Abstract
There are several important challenges in radiomics research; one of them is feature selection. Since many quantitative features are non-informative, feature selection becomes essential. Feature selection methods have been mixed with filter, wrapper, and embedded methods without a rule of thumb. This study aims to develop a framework for optimal feature selection in radiomics research. We developed the framework that the optimal features were selected to quickly through controlling relevance and redundancy among features. A ‘FeatureMap’ was generated containing information for each step and used as a platform. Through this framework, we can explore the optimal combination of radiomics features and evaluate the predictive performance using only selected features. We assessed the framework using four real datasets. The FeatureMap generated 6 combinations, with the number of features selected varying for each combination. The predictive models obtained high performances; the highest test area under the curves (AUCs) were 0.792, 0.820, 0.846 and 0.738 in the cross-validation method, respectively. We developed a flexible framework for feature selection methods in radiomics research and assessed its usefulness using various real-world data. Our framework can assist clinicians in efficiently developing predictive models based on radiomics.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-80863-8.
Keywords: Radiomics, Feature selection, FeatureMap, Prediction model, Machine learning
Subject terms: Cancer, Medical research
Introduction
Radiomic features are radiologic biomarkers that are quantitatively assessed and extracted from medical images of one or more modalities, including computed tomography (CT), magnetic resonance imaging (MRI), positron emission tomography (PET), and ultrasound1. These biomarkers include first-order statistical features describing the distribution of voxel intensities, shape-based features describing the shape of the region of interest and its geometric properties, and texture features describing the spatial variations in voxel intensity levels2. Radiomics can mean a research area for radiome (i.e., the whole set of radiomic features), but more likely refer to a wholistic methodology that encompasses six components: (1) image acquisition and pre-processing, (2) image segmentation, (3) feature extraction, (4) feature selection (FS) or dimension reduction, (5) derivation and evaluation of predictive models, and (6) clinical application2,3. Despite several unresolved challenges, this methodology is extensively utilized to facilitate non-invasive clinical decision-making in oncology research4–7.
Like the other radiomics components, FS is an essential step during which unnecessary noise information is reduced by selecting an optimal subset of features that are reliable, likely relevant to a diagnosis or prognosis endpoint, but not redundant to each other. Due to the high-dimensional and complex nature of radiomics feature data derived from a relatively limited set of images8–10, FS prior to predictive modeling is essential to enhance predictive accuracy and mitigate the risk of model overfitting11–13. Current FS methods are categorized into three types: filter, wrapper, and embedded methods. FS have been proceeded with a single FS method, or with a serial combination of multiple methods. Although pros and cans of FS procedures have been discussed, there is a lack of rules of thumb as well as guidelines and tools for choosing a proper FS procedure14–18.
Artificial intelligence (AI) applications have been increased in the medical field, aiming to replace repetitive tasks encountered in routinely clinical workflows and to provide clinical decision support systems (CDSSs). AI techniques have been employed for diagnosis or prognosis prediction in radiomics research19, and it made the cooperation between clinical and AI domains essential20,21. Various technical options have been available for key elements of AI development, including machine learning (ML) algorithms, model building and testing data construction, and FS. It became a challenge to evaluate all available options and choose an optimal one.
Since ML-based methods have been introduced for FS, clinicians and ML experts need to cooperate to establish an optimal FS strategy. It requires an efficient framework that contains easy and fast tools (1) to assess and store feature characteristics, including indices of reliability/robustness, clinical relevance, and redundancy; and (2) to populate and compare candidate FS strategies by applying various FS methods, model-building and evaluation algorithms, and performance measures. Our objective is to develop an efficient and practical framework for FS, particularly in the context of high-dimensional data, that delivers all essential information and facilitates collaboration between clinicians and ML experts.
Materials and methods
Feature selection in radiomics
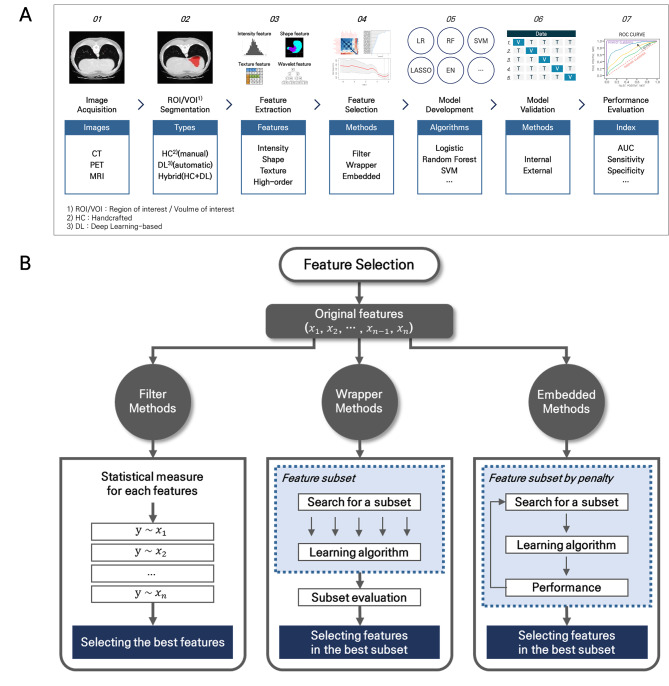
The radiomics analysis process can consist of seven steps, from image acquisition to model evaluation (Fig. 1A). The FS is an important step to remove noise information, improve the predictive performance, and prevent the overfitting risk of the model. Many FS methods predominantly rely on repeatability and reproducibility of individual features, relevance to a clinical outcome, and redundancy among features22,23. The repeatability and reproducibility are often checked to screen-out features with poor data quality prior to the FS steps based on the relevance and redundancy that are considered comprehensively7,11–13,24.
Fig. 1.
The radiomics analysis process. (A) Radiomics workflow. (B) Feature selection methods.
There are three categories of FS methods: filter, wrapper, and embedded approaches (Fig. 1B). These methods can be used individually or in combination22,23. Filter methods assign an independent score to each feature, and select features whose scores satisfy a pre-defined criterion prior to the model building process. The scores are developed by evaluating relevance of each feature to the clinical outcome, with statistical or informational metrics, including correlation coefficients, minimum redundancy maximum relevance, or mutual information. Because filter approaches operate in per-feature level, they are fast and easy to implement while they may miss feature combinations that could be synergetic for prediction.
Wrapper methods search the optimal subset of features with the best model performance among all possible subsets when modeling is based on a specific ML algorithm. Such ML algorithms should provide individual feature ranking. For example, multivariable regressions with p-values for model difference when eliminating or adding a feature, and random forest (RF) and extreme gradient boosting (XGBoost) with feature importance. A wrapper technique requires a search method that iteratively adds high-ranked features of removes low-ranked ones. Examples include backward elimination, recursive feature elimination, forward selection, sequential selection, and Boruta. While the wrapper methods tend to be computationally expensive and susceptible to overfitting, they usually result in better predictive performance than filter methods.
Embedded methods incorporate the FS process directly into the ML-based model building process to encompass benefits of filter and wrapper methods. Examples include Least Absolute Shrinkage and Selection Operator (LASSO) and Elastic-Net (EN). These methods search for the best subset of features with high performance during the learning process while maintaining reasonable computational costs. Embedded methods are likely to suffer from limited interpretability, and cannot be supported by all ML algorithms.
Study Design: Framework for optimal feature selection
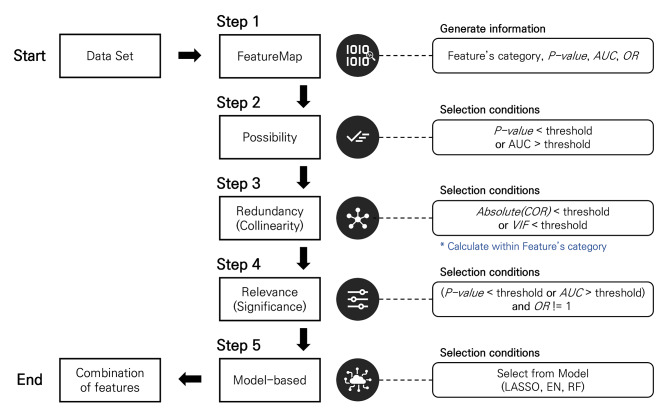
Our framework facilitates an easy and efficient application of FS methods in combination based on ‘FeatureMap’ in Step 1: filter methods in Steps 2 ~ 4 and wrapper or embedded methods in Step 5 (Fig. 2, Supplementary Fig. S1). In Step 1, all metrics necessary for decision-making in Step 2 (e.g., p-value, area under the curve (AUC) and odds ratio (OR)) are calculated by conducting univariable analyses of individual quantitative features extracted from medical images, and saved with basic information (e.g., name, category and category identifier (ID)) as backbone of the FeatureMap (Supplementary Fig. S2). The category describes morphologic, intensity- or texture-based characteristics of a radiomic feature. An identical category ID signifies that the features belong to the same category. All other information required for decision in each step and the corresponding results are appended into the FeatureMap throughout the framework.
Fig. 2.
The framework for feature selection in radiomics research.
In Step 2, a single or a set of thresholds for the metrics can be considered for fast screening and dimensionality reduction. The filtering status of each feature is recorded in the ‘FeatureMap’ (Supplementary Fig. S3). The Step 2 was designed to reduce the ultrahigh dimensionality in radiomics data by applying on the sure independence screening (SIS)25,26, in which every feature with ‘weak’ relevance to the outcome (e.g., univariable association or predictability) should remain for the next filter steps. Because it is necessary to avoid unnecessary elimination of useful features for prediction in this step, a threshold(s) need to be too stringent, and FDR correction may not be of concern. The Step 2 is optional and can be skipped when data do not suffer from the ultrahigh dimensionality.
The core of this framework generates a ‘FeatureMap’ containing information for decision-making in the first step, and then provides various combinations of features in the last step. The FeatureMap saves all information at each framework step, thus reducing unnecessary calculations and minimizing resources though recycling saved information. This makes it possible to perform FS efficiently and quickly.
Step 3 removes the excessive redundancy (or collinearity) between selected features in Step 2, using the absolute value of spearman correlation coefficient (COR) and variance inflation factor (VIF) as the selection criterion (Supplementary Fig. S4). To densify the dataset and remove highly correlated features in Step 3, we employed a high-correlation filter method: (1) calculate pairwise correlations; (2) eliminate the feature with weaker relevance from the most highly correlated pair; (3) repeat until all remaining pairs have a correlation below a predefined threshold. This method can be applied to all features whose categories are same. There are three options for selecting the criterion for weaker relevance: statistical significance (p-value), effect size (OR), and predictability (AUC). For example, the representative feature with the lowest p-value in the FeatureMap is selected when using the p-value as the criteria.
Step 4 confirms the relevance (or significance) between the outcome and the selected features from the Step 3, and also separates into two tracks in order to consider redundancy (COR, VIF) at the same time. In this step, the candidate features can be selected that the p-value or AUC is satisfied the criterion and max(OR, 1/OR)-1 is greater than threshold (Supplementary Fig. S5).
Step 5 selects the final features using automatic embedded FS algorithms which LASSO, EN and RF. In the RF, the top-ranking with a variable importance can be used as the criterion for feature selection. As a result, maximum 6 combination of features can be generated through two tracks in Step 4 and three embedded algorithms in Step 5 (Supplementary Fig. S6).
In this paper, thresholds were set to 0.5 of p-value in Step 2, 0.8 of COR and 4 of VIF in Step 3, 0.2 of p-value, 0.55 of AUC and 1e-7 of OR in Step 4, 10 of the top-ranking in Step 5. These thresholds can be changed by researcher.
Performance assessment using real datasets
To assess and validate the framework, multiple real clinical datasets obtained for other retrospective studies with binary clinical outcome were used. The Institutional Review Boards (IRBs) of the Samsung Medical Center and the Soonchunhyang University Seoul Hospital approved this study (IRB No. 2020-03-086 and IRB No. 2019-05-032, respectively) and waived the requirement for informed consent given the retrospective nature of the study and all methods were performed in accordance with the relevant guidelines and regulations. All patient information was anonymized and de-identified.
Dataset 1 is metabolic syndrome data with PET-CT images in Soonchunhyang University Seoul Hospital. The objective is to develop a predictive model for the improvement of metabolic syndrome after bariatric surgery (yes, no). The sample size is 79, with 854 features and 55 categories (IRB No. 2019-05-032).
Dataset 2 is gastric cancer data with CT images in Samsung Medical Center. The objective is to develop a predictive model for diagnostic EMT subtype (yes, no). The sample size is 125, with 1223 features and 79 categories (IRB No. 2020-03-086).
Dataset 3 is low-grade gliomas public-data with MRI images in the cancer genome atlas and the cancer imaging archive. The objective is to develop a predictive model for mutation of the 1p19q gene (wild gene, mutated gene)27. The sample size is 105, with 640 features and 38 categories.
Dataset 4 is prostate imaging cancer AI (PI-CAI) public-data with T2-weighted MRI images28. The objective is to develop a predictive model for a malignancy of prostatic tissue (yes, no). The sample size is 969, with 1015 features and 67 categories.
We assessed the utility as the perspective of screening to confirm the predictive performance using only the selected features from the framework. The procedures for the predictive model were set as follows, and these settings can be flexibly changed by the researcher. To develop the predictive models, the full data was randomly divided into the model development (70%) and the model validation (30%) data. To improve the predictive performance, we performed normalization, standardization, and a random search using 5 repeated 5-fold cross-validation for optimization. On the other hand, we considered a bootstrap method with 500 iterations for reducing partition bias and used default values of hyperparameter without optimization. We used three popular algorithms: logistic regression (LR), RF and support vector machine (SVM). To assess the predictive performance of the model, we used accuracy, sensitivity, specificity, balanced accuracy (BA), and AUC. The BA was used to determine the optimal cutoff for the predictive model.
We compared the number of features in various combinations selected through the framework and identified the effect on the predictive performance according to the number of features. To evaluate the effectiveness of our framework, we considered three commonly-used FS methods (i.e., LASSO, XGBoost, and Boruta) as reference FS methods. Calibration curves were compared between framework method and reference FS methods for each of the final models with the highest test AUC. All procedures of the framework and predictive model from development to validation were implemented using R (version 4.1.3).
Results
Though the framework, the optimal combinations of features were different depending on the data (Table 1). In addition, the predictive models each data showed overall high performance in various combinations rather than in a specific combination.
Table 1.
Overview of the datasets and comparison of the predictive performances between FS methods.
| Dataset | Sample Size | Number of features | Class balance (%) | FS method | Number of selected features | Predictive algorithm | Predictive Performancesa | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AUC | Accuracy | Sensitivity | Specificity | Balanced Accuracy |
|||||||
| 1 | 79 | 854 | 36.7% | Framework (VIF-RF) | 10 | LR | 0.792 | 0.870 | 0.625 | 1.000 | 0.812 |
| Reference (XGBoost) | 10 | SVM | 0.792 | 0.783 | 0.625 | 0.867 | 0.746 | ||||
| 2 | 125 | 1223 | 38.4% | Framework (COR-RF) | 10 | LR | 0.820 | 0.811 | 0.929 | 0.739 | 0.834 |
| Reference (Boruta) | 1 | LR | 0.618 | 0.622 | 0.643 | 0.609 | 0.626 | ||||
| 3 | 105 | 640 | 63.8% | Framework (COR-RF) | 10 | RF | 0.846 | 0.871 | 0.950 | 0.727 | 0.839 |
| Reference (XGBoost) | 10 | RF | 0.827 | 0.871 | 0.950 | 0.727 | 0.839 | ||||
| 4 | 969 | 1015 | 65.7% | Framework (COR-LASSO) | 24 | LR | 0.738 | 0.738 | 0.801 | 0.616 | 0.709 |
| Reference (LASSO) | 41 | LR | 0.744 | 0.755 | 0.801 | 0.667 | 0.734 | ||||
FS, feature selection; AUC, area under the curve; LR, logistic regression; RF, random forest; SVM, support vector machine; COR, correlation; VIF, variance inflation factor; LASSO, least absolute shrinkage and selection operator; XGBoost, extreme gradient boosting.
aFinally selected predictive model for each test data.
Dataset 1: predictive model for improvement metabolic syndrome after bariatric surgery with metabolic syndrome data
The FeatureMap generated through the framework consisted of 6 combinations as shown in Supplementary Fig. S1. The number of features selected based on the COR were 43 with LASSO, 44 with EN and 10 with RF, respectively. On the other hand, the number of features selected based on the VIF were 26 with LASSO, 30 with EN and 10 with RF, respectively.
The predictive performances of each combination with cross-validation and bootstrap methods are shown in Supplementary Table S1. Also, the AUCs were represented using a heatmap according to each combination (Supplementary Fig. S7).
In the cross-validation method, the LR model obtained the best performance at combination of VIF and RF, with the train AUC of 0.833 and the test AUC of 0.792. The RF model obtained the train AUC of 1.000 and test AUC of 0.692 at combination of COR and RF. The SVM model obtained the train AUC of 0.819 and test AUC of 0.783 at combination of VIF and RF. Similarly, in the bootstrap method, the LR and RF models obtained the best performances at combination of VIF and RF, with the train AUCs of 0.890 and 0.900, the test AUCs of 0.641 and 0.696, respectively. The SVM model obtained the train AUC of 0.996 and test AUC of 0.695 at combination of COR and EN. As shown in Supplementary Table S1 and Fig. S7, various performances were obtained depending on the various combinations rather than in a specific combination.
Dataset 2: predictive model for diagnostic EMT subtype with gastric cancer data
The number of features selected based on the COR were 2 with LASSO, 71 with EN and 10 with RF, respectively. On the other hand, the number of features selected based on the VIF were 2 with LASSO, 5 with EN and 10 with RF, respectively (Supplementary Fig. S8).
In the cross-validation method, all models obtained the best performances at combination of COR and RF with the test AUC of 0.820 in the LR, the test AUC of 0.758 in the RF and the test AUC of 0.776 in the SVM, respectively (Supplementary Table S2 and Fig. S9A). In the bootstrap method, the LR model obtained the best performance at combination of COR and LASSO with the train AUC of 0.717 and the test AUC of 0.684. The RF model obtained the best performance at combination of VIF and RF with the train AUC of 0.908 and the test AUC of 0.680. On the other hand, the SVM model obtained the best performance at combination of COR and RF, with train AUC of 0.922 and the test AUC of 0.664, respectively (Supplementary Fig. S9B).
Dataset 3: predictive model for the mutation of the 1p19q gene with low-grade gliomas data
The number of features selected based on the COR were 25 with LASSO, 17 with EN and 10 with RF, respectively. On the other hand, the number of features selected based on the VIF were 22 with LASSO, 38 with EN and 10 with RF, respectively (Supplementary Fig. S10).
In the cross-validation method, the LR model obtained the best performance at combination of COR and EN with the train AUC of 1.000 and the test AUC of 0.782. The RF model obtained the best performance at combination of COR and RF with the train AUC of 1.000 and the test AUC of 0.846. On the other hand, the SVM model obtained the best performance at combination of COR and LASSO, with train AUC of 1.000 and the test AUC of 0.836, respectively (Supplementary Table S3 and Fig. S11A). In the bootstrap method, all models obtained the best performances at combination of COR and LASSO with test AUCs of 0.875 in the LR, 0.952 in the RF and 0.962 in the SVM, respectively (Supplementary Fig. S11B).
Dataset 4: predictive model for the malignancy of prostatic tissue with PI-CAI data
The number of features selected based on the COR were 24 with LASSO, 28 with EN and 10 with RF, respectively. On the other hand, the number of features selected based on the VIF were 17 with LASSO, 11 with EN and 10 with RF, respectively (Supplementary Fig. S12).
In the cross-validation method, the LR model obtained the best performance at combination of COR and LASSO with the train AUC of 0.735 and the test AUC of 0.738. The RF model obtained the best performance at combination of COR and EN with the train AUC of 1.000 and the test AUC of 0.707. On the other hand, the SVM model obtained the best performance at combination of COR and LASSO, with train AUC of 0.762 and the test AUC of 0.718, respectively (Supplementary Table S4 and Fig. S13A).
Comparisons of the predictive performance between FS framework and reference FS methods
The predictive performances for each reference FS method are shown in Supplementary Table S5, and the final models were selected at high test AUC. In datasets 1 and 3, XGBoost selected 10 features, with test AUCs of 0.792 and 0.827, respectively. For datasets 2 and 4, the Boruta and LASSO algorithms selected 1 and 41 features, with test AUCs of 0.618 and 0.744, respectively. Calibration performance of our framework was comparable to reference FS methods (Supplementary Fig. S14). In terms of the integrated calibration index, our framework showed good calibration for datasets 2, 3, and 4, with values of 0.041, 0.091, and 0.049, respectively.
Discussion
In the present study, we utilized four different real datasets to develop the framework for radiomics research. The FeatureMap generated 6 combinations, with the number of features selected varying for each combination. The threshold values were set the same for each step of the framework, and the predictive performances were compared using only selected features. Compared to reference FS methods, our framework showed mostly superior performance (Table 1). In dataset 4, the framework’s AUC was slightly lower by 0.006 (0.738 vs. 0.744) but obtained this with 17 fewer features. Despite the slight reduction in discrimination performance, calibration performance remained high (Supplementary Fig. S14). By the principle of parsimony (called Occam’s razor), our framework is more efficient. When developing clinical prediction models, various factors should be considered, such as data size, the number of features, cost, and resources in the clinical environment29,30. Thus, when model performances are comparable, selecting a simpler model is advantageous. Through these findings, we have validated the usefulness of our framework. If researchers have radiomics feature data, they can select the optimal features easily and quickly by setting thresholds. The options of ML were simply applied for quick model development as the perspective of screening. These tasks can be flexibly applied, for example, if resources such as computing power, time or cost are abundant, the optimization process can be added in the bootstrap method to improve the predictive performance. Moreover, we expect that a better predictive model can be developed, if the optimal combination of features though the framework is combined with other clinical information.
In radiomics research, it is necessary to standardize methodology based on reproducibility3,5,7–9,19. Our findings confirmed the potential for both standardization and reproducibility for radiomics-based feature selection methods. We can secure evidence of standardization as the redundancy and relevance are considered, and all processes can be transparently provided. For this way, the mixed FS methods can be consistently standardized using the framework. Similarly, reproducibility can also be secured, as all information of the FS process is saved in FeatureMap. If the predictive performance of the selected feature combination is not satisfactory, then it is possible to develop a new predictive model by utilizing the information of another combination saved in the FeatureMap. Our framework is easily expandable to other metrics since any metric can be added into ‘FeatureMap’ because of its scalable nature. Through this process, it is possible to develop the predictive model quickly and efficiently from the screening perspective, and the FeatureMap can serve as a platform within the framework itself. Therefore, it is possible to solve the complexity of radiomics research one step further and make more robust decision-making based on reproducibility and standardization of the framework.
Another feasibility aspect is an Automated Machine Learning (AutoML) that is pipeline for automated machine learning from data preparation to model evaluation, and there are tools using AutoML are Auto-Weka, MLjar, and H2O, etc31,32. The objective of AutoML is to automate and solve the manual and repetitive tasks of ML pipelines by various domain experts when developing the predictive models. Using our framework to implement the process from FS to model evaluation as the pipeline, it is possible to develop the AutoML for radiomics-based FS. Then, the researcher can automatically evaluate the model through iterative tasks by adjusting the threshold of the framework. This can reduce costs and time, while also improving the accessibility and productivity of radiomics research. Furthermore, if we expand it as the pipeline that automates all steps of the radiomics workflow in Fig. 1, we can develop the platform specialized in radiomics research using only medical images.
On the other hand, the ML-based model cannot avoid the “black box” problem, and it is also true in radiomics research. To address this issue, there have been many studies on explainable AI (XAI) to gain insight and explain factors from the predictive model33–36. Especially in clinical settings, since it is directly related to patient care, it is even more important to have the XAI model that provides the interpretation of the results. The use of methods such as LIME or SHAP can improve explanatory power37,38. In addition, if we can reversely float the selected features with high prediction performance in the framework onto the images and validate the clinical meaning by clinicians, it can be utilized as the XAI model.
Our study has several limitations. First, there is a problem applying a numeric and survival outcome because our framework used the binary outcome. A study design that can be applied to survival analysis is needed in the future. Second, it is necessary to research a method of repeating the model-based step using the bootstrap approach within our framework and utilizing the frequency of features selected for each result. This means a stability of radiomics features, which is how reliably features are selected. According to the frequency of features, the number of top-ranking can be selected, or features selected over a certain percentage of the number of iterations can be finally selected. In this way, the framework can be extended, if considering the stability in addition to redundancy and relevance. Third, feature data must be prepared in advance. Since the framework uses extracted feature data, issues such as feature reliability, data imbalance or missing values must be considered before using the framework. Once the data is prepared, the framework can be flexibly applied in radiomics research. Fourth, we tried to demonstrate the effectiveness of our framework using four real datasets. Although these datasets have different characteristics, such as different sample sizes, the number of features, and outcome balances, the demonstration with only four datasets was not enough to confirm the effectiveness of our framework. Additionally, some guidelines need to be suggested so that users can set good thresholds. However, our framework enables users to explore various threshold settings easily and quickly and to choose an optimal setting.
The number of clinical research using radiomics is increasing, and many predictive models are suggested for the response evaluation of new treatment for various diseases. However, the poor validation of the model and inter-observer variability of the analysis are hurdles of clinical utilization of radiomics. Therefore, establishing the standardization of methodology for radiomics research is necessary1. In this study, we developed the framework for feature selection methods in radiomics research and assessed its usefulness using various real-world data. Our framework can assist clinicians in efficiently developing the predictive models based on radiomics.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work is supported by FM 2030 project of Samsung medical center (SMX1240801).
Abbreviations
- CT
Computed tomography
- MRI
Magnetic resonance image
- PET
Positron emission tomography
- AI
Artificial intelligence
- ML
Machine learning
- FS
Feature selection
- COR
Correlation
- VIF
Variance inflation factor
- LR
Logistic regression
- LASSO
Least absolute shrinkage and selection operator
- EN
Elastic-net
- RF
Random forest
- SVM
Support vector machine
- XGBoost
Extreme gradient boosting
- AUC
Area under the curve
- OR
Odds ratio
- AutoML
Automated ML
- XAI
Explainable AI
Author contributions
Study design: S.H., S.H., W.K.J., K.K. Collected the data: W.K.J., E.O. Contributed data or analysis tools: E.O., W.J.L., W.K.J. Performed the analysis: S.H., S.H. Wrote the paper: S.H., S.H. Manuscript editing: W.K.J., K.K.
Data availability
Dataset 1 and 2 generated during and/or analyzed during the current study are not publicly available due to privacy and ethical considerations. However, these datasets can be made accessible to qualified researchers upon reasonable request to the corresponding author (W.K.J.), any specific accession codes or unique identifiers associated with the datasets will be provided upon approval of the request.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Sungsoo Hong and Sungjun Hong contributed equally to this work.
Contributor Information
Woo Kyoung Jeong, Email: jeongwk@gmail.com.
Kyunga Kim, Email: kyunga.j.kim@samsung.com.
References
- 1.Jeong, W. K., Jamshidi, N., Felker, E. R., Raman, S. S. & Lu, D. S. Radiomics and radiogenomics of primary liver cancers. Clin. Mol. Hepatol.25, 21–29. 10.3350/cmh.2018.1007 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang, J., Wolfram, D. & Ma, F. The impact of big data on research methods in information science. Data Inform. Manage.7, 100038 (2023). [Google Scholar]
- 3.Lambin, P. et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat. Reviews Clin. Oncol.14, 749–762 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Lambin, P. et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur. J. Cancer. 48, 441–446. 10.1016/j.ejca.2011.11.036 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gillies, R. J., Kinahan, P. E. & Hricak, H. Radiomics: images are more than pictures, they are data. Radiology278, 563–577. 10.1148/radiol.2015151169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiesa-Estomba, C. M. et al. Radiomics and texture analysis in laryngeal cancer. Looking for new frontiers in precision medicine through imaging analysis. Cancers (Basel). 11, 1409. 10.3390/cancers11101409 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shur, J. D. et al. Radiomics in Oncology: a practical guide. Radiographics41, 1717–1732. 10.1148/rg.2021210037 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demircioğlu, A. Benchmarking feature selection methods in Radiomics. Invest. Radiol.57, 433–443. 10.1097/rli.0000000000000855 (2022). [DOI] [PubMed] [Google Scholar]
- 9.Ligero, M. et al. Selection of Radiomics Features based on their reproducibility. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc.2019, 403–408. 10.1109/embc.2019.8857879 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Yuan, R., Tian, L. & Chen, J. An RF-BFE algorithm for feature selection in radiomics analysis. Med. Imaging 2019: Imaging Inf. Healthc. Res. Appl.10954, 183–188. 10.1117/12.2512045 (2019). [Google Scholar]
- 11.Saeys, Y., Inza, I. & Larranaga, P. A review of feature selection techniques in bioinformatics. Bioinformatics23, 2507–2517. 10.1093/bioinformatics/btm344 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Li, J. et al. Feature selection: a data perspective. ACM Comput. Surv.50, 1–45. 10.1145/3136625 (2017). [Google Scholar]
- 13.Kumar, V. & Minz, S. Feature selection: a literature review. SmartCR4, 211–229 (2014). [Google Scholar]
- 14.Wang, J., Zeng, J., Li, H. & Yu, X. A deep learning Radiomics Analysis for Survival Prediction in Esophageal Cancer. J. Healthc. Eng.2022 (4034404). 10.1155/2022/4034404 (2022). [DOI] [PMC free article] [PubMed]
- 15.Xu, L. et al. A radiomics approach based on support vector machine using MR images for preoperative lymph node status evaluation in intrahepatic cholangiocarcinoma. Theranostics9, 5374–5385. 10.7150/thno.34149 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang, M. et al. Computed-tomography-based Radiomics Model for Predicting the malignant potential of gastrointestinal stromal tumors preoperatively: a Multi-classifier and Multicenter Study. Front. Oncol.11, 582847. 10.3389/fonc.2021.582847 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ren, J., Qi, M., Yuan, Y., Duan, S. & Tao, X. Machine learning-based MRI texture analysis to predict the histologic Grade of oral squamous cell carcinoma. AJR Am. J. Roentgenol.215, 1184–1190. 10.2214/AJR.19.22593 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Nagawa, K. et al. Texture analysis of muscle MRI: machine learning-based classifications in idiopathic inflammatory myopathies. Sci. Rep.11, 9821. 10.1038/s41598-021-89311-3 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Timmeren, J. E., Cester, D., Tanadini-Lang, S., Alkadhi, H. & Baessler, B. Radiomics in medical imaging—how-to guide and critical reflection. Insights into Imaging. 11, 1–16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mayerhoefer, M. E. et al. Introduction to Radiomics. J. Nucl. Med.61, 488–495. 10.2967/jnumed.118.222893 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laajili, R., Said, M. & Tagina, M. Application of radiomics features selection and classification algorithms for medical imaging decision: MRI radiomics breast cancer cases study. Inf. Med. Unlocked. 27, 100801 (2021). [Google Scholar]
- 22.Zhang, W., Guo, Y. & Jin, Q. Radiomics and its feature selection: a review. Symmetry15, 1834 (2023). [Google Scholar]
- 23.Wang, K., An, Y., Zhou, J., Long, Y. & Chen, X. A novel multi-level feature selection method for radiomics. Alexandria Eng. J.66, 993–999 (2023). [Google Scholar]
- 24.Khaire, U. M. & Dhanalakshmi, R. Stability of feature selection algorithm: a review. J. King Saud University-Computer Inform. Sci.34, 1060–1073 (2022). [Google Scholar]
- 25.Fan, J. & Lv, J. Sure independence screening for ultrahigh dimensional feature space. J. R Stat. Soc. Ser. B Methodol.70, 849–911 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chowdhury, M. Z. I. & Turin, T. C. Variable selection strategies and its importance in clinical prediction modelling. Family Med. Community Health8, e000262 (2020). [DOI] [PMC free article] [PubMed]
- 27.Mosquera, C. Radiomics for LGG dataset. Kaggle. (2019). https://kaggle.com/competitions/glioma-radiomics
- 28.Demircioğlu, A. & radMLBench A dataset collection for benchmarking in radiomics. Comput. Biol. Med.182, 109140 (2024). [DOI] [PubMed] [Google Scholar]
- 29.Efthimiou, O. et al. Developing clinical prediction models: a step-by-step guide. Bmj386, e078276. 10.1136/bmj-2023-078276 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanchez-Pinto, L. N. & Bennett, T. D. Evaluation of machine learning models for clinical prediction problems. Pediatr. Crit. Care Med.23, 405–408. 10.1097/pcc.0000000000002942 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He, X., Zhao, K. & Chu, X. AutoML: a survey of the state-of-the-art. Knowl-Based Syst.212, 106622 (2021). [Google Scholar]
- 32.Truong, A. et al. Towards automated machine learning: Evaluation and comparison of AutoML approaches and tools. 2019 IEEE 31st international conference on tools with artificial intelligence (ICTAI) 2019:1471–1479. 10.1109/ICTAI.2019.00209
- 33.Lundberg, S. M. & Lee, S. I. A unified approach to interpreting model predictions. NIPS’17: Proc. 31st Int. Conf. Neural Inform. Process. Syst.4768–477710.5555/3295222.3295230 (2017).
- 34.Covert, I. C., Lundberg, S. & Lee, S. I. Explaining by removing: a unified framework for model explanation. J. Mach. Learn. Res.22, 9477–9566 (2021). [Google Scholar]
- 35.Ribeiro, M. T., Singh, S. & Guestrin, C. Why should i trust you? Explaining the predictions of any classifier. Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining 2016:1135–1144. 10.1145/2939672.2939778
- 36.Severn, C. et al. A Pipeline for the implementation and visualization of Explainable Machine Learning for Medical Imaging using Radiomics features. Sens. (Basel). 22, 5205. 10.3390/s22145205 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lundberg, S. M. et al. Explainable machine-learning predictions for the prevention of hypoxaemia during surgery. Nat. Biomed. Eng.2, 749–760. 10.1038/s41551-018-0304-0 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumarakulasinghe, N. B., Blomberg, T., Liu, J., Leao, A. S. & Papapetrou, P. Evaluating local interpretable model-agnostic explanations on clinical machine learning classification models. IEEE 33rd Int. Symp. Computer-Based Med. Syst. (CBMS). 10.1109/CBMS49503.2020.00009 (2020). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Dataset 1 and 2 generated during and/or analyzed during the current study are not publicly available due to privacy and ethical considerations. However, these datasets can be made accessible to qualified researchers upon reasonable request to the corresponding author (W.K.J.), any specific accession codes or unique identifiers associated with the datasets will be provided upon approval of the request.