Abstract
Understanding the intricate interplay between nanoparticle-mediated cyanobacterial interactions is pivotal in elucidating their impact on the transcriptome and lipidome. In the present study, total fatty acid methyl esters (FAMEs) in the wild-type (B481-WT) and transformant (B481-SD) Fremyella diplosiphon strains treated with nanoscale zero-valent iron nanoparticles (nZVIs) were characterized, and transcriptome changes analyzed. Comprehensive two-dimensional gas chromatography-time-of-flight mass spectrometry revealed a 20–25% higher percentage of FAMEs in nZVI-treated F. diplosiphon strain B481-SD compared to B481-WT. Accumulation of alkanes was significantly higher (> 1.4 times) in both strains treated with 25.6 mg L−1 nZVIs compared to the untreated control. In addition, we observed significantly higher levels of monounsaturated FAMEs (11%) in B481-WT in 3.2 (11.34%) and 25.6 mg L−1 (11.22%) nZVI-treated cells when compared to the untreated control (7%). Analysis of the F. diplosiphon transcriptome treated with 3.2 mg L−1 revealed a total of 1811 and 1651 genes that were differentially expressed in B481-SD and B481-WT respectively. While the expression of iron uptake and ion channel genes was downregulated, genes coding for photosynthesis, pigment, and antioxidant enzymes were significantly (p < 0.05) upregulated in B481-SD treated with 3.2 mg L−1 nZVIs compared to the untreated control. This study on essential FAMEs and regulation of genes in nZVI-treated F. diplosiphon strains provides a molecular framework for optimization of metabolic pathways in this model species.
Keywords: Cyanobacteria, Lipidome, Nanoparticles, Transcriptome
Subject terms: Nanobiotechnology, Transcriptomics
Introduction
Only a few microorganisms, including yeasts, molds, algae, and cyanobacteria can accumulate lipids to more than 20% of their dry cell weight and are thus termed as oleaginous1. Of these, cyanobacteria are ideal third and fourth-generation organisms for biotechnological advancements, particularly in the fields of biofuel production and carbon capture2. Although cyanobacteria-derived lipids store twice as much energy per carbon atom as carbohydrates, offering a two fold increase in fuel energy content, achieving commercial viability for cyanobacterial fuel remains a significant challenge3,4. Of the various species, Fremyella diplosiphon, a model organism renowned for its short generation cycle and adaptability to a wide range of light, offers a unique platform for unraveling the intricate molecular responses induced by environmental stressors such as salinity and light5. With its abundant high-value fatty acid methyl esters (FAMEs) in transesterified lipids, including value-added 7-hexadecanoic acid (C16:1) and 9-octadecenoic acid (C18:1) methyl esters, F. diplosiphon has garnered significant attention as a promising candidate for bioenergy production6.
Although the impact of abiotic stressors such as heat, cold, salinity, nitrogen starvation, photo-oxidation, anaerobiosis, and osmotic changes on microorganisms are well established7–9, chemical stressors such as nanoparticles and their consequences are yet to be fully understood. Specifically, metallic nanoparticles have started to gain preeminent importance due to their distinctive optical and magnetic characteristics that significantly impact biological processes such as growth, cellular physiology, photosynthetic activity, pigmentation, gene expression, and macromolecule synthesis, including lipid content10–12. This impact is particularly evident in photosynthesis and pigmentation, where the interaction of nanoparticles with cellular structures can enhance or inhibit these processes depending on the concentration, known as the “hormetic effect”13,14. Several studies, such as those by Du et al.15 using Zn nanoparticles on Microcystis aeruginosa, and Bytešníková et al.16 using graphene oxide nanoparticles on Chlamydomonas reinhardtii, have explored these interactions, providing valuable insights into the mechanisms by which nanoparticles influence cellular functions. As vital components of cell membranes, lipids are crucial for maintaining structural integrity and functionality and regulating the pores that control the movement of substances in and out of the cell17. Furthermore, lipids such as sterols are essential in modulating intracellular signaling pathways18, and can alter gene expression involved in the formation of lipid rafts that compartmentalize cellular processes along with the synthesis of secondary messengers in signal transduction and the provision of energy storage through fatty acid metabolism19,20. Therefore, nanoparticles can influence lipid metabolism by altering enzymatic activities involved in lipid synthesis and degradation. Additionally, the interaction of nanoparticles with lipid bilayers can alter membrane properties such as fluidity, permeability, and phase behavior21, since unsaturated fatty acids are essential constituents of polar glycerolipids in biological membranes, and the unsaturation level of membrane lipids is crucial for controlling membrane fluidity22. In living organisms, the regulation of membrane fluidity, which is critical for the normal functioning of biological membranes, plays a significant role in tolerance and acclimatization to environmental stresses through a process known as “homeoviscous adaptation”, which involves changes in membrane fatty acid composition as an adaptive response to environmental changes23. For instance, in a study on the cyanobacterium Arthrospira maxima treated with 5.1 mg L⁻¹ nZVIs, notable changes in lipid composition, including a decrease in saturated and monounsaturated fatty acids, and an increase in polyunsaturated fatty acids was reported24. Consequently, analyzing these interactions is crucial for evaluating the potential benefits and risks associated with nanoparticle exposure in biological systems, as they can have profound implications for cellular physiology, stress response, and overall organismal health.
In addition to lipid analysis, understanding the impact of nanoparticles on the transcriptome of cyanobacteria can provide crucial insights into the molecular mechanisms underlying their responses to these environmental stressors. Depending on the type of nanoparticle exposure, altered gene expression, metabolism, and homeostasis to metals have been documented24. By analyzing changes in gene expression patterns, researchers can elucidate the adaptive strategies cyanobacteria employ when exposed to nanoparticles, potentially uncovering novel pathways for biotechnological applications. A study investigating the impact of 200 mg L−1 nZVIs on the Dictyosphaerium sp. transcriptome revealed 2,065 differentially expressed genes, with 400 upregulated and 1,665 downregulated compared to the untreated control25. This significant alteration in gene expression highlights the profound influence of nanoparticles on cellular processes such as pigment synthesis, photosynthetic efficiency, and stress response pathways, underscoring the importance of understanding these interactions in advancing both environmental and biotechnological research. In addition to profiling genes using the high-throughput transcriptome sequencing (RNA-seq) approach, low-abundance transcripts, which account for approximately 30% of most transcriptomes and novel exons and splice junctions can be quantified26. Moreover, the choice of nanoparticles used in culture treatment can profoundly impact differential gene expression, offering valuable insights into their biological effects. In a study by Mortimer et al.27, carbon and boron nitride-based nanoparticles induced significant transcriptomic responses in bacteria at non-growth-inhibitory concentrations of 10 mg L−1. Specifically, multiwall carbon nanotubes induced the expression of 111 genes, while graphene, boron nitride, and carbon black differentially regulated 44, 26, and 25 genes, respectively.
Despite a report on the impact of optimal nZVI concentration on lipid production in F. diplosiphon, there have been no studies to examine their impact under moderate and high ROS levels in this model species. In addition, there exists a gap in understanding the impact of nZVIs on the differentially expressed targeted genes in this species. Therefore, in the present investigation, we aimed to explore the impact of moderate and sub-optimal concentrations of nZVIs on F. diplosiphon FAME profiles using using comprehensive two-dimensional gas chromatography coupled with time-of-flight mass spectrometry (GCxGC/TOF-MS). In addition, differential expression of targeted genes and signaling pathways in F. diplosiphon under moderate ROS induced by nZVIs was examined using whole transcriptome sequencing.
Materials and methods
Strains, culture conditions, and nanoparticles
F. diplosiphon strains, B481-WT from the UTEX algal culture collection repository (Austin, TX, United States) and B481-SD (B481-WT strain overexpressed with the sterol desaturase gene; accession MH329183) were used in this study. Cultures were grown in liquid BG-11 medium containing 20 mM HEPES to an exponential growth phase (~ optical density 750 nm of 0.8) under continuous shaking at 170 rpm and 28 °C in an Innova 44R incubator shaker (Eppendorf, Hamburg, Germany). Light wavelengths in the shaker were adjusted to 30 µmol m−2 s−1 using the model LI-190SA quantum sensor (Li-Cor, Lincoln, NE, USA). Nanofer 25s zero-valent iron nanoparticles (nZVIs), sourced from Nano Iron Company (Rajhrad, Czech Republic), were added to liquid media at final concentrations of 3.2 mg L−1 and 25.6 mg L−1. These concentrations were selected based on a previous report of their moderate and high levels of ROS in F. diplosiphon28.
Quantification of total lipids in nZVI-treated Fremyella diplosiphon by gravimetric analysis
F. diplosiphon B481-WT and B481-SD strains were grown in BG-11 media containing 3.2 and 25.6 mg L−1 nZVIs for 15 days in conditions mentioned in Sect. 2.1. Cultures grown in the absence of nZVIs under similar conditions served as control. Cells were pelleted, lyophilized, and total lipids extracted using a chloroform: methanol method based on Folch et al.29. Briefly, a 15 ml methanol/chloroform (2:1, v/v) solvent was added to 100 mg of lyophilized cells. The mixture was homogenized, incubated on ice, and centrifuged at 3000 rpm for 10 min to collect the organic phase. The residue was re-extracted thrice with 4 ml of methanol and 2 ml of chloroform, and the organic phase dried in a rotary evaporator to estimate the total lipid content.
GCxGC/TOF-MS analysis of transesterified lipids in nZVI-treated F. diplosiphon
F. diplosiphon lipids were converted to FAMEs through direct transesterification as described by Wahlen et al.30. Lyophilized cells (100 mg) were dissolved in 3 ml methanol containing 1.8% (v/v) sulfuric acid and subjected to 80 °C for 20 min in a commercial multimode scientific microwave (CEM Corporation, USA) with a maximum power output set at 25 W per sample. The reaction was quenched with 4 ml chloroform, washed in distilled water, and centrifuged at 2000 rpm for phase separation. The chloroform phase containing FAMEs and lipids was removed, and the remaining biomass was washed twice with 2 ml chloroform. The transesterified lipids were dried under nitrogen gas and then reconstituted in 2 ml of dichloromethane, with cholestane (50 µg ml−1) added as an internal standard. FAMEs and alkanes were characterized and quantified by GCxGC/TOF-MS as described by Tabatabai et al.31.
Differential gene expression in wild-type and nano-treated F. diplosiphon variants
Total RNA from F. diplosiphon B481-WT and B481-SD strains grown in 3.2 mg L−1 nZVIs and the untreated control was extracted according to the modified method of Fathabad et al.32. RNA integrity was verified prior to library preparation. After removing ribosomal RNA and fragmentation, the first strand of cDNA was synthesized with random hexamer primers, using a reaction buffer containing dTTPs instead of dUTPs for the second strand. The directional library was subjected to end repair, A-tailing, adapter ligation, size selection, USER enzyme digestion, amplification, and purification. Qubit and real-time PCR were used to quantify the library, and a bioanalyzer was used to detect size distribution. Sequencing was performed on the Illumina platform using paired-end reads after cluster generation (Novogene Corporation Inc., CA). Raw data reads of FASTQ format were processed by removing poly-N sequences and low-quality reads, and Q20, Q30, and GC content calculated. Reference genome and gene model annotation files were downloaded from JGI genome portal. Building the index of the reference genome and aligning clean reads were performed using Bowtie 2 software (v23.4.3)33.
Statistical analysis
Total lipid and FAME analysis
Total lipids and FAME content were reported as a cumulative treatment mean ± standard error. Statistical significance was determined using one-way analysis of variance and Tukey’s honest significant difference post-hoc test at 95% confidence intervals (p < 0.05). The single-factor, fixed-effect ANOVA model, Yij = µ + αSi + εij, was used where Y is the total lipid content in strain i and biological replicate j. The µ represents the overall total lipid content with adjustments from the effects of strain (αS), and εij is the experimental error from strain i and biological replicate j.
Gene structure analysis and quantification of gene expression level
Novel genes, operon, and transcription start sites were identified using Rockhopper. The upstream 700 bp sequence of the transcription start site for predicting the promoter using a Time-Delay Neural Network was extracted. Fragments per kilobase of transcript per million mapped reads (FPKM) of each gene were calculated based on gene length and the read count mapped to this gene. The expected number of FPKM transcript sequences was used to determine the effect of sequencing depth and gene length for the read count at the same time.
Differential expression analysis
Differential expression analysis consisting of two biological replicates was performed using the RNA-seq data DESeq2 R package. Negative binomial distribution was employed to determine differential expression in digital gene expression data and resultant p-values were adjusted using Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with an adjusted p-value < 0.05 determined by DESeq were assigned as differentially expressed. Prior to differential gene expression analysis, the read counts were adjusted for each sequenced library by Trimmed Mean of M- values through one scaling normalized factor. The edgeR package (v3.24.3) was used to analyze the differential expression of two conditions. The corrected p-value of 0.05 and |Log2 (fold change) | of 1 were set as the threshold for significantly differential expression.
GO and KEGG enrichment analyses of differentially expressed genes
Gene Ontology (GO) enrichment analysis of differentially expressed genes, incorporating a correction for gene length, was performed by the cluster-Profiler R package (v3.8.1). We considered GO terms with corrected p values less than 0.05 to be significantly enriched by differentially expressed genes. The cluster-Profiler R package was used to test the differential expression of genes in KEGG pathways for statistical enrichment.
Results and discussion
With a predicted proteome to produce and secrete fatty acids, cyanobacteria offer diverse biological and industrial implications. Their inherent attributes, such as environmental sustainability and enhanced adaptive capability of phycobiliproteins, make these organisms a viable source of bioproducts. The present research elucidates new approaches to studying total lipid composition and transcriptome changes in F. diplosiphon strains B481-WT and B481-SD exposed to varying nZVI concentrations.
Total lipids in nZVI-treated F. diplosiphon B481-WT and B481-SD
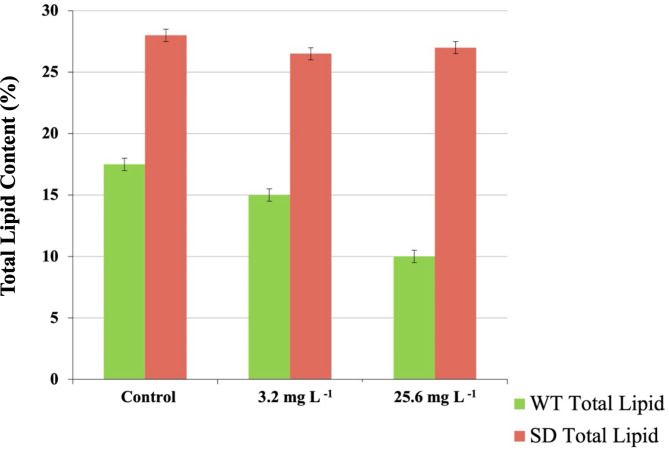
Quantitative analysis by gravimetry offers highly precise and reproducible measurements of total lipid content in various organisms, including cyanobacteria and microalgae32,34. Comparison of total extracted lipids in B481-SD treated with 3.2 and 25.6 mg L−1 nZVIs revealed significantly higher (p < 0.05) total lipid content compared to B481-WT at similar concentrations (Fig. 1). In addition to the cultures treated with nZVIs at both the concentrations, total lipid content in B481-SD was significantly higher than lipids in the untreated B481-WT (Fig. 1). These findings suggest that overexpression of the sterol desaturase gene in B481-SD could have enhanced lipid production by catalyzing an intermediate step in the synthesis of major sterols, in addition to influencing cellular signaling and membrane lipid distribution32. In addition, enhanced tolerance to stressors such as β-lactam (ampicillin, cefotaxime) and aminoglycoside (kanamycin) antibiotics in B481-SD compared to the B481-WT strain has been reported35.
Fig. 1.
Total lipid content in Fremyella diplosiphon grown in 3.2 and 25.6 mg L−1 nanoscale zero-valent iron nanoparticles over a period of 15 days. The average % lipid content of total cellular dry weight (± standard error) for three biological replicates is shown. Different letters above bars indicate significance among treatment means (p < 0.05).
F. diplosiphon FAMEs in optimal and sub-optimal nZVI concentrations
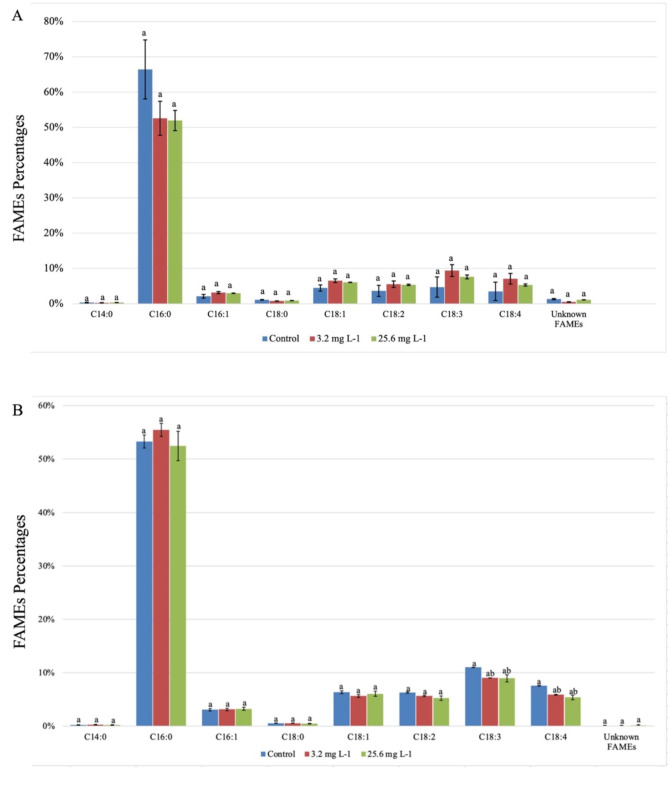
In addition to the total lipid content, we analyzed FAME composition, which is a crucial indicator in determining cyanobacterial lipid quality. As the most abundant type of FAMEs in F. diplosiphon, we observed hexadecanoic acid methyl ester (C16:0) to comprise 66.4% and 53.3% of the total FAMEs in the untreated B481-WT and B481-SD strains respectively. In B481-WT, the percentage of hexadecanoic acid was lower than the untreated control, at 52.6% and 51.9% in 3.2 mg L⁻¹ and 25.6 mg L⁻¹ nZVIs, respectively. The alteration in the predominant saturated fatty acid profile of B481-WT could be attributed to its vulnerability to nZVIs, which could have altered the FAMEs. It should be noted that the heightened sensitivity in B481-WT is possible due to the lack of the overexpressed sterol desaturase gene, unlike B481-SD. In a similar study by Fazelian et al.36, a significant decrease in the percentage of hexadecanoic acid methyl ester (C16:0), from 22.84% in the untreated control to 13.73% in Nannochloropsis oculata treated with Fe2O3-NPs was reported. However, we observed the highest percentage of C16:0 in B481-SD at 3.2 mg L−1 nZVI concentration (55.71%) compared to the untreated control (52.47%) and 25.6 mg L−1 nZVIs (51.78%) (Fig. 2). Since hexadecanoic acid is present in cyanobacterial/microalgal strains ranging from 23 to 43% and the composition of fatty acids is influenced by several parameters37,38, it is not surprising to observe variations in C16:0 between the two strains tested in this study. Thus, alterations in fatty acid saturation can influence the resilience of the species to abiotic stressors such as nZVIs. Similar observations have been reported in other cyanobacterial species, where an increase in unsaturated fatty acid content occurred as an adaptive response for maintaining membrane fluidity under stress conditions and is attributed to the lower melting point of unsaturated fatty acids compared to saturated fatty acids39. This activation could serve as a protective mechanism, where elevated SFAs and MUFAs stabilize cell membranes against oxidative stress induced by nZVIs. In addition, we observed a 1.3X increase in unsaturated fatty acids in untreated B481-SD compared to the untreated B481-WT (Table 1). These results are in accordance with a previous finding where overexpression of the sterol desaturase gene resulted in higher unsaturated fatty acids, possibly due to enhanced intracellular signal activation19. In addition, the genes encoding acyl-CoA desaturase (2.7X) and fatty acid desaturase (1.2X) in B481-SD were significantly enhanced compared to untreated B481-WT (Table 2A).
Fig. 2.
Comparison of total fatty acid methyl esters (FAMEs) in zero-valent iron nanoparticle-treated Fremyella diplosiphon (A) B481-WT and (B) B481-SD. Different letters above error bars indicate significance differences (p < 0.05).
Table 1.
Comparison of saturated and unsaturated fatty acid methyl esters in Fremyella diplosiphon B481-WT and B481-SD strains treated with 3.2 and 25.6 mg L−1 nanoscale zero-valent iron nanoparticles (nZVIs).
| Fatty acid methyl esters (FAME %) | Ratio (B481-WT) | Ratio (B481-SD) | |||
|---|---|---|---|---|---|
| Treatment | FAME type | B481-WT | B481-SD | Saturated/ unsaturated | Saturated/ unsaturated |
| Control (untreated) | Saturated | 78.63% | 61.18% | 3.7 | 1.6 |
| Monounsaturated | 7.61% | 10.67% | |||
| Polyunsaturated | 13.76% | 28.15% | |||
| 3.2 mg L−1 nZVIs | Saturated | 62.82% | 65.72% | 1.7 | 1.9 |
| Monounsaturated | 11.34% | 10.29% | |||
| Polyunsaturated | 25.84% | 24.00% | |||
| 25.6 mg L−1 nZVIs | Saturated | 66.05% | 64.77% | 1.9 | 1.8 |
| Monounsaturated | 11.22% | 11.35% | |||
| Polyunsaturated | 22.73% | 23.89% | |||
Table 2.
Comparison of Fold change values in lipid metabolism genes of untreated Fremyella diplosiphon B481-SD versus untreated B481-WT (A), and pigmentation genes in B481-WT treated with 3.2 mg L−1 compared to B481-SD treated with 3.2 mg L−1 nanoscale zero-valent iron nanoparticles (nZVIs) (B).
| A. B481-SD untreated vs. B481-WT untreated control | ||
| Gene ID | Gene | Fold change |
| FDUTEX481_RS12380 | sterol desaturase | 1.31934689 |
| FDUTEX481_RS13295 | fatty acid desaturase | 1.22612004 |
| FDUTEX481_RS23510 | acyl-CoA desaturase | 2.72771528 |
| B. B481-WT treated with 3.2 mg L−1 nZVIs vs. B481-SD treated with 3.2 mg L−1 nZVIs | ||
| Gene ID | Gene | Fold change |
| FDUTEX481_RS17975 | C-phycoerythrin subunit beta (cpeB) | -0.0305466 |
| FDUTEX481_RS06080 | allophycocyanin alpha-B chain (apcD) | 1.14585931 |
| FDUTEX481_RS16175 | phycocyanin subunit alpha (cpcA) | 0.99662662 |
| FDUTEX481_RS10560 | allophycocyanin subunit alpha 2 (apcA2) | 1.85575686 |
Our results on lipid profiling revealed a ~ 1.5X increase in monounsaturated fatty acids in B481-WT treated with 3.2 and 25.6 mg L−1 nZVIs compared to the untreated control, while no significant changes were observed in B481-SD (Table 1). Interestingly, we noted ~ 1.4X more MUFAs in the untreated B481-SD compared to untreated B481-WT (Table 1). These results indicate that nZVIs at both 3.2 and 25.6 mg L−1 concentrations could significantly impact MUFA production in B481-WT; however, in comparison to the untreated cultures, B481-SD (10.67%) exhibited higher MUFAs than B481-WT (7.61%), possibly due to alterations in lipid saturation linked to sterol desaturase. In a similar study by Lu et al.40, exposure of Chlamydomonas sp. to isobutyl cyanoacrylate nanoparticles resulted in increased MUFAs, including C18: 1. These changes were attributed to intracellular stress-associated alterations in cyanobacterial lipid profile distribution. Additionally, a strong connection between antioxidant activity and MUFAs was demonstrated in a comparative study, in which Gloeocapsa sp. had the highest MUFA content (27.01 ± 0.15) corresponding to the highest antioxidant DPPH activity (88.84 ± 10.26), surpassing Chroococcidiopsis sp. (19.71 ± 0.93) and Merismopedia sp. (27.32 ± 2.83)41. These data support our findings that the increased MUFA levels observed in B481-WT and B481-SD treated with 3.2 mg L−1 nZVIs relate to the antioxidant-related benefits of MUFAs and improved oxidative stress resilience.
Further, our findings revealed that 61.2 to 80.5% of the extracted FAMEs consisted of SFAs (Table 1). It was interesting to note fewer SFAs in B481-WT treated with 3.2 (62.82%) and 25.6 (66.05%) mg L−1 nZVIs compared to the untreated control (78.63%). In a similar study on Nannochloropsis oculata treated with Fe2O3-NPs, an 8.69% decrease in SFAs compared to the untreated control was reported36. However, we observed 65.72% and 64.77% SFAs in B481-SD treated with 3.2 and 25.6 mg L−1 nZVIs compared to 61.18% in the control, suggesting the impact of the overexpressed sterol desaturase gene on B481-SD fatty acid composition. It is important to note the role and benefits of SFAs for industrial applications, including their use in renewable energy production. Despite their poor stability at low temperatures, SFAs are essential constituents of FAMEs like the MUFAs, contributing to high combustion properties such as higher cetane number and greater calorific value40. Thus, a balanced enhancement of SFAs and MUFAs in response to nZVI treatment is crucial for achieving optimal bioproducts.
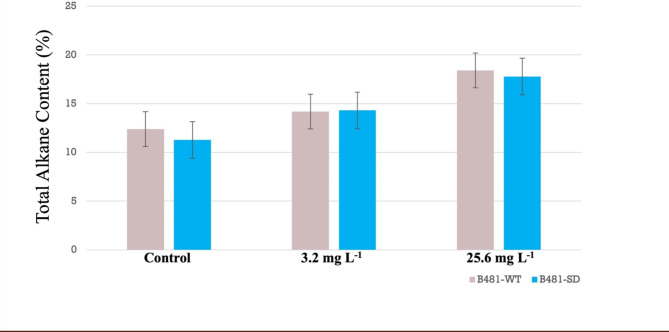
As energy-dense sources, alkanes represent another vital group of key infrastructure-compatible biofuel components synthesized by cyanobacteria. Our findings indicated significantly high (p < 0.05) alkane content of 1.4X in B81-WT and 1.5X in B481-SD strains treated with 25.6 mg L−1 nZVIs (Fig. 3). The observed increase in alkane content can be attributed to the potential role of nZVIs in altering the activity of enzymes such as acyl-ACP reductase and aldehyde deformylating oxygenase, which are key in converting fatty acyl-ACP to alkanes, thus boosting the overall energy content42. This increase could be attributed to modulations in redox balance, which aid in regulating cyclic electron flow and suppressing ROS43, thus enabling the organism to withstand stress while maintaining optimal growth and productivity. Similar findings were observed in a previous study in F. diplosiphon B481-SD treated with 3.2 mg L−1 nZVIs and 0.8 mg L−1 ampicillin, which resulted in a 2–4 fold augmentation in alkanes compared to the untreated control44. Furthermore, in a study by Schirmer et al.45, a total of 17 genes were identified by subtractive genome analysis in alkane-producing cyanobacteria, while these genes were notably absent in non-alkane-producing species. Of the seventeen, two genes (AAR and ADO), were found to code for crucial enzymes (acyl-ACP reductase and aldehyde-deformylating oxygenase), which led to the discovery of the alkane biosynthesis pathway using fatty acyl ACP as its substrate.
Fig. 3.
Comparison of alkane content in Fremyella diplosiphon strains B481-WT and B481-SD grown in 3.2 and 25.6 mg L−1 zero-valent iron nanoparticles over a period of 15 days. The average % alkane content of total cellular dry weight (± standard error) for three biological replicates is shown.
Differential gene expression in nZVI-treated F. diplosiphon
To gain insight into the transcriptome changes in F. diplosiphon treated with 3.2 mg L−1 nZVIs, we compared the differential gene expression of treated cells to the untreated control. All comparisons were made using non-redundant lists of genes after the removal of gene IDs without an entire number. When more than one gene (pseudogenes) was recognized, one was selected, and in the case of redundant gene ID numbers, the one that was most altered was chosen.
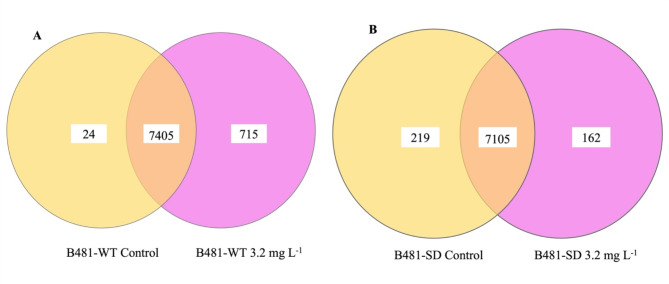
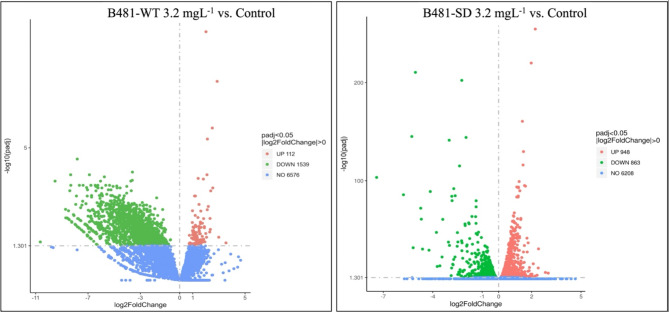
Our results indicated specific cellular functions to be regulated in nZVI-treated F. diplosiphon, in addition to the function of genes and the molecular mechanisms underlying specific biological processes. The reads from Tolypothrix sp. PCC 7601 were aligned to the F. diplosiphon reference genome and used as a reference. Using the methodology and filters outlined above, we generated lists of changes in 2-fold altered genes. In total, about 30 million raw paired-end reads were generated for each sample, with an average of 29,625,787 for B481-WT and 31,608,579 for B481-SD. A comparison of expressed genes in the two strains revealed a higher number of genes in the F. diplosiphon B481-SD. As illustrated in the Venn diagram (Fig. 4), a total of 715 genes were uniquely expressed in B481-WT treated with 3.2 mg L−1 nZVIs, 24 genes in the untreated control, and 7405 genes were commonly expressed. On the contrary, strain B481-SD treated with 3.2 mg L−1 nZVIs revealed 162 genes while 219 genes were detected in the control; however, 7105 genes were commonly expressed (Fig. 4). Analysis of differentially expressed genes (DEG) Log2 fold change of B481-WT treated with 3.2 mg L−1 nZVIs revealed 112 up-regulated, 1539 down-regulated genes, and 6576 unaltered genes compared to the untreated control (Fig. 4). Interestingly, we observed fewer downregulated but more upregulated genes in F. diplosiphon B481-SD, in which Log2 fold changes in 3.2 mg L−1 nZVI-treated cells vs. B481-SD control indicated 863 down-regulated genes and 948 up-regulated genes as depicted in the differential gene count and volcano plot (Fig. 5). These results suggest that iron levels are pivotal to ensure the balance in genetic expression changes. Alternatively, it is also possible that iron limitation can be crucial in both the up-regulation and down-regulation of genes. A previous study has shown that severe iron limitation caused the downregulation of 36 genes and the upregulation of 35 genes, while mild iron limitation resulted in the downregulation of 27 genes and the upregulation of 62 genes in Synechococcus sp. PCC700246.
Fig. 4.
Venn diagrams showing the number of genes expressed in Fremyella diplosiphon strains (A) B481-WT and (B) B481-SD grown in 3.2 mg L−1 zero-valent iron nanoparticles compared to the untreated control.
Fig. 5.
Volcanic representation of differentially expressed genes in Fremyella diplosiphon strains (A) B481-WT and (B) B481-SD treated with 3.2 mg L−1 zero-valent iron nanoparticles compared to untreated control.
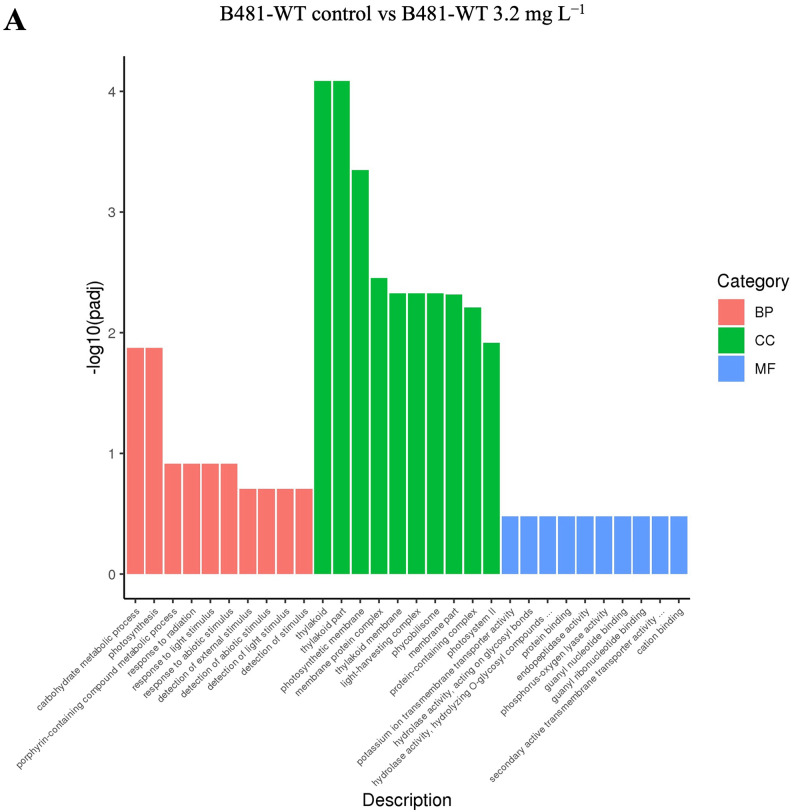
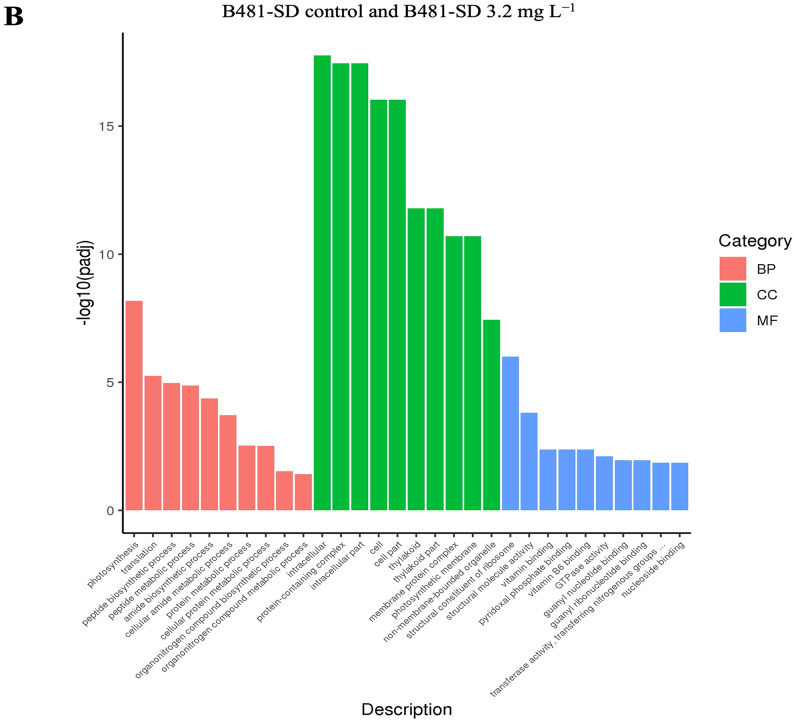
The gene ontology (GO) enrichment histogram of co-upregulated and downregulated genes is shown in Fig. 6. In the three groups (BP, blue; MF, red; and CC, green), the EC score was calculated for each GO category and compared with that of random sampling (purple) to estimate the statistical significance level. The histogram intuitively reflects the distribution of the number of DEGs in GO terms enriched in BP, CC, and MF. The ordinate is the enriched GO term, and the abscissa is the number of differentially expressed genes in the term.
Fig. 6.
Biological Process (BP), Molecular Function (MF), and Cellular Component (CC) groups of GO (gene ontology) in Fremyella diplosiphon (A) B481-WT untreated control versus B481-WT treated with 3.2 mg L−1 nanofer 25s zerovalent nanoparticles-treated (nZVIs), (B) B481-SD control versus B481-SD 3.2 mg L−1 nZVI-treated cultures. The total number of categories in each GO is indicated in parentheses. Fraction of categories (y axis) indicates the ratio of the number of GO categories with higher EC score than each threshold EC score (x axis) to the total number of GO categories.
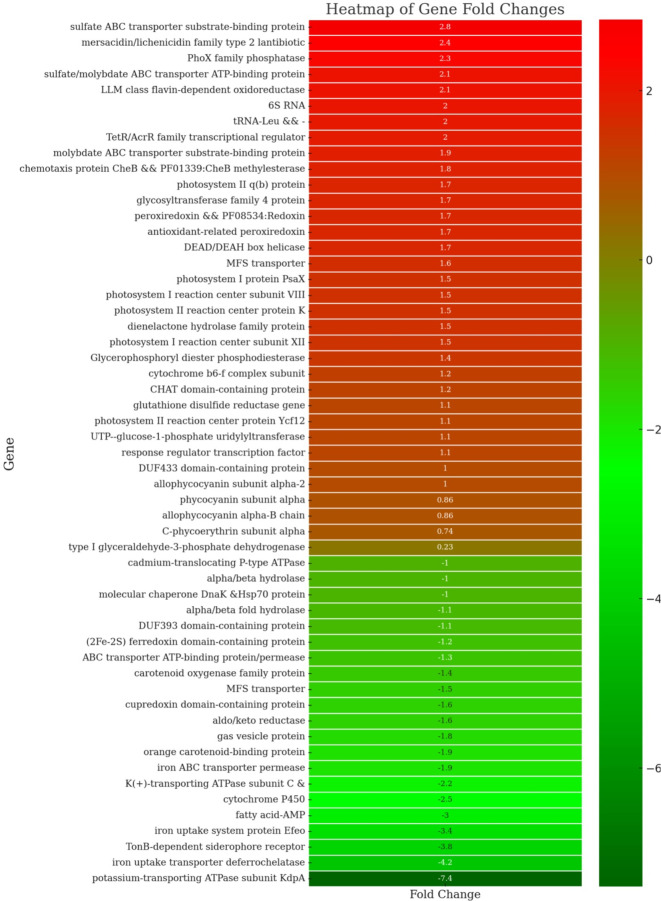
The application of nZVIs can significantly influence the photosynthetic efficiency of cyanobacteria by interacting with photosystem I (PSI) and photosystem II (PSII). A previous study has shown that nZVIs can enhance electron transport rates in PSI and PSII47, improving light absorption and energy conversion. Furthermore, experiments have demonstrated that nanoparticles, by eliminating unwanted wavelengths, can increase the target wavelengths, thereby promoting pigment content and enhancing the expression of photosynthetic genes48. In this study, we observed an upregulation of the genes involved in photosynthesis, including psaI, psaX, psaM, psbM, psbK, psbA, and psb30 in B481-SD treated with 3.2 mg L−1 nZVIs compared to the untreated control, as indicated by a Log2 fold increase between 1.1X and 1.7X (Fig. 7). These findings indicate the impact of nZVIs on F. diplosiphon PSI and PSII, enhancing the photosynthetic capacity by upregulating the target genes. This effect may be caused by the uptake of zerovalent iron molecules by the cells, which can be involved as a cofactor in enzymatic pathways to enhance photosynthesis49. Similarly, a study by Zhu et al.47 reported that Chlorella pyrenoidosa treated with 15 mg L−1 of TiO2 nanoparticles increased PSI and PSII gene expression, including psaI, psbA, psbM, and psbK, with a Log2 fold change between 3.56 and 5.51. Moreover, given that nZVI produces Fe(II) through oxidation50, both nZVI and iron oxides could be responsible for the growth and physiological responses of treated cells. The Fe(II) generated can further participate in redox reactions, facilitating electron transfer processes crucial for photosynthesis and other metabolic activities51.
Fig. 7.
Heat map representing differential gene expression between Fremyella diplosiphon strains B481-SD and B481-WT treated with 3.2 mg L−1 zero-valent iron nanoparticles. The heat map displays fold changes, with upregulated genes shown in red and downregulated genes in green. The intensity of the color corresponds to the magnitude of the fold change, with darker shades indicating more significant changes.
In addition, phycobilisomes and carotenoids in cyanobacteria absorb and transfer excitation energy with high quantum efficiency to photosystem II and I in the photosynthetic lamellae52. Comparison of the pigment-coding genes cpeA, apcD, cpcA, and apcA2 in 3.2 mg L−1 nZVIs and untreated B481-SD showed a significant upregulation only in the apcA2 gene (Fig. 7). In contrast, B481-WT treated with 3.2 mg L−1 showed upregulation of both apcD (Log2 Fc 1.1) and apcA2 genes (Log2 Fc 1.86) (Table 2B). The upregulation of the pigment coding genes in both strains treated with 3.2 mg L−1 nZVIs indicated cellular protection against nanoparticle-associated cellular stress. In a similar study by Ruan et al.53, Synechocystis sp. PCC 6803 exposed to 0.25 mg L−1 Cadmium nanoparticles for 72 h, upregulation of allophycocyanin subunit alpha 2 and phycocyanin alpha subunit compared to the untreated control group due to a 3–4 fold increase in ROS was observed.
Although nZVIs can be beneficial for gene expression studies related to photosynthesis, pigment production, and lipid synthesis, one of the most important mechanisms underlying these effects is oxidative stress54. The generation of ROS through oxidative stress can enhance these processes by promoting cellular activities and metabolic pathways55. However, maintaining a delicate balance between beneficial oxidative stress and detrimental oxidative damage is crucial for cellular survival. Therefore, the regulation of ROS levels is essential to harness the positive effects of nZVIs while preventing potential negative impacts on cell health. Antioxidant systems play a vital role in this regulation by scavenging excess ROS and protecting cellular components from oxidative damage. Interestingly, the Log2 FC for the glutathione reductase gene in B481-SD treated with 3.2 mg L−1 nZVIs was 1.12 fold times higher compared to the untreated control (Fig. 7). As an essential component of the antioxidant defense system in both cyanobacteria and plants, this gene has been reported to maintain cellular redox homeostasis by reducing glutathione disulfide to the sulfhydryl form antioxidant56. Another notable upregulated gene enhancing antioxidant capacity in nZVI-treated B481-SD was peroxiredoxin (Log2 FC 1.7), which is well-known for maintaining redox homeostasis in cyanobacteria57. This is supported by a study on Anabaena PCC 7120, which demonstrated that exposure to H2O2 could induce overexpression of peroxiredoxin Q proteins, thereby reducing oxidative stress in intact photosynthetic pigments58. Thus, the better survival rate of nZVI-treated B481-SD compared to B481-WT could be attributed to the elevated expression of genes coding for antioxidant enzymes.
In addition, differential expression of multiple iron transmembrane transporter protein genes was observed in both strains treated with nZVIs. While the genes coding for iron uptake transporter deferrochelatase, TonB-dependent siderophore receptor, and the iron uptake system protein Efeo were down-regulated by Log2 fold changes ranging from 3.4 to 4.2 in B481-SD, expression of these genes were not significant (p > 0.05) in B481-WT (Fig. 7). It is well known that several iron-uptake channels identified in cyanobacteria enable their survival in environments with scarce iron supply59,60. However, maintaining intracellular iron levels is pivotal for cellular homeostasis, and the iron channels located on the cellular membrane in cyanobacteria play a crucial role in ensuring this vital balance. Thus, it is possible that genes encoding these specific channels could be downregulated as a result of negative regulation systems due to the increased intracellular iron levels. Consequently, we hypothesize that B481-SD has a delicate mechanism to maintain intracellular iron homeostasis in contrast to B481-WT. Besides, the regulation of other ion channels, such as potassium ions (K+), can be vital considering the impact of nZVIs on total intracellular ion balance. A low concentration of K+ was reported to increase cellular survival61, and various cyanobacterial cells such as Microcystis were found to be much more sensitive to K+ than other cations62. Additionally, we observed that the potassium-transporting ATPase subunit C and the potassium-transporting ATPase subunit KdpA in B481-SD were downregulated by Log2 fold changes of 2.2 and 7.4, respectively, when compared to the untreated culture (Fig. 7). Given the level of transcriptome changes observed in the present study, we infer that a number of genes were differentially regulated by exposure of F. diplosiphon to nZVIs.
Conclusion
The present study demonstrated significant alterations in the total lipid content of both the wild-type and transformant strains of F. diplosiphon treated with nZVIs. This effect was corroborated by the increased percentages of monounsaturated fatty acids and the distinctive FAME profiles observed, particularly hexadecanoic acid. Transcriptome analysis further illuminated the regulatory shifts in gene expression associated with iron uptake, photosynthetic activity, and fatty acid metabolism, underscoring the complex interplay between metabolic pathways and nZVIs as external stressors. These findings not only deepen our understanding of the metabolic alterations in F. diplosiphon as a response to nZVI exposure, but also highlight the improved potential of the transformant strain for biofuel production.
Acknowledgements
This research was funded by the National Science Foundation’s Nanoscale Interactions Program (1900966) and co-supported by the Excellence in Research. Work conducted at the National High Magnetic Field Laboratory was supported by the NSF Division of Chemistry and Division of Materials Research through DMR-2128556, as well as the State of Florida.
Author contributions
Conceptualization, H.C., A.S.A., V.S.; design S.G., H.C., A.S.A., V.S.; investigation S.G., H.C., Y.Y.; validation, S.G., Y.Y., H.C., S.M.M.; writing and editing, Y.Y., S.G., H.C., A.S.A., S.M.M., V.S.; funding, V.S., H.C.; and project administration, H.C., V.S. All authors have read and agreed to the final version of the manuscript.
Data availability
Sequence data that support the findings of this study have been deposited in the National Center for Biotechnology Information Archive with the primary accession code MH329183.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
The original online version of this Article was revised: In the original HTML version of this Article, the citation for Reference 11 was omitted in the Introduction and inadvertently cited as Reference 13. Additionally, in the Materials and methods section, under the subheading ‘Differential gene expression in wild-type and nano-treated F. diplosiphon variants’, the citation for ‘Fathabad et al.’ was incorrectly linked to ‘Folch et. al.’ (Reference 29). Full information regarding the corrections made can be found in the correction for this Article.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
1/30/2025
A Correction to this paper has been published: 10.1038/s41598-025-87330-y
References
- 1.Patel, A. et al. An overview of potential oleaginous microorganisms and their role in biodiesel and omega-3 fatty acid-based industries. Microorganisms8, 434 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koberg, M. & Gedanken, A. Optimization of bio-diesel production from oils, cooking oils, microalgae, and castor and jatropha seeds: probing various heating sources and catalysts. Energy Environ. Sci.5, 7460–7469 (2012). [Google Scholar]
- 3.Nozzi, N. E., Oliver, J. W. K. & Atsumi, S. Cyanobacteria as a platform for biofuel production. Front. Bioeng. Biotechnol.1, 7 (2013). [DOI] [PMC free article] [PubMed]
- 4.Dismukes, G. C., Carrieri, D., Bennette, N., Ananyev, G. M. & Posewitz, M. C. Aquatic phototrophs: efficient alternatives to land-based crops for biofuels. Curr. Opin. Biotechnol.19, 235–240 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Hannon, M., Gimpel, J., Tran, M., Rasala, B. & Mayfield, S. Biofuels from algae: challenges and potential. Biofuels1, 763–784 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knoot, C. J., Ungerer, J., Wangikar, P. P. & Pakrasi, H. B. Cyanobacteria: promising biocatalysts for sustainable chemical production. J. Biol. Chem.293, 5044–5052 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh, P., Srivastava, A. & Shukla, E. Response of cyanobacteria during abiotic stress with special reference to membrane biology: an overview. in Cyanobacterial Biotechnology in the 21st Century (eds Neilan, B., Passarini, M. R. Z., Singh, P. K. & Kumar, A.) 63–84 (Springer Nature, Singapore (2023). [Google Scholar]
- 8.Yang, W., Wang, F., Liu, L. N. & Sui, N. Responses of membranes and the photosynthetic apparatus to salt stress in cyanobacteria. Front. Plant. Sci.11, 713 (2020). [DOI] [PMC free article] [PubMed]
- 9.Wu, M. et al. Effects of different abiotic stresses on carotenoid and fatty acid metabolism in the green microalga Dunaliella salina Y6. Ann. Microbiol.70, 48 (2020). [Google Scholar]
- 10.Shakeel, T. et al. A consensus-guided approach yields a heat-stable alkane-producing enzyme and identifies residues promoting thermostability. J. Biol. Chem.293, 9148–9161 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sekoai, P. T. et al. Application of nanoparticles in biofuels: an overview. Fuel237, 380–397 (2019). [Google Scholar]
- 12.Klähn, S. et al. Alkane biosynthesis genes in cyanobacteria and their transcriptional organization. Front. Bioeng. Biotechnol.2, 24 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuo, S., Yang, H., Jiang, X. & Ma, Y. Magnetic Fe3O4 nanoparticles enhance cyanobactericidal effect of allelopathic p-hydroxybenzoic acid on Microcystis aeruginosa by enhancing hydroxyl radical production. Sci. Total Environ.770, 145201 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Yalcin, Y. S., Aydin, B. N. & Sitther, V. Impact of zero-valent iron nanoparticles and ampicillin on adenosine triphosphate and lactate metabolism in the cyanobacterium Fremyella diplosiphon. Microorganisms12, 612 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du, J. et al. Contributions of Zn ions to ZnO nanoparticle toxicity on Microcystis aeruginosa during chronic exposure. Bull. Environ. Contam. Toxicol.103, 802–807 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Bytešníková, Z. et al. New insight into the biocompatibility/toxicity of graphene oxides and their reduced forms on Chlamydomonas reinhardtii. NanoImpact31, 100468 (2023). [DOI] [PubMed] [Google Scholar]
- 17.Hori, K. et al. Recent advances in research on biointerfaces: from cell surfaces to artificial interfaces. J. Biosci. Bioeng.133, 195–207 (2022). [DOI] [PubMed] [Google Scholar]
- 18.Shah, A. M. et al. A hidden treasure of polyunsaturated fatty acids. Front. Nutr.9, 827837 (2022). [DOI] [PMC free article] [PubMed]
- 19.Eungrasamee, K., Incharoensakdi, A., Lindblad, P. & Jantaro, S. Synechocystis sp. PCC 6803 overexpressing genes involved in CBB cycle and free fatty acid cycling enhances the significant levels of intracellular lipids and secreted free fatty acids. Sci. Rep.10, 4515 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eungrasamee, K., Zhu, Z., Liu, X., Jantaro, S. & Lindblad, P. in Chapter 5 - Lipid Metabolism in cyanobacteria: Biosynthesis and Utilization. 85–116 (eds Cyanobacteria, A. K. & Singh, S. S.) (Academic, 2024).
- 21.Yadav, G. et al. Lipid content, biomass density, fatty acid as selection markers for evaluating the suitability of four fast growing cyanobacterial strains for biodiesel production. Bioresour Technol.325, 124654 (2021). [DOI] [PubMed] [Google Scholar]
- 22.Ishikawa-Ishiwata, Y., Nosaka, Y., Usui, T. & Sasaki, H. Growth characteristics and fatty acids of the thermotolerant green alga Desmodesmus sp. isolated from a freshwater puddle in Ibaraki, Japan. Plankton Benthos Res.18, 74–83 (2023). [Google Scholar]
- 23.de la Rosa, F., De Troch, M., Malanga, G. & Hernando, M. Differential sensitivity of fatty acids and lipid damage in Microcystis aeruginosa (cyanobacteria) exposed to increased temperature. Comp. Biochem. Physiol. Part. C Toxicol. Pharmacol.235, 108773 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Pádrová, K. et al. Trace concentrations of iron nanoparticles cause overproduction of biomass and lipids during cultivation of cyanobacteria and microalgae. J. Appl. Phycol.27, 1443–1451 (2015). [Google Scholar]
- 25.Jiang, Y. et al. Nanoscale zero-valent iron alters physiological, biochemical, and transcriptomic response of nonylphenol-exposed algae (Dictyosphaerium Sp). Environ. Sci. Pollut Res.29, 20711–20720 (2022). [DOI] [PubMed] [Google Scholar]
- 26.Van Aken, B. Gene expression changes in plants and microorganisms exposed to nanomaterials. Curr. Opin. Biotechnol.33, 206–219 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Mortimer, M., Devarajan, N., Li, D. & Holden, P. A. Multiwall carbon nanotubes induce more pronounced transcriptomic responses in Pseudomonas aeruginosa PG201 than graphene, exfoliated boron nitride, or carbon black. ACS Nano. 12, 2728–2740 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Fathabad, S. G. et al. Impact of zero-valent iron nanoparticles on Fremyella diplosiphon transesterified lipids and fatty acid methyl esters. ACS Omega. 5, 12166–12173 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Folch, J., Lees, M. & Stanley, G. H. S. A simple method for isolation and purification of total lipides from animal tissues. J. Biol. Chem.226, 497–509 (1957). [PubMed] [Google Scholar]
- 30.Wahlen, B. D., Willis, R. M. & Seefeldt, L. C. Biodiesel production by simultaneous extraction and conversion of total lipids from microalgae, cyanobacteria, and wild mixed-cultures. Bioresour Technol.102, 2724–2730 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Tabatabai, B. et al. Fremyella diplosiphon as a biodiesel agent: Identification of fatty acid methyl esters via microwave-assisted direct in situ transesterification. BioEnergy Res.11, 528–537 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gharaie Fathabad, S. et al. Augmenting Fremyella diplosiphon cellular lipid content and unsaturated fatty acid methyl esters via sterol desaturase gene overexpression. Appl. Biochem. Biotechnol.189, 1127–1140 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goldoost, H., Vahabzadeh, F. & Fallah, N. Lipids productivity of cyanobacterium Anabaena Vaginicola in an internally illuminated photobioreactor using LED bar lights. Sci. Rep.14, 6857 (2024). [DOI] [PMC free article] [PubMed]
- 35.Yalcin, Y. S. et al. Lipid production and cellular changes in Fremyella diplosiphon exposed to nanoscale zerovalent iron nanoparticles and ampicillin. Microb. Cell. Factor.22, 108 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fazelian, N., Yousefzadi, M. & Movafeghi, A. Algal response to metal oxide nanoparticles: analysis of growth, protein content, and fatty acid composition. BioEnergy Res.13, 944–954 (2020). [Google Scholar]
- 37.Ševců, A., El-Temsah, Y. S., Joner, E. J. & Černík, M. Oxidative stress induced in microorganisms by zero-valent iron nanoparticles. Microbes Environ.26, 271–281 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Kramm, A., Kisiela, M., Schulz, R. & Maser, E. Short-chain dehydrogenases/reductases in cyanobacteria. FEBS J.279, 1030–1043 (2012). [DOI] [PubMed] [Google Scholar]
- 39.de la Rosa, F., Pezzoni, M., De Troch, M., Costa, C. S. & Hernando, M. Effects of temperature up-shift and UV-A radiation on fatty acids content and expression of desaturase genes in cyanobacteria Microcystis aeruginosa: stress tolerance and acclimation responses. Photochem. Photobiol Sci.23, 1167–1178 (2024). [DOI] [PubMed] [Google Scholar]
- 40.Lu, H. et al. Enhanced triacyclglycerols and starch synthesis in Chlamydomonas stimulated by the engineered biodegradable nanoparticles. Appl. Microbiol. Biotechnol.107, 971–983 (2023). [DOI] [PubMed] [Google Scholar]
- 41.Wickramasinghe, M. et al. Exploration of antioxidant activities, microstructural properties, and fatty acid composition of three cyanobacteria species. Biocatal. Agric. Biotechnol.56, 103015 (2024). [Google Scholar]
- 42.Hayashi, Y. & Arai, M. Recent advances in the improvement of cyanobacterial enzymes for bioalkane production. Microb. Cell. Factories. 21, 256 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berla, B. M., Saha, R., Maranas, C. D. & Pakrasi, H. B. Cyanobacterial alkanes modulate photosynthetic cyclic electron flow to assist growth under cold stress. Sci. Rep.5, 14894 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ling, H., Chen, B., Kang, A., Lee, J. M. & Chang, M. W. Transcriptome response to alkane biofuels in Saccharomyces cerevisiae: identification of efflux pumps involved in alkane tolerance. Biotechnol. Biofuels. 6, 95 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schirmer, A., Rude, M. A., Li, X. & Popova, E. Del Cardayre, S. B. Microbial biosynthesis of alkanes. Science329, 559–562 (2010). [DOI] [PubMed] [Google Scholar]
- 46.Blanco-Ameijeiras, S., Cosio, C. & Hassler, C. S. Long-term acclimation to iron limitation reveals new insights in metabolism regulation of Synechococcus sp. PCC7002. Front. Mar. Sci.4, 247 (2017).
- 47.Zhu, L. et al. Physiological and transcriptomic analysis reveals the toxic and protective mechanisms of marine microalga Chlorella pyrenoidosa in response to TiO2 nanoparticles and UV-B radiation. Sci. Total Environ.912, 169174 (2024). [DOI] [PubMed] [Google Scholar]
- 48.Salehi, B. & Wang, L. Critical review on nanomaterials for enhancing bioconversion and bioremediation of agricultural wastes and wastewater. Energies15, 5387 (2022). [Google Scholar]
- 49.Gichuki, S. M. et al. Zero-valent iron nanoparticles induce reactive oxygen species in the cyanobacterium, Fremyella diplosiphon. ACS Omega. 6, 32730–32738 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodríguez-Rasero, C. et al. Use of zero-valent iron nanoparticles (nZVIs) from environmentally friendly synthesis for the removal of dyes from water—A Review. Water16, 1607 (2024). [Google Scholar]
- 51.Song, Y. et al. Nano zero-valent iron harms methanogenic archaea by interfering with energy conservation and methanogenesis. Environ. Sci. Nano. 8, 3643–3654 (2021). [Google Scholar]
- 52.Harke, M. J. & Gobler, C. J. Daily transcriptome changes reveal the role of nitrogen in controlling microcystin synthesis and nutrient transport in the toxic cyanobacterium, Microcystis aeruginosa. BMC Genom.16, 1068 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ruan, G., Mi, W., Yin, X., Song, G. & Bi, Y. Molecular responses mechanism of Synechocystis sp. PCC 6803 to cadmium stress. Water14, 4032 (2022). [Google Scholar]
- 54.Vargas-Estrada, L. et al. Role of nanoparticles on microalgal cultivation: A review. Fuel280, 118598 (2020). [Google Scholar]
- 55.Yalcin, Y. S., Aydin, B. N., Sayadujjhara, M. & Sitther, V. Antibiotic-induced changes in pigment accumulation, photosystem II, and membrane permeability in a model cyanobacterium. Front. Microbiol.13, 930357 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chakdar, H., Thapa, S., Srivastava, A. & Shukla, P. Genomic and proteomic insights into the heavy metal bioremediation by cyanobacteria. J. Hazard. Mater.424, 127609 (2022). [DOI] [PubMed] [Google Scholar]
- 57.Liu, X. et al. Anti-oxidant mechanisms of Chlorella pyrenoidosa under acute GenX exposure. Sci. Total Environ.797, 149005 (2021). [DOI] [PubMed] [Google Scholar]
- 58.Hrdlickova, R., Toloue, M. & Tian, B. RNA -Seq methods for transcriptome analysis. WIREs RNA. 8, e1364 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mirus, O., Strauss, S., Nicolaisen, K., Von Haeseler, A. & Schleiff, E. TonB-dependent transporters and their occurrence in cyanobacteria. BMC Biol.7, 68 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lau, C. K. Y., Krewulak, K. D. & Vogel, H. J. Bacterial ferrous iron transport: the Feo system. FEMS Microbiol. Rev.40, 273–298 (2016). [DOI] [PubMed] [Google Scholar]
- 61.Shukla, B. & Rai, L. C. Potassium-induced inhibition of nitrogen and phosphorus metabolism as a strategy of controlling Microcystis blooms. World J. Microbiol. Biotechnol.23, 317–322 (2007). [Google Scholar]
- 62.Sandrini, G., Huisman, J. & Matthijs, H. C. P. Potassium sensitivity differs among strains of the harmful cyanobacterium Microcystis and correlates with the presence of salt tolerance genes. FEMS Microbiol. Lett.362, fnv121 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequence data that support the findings of this study have been deposited in the National Center for Biotechnology Information Archive with the primary accession code MH329183.