Abstract
Renal cell carcinoma (RCC) ranks as a prevalent malignant neoplasm, with clear cell renal cell carcinoma (ccRCC, also known as KIRC) accounting for approximately 75% of all RCC cases. The farnesoid X receptor (FXR, encoded by NR1H4), functioning as a nuclear receptor, plays a crucial role in regulating gene transcription. Although the involvement of FXR in tumors of the digestive system and in acute kidney injury has been extensively studied, its specific role in the pathogenesis of ccRCC has yet to be thoroughly investigated. Consequently, the objective of our current investigation is to uncover the functional roles of FXR in ccRCC. In this study, plasmids for the overexpression of FXR were constructed, and small interfering RNA (siRNA) constructs were designed. Dual-luciferase reporter assays confirmed a direct binding interaction between FXR and the promoter of the matrix metalloproteinase 7 (MMP-7) gene. Additionally, a mouse xenograft model elucidated the regulatory effect of FXR on MMP-7 in the context of tumor growth. This study elucidates how FXR regulates the promotion of ccRCC through the MMP-7-mediated EMT pathway. Interestingly, FXR is typically regarded as a tumor suppressor gene that affects gastrointestinal tumors, providing a potential new therapeutic direction for ccRCC.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-80368-4.
Keywords: FXR, MMP-7, EMT, ccRCC
Subject terms: Oncogenes, Cancer, Cancer genetics, Cancer epigenetics
Introduction
In the year 2022, RCC was positioned as the 14th leading cause of cancer burden among 36 major malignancies worldwide, with a reported incidence of 434,419 new cases and a mortality rate of 155,702 deaths globally1. RCC is predominantly classified into three principal subtypes: ccRCC, papillary (pRCC), and chromophobe (chRCC), with additional rare variants including those of the collecting duct, medullary, and those related to hereditary leiomyomatosis and renal cell cancer (HLRCC) syndromes2–4. The ccRCC subtype is so named due to the characteristic lipid-clearing appearance observed during histological slide preparation, and RCC constitutes the most frequent histological variant of kidney cancer5. RCC constitutes a considerable health hazard to the global population.
FXR is a member of the nuclear receptor superfamily of ligand-activated transcription factors, primarily expressed in the liver, intestines, kidneys, and adrenal glands6. As a bile acid-activated nuclear receptor, FXR plays a pivotal role in regulating bile acid metabolism by modulating synthesis, transport, and facilitating bile excretion7,8. FXR exerts crucial functions in the pathogenesis of various tumors. For example, in colorectal cancer, it impedes tumor progression through the activation of dehydrogenoreductase 9 (DHRS9)9, thereby suppressing cellular oxidative phosphorylation, or by engaging the suppressor of cytokine signaling 3 (SOCS3) gene to inhibit the (JAK2/STAT3) signaling pathway10. In esophageal squamous cell carcinoma, FXR acts as a tumor suppressor by countering the (ERK1/2) signaling pathway11. Interestingly, the role of FXR is context-dependent and can exhibit paradoxical effects in certain cancers. In breast cancer, for instance, it promotes bone metastasis by mimicking the bone microenvironment and upregulating the expression of runt-related transcription factor 2 (RUNX2)12. Furthermore, in pancreatic cancer with lymph node metastasis, FXR augments the migratory and invasive capabilities of tumor cells13.
In the urogenital system, investigations into the FXR have garnered significant attention. In the context of bladder cancer, the FXR agonist GW4064 has been shown to suppress the migratory and invasive capacities of human bladder cancer cells by downregulating the expression of tissue plasminogen activator B and matrix metalloproteinase 2 (MMP2)14. Concerning renal function, evidence suggests that FXR plays a crucial role in promoting water reabsorption under physiological conditions and in maintaining cell viability under conditions of high osmotic pressure. Moreover, in pathological states, FXR has been implicated in the attenuation of apoptosis, autophagy, and iron accumulation within renal tubular epithelial cells15. Notably, the role of FXR in ccRCC remains unexplored, highlighting a significant gap in current research.
The present investigation elucidates that FXR plays a pivotal role in the regulation of MMP-7 expression in ccRCC. Matrix metalloproteinases (MMPs) constitute a family of zinc-dependent endopeptidases, encompassing over 21 homologs in humans and a diverse array of orthologs across different species. Numerous MMPs, such as MMP-2 and MMP-9, are produced by stromal cells within the tumor microenvironment, including fibroblasts, myofibroblasts, inflammatory cells, and endothelial cells. In contrast, certain MMPs, including MMP-7, are predominantly expressed by the cancer cells themselves16. MMP7, the smallest member of the MMP family17, exhibits specific physiological functions in the kidney18, including the proteolysis of podocyte slit diaphragm proteins, which can result in proteinuria19, and the promotion of Fas ligand (FasL) expression in interstitial fibroblasts, thereby facilitating apoptosis of these cells20. Furthermore, MMP-7 is implicated in a variety of biological processes, including the epithelial-mesenchymal transition (EMT), by virtue of its capacity to induce FasL expression21. These observations highlight the critical importance of FXR-mediated regulation of MMP-7 in the pathogenesis of ccRCC, and they suggest potential therapeutic implications for understanding tumor progression and developing novel treatment strategies.
The present study elucidates the pivotal role of FXR in the pathogenesis of ccRCC by modulating the EMT pathway through MMP-7, which facilitates cancer progression. Initially, the expression levels of FXR in RCC tissues were comprehensively examined, along with the impact of altered FXR expression on the migratory and invasive capabilities of cancer cells. Subsequently, similar assessments of MMP-7 expression and function were performed. To validate the regulatory relationship between FXR and MMP-7, dual-luciferase reporter gene assays were employed to confirm the molecular interactions. Additionally, in vivo tumor formation studies in murine models were conducted to corroborate these findings. This research provides novel insights into the functional role of FXR in RCC and suggests potential therapeutic avenues for its clinical management.
Materials and methods
Cell culture and treatments
786-O, 769-P, HK-2, and 293 T cells were obtained from the American Type Culture Collection (ATCC), where they are authenticated by STR analysis and routinely tested for mycoplasma contamination. These cells were cultured in a humidified atmosphere at 37 °C with 5% CO2. 786-O, and 769-P cells were cultured in RPMI-1640 medium (Invitrogen, USA), while HK-2 and 293 T cells were cultured in DMEM medium (Invitrogen, USA). All culture media were supplemented with 10% fetal bovine serum (GIBCO, USA), 1% penicillin/streptomycin (Hyclone).
Lentiviral vectors and transfection
The coding sequence of FXR was cloned into the PMX vector to generate the FXR overexpression plasmid. siRNAs targeting FXR and MMP-7 were purchased from Sangon Biotech. The overexpression plasmid for MMP-7, pCMV-MMP-7, was obtained from Miaoling Biology. All transfections were conducted according to the manufacturer’s instructions using the Lipofectamine 6000 reagent from Beyotime Biotechnology.
RNA isolation, quantitative real-time PCR (qRT-PCR)
Cellular RNA was initially extracted using TRIzol reagent (Invitrogen, USA), followed by reverse transcription performed with the PrimeScript™ RT reagent kit (TakaRa, Japan). Subsequently, qRT‐PCR was conducted using the LightCycler 480 detection system. The relative mRNA levels of the gene of interest were determined using the 2^-ΔΔCt method and normalized to GAPDH expression. Specific primer sequences are listed in Supplementary Table S1.
Protein extraction and Western blot
Cells were lysed using RIPA buffer, and protein concentrations were determined with the BCA protein assay kit (Beyotime, Shanghai, China). Subsequently, cell proteins were separated by SDS-PAGE and transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, USA). Membranes were blocked with 5% skim milk for one hour at room temperature, followed by overnight incubation at 4 °C with primary antibodies. After washing, membranes were then incubated with appropriate secondary antibodies for one hour at room temperature. Protein bands were detected using the ChemiDocTM XRS + system, and protein abundance was quantified using Image J software. Antibody details are provided in Supplementary Table S2.
Immunohistochemistry (IHC) and immunofluorescence staining
Three pairs of kidney renal clear cell carcinoma (KIRC) tissues were obtained from patients undergoing urological surgery at Renmin Hospital of Wuhan University. The privacy rights of human subjects were always respected. All participants, human data, and tissues were acquired after obtaining informed consent, and this study was approved by the Ethics Committee of Renmin Hospital of Wuhan University. All methods were carried out in accordance with the principles outlined in the Declaration of Helsinki. In the IHC experiment, tissue sections were initially subjected to standard dewaxing followed by hydration using citrate buffer. After blocking with 3% hydrogen peroxide and 10% goat serum, tissue sections were incubated overnight at 4 °C with a specific antibody against FXR (dilution 1:100). Subsequently, sections were incubated at 37 °C for 1 h with a secondary antibody. Visualization was achieved using diaminobenzidine (DAB, Servicebio, G1212-200 T), and tissue sections were observed under a microscope by two researchers. For immunofluorescence staining, antibodies against FXR (1:100) and MMP-7 (1:100) were used simultaneously in the staining procedure. All research involving human participants complies with the Declaration of Helsinki.
Luciferase reporter assay
For the Luciferase reporter assay, the upstream 2000 bp region of the MMP-7 promoter was cloned into the pGL3 luciferase reporter plasmid. Two potential binding sites of FXR with MMP-7 were predicted using the JASPAR website, and the plasmid construction was carried out by Miaoling Biology. In the experiment, the plasmid containing the MMP-7 promoter-driven Firefly luciferase reporter gene was co-transfected with the pTK-RL plasmid into cells. Additionally, siFXR was introduced. The experiment was conducted using the Dual Luciferase Reporter Gene Assay Kit (Beyotime, Shanghai, China). This setup allows for the assessment of FXR’s regulatory effect on the MMP-7 promoter activity through monitoring Firefly luciferase activity normalized to Renilla luciferase activity as an internal control.
Wound-healing assays
Cells were cultured in six-well plates until reaching 90% confluence. Using a 10 μL pipette tip, artificial vertical scratches were created on the cell monolayer. Subsequently, cells were maintained in serum-free medium for an additional 24 h. Images of wound closure were captured under a microscope at 0, 12, and 24 h, respectively.
Transwell assays
In the Transwell assay, the lower chamber was filled with 600 μL of RPMI-1640 medium containing 30% FBS. The upper chamber filters were pre-coated with 50 μL of Matrigel (Servicebio, G4130), and 10 × 104 cells were seeded into each upper chamber. The cells were then incubated at 37 °C for 24 h. After incubation, non-migratory cells on the upper surface of the Transwell membrane were washed off with PBS. The migrated or invaded cells underneath the membrane were fixed with 4% paraformaldehyde (Beyotime, P0099) and stained with 1% crystal violet (Beyotime, C0121) . Subsequently, these cells were observed under a microscope.
Cell proliferation assays
A total of 4,000 corresponding treated cells were seeded into a 96-well plate. After 36 or 48 h, 10 μl of CCK-8 solution was added to each well. The plate was then incubated in a 37 °C incubator for 2 h, and the absorbance at 450 nm was measured.
Animal experiments
6-week-old BALB/c nude mice were randomly divided into 3 groups (n = 5). Group 1 received subcutaneous injections of 5 × 106 786-O siRNA Negative Control (siNC) cells (left flank) and 786-O siFXR cells (right flank). Group 2 received injections of 5 × 106 786-O siNC cells + pCMV-T7 (left flank) and 786-O siNC cells + pCMV-MMP-7 (right flank). Group 3 received injections of 5 × 106 786-O siFXR cells + pCMV-T7 (left flank) and 786-O siFXR cells + pCMV-MMP-7 (right flank). Tumor volumes were assessed every 3 days, Euthanize the mice two weeks later by inhaling 5% isoflurane and collect the tumors. All procedures followed ethical guidelines for animal experiments.
Bioinformatical analysis
Online analysis of FXR mRNA and protein expression levels was conducted using data from The Cancer Genome Atlas (TCGA) and the Clinical Proteomic Tumor Analysis Consortium (CPTAC) available on the UALCAN website. The GSE53757 dataset from the GEO database was divided into two groups based on FXR expression levels, followed by differential analysis and the creation of a volcano plot.
Statistical analysis
All experiments were conducted at least three times. Data in this study are presented as mean ± SD and were analyzed using SPSS 22.0 and GraphPad Prism 10 software. Differences among groups were compared using Student’s t-test or one-way ANOVA. A p-value less than 0.05 was considered statistically significant.
Results
FXR expression is elevated in ccRCC tissues and cell lines
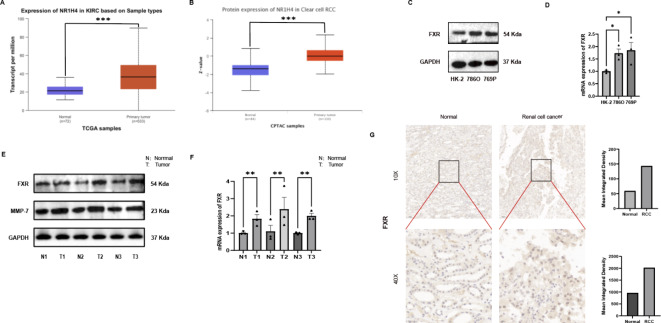
A comprehensive analysis of data from TCGA and CPTAC revealed significant upregulation of FXR mRNA and protein levels within RCC tumor tissues (Fig. 1A,B)22,23. To corroborate these findings, a comparative assessment of FXR protein and mRNA levels was conducted across renal tubular epithelial cells (HK-2) and a panel of renal cancer cell lines (786-O, 769-P). Our analysis revealed that the 786-O and 769-P cell lines exhibited substantially elevated expression of FXR when compared with HK-2 cells (Fig. 1C,D). Furthermore, mRNA and protein expression profiles were isolated and analyzed from three pairs of RCC tissues, which further validated the heightened expression of FXR observed initially (Fig. 1E,F). Finally, immunohistochemical staining of RCC tumor tissues, when compared to adjacent normal renal tissues, confirmed a marked increase in protein levels of FXR within the tumor samples (Fig. 1G). Collectively, these findings provide robust evidence for the overexpression of FXR in both RCC tissues and established cell lines, suggesting a potential pivotal role for FXR in the pathogenesis of RCC.
Fig. 1.
FXR expression is elevated in ccRCC tissues and cell lines. (A) The Ualcan website utilized the TCGA database to assess elevated RNA expression levels of FXR in tumor tissues. (p = 1.62458935193399E-12) (B) The Ualcan website utilized the CPTAC database to examine increased protein expression levels of FXR in tumor tissues. (p = 1.10805474525263E-14). (C) FXR protein expression levels in ccRCC cell lines. (D) FXR mRNA expression levels in ccRCC cell lines. (E) FXR protein expression levels in normal tissues (N) and ccRCC tissues (T). (F) FXR RNA expression levels in normal tissues (N) and ccRCC tissues (T). (G) Immunohistochemical findings comparing tumor and normal tissues.
FXR promotes migration and invasion of ccRCC cells in vitro
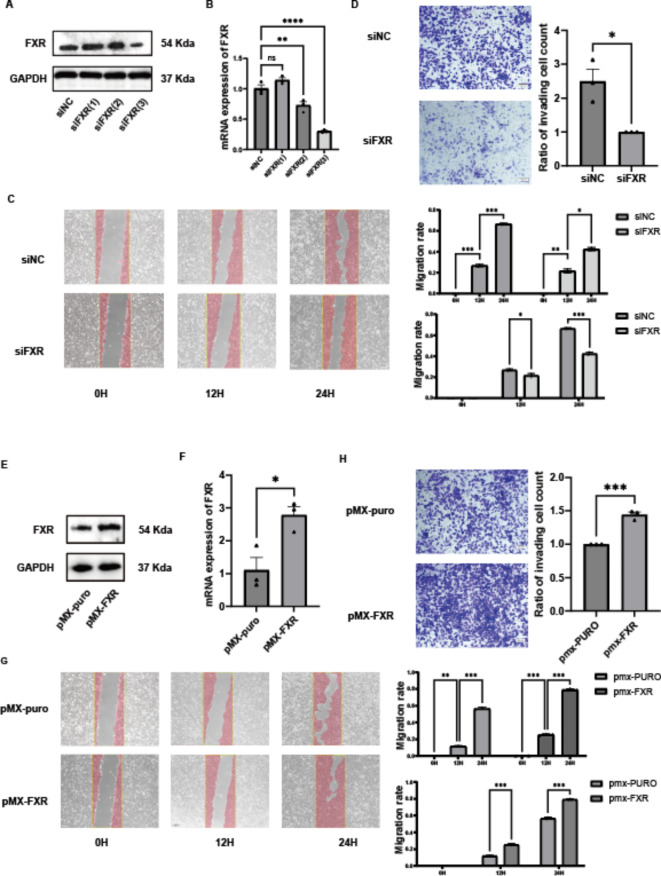
Migration and invasion are critical phenotypes of cancer cells. To investigate the effects of FXR on the migration and invasion abilities of ccRCC cells, FXR expression was manipulated through knockdown and overexpression experiments. For FXR knockdown, 786-O cells were transfected with three different FXR-specific small interfering RNA (siRNAs), and protein levels of FXR (Fig. 2A) as well as mRNA levels (Fig. 2B) were evaluated. Results showed that siFXR(3) was the most effective, therefore, siFXR(3) was selected for subsequent experiments . Transwell assays were conducted where 10 × 104 cells were seeded into the upper chamber with Matrigel-coated membranes. After 24 h of incubation at 37 °C, cells that migrated through the membrane were fixed, stained, and observed under a microscope. FXR knockdown resulted in a decrease in the number of cells that migrated through the Matrigel in Transwell assays (Fig. 2C). Similarly, FXR knockdown significantly reduced the migration rate of 786-O cells at 12 and 24 h (Fig. 2D). Overall, these findings indicate that knockdown of FXR reduces the migration and invasion capabilities of ccRCC cells.
Fig. 2.
FXR promotes migration and invasion of ccRCC cells in vitro. (A) Protein expression levels of FXR after FXR knockdown. (B) mRNA expression levels of FXR after FXR knockdown. (C) Scratch assay of 786O cells after FXR knockdown. (D) Transwell assay of 786O cells after FXR knockdown. (E) Protein expression levels of FXR after FXR overexpression. (F) mRNA expression levels of FXR after FXR overexpression. (G) Scratch assay of 786O cells after FXR overexpression. (H) Transwell assay of 786O cells after FXR overexpression.
To further validate the impact of FXR on migration and invasion abilities, FXR was overexpressed in ccRCC cells.786-O cells were transfected with PMX-FXR plasmid containing the FXR coding sequence, and protein (Fig. 2E) and mRNA levels (Fig. 2F) of FXR were assessed. Transfection with PMX-FXR plasmid led to increased levels of FXR mRNA and protein in 786-O cells, confirming effective overexpression. Transwell assays showed that overexpression of FXR increased the number of 786-O cells that migrated through the Matrigel Similarly, Scratch assays revealed that overexpression of FXR increased the migration rate of 786-O cells at 12 and 24 h. (Fig. 2G,H). In conclusion, FXR promotes migration and invasion of ccRCC cells, as evidenced by both knockdown and overexpression experiments.
MMP-7 expression is elevated in ccRCC tissues and cell lines and its expression correlates with FXR expression
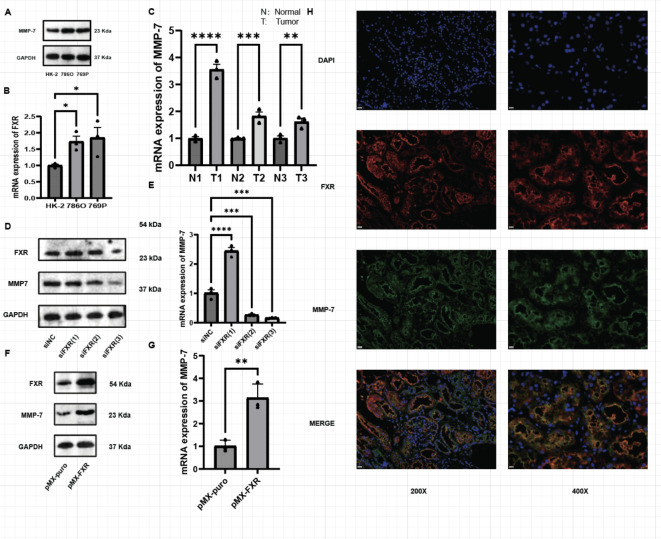
Existing studies have demonstrated that FXR regulates many genes in various cancers9,24–26, including members of the MMP family such as MMP-2, MMP-7, and MMP-9, which are associated with FXR. Furthermore, MMP family genes play significant roles in the proliferation and migration of RCC27. PCR results indicate that the expression levels of MMP-2, MMP-7, and MMP-9 are elevated in RCC cell lines (Fig. S1A–C). To further investigate, the dataset GSE53757 from the GEO database was categorized into two groups based on FXR expression levels, followed by differential analysis. The results revealed that MMP-7 exhibited the highest correlation with FXR (Fig. S1D). Thus, MMP-7 has been identified as a potential target gene of FXR, warranting further investigation in subsequent experiments. Initially, experiments were conducted to explore MMP-7 expression in ccRCC tissues using protein and mRNA extracted from three pairs of tumor tissues. Similarly, MMP-7 protein (Fig. 3A) and mRNA (Fig. 3B) levels were also higher in ccRCC cell lines compared with HK-2 cell line. Our results indicate elevated levels of MMP-7 protein (Fig. 1E) and mRNA (Fig. 3C) in ccRCC tumor tissues. Thus, MMP-7 is indeed highly expressed in both ccRCC tissues and cell lines.
Fig. 3.
MMP-7 expression is elevated in ccRCC tissues and cell lines and its expression correlates with FXR expression. (A) MMP-7 mRNA expression levels in ccRCC cell lines. (B) MMP-7 mRNA expression levels in ccRCC tissues. (C) Protein expression levels of MMP-7 and FXR after FXR knockdown. (D) mRNA expression levels of MMP-7 after FXR knockdown. (E) Protein expression levels of MMP-7 and FXR after FXR overexpression. (F) mRNA expression levels of MMP-7 after FXR overexpression. (G) Immuno-fluorescence of FXR and MMP-7 in ccRCC tissues.
To validate the correlation between FXR and MMP-7 expression, MMP-7 expression was examined after knocking down or overexpressing FXR. Initially, 786-O cells were transfected with the three siFXR variants mentioned earlier, revealing that MMP-7 protein expression levels paralleled changes in FXR expression (Fig. 3D), with consistent trends observed in mRNA levels (Fig. 3E). Overexpression studies of FXR also indicated increased MMP-7 protein (Fig. 3F) and mRNA (Fig. 3G) expression levels. Furthermore, immunofluorescence staining of FXR and MMP-7 (Fig. 3H) demonstrated overlapping expression patterns, further supporting the correlation between FXR and MMP-7 expression.
Therefore, our findings strongly suggest a regulatory relationship between FXR and MMP-7 expression in ccRCC.
MMP-7 promotes migration and invasion of CcRCC cells
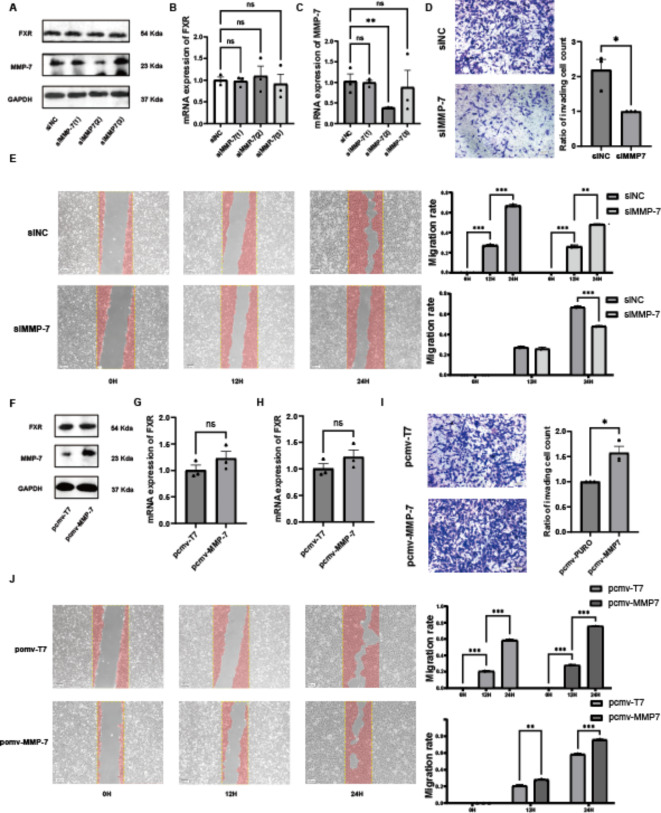
To explore the effects of MMP-7 on migration and invasion in ccRCC, knockdown and overexpression experiments targeting MMP-7 were conducted. Three different MMP-7 siRNAs were transfected into 786-O cells, and protein levels (Fig. 4A) and mRNA levels (Fig. 4B) were assessed. The results showed that siMMP-7 (2) had the most effective knockdown efficiency. Knockdown of MMP-7 did not significantly affect the mRNA and protein expression of FXR (Fig. 4C). Subsequent experiments used siMMP-7 (2).
Fig. 4.
MMP-7 Promotes Migration and Invasion of CcRCC Cells. (A) Protein expression levels of FXR and MMP-7 after MMP-7 knockdown. (B) mRNA expression levels of FXR after MMP-7 knockdown. (C) mRNA expression levels of MMP-7 after MMP-7 knockdown. (D) Scratch assay of 786O cells after MMP-7 knockdown. (E) Transwell assay of 786O cells after MMP-7 knockdown. (F) Protein expression levels of FXR and MMP-7 after MMP-7 overexpression. (G) mRNA expression levels of FXR after MMP-7 overexpression. (H) mRNA expression levels of MMP-7 after MMP-7 overexpression. (I) Scratch assay of 786O cells after MMP-7 overexpression. (J) Transwell assay of 786O cells after MMP-7 overexpression.
Transwell assays were conducted, with 10 × 104 cells plated in each upper chamber and incubated for 24 h at 37 °C. Following fixation and staining, a decreased number of cells migrated through the Matrigel after MMP-7 knockdown was observed (Fig. 4D). Similarly, Scratch assays were performed using 786-O cells. After MMP-7 knockdown, the migration rate of 786-O cells significantly decreased at 12 and 24 h (Fig. 4E). These findings indicate that knockdown of MMP-7 reduces the migration and invasion capabilities of ccRCC cells. To further verify the impact on migration and invasion in ccRCC, MMP-7 was overexpressed. The pCMV-MMP-7 plasmid, containing the FXR coding gene, was transfected into 786-O cells, and protein levels (Fig. 4F) and mRNA levels (Fig. 4G) were measured. Transfection with pCMV-MMP-7 increased both mRNA and protein levels of MMP-7 in 786-O cells, confirming the effectiveness of the pCMV-MMP-7 plasmid. Importantly, FXR mRNA and protein expression remained unaffected (Fig. 4H). Transwell assays showed an increased number of cells migrating through Matrigel after MMP-7 overexpression (Fig 4I). Similarly, Scratch assays revealed increased migration rates of 786-O cells at 12 and 24 h post overexpression of MMP-7 (Fig. 4J). These results indicate that overexpression of MMP-7 enhances the migration and invasion capabilities of ccRCC cells.
In summary, MMP-7 promotes migration and invasion of ccRCC cells. Furthermore, overexpression or knockdown of MMP-7 does not affect FXR expression. Previous studies have suggested that overexpression or knockdown of FXR significantly affects MMP-7 expression. Thus, it can be concluded that FXR regulates MMP-7 expression.
Overexpression of MMP-7 rescues the effects of FXR knockdown on migration and invasion capabilities in ccRCC
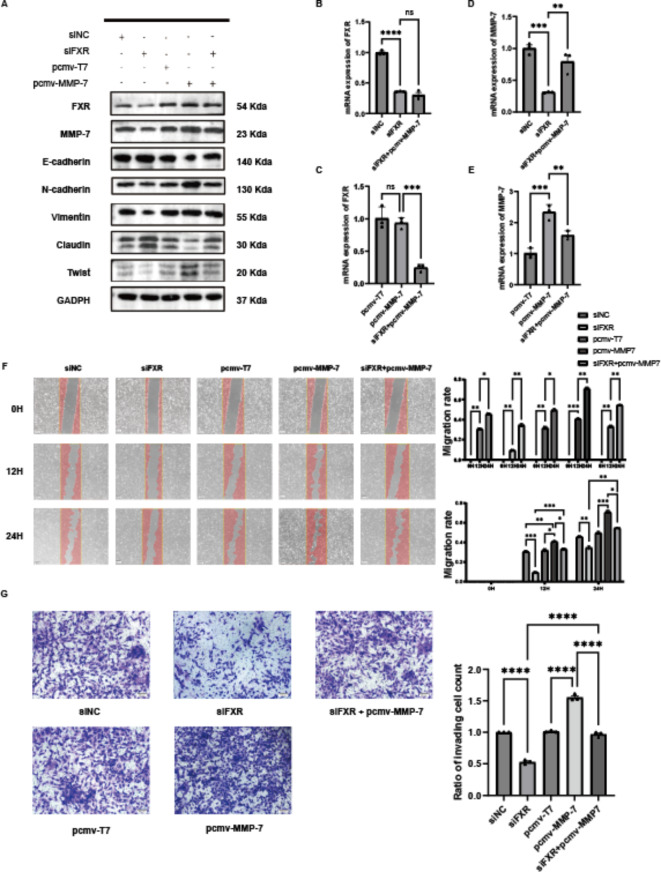
Through literature review and bioinformatics analysis, it was found that FXR is associated with the EMT pathway in ccRCC. The EMT pathway is closely linked to tumor invasion and metastasis, and the association of MMP-7 with the EMT pathway has been confirmed in another study 24 Antibodies targeting the EMT pathway were purchased, and protein extracts from cells treated with siFXR and pCMV-MMP-7 were analyzed (Fig. 5A). It was observed that after FXR knockdown, both FXR and MMP-7 protein expression decreased. Concurrently, the expression levels of Twist, Vimentin, and E-cadherin in the EMT pathway decreased, while Claudin and N-cadherin increased. Following MMP-7 overexpression, FXR protein expression remained unchanged, and MMP-7 expression increased. Additionally, the levels of Twist, Vimentin, and E-cadherin in the EMT pathway increased, while Claudin and N-cadherin decreased. Importantly, co-treatment with siFXR and pCMV-MMP-7 reversed the EMT-related proteins expression changes induced by siFXR alone.
Fig. 5.
Overexpression of MMP-7 rescues the effects of FXR knockdown on migration and invasion capabilities in ccRCC. (A) Protein expression of FXR, MMP-7, GAPDH, and EMT-related proteins in rescue experiments overexpressing MMP-7 after FXR knockdown. (B) mRNA expression of FXR, MMP-7, and GAPDH in rescue experiments overexpressing MMP-7 after FXR knockdown. (C) Scratch assay and quantitative analysis in rescue experiments. (D) Transwell assay and quantitative analysis in rescue experiments.
Furthermore, the mRNA levels of FXR and MMP-7 in cells were examined following the aforementioned treatments (Fig. 5B–E). The trends in mRNA levels were consistent with those observed for protein levels: FXR and MMP-7 mRNA levels decreased after FXR knockdown, remained unchanged for FXR after MMP-7 overexpression, and the mRNA expression levels of MMP-7 are elevated. Co-overexpression of MMP-7 on the background of FXR knockdown reduced the effects caused by FXR knockdown. Later, phenotypic experiments were performed based on the aforementioned grouping, including scratch assays and Transwell assays. In the scratch assay (Fig. 5F), quantitative analysis of migration rates indicated that knocking down FXR reduced the migration capability of 786-O cells, whereas overexpression of MMP-7 increased their migration capability. Moreover, co-overexpression of MMP-7 on the background of FXR knockdown reversed the effects induced by FXR knockdown. Similarly, the Transwell assay (Fig. 5G) showed consistent results: FXR knockdown reduced the invasive and migratory abilities of 786-O cells, whereas MMP-7 overexpression enhanced these abilities. Co-overexpression of MMP-7 on the background of FXR knockdown reversed the effects of FXR knockdown. Moreover, experiments were conducted in which FXR was overexpressed and MMP-7 was subsequently knocked down. The expression of EMT-related proteins (Figure S2A), along with results from Transwell assays (Figure S2B) and scratch assays (Figure S2C), showed opposite results compared to those observed previously. To validate that the results of the Transwell and scratch assays were not influenced by cell proliferation, it was necessary to conduct CCK8 assays using cells subjected to the same treatment conditions. The results indicated that there were no significant differences in cell proliferation among the groups at 36 h (Figure S3A–F), with notable differences emerging only at 48 h (Figure S3G–H). Since all Transwell and scratch assays were completed within 24 h, the impact of cell proliferation on these results can be excluded. These results further demonstrate that FXR regulates the progression of clear cell renal cell carcinoma via MMP-7-mediated modulation of the EMT pathway.
Dual-luciferase reporter assays indicated that FXR directly interacts with the promoter region of MMP-7
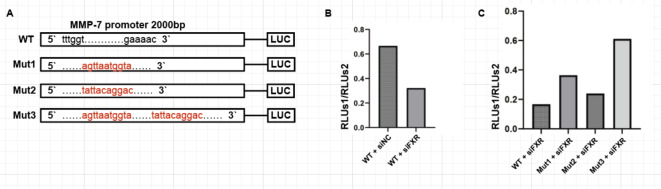
To further validate the regulatory role of FXR on MMP-7, dual-luciferase reporter assays were conducted using 293 T cells. The promoter region of MMP-7 encompassing the first 2000 base pairs was cloned into the pGL3 luciferase reporter vector. Based on the predictions from JASPAR database, two potential FXR binding sites within MMP-7 were identified at positions -1363 and -279 (Fig. 6A). Based on these sites, four plasmids were designed: wild-type (WT), single-point mutant Mut1 and Mut 2, and double-point mutant Mut 3, constructed by Miao Ling Corporation. Upon obtaining the plasmids, the wild-type plasmid W was initially co-transfected with the pTK-RL plasmid into cells in two wells of a 6-well plate. Simultaneously, siFXR and siNC were added separately to these wells. The experiments were performed according to the instructions of the Promega Dual-Luciferase Reporter Assay System. The results indicated a decrease in the ratio of firefly luciferase to Renilla luciferase activity upon FXR knockdown (Fig. 6B), suggesting that FXR activates the wild-type MMP-7 promoter plasmid.
Fig. 6.
Dual-luciferase reporter assays indicated that FXR directly interacts with the promoter region of MMP-7. (A) Schematic diagrams of four luciferase expression plasmids: WT, Mut1, Mut2, Mut3. (B) Ratio of luciferase activity in 293 T cells transfected with WT and then knocked down for FXR. (C) Ratio of luciferase activity in 293 T cells knocked down for FXR and subsequently transfected with the four luciferase expression plasmids.
Subsequently, plasmids WT, Mut 1, Mut 2, and Mut 3 containing the MMP-7 promoter-driven firefly luciferase reporter gene were separately co-transfected with pTK-RL plasmid into cells, along with siFXR. The Dual-Luciferase Reporter Gene Assay Kit from Promega was used for these experiments (Fig. 6C). Results showed a significant increase in the ratio of firefly luciferase to Renilla luciferase activity in Mut 1 and Mut 3 groups, indicating that mutation site 1, located at position -1363 within the MMP-7 promoter region, is indeed the FXR binding site.The dual-luciferase reporter assays conclusively demonstrate that FXR directly interacts with the promoter region of MMP-7.
Subcutaneous tumor experiments in mice further confirm the effects of FXR and MMP-7 on ccRCC
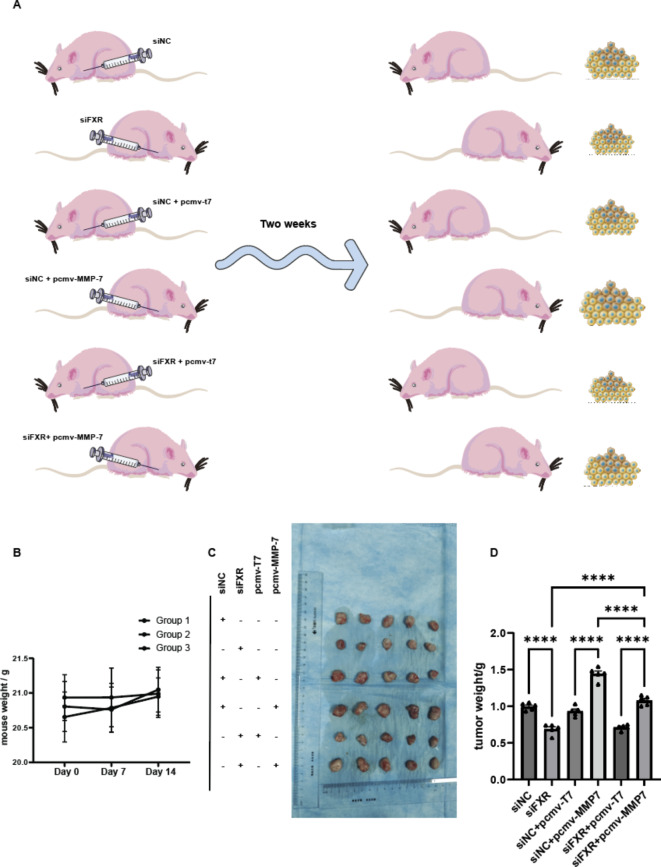
To validate the roles of FXR and MMP-7 in vivo, 6-week-old BALB/c nude mice were randomly divided into three groups. The first group received subcutaneous injections of 5 × 106 786-O siNC cells (left) and 786-O siFXR cells (right). The second group received injections of 5 × 106 786-O siNC + pCMV-T7 cells (left) and 786-O siNC + pCMV-MMP-7 cells (right). The third group received injections of 5 × 106 786-O siFXR + pCMV-T7 cells (left) and 786-O siFXR + pCMV-MMP-7 cells (right) (Fig. 7A). During these two weeks, the mice’s body weight remained essentially unchanged (Fig. 7B). Tumor volumes were evaluated every 3 days, and mice were euthanized after 2 weeks to collect tumors (Fig. 7C). Analysis of tumor weight data (Fig. 7D) showed that in the first group, tumor growth was slower after FXR knockdown. In the second group, tumor growth was accelerated after MMP-7 overexpression. In the third group, tumors grew even faster when MMP-7 was overexpressed in the context of FXR knockdown, and relative to the FXR knockdown group (Group 1), growth was faster, while compared to the MMP-7 overexpression group (Group 2), growth was slower. In conclusion, FXR and MMP-7 promote in vivo growth of ccRCC cells.
Fig. 7.
Subcutaneous tumor experiments in mice further confirm the effects of FXR and MMP-7 on ccRCC. (A) Three groups of nude mice were injected subcutaneously with 786O cells treated accordingly in the left and right axilla. Tumors were harvested two weeks later. (B) Weight changes of the three groups of nude mice. (C) Harvested tumors. (D) Weights of tumors from different groups.
Discussion
Previous investigations into the FXR have primarily concentrated on diseases of the digestive system28. However, accumulating evidence suggests that FXR also exerts pivotal functions within the renal system. In the context of acute kidney injury, several studies have demonstrated that activation of FXR can potently mitigate renal inflammation and cell apoptosis, as well as diminish the levels of reactive oxygen species29. In the realm of chronic kidney diseases, such as diabetic nephropathy, FXR agonists have been shown to enhance cholesterol metabolism30, and to ameliorate renal tubulointerstitial fibrosis31, thereby preventing the progression of diabetic nephropathy. Regarding renal cancer, the literature has highlighted that FXR can promote the differentiation of renal adenocarcinoma cells through the downregulation of Oct3/4 expression15,25. Our previous research has implicated FXR as a potential tumor marker for ccRCC32. Consequently, to elucidate the molecular mechanisms underlying its role in RCC, a more comprehensive investigative approach has been undertaken.
This study reports that FXR promotes invasion and migration of ccRCC through MMP-7-mediated regulation of the EMT pathway. Initially, the impact of FXR on ccRCC migration and invasion was corroborated through knockdown and overexpression experiments. Subsequent studies utilizing similar methodologies confirmed the role of MMP-7 in promoting ccRCC migration and invasion, with evidence indicating that MMP-7 is modulated by FXR to a certain degree. Moreover, two rescue experiments elucidated the regulatory axis of FXR-MMP-7-EMT, and a dual luciferase reporter assay substantiated direct interaction between FXR and the MMP-7 promoter region. In vivo mouse xenograft experiments further validated the in situ significance of FXR in ccRCC pathogenesis.
FXR has been implicated in the pathogenesis of various cancers. In the context of colorectal cancer, it has been shown to synergize with EZH2 inhibitors to facilitate nuclear localization of FXR, leading to increased expression of CDX2 and subsequent inhibition of migration and invasion of cancer cells26. Additionally, FXR agonists have been reported to induce cell cycle arrest in colorectal cancer cells via the miR-135A1/CCNG2 pathway, affecting tumor proliferation33. In bladder cancer, the overexpression of FXR has been associated with reduced levels of vascular endothelial growth factor (VEGF), thus mitigating the malignant phenotype of bladder cancer cells34. Conversely, FXR has also been implicated in promoting tumor progression in other malignancies, such as breast cancer and pancreatic cancer with lymph node metastasis12,13. Notably, a recent report has highlighted that in colorectal cancer, FXR represses MMP-7 transcription by binding to a negative FXR response element within the 5’ region of the MMP-7 promoter35. In contrast, our findings suggest that FXR facilitates MMP-7 transcription. This discrepancy may be due to variations in the binding sites of FXR, which warrants further exploration and detailed mechanistic studies.
MMP-7, a member of the zinc-dependent endopeptidase family termed MMPs, exhibits the capacity to degrade extracellular matrix (ECM) proteins. It is integral in the tissue remodeling processes underpinning a variety of physiological and pathological conditions36. Moreover, MMP-7 is not only involved in ECM protein degradation but also in the breakdown of molecules such as E-cadherin, Fas ligand, and renin19,27. Elevated expression levels of MMP-7 have been correlates with unfavorable prognostic outcomes across a spectrum of cancers, including colorectal, bladder, gastric, pancreatic, and others37–40. MMP-7 has been implicated in the EMT pathway. A previous investigation demonstrated that MMP-7 can proteolytically cleave E-cadherin into soluble E-cadherin (sE-cadherin) within prostate cancer cells, disrupting the E-cadherin-β-catenin complex and facilitating the release of β-catenin, which in turn activates EMT24. In the present study, the overexpression of FXR or MMP-7 was found to be associated with an increase in the expression of EMT biomarkers, including Twist, Vimentin, and N-cadherin, while the levels of Claudin and E-cadherin were diminished. These findings underscore the presence of an FXR-MMP-7-EMT axis. Furthermore, dual-luciferase reporter assays were utilized to validate the direct transcriptional regulation of MMP-7 by FXR, acting on its promoter region.
RCC is highly associated with metabolism41. For instance, MUC1 influences RCC proliferation and angiogenesis by affecting metabolic pathways42. In addition, MUC1 is closely linked to EMT. Adipocytes are a notable feature of RCC43, prompting the question of whether FXR plays a role in lipid accumulation in RCC due to its regulatory effect on fat metabolism. Additionally, FXR has a significant relationship with the immune microenvironment in gastrointestinal tumors44. As a highly aggressive tumor reliant on angiogenesis, immune-related processes are crucial in the metastasis and invasion of RCC45.
This investigation harbors several intrinsic limitations, including the utilization of a relatively modest sample size of patient-derived tissues and a restricted array of ccRCC cell lines. Consequently, there is ample room for further inquiry into the intricate regulatory mechanisms governing the FXR-MMP-7-EMT axis. Despite these constraints, our study contributes compelling evidence in support of the involvement of the FXR-MMP-7-EMT axis in ccRCC pathogenesis. Specifically, it has been demonstrated that activation of FXR not only promotes disease progression but also operates through specific molecular mechanisms. These mechanisms were elucidated through rescue experiments, dual-luciferase reporter assays, and murine tumorigenesis studies, which collectively and robustly validated the presence and significance of the FXR-MMP-7-EMT signaling axis.
Conclusion
In conclusion, our study demonstrates that FXR regulates the EMT pathway through MMP-7 to promote invasion and migration in ccRCC. Subsequent animal experiments strongly indicate that the regulation of FXR and MMP-7 significantly influences tumor progression in ccRCC. This suggests that the FXR-MMP-7-EMT axis could serve as a novel therapeutic and preventive target for ccRCC.
Supplementary Information
Acknowledgements
The authors sincerely thank the Central Laboratory and Urologic Disease Institute of Renmin Hospital of Wuhan University for their support with instruments and equipment. We also appreciate the helpful comments provided by the reviewers on this manuscript.
Abbreviations
- RCC
Renal cell carcinoma
- ccRCC
Clear cell renal cell carcinoma
- FXR, encoded by NR1H4
Farnesoid X receptor
- siRNA
Small interfering RNA
- MMP-7
Matrix metalloproteinase-7
- JAK2/STAT3
Janus kinase 2/signal transducer and activator of transcription 3)
- ERK1/2
Extracellular signal-regulated kinase 1/2
- FasL
Fas ligand
- MUC1
Mucin1
Author contributions
Jiachen Liu: Conceptualization (lead); data curation (lead); writing – original draft (lead);formal analysis (equal); software (equal); validation (equal). Shiyu Huang: Data curation (equal); methodology (equal); supervision (equal); investigation (equal); writing – original draft (equal). Yanguang Hou: methodology (equal); visualization (equal); software (equal); Conceptualization (equal); Shujie Fu: Data curation (equal); software (equal); investigation (equal); methodology (equal); visualization (equal) .Lei Wang: methodology (equal); software (equal); Juncheng Hu: Formal analysis (equal); funding acquisition (equal); project administration (equal); supervision (equal); validation (equal); writing – review and editing (equal) Cheng Liu : writing – review and editing (equal) ;Conceptualization (equal); methodology (equal); software (equal) Xiuheng Liu: supervision (equal); investigation (equal); methodology (equal); validation (equal). All authors read and approved the final manuscript.
Funding
Grants from National Natural Science Foundation of China (No. 82203258).
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.The data that support the findings of this study are available from the corresponding author upon reasonable request. For further information or to request data, please contact the author at: drliuxh@hotmail.com.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All experimental protocols were approved by the Ethics Committee of Wuhan University People’s Hospital and strictly followed the recommendations of the National Institutes of Health Guide for the Care and Use of Laboratory Animals. The study is reported in accordance with the ARRIVE guidelines.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jiachen Liu and Shiyu Huang made equal contributions to this work.
Contributor Information
Juncheng Hu, Email: hujc@whu.edu.cn.
Cheng Liu, Email: 2013103020057@whu.edu.cn.
Xiuheng Liu, Email: drliuxh@hotmail.com.
References
- 1.Bray, F. et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin.74, 229–263. 10.3322/caac.21834 (2024). [DOI] [PubMed] [Google Scholar]
- 2.Moch, H. et al. The 2022 world health organization classification of tumours of the urinary system and male genital organs-part A: Renal, penile, and testicular tumours. Eur. Urol.82, 458–468. 10.1016/j.eururo.2022.06.016 (2022). [DOI] [PubMed] [Google Scholar]
- 3.Linehan, W. M. & Ricketts, C. J. The Cancer Genome Atlas of renal cell carcinoma: Findings and clinical implications. Nat Rev Urol16, 539–552. 10.1038/s41585-019-0211-5 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Rini, B. I., Campbell, S. C. & Escudier, B. Renal cell carcinoma. Lancet373, 1119–1132. 10.1016/s0140-6736(09)60229-4 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Fuhrman, S. A., Lasky, L. C. & Limas, C. Prognostic significance of morphologic parameters in renal cell carcinoma. Am. J. Surg. Pathol.6, 655–663. 10.1097/00000478-198210000-00007 (1982). [DOI] [PubMed] [Google Scholar]
- 6.Forman, B. M. et al. Identification of a nuclear receptor that is activated by farnesol metabolites. Cell81, 687–693. 10.1016/0092-8674(95)90530-8 (1995). [DOI] [PubMed] [Google Scholar]
- 7.Li, T. & Chiang, J. Y. Bile acid signaling in metabolic disease and drug therapy. Pharmacol. Rev.66, 948–983. 10.1124/pr.113.008201 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kainuma, M., Takada, I., Makishima, M. & Sano, K. Farnesoid X receptor activation enhances transforming growth factor β-induced Epithelial-Mesenchymal transition in hepatocellular carcinoma cells. Int. J. Mol. Sci.10.3390/ijms19071898 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao, J. et al. Transcription factor FXR activates DHRS9 to inhibit the cell oxidative phosphorylation and suppress colon cancer progression. Anal. Cell Pathol. (Amst.)2022, 8275574. 10.1155/2022/8275574 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li, S. et al. Farnesoid X receptor activation induces antitumour activity in colorectal cancer by suppressing JAK2/STAT3 signalling via transactivation of SOCS3 gene. J. Cell Mol. Med.24, 14549–14560. 10.1111/jcmm.16083 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng, Q. et al. Activation of FXR suppresses esophageal squamous cell carcinoma through antagonizing ERK1/2 signaling pathway. Cancer Manag. Res.13, 5907–5918. 10.2147/cmar.S243317 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Absil, L. et al. Farnesoid X receptor as marker of osteotropism of breast cancers through its role in the osteomimetism of tumor cells. BMC Cancer20, 640. 10.1186/s12885-020-07106-7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee, J. Y. et al. Farnesoid X receptor, overexpressed in pancreatic cancer with lymph node metastasis promotes cell migration and invasion. Br. J. Cancer104, 1027–1037. 10.1038/bjc.2011.37 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kao, C. C. et al. GW4064 inhibits migration and invasion through cathepsin B and MMP2 downregulation in human bladder cancer. Chem. Biol. Interact.389, 110869. 10.1016/j.cbi.2024.110869 (2024). [DOI] [PubMed] [Google Scholar]
- 15.Guo, Y., Xie, G. & Zhang, X. Role of FXR in renal physiology and kidney diseases. Int. J. Mol. Sci.10.3390/ijms24032408 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egeblad, M. & Werb, Z. New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer2, 161–174. 10.1038/nrc745 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Kessenbrock, K., Plaks, V. & Werb, Z. Matrix metalloproteinases: Regulators of the tumor microenvironment. Cell141, 52–67. 10.1016/j.cell.2010.03.015 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng, C. M. et al. Matrix metalloproteinase-7 promotes chronic kidney disease progression via the induction of inflammasomes and the suppression of autophagy. Biomed. Pharmacother.154, 113565. 10.1016/j.biopha.2022.113565 (2022). [DOI] [PubMed] [Google Scholar]
- 19.Tan, R. J. et al. Tubular injury triggers podocyte dysfunction by β-catenin-driven release of MMP-7. JCI Insight10.1172/jci.insight.122399 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan, R. J., Zhou, D., Zhou, L. & Liu, Y. Wnt/β-catenin signaling and kidney fibrosis. Kidney Int. Suppl.4, 84–90. 10.1038/kisup.2014.16 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu, H. et al. Matrix metalloproteinase-7 protects against acute kidney injury by priming renal tubules for survival and regeneration. Kidney Int.95, 1167–1180. 10.1016/j.kint.2018.11.043 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chandrashekar, D. S. et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia25, 18–27. 10.1016/j.neo.2022.01.001 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chandrashekar, D. S. et al. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia19, 649–658. 10.1016/j.neo.2017.05.002 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang, Q. et al. Interleukin-17 promotes prostate cancer via MMP7-induced epithelial-to-mesenchymal transition. Oncogene36, 687–699. 10.1038/onc.2016.240 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fujino, T. et al. Farnesoid X receptor and liver X receptors regulate Oct3/4 expression by multiple feedback regulating system in normal renal-derived cells and renal adenocarcinoma cells. J. Toxicol. Sci.45, 25–35. 10.2131/jts.45.25 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Yu, J. et al. Activation of FXR and inhibition of EZH2 synergistically inhibit colorectal cancer through cooperatively accelerating FXR nuclear location and upregulating CDX2 expression. Cell Death Dis.13, 388. 10.1038/s41419-022-04745-5 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan, R. J. & Liu, Y. Matrix metalloproteinases in kidney homeostasis and diseases. Am. J. Physiol. Renal. Physiol.302, F1351-1361. 10.1152/ajprenal.00037.2012 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ding, L., Yang, L., Wang, Z. & Huang, W. Bile acid nuclear receptor FXR and digestive system diseases. Acta Pharm. Sin B5, 135–144. 10.1016/j.apsb.2015.01.004 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gai, Z. et al. Farnesoid X receptor activation protects the kidney from ischemia-reperfusion damage. Sci. Rep.7, 9815. 10.1038/s41598-017-10168-6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marquardt, A. et al. Farnesoid X receptor agonism protects against diabetic tubulopathy: Potential add-on therapy for diabetic nephropathy. J. Am. Soc. Nephrol.28, 3182–3189. 10.1681/asn.2016101123 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, X. X. et al. Diabetic nephropathy is accelerated by farnesoid X receptor deficiency and inhibited by farnesoid X receptor activation in a type 1 diabetes model. Diabetes59, 2916–2927. 10.2337/db10-0019 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang, S., Hou, Y., Hu, M., Hu, J. & Liu, X. Clinical significance and oncogenic function of NR1H4 in clear cell renal cell carcinoma. BMC Cancer22, 995. 10.1186/s12885-022-10087-4 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiao, P., Li, S., Zhang, H., Yao, L. & Wang, F. Farnesoid X receptor inhibits proliferation of human colorectal cancer cells via the miR-135A1/CCNG2 signaling pathway. Oncol. Rep.40, 2067–2078. 10.3892/or.2018.6636 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Lai, C. R. et al. Enhancement of farnesoid X receptor inhibits migration, adhesion and angiogenesis through proteasome degradation and VEGF reduction in bladder cancers. Int. J. Mol. Sci.10.3390/ijms23095259 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng, Z., Chen, J., Drachenberg, C. B., Raufman, J. P. & Xie, G. Farnesoid X receptor represses matrix metalloproteinase 7 expression, revealing this regulatory axis as a promising therapeutic target in colon cancer. J. Biol. Chem.294, 8529–8542. 10.1074/jbc.RA118.004361 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edman, K. et al. The discovery of MMP7 inhibitors exploiting a novel selectivity trigger. ChemMedChem6, 769–773. 10.1002/cmdc.201000550 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Masaki, T. et al. Matrilysin (MMP-7) as a significant determinant of malignant potential of early invasive colorectal carcinomas. Br. J. Cancer84, 1317–1321. 10.1054/bjoc.2001.1790 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szarvas, T. et al. Matrix metalloproteinase-7 as a marker of metastasis and predictor of poor survival in bladder cancer. Cancer Sci.101, 1300–1308. 10.1111/j.1349-7006.2010.01506.x (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee, K. H. et al. Relationship between E-cadherin, matrix metalloproteinase-7 gene expression and clinicopathological features in gastric carcinoma. Oncol. Rep.16, 823–830 (2006). [PubMed] [Google Scholar]
- 40.Kuhlmann, K. F. et al. Evaluation of matrix metalloproteinase 7 in plasma and pancreatic juice as a biomarker for pancreatic cancer. Cancer Epidemiol. Biomark. Prev.16, 886–891. 10.1158/1055-9965.Epi-06-0779 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.di Meo, N. A. et al. Renal cell carcinoma as a metabolic disease: An update on main pathways, potential biomarkers, and therapeutic targets. Int. J. Mol. Sci.10.3390/ijms232214360 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lucarelli, G. et al. MUC1 tissue expression and its soluble form CA15–3 identify a clear cell renal cell carcinoma with distinct metabolic profile and poor clinical outcome. Int. J. Mol. Sci.10.3390/ijms232213968 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.di Meo, N. A. et al. The dark side of lipid metabolism in prostate and renal carcinoma: Novel insights into molecular diagnostic and biomarker discovery. Expert Rev. Mol. Diagn.23, 297–313. 10.1080/14737159.2023.2195553 (2023). [DOI] [PubMed] [Google Scholar]
- 44.Jia, W., Xie, G. & Jia, W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat. Rev. Gastroenterol. Hepatol.15, 111–128. 10.1038/nrgastro.2017.119 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vuong, L., Kotecha, R. R., Voss, M. H. & Hakimi, A. A. Tumor microenvironment dynamics in clear-cell renal cell carcinoma. Cancer Discov.9, 1349–1357. 10.1158/2159-8290.Cd-19-0499 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.The data that support the findings of this study are available from the corresponding author upon reasonable request. For further information or to request data, please contact the author at: drliuxh@hotmail.com.