrix of Stereo-seq and scRNAseq was deposited on STOmicsDB69 of China National GenBank Database (accession number: STT0000036, https://db.cngb.org/stomics/project/STT0000036). The publicly available TCGA with Z-scored gene expression matrix and the MSI scores could be downloaded through https://www.cbioportal.org/study/summary?id=coadread_tcga_pan_can_atlas_2018. The publicly available single cell RNA-seq data could be downloaded through the following links: CRC (gene expression omnibus: GSE205506): https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE20550616; RCC (single cell portal: SCP1288): https://singlecell.broadinstitute.org/single_cell/study/SCP1288/tumor-and-immune-reprogramming-during-immunotherapy-in-advanced-renal-cell-carcinoma#study-download70; melanoma (gene expression omnibus: GSE115978): https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE11597871. Source data are provided as a Source Data file. The remaining data are available within the Article, Supplementary Information or Source Data file. Source data are provided with this paper.
Competing interests
Employees of BGI Research Hangzhou/Shenzhen/Chongqing have stock holdings in BGI. All other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yu Feng, Wenjuan Ma.
Contributor Information
Gong Chen, Email: chengong@sysucc.org.cn.
Longqi Liu, Email: liulongqi@genomics.cn.
Jingying Zhou, Email: zhoujy@cuhk.edu.hk.
Rongxin Zhang, Email: zhangrx@sysucc.org.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-54710-3.
References
- 1.Arora, S. et al. Existing and emerging biomarkers for immune checkpoint immunotherapy in solid tumors. Adv. Ther.36, 2638–2678 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.André, T. et al. Pembrolizumab in microsatellite-instability–high advanced colorectal cancer. N. Engl. J. Med.383, 2207–2218 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Le, D. T. et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science357, 409–413 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le, D. T. et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med.372, 2509–2520 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, G. et al. Neoadjuvant PD-1 blockade with sintilimab in mismatch-repair deficient, locally advanced rectal cancer: an open-label, single-centre phase 2 study. Lancet Gastroenterol. Hepatol.8, 422–431 (2023). [DOI] [PubMed] [Google Scholar]
- 6.Chalabi, M. et al. Neoadjuvant immunotherapy leads to pathological responses in MMR-proficient and MMR-deficient early-stage colon cancers. Nat. Med. 26, 566–576 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Yaghoubi, N., Soltani, A., Ghazvini, K., Hassanian, S. M. & Hashemy, S. I. PD-1/ PD-L1 blockade as a novel treatment for colorectal cancer. Biomed. Pharmacother.110, 312–318 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Chen, J. et al. Reprogramming immunosuppressive myeloid cells by activated T cells promotes the response to anti-PD-1 therapy in colorectal cancer. Signal Transduct. Target Ther.6, 4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho, W. W. et al. Dendritic cell paucity in mismatch repair-proficient colorectal cancer liver metastases limits immune checkpoint blockade efficacy. Proc. Natl. Acad. Sci. USA118, 10.1073/pnas.2105323118 (2021). [DOI] [PMC free article] [PubMed]
- 10.Pelka, K. et al. Spatially organized multicellular immune hubs in human colorectal cancer. Cell184, 4734–4752.e4720 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qi, J. et al. Single-cell and spatial analysis reveal interaction of FAP+ fibroblasts and SPP1+ macrophages in colorectal cancer. Nat. Commun.13, 1–20 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang, R. et al. Spatial transcriptome unveils a discontinuous inflammatory pattern in proficient mismatch repair colorectal adenocarcinoma. Fundam. Res.3, 640–646 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Traag, V. A., Waltman, L. & Van Eck, N. J. From Louvain to Leiden: Guaranteeing well-connected communities. Sci. Rep.9, 1–12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joanito, I. et al. Single-cell and bulk transcriptome sequencing identifies two epithelial tumor cell states and refines the consensus molecular classification of colorectal cancer. Nat. Genet. 54, 963–975 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen, K. et al. Resolving cancer-stroma interfacial signalling and interventions with micropatterned tumour-stromal assays. Nat. Commun.5, 5662 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, J. et al. Remodeling of the immune and stromal cell compartment by PD-1 blockade in mismatch repair-deficient colorectal cancer. Cancer Cell41, 1152–1169.e1157 (2023). [DOI] [PubMed] [Google Scholar]
- 17.Polanski, K. et al. BBKNN: fast batch alignment of single cell transcriptomes. Bioinformatics36, 964–965 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cable, D. M. et al. Robust decomposition of cell type mixtures in spatial transcriptomics. Nat. Biotechnol.40, 517–526 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xun, Z. et al. Reconstruction of the tumor spatial microenvironment along the malignant-boundary-nonmalignant axis. Nat. Commun.14, 933 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, C. et al. Neuropilin-1 is a T cell memory checkpoint limiting long-term antitumor immunity. Nat. Immunol.21, 1010–1021 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horton, B. L. et al. Lack of CD8(+) T cell effector differentiation during priming mediates checkpoint blockade resistance in non-small cell lung cancer. Sci. Immunol.6, eabi8800 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang, Q. et al. Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell179, 829–845.e820 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Lee, A. H. et al. Neoadjuvant PD-1 blockade induces T cell and cDC1 activation but fails to overcome the immunosuppressive tumor associated macrophages in recurrent glioblastoma. Nat. Commun.12, 6938 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Magen, A. et al. Intratumoral dendritic cell-CD4(+) T helper cell niches enable CD8(+) T cell differentiation following PD-1 blockade in hepatocellular carcinoma. Nat. Med. 29, 1389–1399 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sahai, E. et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat. Rev. Cancer20, 174–186 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Dijk, N. et al. Preoperative ipilimumab plus nivolumab in locoregionally advanced urothelial cancer: the NABUCCO trial. Nat. Med26, 1839–1844 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Niogret, J. et al. Follicular helper-T cells restore CD8(+)-dependent antitumor immunity and anti-PD-L1/PD-1 efficacy. J. Immunother. Cancer9, 10.1136/jitc-2020-002157 (2021). [DOI] [PMC free article] [PubMed]
- 28.Yang, W. et al. A selective HDAC8 inhibitor potentiates antitumor immunity and efficacy of immune checkpoint blockade in hepatocellular carcinoma. Sci. Transl. Med.13, eaaz6804 (2021). [DOI] [PubMed] [Google Scholar]
- 29.Yang, M. et al. CXCL13 shapes immunoactive tumor microenvironment and enhances the efficacy of PD-1 checkpoint blockade in high-grade serous ovarian cancer. J. Immunother. Cancer9, 10.1136/jitc-2020-001136 (2021). [DOI] [PMC free article] [PubMed]
- 30.Groom, J. R. & Luster, A. D. CXCR3 ligands: redundant, collaborative and antagonistic functions. Immunol. Cell Biol.89, 207–215 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Longo, S. K., Guo, M. G., Ji, A. L. & Khavari, P. A. Integrating single-cell and spatial transcriptomics to elucidate intercellular tissue dynamics. Nat. Rev. Genet.22, 627–644 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thommen, D. S. et al. A transcriptionally and functionally distinct PD-1+ CD8+ T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat. Med.24, 994–1004 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caushi, J. X. et al. Transcriptional programs of neoantigen-specific TIL in anti-PD-1-treated lung cancers. Nature596, 126–132 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng, L. et al. Pan-cancer single-cell landscape of tumor-infiltrating T cells. Science374, abe6474 (2021). [DOI] [PubMed] [Google Scholar]
- 35.Qiu, X. et al. Single-cell mRNA quantification and differential analysis with Census. Nat. Methods14, 309–315 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qiu, X. et al. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods14, 979–982 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol.32, 381–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fu, J. et al. Large-scale public data reuse to model immunotherapy response and resistance. Genome Med.12, 21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang, P. et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med.24, 1550–1558 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feig, C. et al. Targeting CXCL12 from FAP-expressing carcinoma-associated fibroblasts synergizes with anti-PD-L1 immunotherapy in pancreatic cancer. Proc. Natl Acad. Sci. USA110, 20212–20217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Izumi, D. et al. CXCL12/CXCR4 activation by cancer-associated fibroblasts promotes integrin beta1 clustering and invasiveness in gastric cancer. Int J. Cancer138, 1207–1219 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Verginadis, I. I. et al. A stromal Integrated Stress Response activates perivascular cancer-associated fibroblasts to drive angiogenesis and tumour progression. Nat. Cell Biol.24, 940–953 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buechler, M. B. et al. Cross-tissue organization of the fibroblast lineage. Nature593, 575–579 (2021). [DOI] [PubMed] [Google Scholar]
- 44.Street, K. et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics19, 477 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu, T., Zhou, L., Li, D., Andl, T. & Zhang, Y. Cancer-associated fibroblasts build and secure the tumor microenvironment. Front Cell Dev. Biol.7, 60 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pham, D. et al. Robust mapping of spatiotemporal trajectories and cell-cell interactions in healthy and diseased tissues. Nat. Commun.14, 7739 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Diao, Y. et al. Identification of novel GLI1 target genes and regulatory circuits in human cancer cells. Mol. Oncol.12, 1718–1734 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skoda, A. M. et al. The role of the Hedgehog signaling pathway in cancer: A comprehensive review. Bosn. J. Basic Med. Sci.18, 8–20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jacob, S., Aguado, M., Fallik, D. & Praz, F. The role of the DNA mismatch repair system in the cytotoxicity of the topoisomerase inhibitors camptothecin and etoposide to human colorectal cancer cells. Cancer Res.61, 6555–6562 (2001). [PubMed] [Google Scholar]
- 50.Tumeh, P. C. et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature515, 568–571 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nirmal, A. J. et al. The spatial landscape of progression and immunoediting in primary melanoma at single cell resolution. Cancer Discov12, 1518–1541 (2022). [DOI] [PMC free article] [PubMed]
- 52.Liu, Y. et al. Identification of a tumour immune barrier in the HCC microenvironment that determines the efficacy of immunotherapy. J. Hepatol.78, 770–782 (2023). [DOI] [PubMed] [Google Scholar]
- 53.Li, B. et al. Benchmarking spatial and single-cell transcriptomics integration methods for transcript distribution prediction and cell type deconvolution. Nat. Methods19, 662–670 (2022). [DOI] [PubMed] [Google Scholar]
- 54.Sautes-Fridman, C., Petitprez, F., Calderaro, J. & Fridman, W. H. Tertiary lymphoid structures in the era of cancer immunotherapy. Nat. Rev. Cancer19, 307–325 (2019). [DOI] [PubMed] [Google Scholar]
- 55.Thommen, D. S. et al. A transcriptionally and functionally distinct PD-1(+) CD8(+) T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat. Med24, 994–1004 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun, X. et al. Tumour DDR1 promotes collagen fibre alignment to instigate immune exclusion. Nature599, 673–678 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Berman, D. M. et al. Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature425, 846–851 (2003). [DOI] [PubMed] [Google Scholar]
- 58.Hanna, A. & Shevde, L. A. Hedgehog signaling: modulation of cancer properies and tumor mircroenvironment. Mol. Cancer15, 24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Emiloju, O. E. & Sinicrope, F. A. Neoadjuvant immune checkpoint inhibitor therapy for localized deficient mismatch repair colorectal cancer: A review. JAMA Oncol.9, 1708–1715 (2023). [DOI] [PubMed] [Google Scholar]
- 60.Ye, S. B. et al. Association of mismatch repair status with survival and response to neoadjuvant chemo(radio)therapy in rectal cancer. NPJ Precis Oncol.4, 26 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen, W. et al. Live-seq enables temporal transcriptomic recording of single cells. Nature608, 733–740 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen, A. et al. Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays. Cell185, 1777–1792.e1721 (2022). [DOI] [PubMed] [Google Scholar]
- 63.Wei, X. et al. Single-cell Stereo-seq reveals induced progenitor cells involved in axolotl brain regeneration. Science377, eabp9444 (2022). [DOI] [PubMed] [Google Scholar]
- 64.Chen, A. et al. Single-cell spatial transcriptome reveals cell-type organization in the macaque cortex. Cell, 10.1016/j.cell.2023.06.009 (2023). [DOI] [PubMed]
- 65.Dobin, A. et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell184, 3573–3587.e3529 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pedregosa, F. et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res.12, 2825–2830 (2011). [Google Scholar]
- 68.O’Reilly, E. A. et al. The fate of chemoresistance in triple negative breast cancer (TNBC). BBA Clin.3, 257–275 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu, Z. et al. STOmicsDB: A comprehensive database for spatial transcriptomics data sharing, analysis and visualization. Nucleic Acids Res52, D1053–D1061 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bi, K. et al. Tumor and immune reprogramming during immunotherapy in advanced renal cell carcinoma. Cancer Cell39, 649–661.e645 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jerby-Arnon, L. et al. A cancer cell program promotes T cell exclusion and resistance to checkpoint blockade. Cell175, 984–997.e924 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Information
Data Availability Statement
The raw sequencing FASTQ files of the scRNA-seq and stereo-seq data generated in this study could be accessed on Genome Sequence Archive (accession number: PRJCA020107, https://ngdc.cncb.ac.cn/gsa-human/browse/HRA005647). The processed matrix of Stereo-seq and scRNAseq was deposited on STOmicsDB69 of China National GenBank Database (accession number: STT0000036, https://db.cngb.org/stomics/project/STT0000036). The publicly available TCGA with Z-scored gene expression matrix and the MSI scores could be downloaded through https://www.cbioportal.org/study/summary?id=coadread_tcga_pan_can_atlas_2018. The publicly available single cell RNA-seq data could be downloaded through the following links: CRC (gene expression omnibus: GSE205506): https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE20550616; RCC (single cell portal: SCP1288): https://singlecell.broadinstitute.org/single_cell/study/SCP1288/tumor-and-immune-reprogramming-during-immunotherapy-in-advanced-renal-cell-carcinoma#study-download70; melanoma (gene expression omnibus: GSE115978): https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE11597871. Source data are provided as a Source Data file. The remaining data are available within the Article, Supplementary Information or Source Data file. Source data are provided with this paper.