Abstract
Microplastics (MPs) in aquatic environments easily support biofilm development, which can interact with other environmental pollutants and act as harbors for microorganisms. Recently, numerous studies have investigated the fate and behavior of MP biofilms in aquatic environments, highlighting their roles in the spread of pathogens and antibiotic resistance genes (ARGs) to aquatic organisms and new habitats. The prevalence and effects of MP biofilms in aquatic environments have been extensively investigated in recent decades, and their behaviors in aquatic environments need to be synthesized systematically with updated information. This review aims to reveal the development of MP biofilm and its interactions with antibiotics, ARGs, and pathogens in aquatic environments. Recent research has shown that the adsorption capabilities of MPs to antibiotics are enhanced after the biofilm formation, and the adsorption of biofilms to antibiotics is biased towards chemisorption. ARGs and microorganisms, especially pathogens, are selectively enriched in biofilms and significantly different from those in surrounding waters. MP biofilm promotes the propagation of ARGs through horizontal gene transfer (HGT) and vertical gene transfer (VGT) and induces the emergence of antibiotic-resistant pathogens, resulting in increased threats to aquatic ecosystems and human health. Some future research needs and strategies in this review are also proposed to better understand the antibiotic resistance induced by MP biofilms in aquatic environments.
Keywords: Aquatic environment, Microplastic biofilm, Antibiotic, Antibiotic resistance gene, Pathogen
Graphical abstract
Highlights
-
•
The formation of MP biofilm and associated factors is reviewed.
-
•
The adsorption mechanism of antibiotics by biofilm is summarized.
-
•
The distribution and risk of ARGs and pathogens in biofilm is discussed.
-
•
Perspectives on the biofilm-induced antibiotic resistance are proposed.
1. Introduction
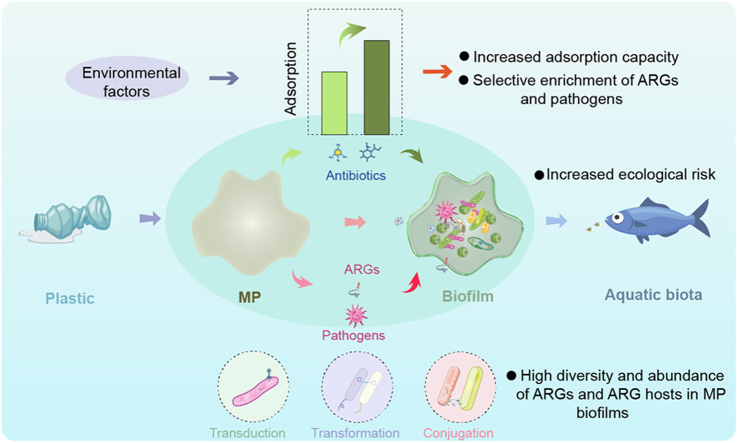
Microplastics (MPs), plastic debris less than 5 mm in size, have been found ubiquitously in aquatic environments due to the high consumption of plastic products and improper plastic waste management [1]. Most MPs are difficult to degrade, easy to transport, and able to accumulate in the food web, posing risks to the health of ecosystems and humans [2]. MPs in aquatic environments provide suitable substrates for microorganisms to colonize and result in the formation of biofilms, known as “plastisphere” [[3], [4], [5]]. The plastisphere constitutes a new ecological niche to strengthen the communications (e.g., transcriptional response and exchange of genes and nutrients) among these microorganisms [6]. Microorganisms colonized on the MP surface can further influence the degradation, sedimentation, transport, adsorption, and other behaviors of MPs in aquatic environments [7]. For example, some studies showed that the adsorption and stabilization of antibiotics were enhanced on biofilm-developed MPs, and the adsorption of antibiotics was biased towards chemisorption [8,9].
As emerging pollutants, antibiotics are widely distributed in aquatic environments, and their distribution is highly associated with anthropogenic activities. Antibiotics are commonly used to prevent or treat bacterial infections in humans or livestock, and up to 200,000 tons of antibiotic-related products are consumed globally each year [10]. Since not all antibiotics can be absorbed or metabolized in the body, 80%–90% are released into the natural environment in the form of maternal, metabolite, or conjugated substances via urine or feces [11]. When antibiotics are absorbed by the MP biofilm, the survival pressure of microorganisms, especially bacteria in the biofilm, increases, leading to the propagation and transmission of antibiotic resistance genes (ARGs) via horizontal gene transfer (HGT) or vertical gene transfer (VGT). Meanwhile, the diversity and abundance of antibiotic-resistant bacteria (ARB) and potential hosts of ARGs may also increase [[12], [13], [14], [15]]. Up to now, more than 400 ARG subtypes mainly belong to multidrug, tetracycline, beta-lactam, sulfonamide, macrolide, aminoglycoside, quinolone, and chloramphenicol classes have been detected in MP biofilms in aquatic environments [5,6,[16], [17], [18]].
In addition to acting as an ideal pool for antibiotics and ARGs, MP biofilm is also an important hub for pathogens such as Pseudomonas monteilii, Salmonella enterica, and Vibrio sp. in aquatic environments [[19], [20], [21]]. Due to the frequent gene exchanges in MP biofilm, increasing numbers of pathogens were found to possess exogenous ARGs in their genomes [6]. After acquiring new ARGs, the previously sensitive pathogens may become resistant to antibiotics, reducing the effectiveness of the antibiotics when treating bacterial diseases [22]. Therefore, it is essential to classify the distribution of pathogens in MP biofilm in aquatic environments.
In this review, the recent publications are collected and analyzed based on the data from Web of Science, Scopus, Science Direct scientific databases, and Google Scholar using the keywords “microplastic” OR “micro-plastic” OR “microplastics” AND “biofilm” OR “biofilms” AND “antibiotic” OR “antibiotics” AND “water” OR “aquatic”. From the search results, only the studies associated with antibiotic adsorption and desorption, ARGs, and pathogens were considered in this review. The purpose of this review was to update knowledge on known impacts of MP biofilms on the fate of antibiotics, ARGs, and pathogens in aquatic environments.
2. Formation of MP biofilm in aquatic environment
2.1. MP as a suitable substrate for biofilm formation
Early in 1972, the colonization of diatoms and hydroids on the surface of plastic particles (pellet, 2.5–5.0 mm) in the western Sargasso Sea was first reported [23]. During the last decades, an increasing number of protozoa (Alveolata), algae (Chlorophyta and Charophyta), fungi, viruses, and bacteria were found in MP biofilms [24]. Field and laboratory experiments both showed that the microorganic community in MP biofilms was distinct from those in the surrounding environments [6,17,19]. Importantly, MP biofilms can act as “protective clothing” for microorganisms in some extreme conditions [25]. In addition to the microorganisms, extracellular polymeric substances (EPS), and other abiotic substrates are also contained in MP biofilms [26], which may further affect the fate of MPs in aquatic environments.
2.2. Formation of MP biofilm
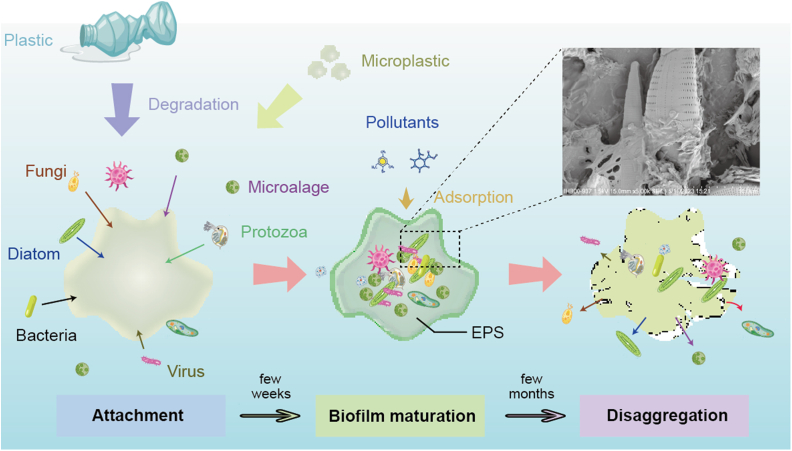
The formation of MP biofilm is a dynamic and complex process. According to published studies, the formation of MP biofilm mainly includes three stages, as shown in Fig. 1: (1) microorganisms, including protozoa, algae, fungi, bacteria, and viruses reversibly and irreversibly adhere on the surface of MP via weak (e.g., random encounters) or strong (e.g., chemotaxis) interactions. Meanwhile, pollutants such as antibiotics, metal ions, and endocrine disruptors are adsorbed into the MP through covalent bonds, hydrophobic interactions, or π–π interactions, etc.; (2) Pollutants accumulate on the biofilm surface to form “ecocorona”, thus facilitating the colonization of microorganisms. EPS is produced and secreted by microorganisms, and the microorganisms are rapidly propagated around the surface of the MP to form a stable three-dimensional community, in which signaling molecules are synthesized and released in large amounts; (3) microorganisms released from the biofilm and return to the surrounding environments as a result of MP degradation or accumulation of harmful molecules [27,28].
Fig. 1.
Formation of biofilm on the surface of MP (elements are collected from Freepik). Scanning electron microscope image, captured by ourselves using a Field-Emission Scanning Electron Microscopy (S-4800, HITACHI, Japan), shows the colonization of diatoms and bacteria in MP biofilm. MP, microplastic; EPS, extracellular polymeric substances.
2.3. Microbial community of MP biofilm
Compared with free-living microorganisms in the surrounding water, microorganisms in MP biofilm are significantly different and highly unique [6,17,[29], [30], [31]]. Microalgae such as Cyanophyta (cyanobacteria) are an important part of MP biofilms both in marine and freshwater environments [[32], [33], [34]]. Besides, Dolichospermum flosaquae forming bundles of filaments also can attach to the surface of plastic debris [35]. In addition to microalgae, bacteria are also an important component of MP biofilms. Bacillus, Mycobacterium, Pseudomonas, Vibrio, and Aeromonas sp. have been found widely distributed in MP biofilms [15,29,30,[36], [37], [38]]. Though some published studies revealed that bacteria were selectively colonized on specific types of MPs [39,40], the core bacteria in MP biofilms are dominated by surrounding environments rather than MP types at large scales [31,37,41]. A field study conducted in the Houxi River demonstrated that bacterial composition was more similar in the biofilms of MPs isolated from the same sampling point regardless of MP types [31].
After being attached to the MP, some of them with quorum sensing (QS) systems can secrete and detect signal molecules, which may alter the original relationships of microorganisms in that micro-environment, thus affecting biofilm formation and antibiotic metabolite production [42]. Due to the frequent communications among microorganisms in MP biofilms, bacteria in MP biofilms may also develop a high tolerance to antibiotics. For instance, Vibrio and Shewanella sp. isolated from polypropylene (PP) and polyvinyl chloride (PVC) MPs are resistant to cefazolin, tetracycline, etc., and the multiple antibiotics resistance (MAR) indexes of these strains are significantly higher than those in surrounding environments [22].
The composition of colonized bacteria in MP biofilms is a result of their adaptation to changes in biofilms, such as the accumulation of harmful molecules. Due to the widespread flagellar motility and metabolic versatility [43], Proteobacteria is always the predominant phylum in MP biofilms at the early stage and is considered to be inchoate colonizers [33,44]. However, the proportion of Proteobacteria in MP biofilm is significantly decreased as the biofilm matures [30,43]. On the contrary, the proportion of the Bacteroidota (secondary biofilm colonizers) is significantly increased, which may be due to most of them having the ability to degrade organic matter and secrete EPS, thus promoting the colonization of other bacteria [45,46]. Some rare phyla, such as Spirochaetes, Synergistetes, and Pacearchaeota which are not found in water, are also presented in biofilms, highlighting the role of MPs as carriers in aquatic environments [30].
2.4. Factors influencing the formation of MP biofilm
Previously, some studies focused on MP biofilms indicated that the formation of biofilm can be affected not only by the environmental conditions (e.g., pH, ultraviolet, salinity, and nutrients) but also by the properties of MPs (e.g., shape, type, color, size, hydrophobicity, and characteristic) [26,[47], [48], [49], [50], [51]]. In addition to the aforementioned influencing factors, some easily overlooked factors also play important roles in the formation of MP biofilms.
(1) Co-effect of aged treatment and MP shape: Aging treatment of MPs does not always promote the formation of MP biofilm. In a previous study, Shan et al. found that microbes tend to adhere to aged PP MPs (fragment, 5 mm × 5 mm), and the relative abundance of Cyanobacteria was higher on the surface of aged MPs [52]. However, a microcosm incubation experiment showed that higher biomass was seemingly formed in virgin PP MPs (particle, 750–1000 μm) than the aged MPs before 30 days of incubation. Compared with virgin PP MPs, the aged MPs have rougher and more cracked surfaces but a lower contact angle, indicating that a hydrophobic surface is more important for microorganism attachment than the role of surface roughness in irregularly spherical MPs, and aging treatment decreases the interaction of MPs with surrounding microorganisms [53].
(2) Depth in water: Water depth has a negative effect on the formation rate of MP biofilms in aquatic environments. Tu et al. previously placed the MPs exposure device containing polyethylene (PE, square, fragment, 4 ± 1 mm) MPs in an offshore aquaculture area at different depths (2–12 m), and results showed that the total biomass of biofilms decreased with water depth, which may be due to the low abundances of microorganisms in deep water [54]. Another study conducted in Qinhuai River also revealed that the biomass of biofilms on the polyethylene terephthalate (PET, 5 mm × 5 mm) MPs and polylactic acid (PLA, fragment, 5 mm × 5 mm) MPs showed depth-decay variations (30, 90, and 200 cm) regardless of the relatively high diversity and abundance of bacteria in deep water [55]. It seems that the low microbial metabolic activities caused by low temperatures and weak sunlight may be the key factors slowing the formation rate of MP biofilms in deep water.
(3) Pollutants: Pollutants such as antibiotics and heavy metals can disturb the nutrient exchange and communications among different microorganisms and put selective pressure on them [56]. Hence, pollutants can inhibit the MP biofilm formation, but the inhibition effects are varied by species. For example, the single treatment of 0.5 mg/L tetracycline, 0.5 mg/L ampicillin, and 1.0 mg/L zinc only inhibited the PVC MPs biofilm formation at the early 28 days. However, by day 84, the single treatment with 1.0 mg/L copper and co-treatment with antibiotics and heavy metals still inhibited biofilm formation [15].
(4) Conjugative plasmids: Conjugative plasmids, such as RP4, are frequently detected in aquatic environments and have been evidenced to promote the colonization of bacteria on the surface of MPs when conjugative pili synthesis genes (e.g., traA and traB) are highly expressed [57]. Although for pili synthesis-repressed plasmids (e.g., RP4 plasmid), the expression of pili genes would be at a high level only when the donor encountered the recipients or activated by some promoters [58], the donors, recipients, and pollutants always coexist in the real aquatic environment.
3. Adsorption and desorption of antibiotics by MP biofilm
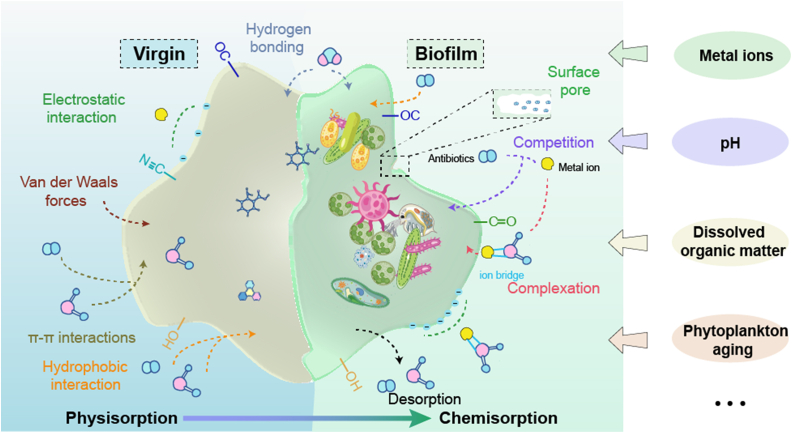
Owing to the highly specific surface areas with porous structures, MPs are important reservoirs for pollutants such as antibiotics [59]. Recently, numerous studies conducted in the laboratory and field investigated the adsorption and desorption behaviors of MPs or MP biofilms on the pollutants (Table 1) [8,9,[60], [61], [62], [63]]. Results from these studies showed that biofilms have a stronger adsorbability to antibiotics than virgin MPs and can alter the transport of pollutants in water. As described in previous studies, the adsorption of antibiotics by MP depends on the type of sorbents and adsorbents, mainly through six mechanisms: (I) Electrostatic attraction; (II) Hydrogen bonding; (III) Pore fitting; (IV) π–π interaction; (V) Hydrophobic interaction; (VI) Van der Waals forces [60,[64], [65], [66]]. The adsorption mechanism of antibiotics in MP biofilms is summarized in Fig. 2.
Table 1.
Influence of microplastic biofilm and metal ions on the adsorption of antibiotics.
| MP types | Size | Shape | AT types and concentration | Metal ions and concentration | Adsorption mechanism | Adsorption amount of biofilm (virgin MPs) | References |
|---|---|---|---|---|---|---|---|
| PE | 60–150 μm | Pellet | TC | 0.5 mg/L Cu2+ | Complexation, hydrophobic interaction, electrostatic interaction, π–π interaction, competition | Bio-PE: qmax = 762.2 μg/g (237.5 μg/g) Bio-PE + Cu: qmax = 950.1 μg/g (911.2 μg/g) |
[60] |
| PS | 0.45–1 mm | Foam | OTC | NA | Electrostatic interaction, hydrogen-bond, π–π interaction | Bio-PS: qmax = 27,500 ± 5120 μg/g (1520 ± 120 μg/g) | [73] |
| PLA | 75–150 μm | NA | OTC | NA | Hydrogen bonding, electrostatic interaction, hydrophobic interaction | Bio-PLA: qmax = 727.825 μg/g (581.187 μg/g) Degraded PLA: qmax = 1193.346 μg/g |
[69] |
| PBAT | <100 μm | NA | OTC | 0.8 mg/L Cu2+ | Complexation, hydrophobic interaction, electrostatic interaction, π–π interaction | Bio-PBAT: qmax = 1396.21 μg/g (692.05 μg/g) Degraded PBAT: qmax = 1869.93 μg/g Bio-PBAT + Cu: qmax = 3713.90 μg/g (3675.88 μg/g) Degraded PBAT + Cu: qmax = 4355.83 μg/g |
[63] |
| PVC, PA, HDPE | PVC: 75 μm PA: 250 μm; HDPE: 74 μm | Pellet | NOR | NA | Hydrophobic interaction, hydrogen-bond, electrostatic interaction | Bio-PVC: qmax = 5889 μg/g (3825 μg/g) Bio-PA: qmax = 2759 μg/g (2837 μg/g) Bio-HDPE: qmax = 10,585 μg/g (5193 μg/g) |
[8] |
| PE, PLA | 250–500 μm | NA | TC-HCl, CIP | NA | Hydrogen-bond interaction, electrostatic interaction, hydrophobic interaction | For TC-HCl, Bio-PE: 427 μg/g (177 μg/g) Bio-PLA: 375 μg/g (145 μg/g); For CIP, Bio-PE: 398 μg/g (98 μg/g) Bio-PLA: 189 μg/g (81 μg/g) |
[9] |
| PE | ∼150 μm | NA | TC-HCl, CFX | NA | Hydrophobic interactions, hydrogen bonding interactions, electrostatic interactions and biodegradation | For TC-HCl, B-PE: 1652 μg/g (1148 μg/g) A-PE: qmax = 2418 μg/g B-A-PE: qmax = 2989 μg/g; For CFX, B-PE: 1192 μg/g (655 μg/g) A-PE: qmax = 1319 μg/g B-A-PE: qmax = 2037 μg/g; |
[71] |
A-PE, aged polyethylene; B-A-PE, aged with biofilm-developed polyethylene; B-PE, biofilm-developed polyethylene; CFX, cefalexin; CIP, ciprofloxacin; HDPE, high-density polyethylene; NA, not available; NOR, norfloxacin; OTC, oxytetracycline; PA, polyamide; PBAT, polyester polybutylene adipate terephthalate; PE, polyethylene; PLA, polylactic acid; PS, polystyrene; PVC, polyvinyl chloride; TC, tetracycline.
Fig. 2.
Mechanism of antibiotics adsorbed by virgin MP and MP biofilms, and factors that influence the adsorption and desorption of antibiotics (elements are collected from Freepik).
3.1. Adsorption and desorption of antibiotics by biofilm
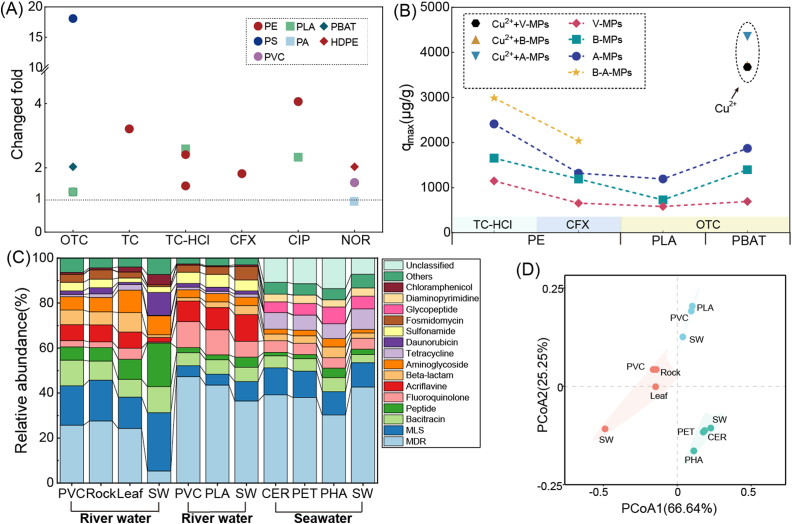
Biofilms are essential for MPs to adsorb high amounts of antibiotics from surrounding environments (Fig. 3A). For example, the adsorption capacities of PBAT MPs on oxytetracycline (OTC) were increased by 101.7% after the formation of biofilms [63]. The abundant oxygen-containing functional groups in EPS can make antibiotics more firmly accumulate on the MP surface, thus promoting the accumulation of antibiotics in MP biofilms. In PE MPs, functional groups at 1033 cm−1 (carbon-oxygen) and 1640 cm−1 (carbanyl) were newly generated after the biofilm formation, which further strengthened the hydrogen-bonding interactions between PE MPs and ciprofloxacin [9]. The surface pores of biofilm also can enhance the adsorption capacity of MPs [67,68]. In addition to these changes, the specific surface area and low zeta potential also make it easier for MPs to adsorb antibiotics [8,[69], [70], [71]]. Compared with aged MPs, the maximum adsorption amount of antibiotics in the biofilm is significantly lower (Fig. 3B), which is because of the enlarged original pores and increased number of small pores (less than 5 nm) on the surface of aged MPs [68]. Besides, the adsorption of antibiotics in biofilm is reversible compared to virgin MPs due to degradation of the biological sites [72]. For virgin MPs, the adsorption of antibiotics by MPs can be divided into three stages, while only two stages were found in biofilm [73]. Regardless of whether the adsorption of antibiotics on virgin MPs is biased towards physisorption, the adsorption of antibiotics on MP biofilms is mainly controlled by chemisorption [8,9,71,74]. The change in adsorption ways is related not only to the changes of MPs themselves but also to microorganisms in biofilms.
Fig. 3.
The changed folds of maximum adsorption capacity of MP biofilms on antibiotics compared to virgin MPs (A), and the maximum adsorption capacity of antibiotics with different forms of MPs (B); the composition of ARG types in MP biofilms, natural substrates (Rock, Leaf, and CER) (C), and the principal co-ordinates analysis of top 15 ARG types among different groups (D). Data in (A) and (B) are collected from Table 1, and data in (C) and (D) are collected from references [6,19,28]. ARG, antibiotic resistance; CER, ceramic pellets; PET, polyethylene terephthalate; PHA, polyhydroxyalkanoate; SW, surrounding water.
Due to the dynamic changes of surrounding environments and metabolic activities of microorganisms in biofilms, the adsorbed antibiotics in MPs may be released into surrounding waters and act as new sources of contaminants. In the desorption process, a phenomenon called “desorption hysteresis” occurred after biofilm formation or aged treatment, which may be highly associated with the reduced hydrophobic and π–π interactions and increased electrostatic interactions [75]. Compared with virgin MPs, the adsorbed and immobilized antibiotics on the biofilms are more reversible, which means a higher possibility of antibiotic desorption in biofilms [63,68]. However, if antibiotics coexisted with heavy metals such as Cu2+, antibiotics would be more stabilized in MP biofilms than in virgin MPs, thus inhibiting the desorption of antibiotics from MP biofilms [60].
3.2. Factors influencing the adsorption and desorption of antibiotics by biofilm
The adsorption and desorption capacities of antibiotics in MP biofilms are not only influenced by the intrinsic characteristics of MPs, but also affected by the changes from surrounding environments and even the microorganisms in MP biofilms [27]. In this section, some influencing factors, such as metal ions, pH, dissolved organic matter, and phytoplankton aging, are discussed.
(1) Metal ions: Metal ions such as copper (Cu) have been confirmed to enhance the adsorption of OTC and tetracycline (TC) on biofilm (Fig. 3B), which may be attributed to the ion bridge formed between OTC (or TC) and Cu2+ [60,63]. After complexing with Cu2+, OTC or TC can be adsorbed by biofilm through electrostatic interaction. In contrast to adsorption, Cu2+ did not promote the desorption of OTC from biofilm [63], but TC could accelerate the release of Cu2+ from MP biofilms [60,76].
(2) pH: As aforementioned, Cu2+ enhanced the adsorption of antibiotics on biofilm. However, Cu2+ is hard to adsorb on positively charged MP surfaces at low pH, and low pH conditions (high H+ concentration) would weaken the ion bridging effect between MPs and antibiotics, thus reducing the adsorption capacity of MPs [60,63]. In addition, at low pH conditions, the H+ in solution may compete with the antibiotics for adsorption and reduce the amount of antibiotics adsorbed on MPs [8].
(3) Dissolved organic matter (DOM): The influence of DOMs, such as fulvic acid (FA), on the adsorption of antibiotics varies with MP types. In previous studies, the author found FA not only competed with OTC to adsorb on the PLA and polyester polybutylene adipate terephthalate (PBAT) MP surface through inter-molecular forces but also competed with OTC for the PLA or PBAT-Cu adsorption sites, thus decreasing the adsorbed amount of OTC on biofilm [63,68]. However, another study revealed that FA could enhance the adsorption of OTC on the biofilm of polystyrene (PS) MPs, which may be attributed to the bridging effect of adsorbed FA with PS MPs and OTC [73].
(4) Phytoplankton aging: The effects of phytoplankton aging on the adsorption of antibiotics on MP biofilms are highly dependent on the species tolerance and biofouling level [62]. When Microcystis aeruginosa is the main species in MP biofilms, the hepatotoxin released by M. aeruginosa itself may be adsorbed by biofilms and then repress the potency on phosphatase-specific binding activity in EPS, which further reduces the adsorbed amount of antibiotics on biofilms. However, a large amount of EPS was observed in MP biofilms after the aging and biofouling of Chlorella vulgaris, which is beneficial for biofilms to adsorb antibiotics mainly through hydrophobic interactions.
4. ARGs in MP biofilm
As “hitchhikers”, MPs can randomly transfer ARGs to new environments, causing potential threats to ecosystems and human health. Previous studies demonstrated that ARGs are selectively enriched on the surface of MPs [6,18,38,43,55], and frequently transferred among different microbes in biofilm through HGT or VGT [77]. Hence, the proliferation and spread of ARGs conferred by MPs are crucial to controlling the ARG problem in aquatic environments. The occurrence of MP-associated ARGs investigated in recent research is summarized in Table 2.
Table 2.
Antibiotic resistance genes and mobile genetic elements in microplastic biofilm in aquatic environments.
| Location | MP types | Sizes | Shapes | Methods | Distribution of ARGs and MGEs | References |
|---|---|---|---|---|---|---|
| Huangpu River | Dominant with PET, PA, PMMA, and PE | 43.47–5765 μm | Fiber, fragment, film, and foam | Metagenomics | 313 ARG subtypes, dominant with multidrug, Bacitracin, and tetracycline, total ARGs abundances ranged from 0.016 to 0.15 copies/16S rRNA; 82 MGE subtypes, dominant with transposase, average abundance of 0.077 copies/16S rRNA; the subtypes and abundances of ARGs and MGEs in biofilms were lower than that in SW | [17] |
| Haihe River | PVC | 3 mm | Pellet | Shotgun metagenomics | 188 ARG subtypes in biofilm, higher than SW (67), dominant with multidrug, MLS, and bacitracin | [19] |
| Zuo River | PP, PE, PS, PET, PC | 100 μm–5 mm | Foam, fiber, fragment, film, pellet | Metagenomics | 1211 ARG subtypes, including multidrug (43.58%), tetracycline (10.88%), MLSB (10.77%), and glycopeptide (8.71%); Among these genes, Among them, the genes macB (MLS), tetA(58) (tetracycline), novA (aminocoumarin), bcrA (peptide), and evgS (multidrug) were consistently detected in high abundance. | [4] |
| Houxi River | PE, PS | ≤75 μm | NA | HT-qPCR | 19–88 ARG subtypes and 7–18 MGE subtypes, both are significantly lower than those in SW; dominant with aminoglycoside and multidrug; the abundance of ARGs in biofilms ranged from 0.002 to 0.12 copies/16S rRNA, and the abundance of ARGs in PE MPs > PS MPs | [31] |
| Qinghuai River | PET, PLA | 5 × 5 mm | Fragment | Quantitative real-time PCR (qPCR) | qnrS and blaNDM-1 were the predominant ARGs in biofilm, the abundances of ARGs in biofilm showed depth-decay variations, and the abundance of ARGs were PLA > PET > water | [55] |
| Ganjiang River | PE, PP, and polybutadiene | NA | Fragment, fiber, pellet, and film | qPCR | The average abundance of total ARGs in MP biofilms was 4.8 × 10−2, higher than that in wood, sediment, and water, but no statistically significant difference was observed among the different media; dominant with sul1, sul2, and ermF | [78] |
| Northern Gulf of Mexico | PET, PHA | 3–4 mm | Pellet | Metagenomics | 339 and 335 ARG subtypes in PET and PHA MPs, higher than SW (310), dominant with multidrug, MLS, tetracycline, and beta-lactam; the ARG abundances in PET and PHA MPs are higher than that in SW | [28] |
| Zhenhai Bay | PP, PE, PET, PHB, PLA | 3–4 mm | Fragment | qPCR | sul1 was the predominant ARG (approximately 0.01 copies/16S rRNA), followed by qnrS and blaTEM, and the abundance of intI1 ranged from 10−4 to 10−2 copies/16S rRNA | [43] |
PMMA, polymethyl methacrylate; PP, polypropylene; PC, polycarbonate; PCL, polycaprolactone.
4.1. Occurrence of ARGs in MP biofilm
The colonization of ARGs in biofilms depends on the surrounding waters rather than on MP types or shapes [12,28,78]. In terms of the antibiotic resistome previously reported in water ecosystems, the composition of ARGs in MP biofilms, detected using metagenomic methods varied greatly in the coastal [28], Zhuo River [4], and Huangpu River [17]. Another study revealed that ARGs in biofilms of PE and PS MPs isolated from the Shidou reservoir were similar to those in their surrounding water but significantly distinct from those in biofilms of PE and PS isolated from the Xinglin Bay [31]. Among these ARGs, multidrug is always the predominant ARG type in biofilms, while the dominance of other ARG types, such as tetracycline [4], beta-lactam [14], aminoglycoside [31], bacitracin [17], and MLS [28], in biofilms depends on the surrounding waters (Fig. 3C). Like the occurrence of bacterial community in MP biofilms, the composition of ARGs in MP biofilms is also dominated by surrounding environments rather than MP types at large scales (Fig. 3D).
According to a previous study, ARGs in natural environments did not always confer antibiotic resistance under low antibiotic residual conditions [79]. For example, sul1 and tetP genes in PLA biofilm were detected with high abundances, but the transcription level of these two genes was very low [6]. Therefore, under that condition, the functions of most ARGs, especially multidrug-related genes, are mainly associated with cell detoxification [80], bacterial virulence [81], and cell-in-cell communication [82], which are beneficial for the frequent communication among bacteria within the biofilm and the adaptation to the adverse environments.
Despite the diversity and richness of ARGs in biofilms being usually lower than that in surrounding water, some ARGs are selectively enriched in biofilms. A previous study performed in the Huangpu River revealed that the abundances of lnuA, mexL, LRA-12, and rosA genes in biofilms were 5.3–172.6 folds higher than that in waters [17]. In addition, the relationship between bacteria and ARGs in MP biofilm was more complex than that in surrounding waters and natural carriers such as quartz sand [83], rock [19], and minerals [29]. More importantly, MP biofilms reduced the degradation rates of antibiotics [29], and the high antibiotic residue in biofilm may further promote the proliferation of ARGs. Therefore, compared with the natural carrier, MP biofilms have great advantages for the propagation and spread of ARGs in water ecosystems.
Some studies have investigated the dynamic evolution of ARGs in MP biofilms [36,43,55,84]. Results from these studies suggested that the evolution of ARGs was random over time. As ARGs are intrinsic in the genomes of bacteria [79], the dynamic of the bacterial community in biofilm may explain the evolution of ARGs. Chen et al. found that the dynamic evolution of mcr-1, qnrA, and sul1 genes in PLA biofilm was consistent with Hydrogenophaga, Nitrospira, and Methyloversatilis [55]. When MPs are transferred to new aquatic environments, the communication between biofilm bacteria and free-living bacteria would further result in the evolution of ARGs and even ARG hosts in the plastisphere and surrounding environments, posing increased threats to the ecosystem and human health [18].
4.2. Mechanisms of the propagation of ARGs in MP biofilm
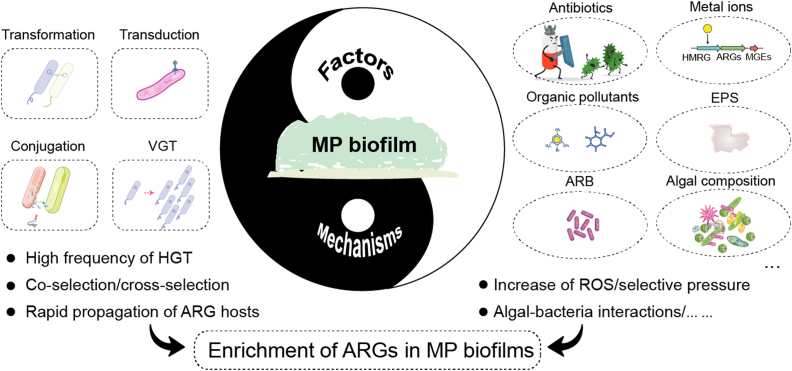
Numerous studies revealed that the HGT process conferred by mobile genetic elements (MGEs) is critical for the propagation of ARGs in biofilms [13,14,38,78,[85], [86], [87], [88], [89]]. Though the frequency of HGT occurring in natural waters is very low [90], the ARG transformation frequencies in MP biofilms are hundreds of times higher [77,85]. Up to now, more than 100 subtypes of MGEs have been identified in MP biofilms, mainly consisting of plasmid, transposase, recombinase, insertion sequence (IS), integrase, and integron [17]. The wide distribution of MGEs in biofilm strengthens the relationship between bacteria and ARGs, thus inducing the emergence of new ARG hosts [31]. In addition, the propagation of ARG hosts through the VGT process also results in the propagation of ARGs in biofilm (Fig. 4) [91]. Considering the complex transmission and propagation of ARGs in biofilms, some factors such as antibiotics, heavy metals, organic pollutants (non-antibiotics), EPS, ARB, and algal composition can also affect these processes (Fig. 4).
Fig. 4.
The mechanisms of ARG enrichment in MP biofilms and the influence of potential environmental factors on ARG propagation (elements are collected from Freepik). ARB, antibiotic-resistant bacteria; HGT, horizontal gene transfer; HMRG, heavy metal resistance gene; MGE, mobile genetic element; ROS, reactive oxygen species; VGT, vertical gene transfer.
(1) Antibiotics: Several studies revealed that MP biofilm can enrich antibiotics from surrounding water, thus resulting in a high residue of antibiotics in biofilms, which may further promote the propagation of ARGs via altering the bacterial community or activating MEGs [12,14,83]. Co-selection and cross-selection are the mechanisms by which antibiotics promote the propagation of ARGs [92,93]. In biofilms, very low concentrations of antibiotics may also have a selective effect if they are bioavailable [94]. In a field experiment, although the concentrations of antibiotics residual in biofilms were lower than 100 ng/g, the residue antibiotics in biofilms increased the abundances of sul1, copA, and intI1 in MP biofilms, and the abundances of ARGs were significantly higher than that in water and sediment, indicating that biofilm can amplify the selective effects of antibiotics on biofilm-bacteria and further promote the propagation of ARGs [37]. In addition, MP biofilms slow the degradation of antibiotics in aquatic environments when compared with natural substrates (e.g., quartzite), and the high residue of antibiotics in MP biofilms maintains the high abundances of ARGs for a prolonged period even if the desorption of antibiotics occurs [29].
(2) Heavy metals: On the one hand, if ARG and heavy metal resistance genes (HMRGs) existed simultaneously on the same MGEs, the selective pressure of heavy metals on microbes may also induce the propagation of ARGs through co-selection [[95], [96], [97]]; on the other hand, low concentrations of heavy metals may increase the contents of reactive oxygen species (ROS) products, thus promoting the HGT of ARGs [15,58,88,98]. Excessive ROS generation could also result in the reduction of ARGs due to decreased HGT of ARGs and even cell death [99,100]. In addition, heavy metals could also promote the co-selection of ARGs with antibiotics [14].
(3) Other organic pollutants: Numerous studies evidenced that non-antibiotic organic pollutants such as antidepressants and polycyclic aromatic hydrocarbons also promoted the spread of ARGs via HGT [[101], [102], [103]]. According to these published studies, the overproduction of ROS and universal stress response were also the main mechanisms that enhanced the efficiency of HGT. In a previous study, the formation of biofilms on triclocarban-contaminated PE MPs was not impeded, and triclocarban promoted the propagation of ARGs via MGEs [14].
(4) Extracellular polymeric substances: EPS is beneficial to promoting the propagation of ARGs in biofilm. In EPS, numerous negatively charged functional groups (e.g., carboxyl, hydroxyl, and phosphate) provided numerous adsorption sites for environmental pollutants and extracellular ARGs [[104], [105], [106], [107]], which further promotes ARG proliferation in biofilms. In previous studies, a positive correlation was observed between the abundance (or expression) of intI1 and EPS [107,108], indicating that EPS positively activated the occurrence of HGT. Another study conducted by Guo et al. also revealed that EPS is essential in controlling the conjugal transfer of ARGs in biofilm [109].
(5) Antibiotic-resistant bacteria: MP can act as an important harbor for ARB and is beneficial for the selective growth of ARB even under the stress of antibiotics [110]. Meanwhile, ARB in MP biofilm can alleviate the pressure of antibiotics on antibiotic-sensitive strains to some extent and finally promote the propagation of ARGs [29]. In a previous study, ARBs, including Pseudomonas, Aeromonas, and Bacillus, were considered to promote the propagation of vanA, sul1, and intI1 in MP biofilms under the stress of antibiotics [111]. Another study conducted in mangroves also showed that the abundances of Vibrio and Shewanella in MP biofilms were increased, and the two genera in biofilms showed stronger resistance to antibiotics [22]. In addition, if ARB harbored conjugative resistant plasmid containing ARGs such as RP4, the high expression of pili synthesis genes would promote ARG conjugative transfer efficiency in the MP biofilms and further promote the propagation of related ARGs [57,112].
(6) Algal composition: Although no relevant studies have been conducted to reveal the contribution of algae to the distribution of ARGs in MP biofilms, in other areas, algae are shown to regulate the propagation of ARGs in the phycosphere, with or without the stress of exogenous pollutants via algal–bacteria interaction [[113], [114], [115], [116]]. Importantly, the core microbiota in the phycosphere is species-specific [117]. According to a previous study, algae may occupy a more dominant ecological niche in large MPs (>330 μm) than bacteria [24], and therefore, the composition of algae in the MP biofilms has unneglected effects on the abundance and diversity of ARG hosts and further affects the composition of ARGs in biofilms.
5. MP biofilm acts as the carrier of pathogens
In addition to acting as a reservoir for selective enrichment of ARGs and environmental pollutants, MPs in aquatic environments are also a hotspot for pathogens [118]. Compared to the surrounding waters, MPs provide better survival conditions (e.g., EPS and nutrients) to pathogens and higher possibilities to transport them to new habitats [19]. Under extreme conditions, MP itself can be used as a source of carbon and energy by pathogens [8]. Numerous studies have investigated the composition of pathogens in biofilms through metagenomic and 16S rRNA sequencing analysis [4,78,119]. Some cultivable taxa of them, such as Klebsiella and Vibrio spp., were previously isolated from MP biofilms [21,120]. The information on pathogens mentioned in recent research is summarized in Table 3.
Table 3.
Pathogens in microplastic biofilm in aquatic environments.
| Location | Sample types | MP types | Sizes | Shapes | Microbe identification strategy | Main pathogens | References |
|---|---|---|---|---|---|---|---|
| Zuo River | Water | PP, PE, PS, PET, PC | 100 μm–5 mm | Foam, fiber, fragment, film, pellet | Metagenomics | P. aeruginosa, Salmonella, Roseomonas sp. and Betaproteobacteria | [4] |
| BeiLun River | Water | PE, PP, PS | NA | Fiber | Metagenomics | Human_herpesvirus_3, Arthrobacter, Salmonella | [119] |
| Ganjiang River | Water | NA | ≥50 μm | Fragment (69.6%), fiber (12.9%), pellet (8.9%), film (8.5%) | 16S rRNA gene sequencing | Streptococcus mitis, Pseudomonas spp., K. pneumoniae, Salmonella enterica and Aeromonas hydrophila | [78] |
| Haihe River | Water | PVC | 3 mm | Pellets | Shotgun metagenomics |
Pseudomonas monteilii, P. Mendocina and P. syringae |
[19] |
| Houxi River | Water | PE, PS | ≤75 μm | NA | 16S rRNA gene sequencing | Mycobacterium spp., M. smegmatis, M. gilvum, M. abscessus, K. pneumoniae and Enterobacter cloacae | [31] |
| Mondego River | Water | PP, PE, PET | 500–1000 μm | Particles | Cultivation and BOX-PCR | E. coli and K. pneumoniae | [127] |
| Lake Maggiore | Water | PP, PET, PBAT, PCL | 4 mm | Fragments (Squares) | 16S rRNA gene sequencing | Flavobacterium, Roseomonas and Legionella | [140] |
| Taihu Lake | Surface water | PET (53.4%), polyester (11.7%), polyamide (6.8%) | 23.0–4238.9 μm | Fibers(92.9%), fragments(3.87%), films (2.38%) | Metagenomics | P. aeruginosa (14.3%), Salmonella enterica (13.6%), Xanthomonas oryzae (10.3%), P. syringae (6.32%), and Burkholderia cenocepacia (3.57%) | [20] |
| East and west coast of Scotland | Water | PE, PS | 2–5 mm | Beads | Cultivation | Campylobacter, E. coli, Intestinal enterococci, Klebsiella, P. aeruginosa, Salmonella and Vibrio sp | [120] |
| Northern Gulf of Mexico | Water | PET, PHA | 3–4 mm | Pellets | Metagenomics | Candida albicans, Enterobacter aerogenes, E. coli and P. aeruginosa | [28] |
| Dongshan Bay | Water | PS, PP, PE, PET, PVC | <0.2 mm (particles) | Particles: PS, PP, PET, PVC films: PE | 16S rRNA gene sequencing | Aeromonas, Escherichia–Shigella, Mycobacterium, Pseudomonas, Staphylococcus, Streptococcus, Vibrio | [61] |
| Coastal beaches | Water | NA | ≥5 μm | NA | 16S rRNA gene sequencing | Clostridium, Aeromonas, Bifidobacterium, Escherichia, Helicobacter, Streptococcus, Vibrio, Collinsella and Photobacterium | [141] |
| Municipal sewage | Sewage | PE | 53–63 μm | Microbeads | 16S rRNA gene sequencing | Arcobacter cryaerophilus, Aeromonas salmonicida, V. areninigrae and V. navarrensis. | [122] |
| Municipal activated sludge | Sludge | PE, PS | 85–106 μm | Spherical | 16S rRNA gene sequencing | Raoultella ornithinolytica Stenotrophomonas maltophilia | [142] |
| Forth Catchment, Scotland | Effluent, water and estuary | PE | 2 mm | Beads | Cultivation | P. aeruginosa and E. coli | [143] |
| Northwestern Mediterranean | Surface water, sediment | NA | NA | NA | Cultivation | Vibrio sp., E. coli, and Pseudomonas sp. | [123] |
| Mah'ebourg fish farm | Water | NA | NA | Ties, buoys, nets and pipes | 16S rRNA sequencing and Cultivation | V. alginolyticus and Photobacterium damselae | [124] |
| Fish ponds | Water | NA | ≥300 μm | NA | 16S rRNA gene sequencing and Cultivation | Vibrio sp. | [21] |
5.1. Pathogens in MP biofilm
According to previous studies, Vibrio sp. is regarded as an opportunistic biofilm former under appropriate growth conditions [121]. Since its first discovery in marine MPs collected from the North Atlantic [34], Vibrio sp. is frequently detected in biofilm in various waters with high abundances [21,61,120,[122], [123], [124]]. For example, the abundance of Vibrio sp. previously reported in PS MPs was up to 6.95 × 104 copies/ng but is lower than that in wood and surrounding waters (1.16 × 105–3.28 × 105 copies/ng) [121]. Compared to Vibrio sp. in surrounding waters, Vibrio sp. in biofilm showed high resistance to antibiotics [22] and highly activated metabolic activity [125], suggesting that Vibrio sp. transferred by MPs is easy to cause outbreaks of diseases and reduces the therapeutic effect of antibiotics.
In addition to Vibrio sp., some pathogens such as Klebsiella, Pseudomonas, and Salmonella spp. are also found to be selectively enriched in biofilm with high antibiotic resistance and possess various ARGs (e.g., gyrA, qnrS, and sul1) [120,126,127]. The acquisition of ARGs by pathogens is mainly through HGT or gene mutation. In Houxi River, Mycobacterium smegmatis not only had a significant co-occurring relationship with ARGs but also significantly and positively with MGEs [31], highlighting the propagation of ARGs in pathogens via HGT. In Mondego River, the ciprofloxacin-resistant phenotype of the Enterobacteriaceae strains in PP, PE, and PET MPs was conferred by two mutant genes-gyrA and parC [127]. The study also revealed that the pollutants, especially enrofloxacin and ciprofloxacin absorbed in biofilms, may be responsible for the emergence of antibiotic-resistant pathogens.
5.2. Threats of pathogen-colonized MPs on aquatic organisms
In aquatic environments, MPs carrying pathogens can later be ingested by aquatic organisms through food chains. Numerous studies have confirmed that aquatic organisms, especially filter feeders, easily take up MP particles from their surrounding water [128]. Although MPs are retained in aquatic organisms within a limited time in most situations, pathogens may be released into the intestine during the digestion process, further causing the outbreak of infections. A recent study showed that the pathogen-colonized plastic debris increased the outbreak frequency of disease in corals from 4% to 89% [129]. Another study revealed that the uptake of Escherichia coli-coated MPs by Ostrea edulis was significantly higher than virgin MPs, and the ingested E. coli-coated MPs further significantly increased the oxygen consumption and respiration rate of oysters over time. However, the virgin MPs didn't induce any measurable significant physiological responses [130]. In addition, a study conducted on Daphnia magna directly demonstrated the pathogen transfer via MPs ingestion [131]. Results from this study showed that Shigella flexneri (a human pathogen with sulfonamides resistance) colonized PS MPs were ingested by D. magna and blocked in their intestine after 24 h exposure. The expression of some functional genes involved in energy and metabolism, oxidative stress response, growth, etc. was changed after treatment. Importantly, the sul1 gene was detected in D. magna after treatment with S. flexneri colonized PS MPs.
6. Environmental implication of MP biofilms in aquatic environments
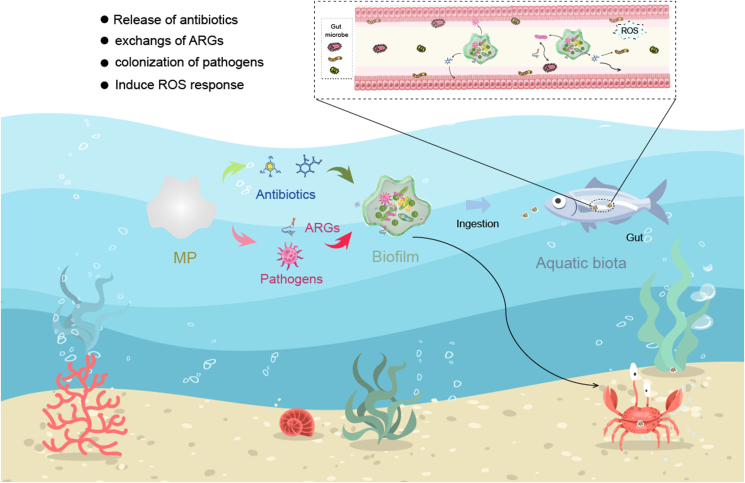
Since MPs were first released into the aquatic environments, the colonization of microorganisms and adsorption of organic pollutants such as antibiotics in MP biofilms have occurred continuously. Within MP biofilms, absorbed antibiotics put selective pressure on bacteria and further promote the propagation of ARGs and pathogens, and microorganisms accelerate the degradation and desorption of antibiotics in return. Though the interactions occurring in biofilms are dynamic and complex, and the compositions of biota and abiotic members vary greatly during the lifecycle of MPs, potential ecological risks to aquatic ecosystems and humans caused by MPs are always increased. In addition to exchanging ARGs with the surrounding bacteria when MPs floated to new environments, antibiotics released from MP biofilms also act as a new source of pollutants.
In water ecosystems, MP biofilms always have adverse effects on aquatic biota [132]. After ingested by aquatic biota, organic pollutants, ARGs, ARG hosts, and pathogens may release from biofilms, and induce the development of antibiotic resistance and even the outbreak of diseases (Fig. 5). As described in a previous study, PS MPs significantly increased the residual concentrations of roxithromycin in the gut of Carassius auratus for a long time, the bioaccumulated roxithromycin further promoted the propagation of ARGs via ARG hosts [133]. A recent study conducted on zebrafish (six months old) revealed that OTC-adsorbed PS MP biofilms induced ROS response and microbiome dysbiosis in the gut [134]. In addition, the abundance of pathogens and ARGs (especially multidrug and macrolide) was also significantly increased. Studies on O. edulis [130], D. magna [131], submerged plants [135,136], free-floating macrophytes [137], and algae [138] also showed the harmful effects of MP biofilms. Therefore, the appropriate management of MP pollution in aquatic environments is an urgent problem for the future.
Fig. 5.
The potential interactions among MP biofilms, aquatic biota, and gut microbes after the ingestion of MPs by aquatic biota. The release of antibiotics and pathogens into the gut and ARGs exchanged between biofilm bacteria and gut microbes, and the induced ROS response of aquatic biota by MPs (elements are collected from Freepik).
7. Challenges and perspectives
Our review summarizes the MP biofilm formation, and its interaction with antibiotics, ARGs, and pathogens in aquatic environments. The biofilm formation of MPs is a complicated process and can be influenced by several factors. Compared to virgin MPs, biofilm enhances the adsorption capacity of MPs to pollutants such as antibiotics. In addition, ARGs and pathogens are selectively enriched in biofilms, which may further threaten the health of aquatic ecosystems and humans. Although the fate and behavior of MP biofilms have been widely investigated in published studies, a deep understanding of the roles that MP biofilms play in aquatic environments is still needed. Challenges in further studies that need to be overcome are described as follows:
(1) The effects of environmental conditions and the properties of MPs on the formation of MP biofilms have been frequently investigated in many studies, but fewer studies focus on the effects of plastic additives (e.g., plasticizers). Actually, plasticizers such as phthalate acid esters are contained in many plastic products. When these plastic products break down into MP in aquatic environments, the phthalate acid esters originating from MPs themselves may affect the colonization of microorganisms in MPs after being released. Therefore, how plasticizers affect the formation of MP biofilms deserves to be investigated in further studies.
(2) Recently published research has primarily focused on the roles of bacteria in MP biofilms, while the roles of microalgae, fungi, and viruses in biofilms are often neglected. In large MPs or plastic debris, algae may occupy a more dominant ecological niche than bacteria. Importantly, algae can control the compositions of bacteria or ARGs in the phycosphere (or surrounding environments) via algal–bacteria interaction, and the core microbiota in the phycosphere is species-specific. Therefore, the roles of these microorganisms, especially microalgae, in ARGs propagation need futher investigations.
(3) The adsorption and desorption of antibiotics in MP biofilms revealed by published studies are almost conducted in laboratory conditions. However, the adsorption and desorption of antibiotics in actual environments can be affected by many unpredictable factors such as temperature and nutrients. Therefore, field studies are needed in the future.
(4) The bacteria and virus communities in MP biofilms have been sufficiently investigated in published studies, but the mechanism by which bacteria and viruses colonized in MP biofilms is still a gap. Most of the present results suggest that the colonization of microbes on MPs is not highly biased; however, whether bacteria with conjugative plasmids or QS systems preferentially colonize on the MPs surface is still worth further investigation.
(5) Numerous studies only investigated the abundances of ARGs in MP biofilms via metagenomic analysis or HT-qPCR, and ARGs with high abundances were considered the predominant ARGs. However, the high abundance of ARGs is not equal to the high expression of ARGs in biofilms, whether these high abundance ARGs are activated in MP biofilms is still unknown. To solve this issue, methods such as meta-transcriptomics should be used jointly to reveal the “true” ARGs in MP biofilms.
(6) In many studies, the bacteria significantly and positively correlated with ARGs would be considered as the hosts of ARGs. Whereas, a high correlation coefficient only indicates a good co-occurrence pattern of bacteria and ARGs, whether the ARGs are located on the genome or plasmid of the target bacteria is still unknown. What's more, there are no uniform standards for R- and P-values in identifying ARG hosts, and the accuracy of the R- and P-values is highly dependent on the number of samples. These aforementioned limitations will further affect the identification of real ARG hosts. In future studies, methods such as the voting mechanism used in metagenomics should be applied to more accurately identify ARG hosts. The detailed information on related methods to identify ARG hosts is listed as follows: ARG relative open reading frames were first annotated with DeepARG software and identified as ARG-like ORFs. The amino acid sequences of ORFs in contigs carrying ARGs (ARC) were subsequently extracted and annotated to the NCBI-NR database to identify the potential ARC hosts (e-value ≤ 1e-5, Diamond, blastp method). The voting mechanism was finally used to determine the taxonomic annotation of ARC hosts: If more than 50% of the ARG-like ORFs on each ARC were assigned to the same taxon, the ARC was assigned to the corresponding taxonomic rank (domain/kingdom/phylum/class/order/family/genus) [139].
CRediT authorship contribution statement
Jia Jia: Conceptualization, Investigation, Supervision, Writing – original draft. Qian Liu: Conceptualization, Investigation, Supervision. E. Zhao: Conceptualization, Investigation, Supervision. Xin Li: Conceptualization, Investigation, Supervision. Xiong Xiong: Conceptualization, Investigation, Supervision. Chenxi Wu: Conceptualization, Funding acquisition, Investigation, Project administration, Resources, Writing – review & editing.
Declaration of competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this article.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (41877394 and 51909012) and the Qinghai Science and Technology Plan (2023-ZJ-905T).
References
- 1.Thompson R.C., Olsen Y., Mitchell R.P., Davis A., Rowland S.J., John A.W., et al. Lost at sea: where is all the plastic? Science. 2004;304:838. doi: 10.1126/science.1094559. [DOI] [PubMed] [Google Scholar]
- 2.Rafa N., Ahmed B., Zohora F., Bakya J., Ahmed S., Ahmed S.F., et al. Microplastics as carriers of toxic pollutants: source, transport, and toxicological effects. Environ. Pollut. 2023;343 doi: 10.1016/j.envpol.2023.123190. [DOI] [PubMed] [Google Scholar]
- 3.Leiser R., Wu G.M., Neu T.R., Wendt-Potthoff K. Biofouling, metal sorption and aggregation are related to sinking of microplastics in a stratified reservoir. Water Res. 2020;176 doi: 10.1016/j.watres.2020.115748. [DOI] [PubMed] [Google Scholar]
- 4.Li R., Zhu L., Wang Y., Zhu Y.G. Metagenomic insights into environmental risk of field microplastics in an urban river. Water Res. 2022;223 doi: 10.1016/j.watres.2022.119018. [DOI] [PubMed] [Google Scholar]
- 5.Song H., Xiao S., Zhou X., Li Y., Tao M., Wu F., et al. Temporal dynamics of bacterial colonization on five types of microplastics in a freshwater lake. Sci. Tot. Environ. 2024;913 doi: 10.1016/j.scitotenv.2023.169697. [DOI] [PubMed] [Google Scholar]
- 6.Wu X., Liu Z., Li M., Bartlam M., Wang Y. Integrated metagenomic and metatranscriptomic analysis reveals actively expressed antibiotic resistomes in the plastisphere. J. Hazard. Mater. 2022;430 doi: 10.1016/j.jhazmat.2022.128418. [DOI] [PubMed] [Google Scholar]
- 7.Jia J., Zhao E., Xiong X., Wu C. In: Micro/Nanoplastics in the Aquatic Environment: Fate Toxicology and Management. Malafaia G., editor. Elsevier; 2024. Chapter Eight - Effects of biofilm on the fate and behavior of microplastics in aquatic environment; pp. 197–225. [Google Scholar]
- 8.He S., Tong J., Xiong W., Xiang Y., Peng H., Wang W., et al. Microplastics influence the fate of antibiotics in freshwater environments: biofilm formation and its effect on adsorption behavior. J. Hazard. Mater. 2023;442 doi: 10.1016/j.jhazmat.2022.130078. [DOI] [PubMed] [Google Scholar]
- 9.Wu Z., Guo Z., Dong D., Wu F., Li J., Yang X. Distinguishable adsorption interaction of virgin and biofilm covered polyethylene and polylactic acid for antibiotics. J. Environ. Chem. Eng. 2023;11 [Google Scholar]
- 10.Omuferen L.O., Maseko B., Olowoyo J.O. Occurrence of antibiotics in wastewater from hospital and convectional wastewater treatment plants and their impact on the effluent receiving rivers: current knowledge between 2010 and 2019. Environ. Monit. Assess. 2022;194:306. doi: 10.1007/s10661-022-09846-4. [DOI] [PubMed] [Google Scholar]
- 11.Kümmerer K. Antibiotics in the aquatic environment – a review – part I. Chemosphere. 2009;75:417–434. doi: 10.1016/j.chemosphere.2008.11.086. [DOI] [PubMed] [Google Scholar]
- 12.Wang S., Xue N., Li W., Zhang D., Pan X., Luo Y. Selectively enrichment of antibiotics and ARGs by microplastics in river, estuary and marine waters. Sci. Tot. Environ. 2020;708 doi: 10.1016/j.scitotenv.2019.134594. [DOI] [PubMed] [Google Scholar]
- 13.Zhang S., Cui L., Zhao Y., Xie H., Song M., Wu H., et al. The critical role of microplastics in the fate and transformation of sulfamethoxazole and antibiotic resistance genes within vertical subsurface-flow constructed wetlands. J. Hazard. Mater. 2023;465 doi: 10.1016/j.jhazmat.2023.133222. [DOI] [PubMed] [Google Scholar]
- 14.Zhao Y., Gao J., Wang Z., Cui Y., Zhang Y., Dai H., et al. Distinct bacterial communities and resistance genes enriched by triclocarban-contaminated polyethylene microplastics in antibiotics and heavy metals polluted sewage environment. Sci. Tot. Environ. 2022;839 doi: 10.1016/j.scitotenv.2022.156330. [DOI] [PubMed] [Google Scholar]
- 15.Zhao Y., Gao J., Wang Z., Dai H., Wang Y. Responses of bacterial communities and resistance genes on microplastics to antibiotics and heavy metals in sewage environment. J. Hazard. Mater. 2021;402 doi: 10.1016/j.jhazmat.2020.123550. [DOI] [PubMed] [Google Scholar]
- 16.Magalhães E.A., de Jesus H.E., Pereira P.H.F., Gomes A.S., Santos H.F.D. Beach sand plastispheres are hotspots for antibiotic resistance genes and potentially pathogenic bacteria even in beaches with good water quality. Environ. Pollut. 2023;344 doi: 10.1016/j.envpol.2023.123237. [DOI] [PubMed] [Google Scholar]
- 17.Xu C., Lu J., Shen C., Wang J., Li F. Deciphering the mechanisms shaping the plastisphere antibiotic resistome on riverine microplastics. Water Res. 2022;225 doi: 10.1016/j.watres.2022.119192. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Q., Zhang J., Fang Q., Zhang M., Wang X., Zhang D., et al. Microplastic biodegradability dependent responses of plastisphere antibiotic resistance to simulated freshwater-seawater shift in onshore marine aquaculture zones. Environ. Pollut. 2023;331 doi: 10.1016/j.envpol.2023.121828. [DOI] [PubMed] [Google Scholar]
- 19.Wu X., Pan J., Li M., Li Y., Bartlam M., Wang Y. Selective enrichment of bacterial pathogens by microplastic biofilm. Water Res. 2019;165 doi: 10.1016/j.watres.2019.114979. [DOI] [PubMed] [Google Scholar]
- 20.Xu C., Hu C., Lu J., Yang T., Shen C., Li F., et al. Lake plastisphere as a new biotope in the Anthropocene: potential pathogen colonization and distinct microbial functionality. J. Hazard. Mater. 2024;461 doi: 10.1016/j.jhazmat.2023.132693. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y., Lu J., Wu J., Wang J., Luo Y. Potential risks of microplastics combined with superbugs: enrichment of antibiotic resistant bacteria on the surface of microplastics in mariculture system. Ecotoxicol. Environ. Saf. 2020;187 doi: 10.1016/j.ecoenv.2019.109852. [DOI] [PubMed] [Google Scholar]
- 22.Tan B., Li Y., Xie H., Dai Z., Zhou C., Qian Z.J., et al. Microplastics accumulation in mangroves increasing the resistance of its colonization Vibrio and Shewanella. Chemosphere. 2022;295 doi: 10.1016/j.chemosphere.2022.133861. [DOI] [PubMed] [Google Scholar]
- 23.Carpenter E.J., Smith K.L., Jr. Plastics on the Sargasso sea surface. Science. 1972;175:1240–1241. doi: 10.1126/science.175.4027.1240. [DOI] [PubMed] [Google Scholar]
- 24.Di Pippo F., Crognale S., Levantesi C., Vitanza L., Sighicelli M., Pietrelli L., et al. Plastisphere in lake waters: microbial diversity, biofilm structure, and potential implications for freshwater ecosystems. Environ. Pollut. 2022;310 doi: 10.1016/j.envpol.2022.119876. [DOI] [PubMed] [Google Scholar]
- 25.Wang L., Luo Z., Zhen Z., Yan Y., Yan C., Ma X., et al. Bacterial community colonization on tire microplastics in typical urban water environments and associated impacting factors. Environ. Pollut. 2020;265 doi: 10.1016/j.envpol.2020.114922. [DOI] [PubMed] [Google Scholar]
- 26.Johansen M.P., Cresswell T., Davis J., Howard D.L., Howell N.R., Prentice E. Biofilm-enhanced adsorption of strong and weak cations onto different microplastic sample types: use of spectroscopy, microscopy and radiotracer methods. Water Res. 2019;158:392–400. doi: 10.1016/j.watres.2019.04.029. [DOI] [PubMed] [Google Scholar]
- 27.He S., Jia M., Xiang Y., Song B., Xiong W., Cao J., et al. Biofilm on microplastics in aqueous environment: physicochemical properties and environmental implications. J. Hazard. Mater. 2022;424 doi: 10.1016/j.jhazmat.2021.127286. [DOI] [PubMed] [Google Scholar]
- 28.Sun Y., Cao N., Duan C., Wang Q., Ding C., Wang J. Selection of antibiotic resistance genes on biodegradable and non-biodegradable microplastics. J. Hazard. Mater. 2021;409 doi: 10.1016/j.jhazmat.2020.124979. [DOI] [PubMed] [Google Scholar]
- 29.Wang J., Peng C., Dai Y., Li Y., Jiao S., Ma X., et al. Slower antibiotics degradation and higher resistance genes enrichment in plastisphere. Water Res. 2022;222 doi: 10.1016/j.watres.2022.118920. [DOI] [PubMed] [Google Scholar]
- 30.Deng H., Fu Q., Li D., Zhang Y., He J., Feng D., et al. Microplastic-associated biofilm in an intensive mariculture pond: temporal dynamics of microbial communities, extracellular polymeric substances and impacts on microplastics properties. J. Clean. Prod. 2021;319 [Google Scholar]
- 31.Li H., Luo Q., Zhao S., Zhao P., Yang X., Huang Q., et al. Watershed urbanization enhances the enrichment of pathogenic bacteria and antibiotic resistance genes on microplastics in the water environment. Environ. Pollut. 2022;313 doi: 10.1016/j.envpol.2022.120185. [DOI] [PubMed] [Google Scholar]
- 32.Dussud C., Hudec C., George M., Fabre P., Higgs P., Bruzaud S., et al. Colonization of non-biodegradable and biodegradable plastics by marine microorganisms. Front. Microbiol. 2018;9:1571. doi: 10.3389/fmicb.2018.01571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sgier L., Freimann R., Zupanic A., Kroll A. Flow cytometry combined with viSNE for the analysis of microbial biofilms and detection of microplastics. Nat. Commun. 2016;7 doi: 10.1038/ncomms11587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zettler E.R., Mincer T.J., Amaral-Zettler L.A. Life in the “plastisphere”: microbial communities on plastic marine debris. Environ. Sci. Technol. 2013;47:7137–7146. doi: 10.1021/es401288x. [DOI] [PubMed] [Google Scholar]
- 35.Yokota K., Waterfield H., Hastings C., Davidson E., Kwietniewski E., Wells B. Finding the missing piece of the aquatic plastic pollution puzzle: interaction between primary producers and microplastics. Limnol. Oceanogr. Lett. 2017;2:91–104. [Google Scholar]
- 36.Zhang S., Liu X., Qiu P., Chen B., Xu C., Dong W., et al. Microplastics can selectively enrich intracellular and extracellular antibiotic resistant genes and shape different microbial communities in aquatic systems. Sci. Tot. Environ. 2022;822 doi: 10.1016/j.scitotenv.2022.153488. [DOI] [PubMed] [Google Scholar]
- 37.Guo X.P., Sun X.L., Chen Y.R., Hou L., Liu M., Yang Y. Antibiotic resistance genes in biofilms on plastic wastes in an estuarine environment. Sci. Tot. Environ. 2020;745 doi: 10.1016/j.scitotenv.2020.140916. [DOI] [PubMed] [Google Scholar]
- 38.Sun R., He L., Li T., Dai Z., Sun S., Ren L., et al. Impact of the surrounding environment on antibiotic resistance genes carried by microplastics in mangroves. Sci. Tot. Environ. 2022;837 doi: 10.1016/j.scitotenv.2022.155771. [DOI] [PubMed] [Google Scholar]
- 39.Liu X., Fang L., Yan X., Gardea-Torresdey J.L., Gao Y., Zhou X., et al. Surface functional groups and biofilm formation on microplastics: environmental implications. Sci. Tot. Environ. 2023;903 doi: 10.1016/j.scitotenv.2023.166585. [DOI] [PubMed] [Google Scholar]
- 40.Li W., Zhang Y., Wu N., Zhao Z., Xu W., Ma Y., et al. Colonization characteristics of bacterial communities on plastic debris influenced by environmental factors and polymer types in the Haihe Estuary of Bohai Bay, China. Environ. Sci. Technol. 2019;53:10763–10773. doi: 10.1021/acs.est.9b03659. [DOI] [PubMed] [Google Scholar]
- 41.Li Y.Q., Zhang C.M., Yuan Q.Q., Wu K. New insight into the effect of microplastics on antibiotic resistance and bacterial community of biofilm. Chemosphere. 2023;335 doi: 10.1016/j.chemosphere.2023.139151. [DOI] [PubMed] [Google Scholar]
- 42.Xu X., Wang S., Li C., Li J., Gao F., Zheng L. Quorum sensing bacteria in microplastics epiphytic biofilms and their biological characteristics which potentially impact marine ecosystem. Ecotoxicol. Environ. Saf. 2023;264 doi: 10.1016/j.ecoenv.2023.115444. [DOI] [PubMed] [Google Scholar]
- 43.Xiao S., Zhang Y., Wu Y., Li J., Dai W., Pang K., et al. Bacterial community succession and the enrichment of antibiotic resistance genes on microplastics in an oyster farm. Mar. Pollut. Bull. 2023;194 doi: 10.1016/j.marpolbul.2023.115402. [DOI] [PubMed] [Google Scholar]
- 44.Roager L., Sonnenschein E.C. Bacterial candidates for colonization and degradation of marine plastic debris. Environ. Sci. Technol. 2019;53:11636–11643. doi: 10.1021/acs.est.9b02212. [DOI] [PubMed] [Google Scholar]
- 45.Dang H., Lovell C.R. Bacterial primary colonization and early succession on surfaces in marine waters as determined by amplified rRNA gene restriction analysis and sequence analysis of 16S rRNA genes. Appl. Environ. Microbiol. 2000;66:467–475. doi: 10.1128/aem.66.2.467-475.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang M., Zhao Y., Qin X., Jia W., Chai L., Huang M., et al. Microplastics from mulching film is a distinct habitat for bacteria in farmland soil. Sci. Tot. Environ. 2019;688:470–478. doi: 10.1016/j.scitotenv.2019.06.108. [DOI] [PubMed] [Google Scholar]
- 47.Bhagwat G., Tran T.K.A., Lamb D., Senathirajah K., Grainge I., O'Connor W., et al. Biofilms enhance the adsorption of toxic contaminants on plastic microfibers under environmentally relevant conditions. Environ. Sci. Technol. 2021;55:8877–8887. doi: 10.1021/acs.est.1c02012. [DOI] [PubMed] [Google Scholar]
- 48.Feng L., He L., Jiang S., Chen J., Zhou C., Qian Z.J., et al. Investigating the composition and distribution of microplastics surface biofilms in coral areas. Chemosphere. 2020;252 doi: 10.1016/j.chemosphere.2020.126565. [DOI] [PubMed] [Google Scholar]
- 49.Miao L., Yu Y., Adyel T.M., Wang C., Liu Z., Liu S., et al. Distinct microbial metabolic activities of biofilms colonizing microplastics in three freshwater ecosystems. J. Hazard. Mater. 2021;403 doi: 10.1016/j.jhazmat.2020.123577. [DOI] [PubMed] [Google Scholar]
- 50.Sun X.L., Xiang H., Xiong H.Q., Fang Y.C., Wang Y. Bioremediation of microplastics in freshwater environments: a systematic review of biofilm culture, degradation mechanisms, and analytical methods. Sci. Tot. Environ. 2023;863 doi: 10.1016/j.scitotenv.2022.160953. [DOI] [PubMed] [Google Scholar]
- 51.Wang J., Guo X., Xue J. Biofilm-developed microplastics as vectors of pollutants in aquatic environments. Environ. Sci. Technol. 2021;55:12780–12790. doi: 10.1021/acs.est.1c04466. [DOI] [PubMed] [Google Scholar]
- 52.Shan E., Zhang X., Li J., Sun C., Teng J., Yang X., et al. Incubation habitats and aging treatments affect the formation of biofilms on polypropylene microplastics. Sci. Tot. Environ. 2022;831 doi: 10.1016/j.scitotenv.2022.154769. [DOI] [PubMed] [Google Scholar]
- 53.Liu P., Dai J., Bie C., Li H., Zhang Z., Guo X., et al. Bioaccessibility of microplastic-associated antibiotics in freshwater organisms: highlighting the impacts of biofilm colonization via an in vitro protocol. Environ. Sci. Technol. 2022;56:12267–12277. doi: 10.1021/acs.est.2c02782. [DOI] [PubMed] [Google Scholar]
- 54.Tu C., Chen T., Zhou Q., Liu Y., Wei J., Waniek J.J., et al. Biofilm formation and its influences on the properties of microplastics as affected by exposure time and depth in the seawater. Sci. Tot. Environ. 2020;734 doi: 10.1016/j.scitotenv.2020.139237. [DOI] [PubMed] [Google Scholar]
- 55.Chen Y., Yan Z., Zhou Y., Zhang Y., Jiang R., Wang M., et al. Dynamic evolution of antibiotic resistance genes in plastisphere in the vertical profile of urban rivers. Water Res. 2024;249 doi: 10.1016/j.watres.2023.120946. [DOI] [PubMed] [Google Scholar]
- 56.You X., Xu N., Yang X., Sun W. Pollutants affect algae-bacteria interactions: a critical review. Environ. Pollut. 2021;276 doi: 10.1016/j.envpol.2021.116723. [DOI] [PubMed] [Google Scholar]
- 57.Zhang G., Chen J., Li W. Conjugative antibiotic-resistant plasmids promote bacterial colonization of microplastics in water environments. J. Hazard. Mater. 2022;430 doi: 10.1016/j.jhazmat.2022.128443. [DOI] [PubMed] [Google Scholar]
- 58.Qiu Z., Yu Y., Chen Z., Jin M., Yang D., Zhao Z., et al. Nanoalumina promotes the horizontal transfer of multiresistance genes mediated by plasmids across genera. Proc. Natl. Acad. Sci. U. S. A. 2012;109:4944–4949. doi: 10.1073/pnas.1107254109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Atugoda T., Wijesekara H., Werellagama D.R.I.B., Jinadasa K.B.S.N., Bolan N.S., Vithanage M. Adsorptive interaction of antibiotic ciprofloxacin on polyethylene microplastics: implications for vector transport in water. Environ. Technol. Innov. 2020;19 [Google Scholar]
- 60.Wang Y., Wang X., Li Y., Li J., Wang F., Xia S., et al. Biofilm alters tetracycline and copper adsorption behaviors onto polyethylene microplastics. Chem. Eng. J. 2020;392 [Google Scholar]
- 61.Yu X., Du H., Huang Y., Yin X., Liu Y., Li Y., et al. Selective adsorption of antibiotics on aged microplastics originating from mariculture benefits the colonization of opportunistic pathogenic bacteria. Environ. Pollut. 2022;313 doi: 10.1016/j.envpol.2022.120157. [DOI] [PubMed] [Google Scholar]
- 62.Kiki C., Adéoyé A.B.E., Li X., Yan X., Feng J., Yu C.P., et al. Contrasting effects of phytoplankton aging on microplastic antibiotic adsorption depending on species tolerance, and biofouling level. Water Res. 2023;237 doi: 10.1016/j.watres.2023.119992. [DOI] [PubMed] [Google Scholar]
- 63.Sun Y., Peng B.Y., Wang X., Li Y., Wang Y., Zhang Y., et al. Adsorption and desorption mechanisms of oxytetracycline on poly(butylene adipate-co-terephthalate) microplastics after degradation: the effects of biofilms, Cu(II), water pH, and dissolved organic matter. Sci. Environ. 2023;863 doi: 10.1016/j.scitotenv.2022.160866. [DOI] [PubMed] [Google Scholar]
- 64.Ainali N.M., Kalaronis D., Evgenidou E., Kyzas G.Z., Bobori D.C., Kaloyianni M., et al. Do poly(lactic acid) microplastics instigate a threat? A perception for their dynamic towards environmental pollution and toxicity. Sci. Tot. Environ. 2022;832 doi: 10.1016/j.scitotenv.2022.155014. [DOI] [PubMed] [Google Scholar]
- 65.Magadini D.L., Goes J.I., Ortiz S., Lipscomb J., Pitiranggon M., Yan B. Assessing the sorption of pharmaceuticals to microplastics through in-situ experiments in New York City waterways. Sci. Tot. Environ. 2020;729 doi: 10.1016/j.scitotenv.2020.138766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guo X., Pang J., Chen S., Jia H. Sorption properties of tylosin on four different microplastics. Chemosphere. 2018;209:240–245. doi: 10.1016/j.chemosphere.2018.06.100. [DOI] [PubMed] [Google Scholar]
- 67.Fu J., Li Y., Peng L., Gao W., Wang G. Distinct chemical adsorption behaviors of sulfanilamide as a model antibiotic onto weathered microplastics in complex systems. Colloids Surf. A Physicochem. Eng. Asp. 2022;648 [Google Scholar]
- 68.Sun Y., Wang X., Xia S., Zhao J. New insights into oxytetracycline (OTC) adsorption behavior on polylactic acid microplastics undergoing microbial adhesion and degradation. Chem. Eng. J. 2021;416 [Google Scholar]
- 69.Fan X., Zou Y., Geng N., Liu J., Hou J., Li D., et al. Investigation on the adsorption and desorption behaviors of antibiotics by degradable MPs with or without UV ageing process. J. Hazard. Mater. 2021;401 doi: 10.1016/j.jhazmat.2020.123363. [DOI] [PubMed] [Google Scholar]
- 70.Guo X., Wang X., Zhou X., Kong X., Tao S., Xing B. Sorption of four hydrophobic organic compounds by three chemically distinct polymers: role of chemical and physical composition. Environ. Sci. Technol. 2012;46:7252–7259. doi: 10.1021/es301386z. [DOI] [PubMed] [Google Scholar]
- 71.Ji H., Wan S., Liu Z., Xie X., Xiang X., Liao L., et al. Adsorption of antibiotics on microplastics (MPs) in aqueous environments: the impacts of aging and biofilms. J. Environ. Chem. Eng. 2024 [Google Scholar]
- 72.Guan J., Qi K., Wang J., Wang W., Wang Z., Lu N., et al. Microplastics as an emerging anthropogenic vector of trace metals in freshwater: significance of biofilms and comparison with natural substrates. Water Res. 2020;184 doi: 10.1016/j.watres.2020.116205. [DOI] [PubMed] [Google Scholar]
- 73.Zhang H., Wang J., Zhou B., Zhou Y., Dai Z., Zhou Q., et al. Enhanced adsorption of oxytetracycline to weathered microplastic polystyrene: kinetics, isotherms and influencing factors. Environ. Pollut. 2018;243:1550–1557. doi: 10.1016/j.envpol.2018.09.122. [DOI] [PubMed] [Google Scholar]
- 74.Wang L., Yang H., Guo M., Wang Z., Zheng X. Adsorption of antibiotics on different microplastics (MPs): behavior and mechanism. Sci. Tot. Environ. 2023;863 doi: 10.1016/j.scitotenv.2022.161022. [DOI] [PubMed] [Google Scholar]
- 75.Liu P., Wu X., Liu H., Wang H., Lu K., Gao S. Desorption of pharmaceuticals from pristine and aged polystyrene microplastics under simulated gastrointestinal conditions. J. Hazard. Mater. 2020;392 doi: 10.1016/j.jhazmat.2020.122346. [DOI] [PubMed] [Google Scholar]
- 76.Parolo M.E., Savini M.C., Vallés J.M., Baschini M.T., Avena M.J. Tetracycline adsorption on montmorillonite: pH and ionic strength effects. Appl. Clay Sci. 2008;40:179–186. [Google Scholar]
- 77.Wang H., Xu K., Wang J., Feng C., Chen Y., Shi J., et al. Microplastic biofilm: an important microniche that may accelerate the spread of antibiotic resistance genes via natural transformation. J. Hazard. Mater. 2023;459 doi: 10.1016/j.jhazmat.2023.132085. [DOI] [PubMed] [Google Scholar]
- 78.Hu H., Jin D., Yang Y., Zhang J., Ma C., Qiu Z. Distinct profile of bacterial community and antibiotic resistance genes on microplastics in Ganjiang River at the watershed level. Environ. Res. 2021;200 doi: 10.1016/j.envres.2021.111363. [DOI] [PubMed] [Google Scholar]
- 79.Jia J., Guan Y., Li X., Fan X., Zhu Z., Xing H., et al. Phenotype profiles and adaptive preference of Acinetobacter johnsonii isolated from Ba River with different environmental backgrounds. Environ. Res. 2021;196 doi: 10.1016/j.envres.2021.110913. [DOI] [PubMed] [Google Scholar]
- 80.Martinez J.L. The role of natural environments in the evolution of resistance traits in pathogenic bacteria. Proc. Biol. Sci. 2009;276:2521–2530. doi: 10.1098/rspb.2009.0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lewinson O., Bibi E. Evidence for simultaneous binding of dissimilar substrates by the Escherichia coli multidrug transporter MdfA. Biochemistry. 2001;40:12612–12618. doi: 10.1021/bi011040y. [DOI] [PubMed] [Google Scholar]
- 82.Heurlier K., Dénervaud V., Haas D. Impact of quorum sensing on fitness of Pseudomonas aeruginosa. Int. J. Med. Microbiol. 2006;296:93–102. doi: 10.1016/j.ijmm.2006.01.043. [DOI] [PubMed] [Google Scholar]
- 83.Liu X., Wang H., Li L., Deng C., Chen Y., Ding H., et al. Do microplastic biofilms promote the evolution and co-selection of antibiotic and metal resistance genes and their associations with bacterial communities under antibiotic and metal pressures? J. Hazard. Mater. 2022;424 doi: 10.1016/j.jhazmat.2021.127285. [DOI] [PubMed] [Google Scholar]
- 84.Yang K., Chen Q.L., Chen M.L., Li H.Z., Liao H., Pu Q., et al. Temporal dynamics of antibiotic resistome in the plastisphere during microbial colonization. Environ. Sci. Technol. 2020;54:11322–11332. doi: 10.1021/acs.est.0c04292. [DOI] [PubMed] [Google Scholar]
- 85.Arias-Andres M., Klümper U., Rojas-Jimenez K., Grossart H.P. Microplastic pollution increases gene exchange in aquatic ecosystems. Environ. Pollut. 2018;237:253–261. doi: 10.1016/j.envpol.2018.02.058. [DOI] [PubMed] [Google Scholar]
- 86.Zhou Q., Zhang J., Zhang M., Wang X., Zhang D., Pan X. Persistent versus transient, and conventional plastic versus biodegradable plastic? – two key questions about microplastic-water exchange of antibiotic resistance genes. Water Res. 2022;222 doi: 10.1016/j.watres.2022.118899. [DOI] [PubMed] [Google Scholar]
- 87.Zhang Y., Tao J., Bai Y., Wang F., Xie B. Incomplete degradation of aromatic-aliphatic copolymer leads to proliferation of microplastics and antibiotic resistance genes. Environ. Int. 2023;181 doi: 10.1016/j.envint.2023.108291. [DOI] [PubMed] [Google Scholar]
- 88.Shi J., Lv B., Wang B., Xie B. Insight into the responses of antibiotic resistance genes in microplastic biofilms to zinc oxide nanoparticles and zinc ions pressures in landfill leachate. J. Hazard. Mater. 2023;459 doi: 10.1016/j.jhazmat.2023.132096. [DOI] [PubMed] [Google Scholar]
- 89.Xia H., Yang J., Huang K., Nie C. Microplastics into vermi-wetland lower the treatment performance of organic substances and antibiotic resistance genes in excess sludge. J. Environ. Chem. Eng. 2023;11 [Google Scholar]
- 90.Zha Y., Li Z., Zhong Z., Ruan Y., Sun L., Zuo F., et al. Size-dependent enhancement on conjugative transfer of antibiotic resistance genes by micro/nanoplastics. J. Hazard. Mater. 2022;431 doi: 10.1016/j.jhazmat.2022.128561. [DOI] [PubMed] [Google Scholar]
- 91.Luo T., Dai X., Wei W., Xu Q., Ni B.J. Microplastics enhance the prevalence of antibiotic resistance genes in anaerobic sludge digestion by enriching antibiotic-resistant bacteria in surface biofilm and facilitating the vertical and horizontal gene transfer. Environ. Sci. Technol. 2023;57:14611–14621. doi: 10.1021/acs.est.3c02815. [DOI] [PubMed] [Google Scholar]
- 92.Jia J., Guan Y., Cheng M., Chen H., He J., Wang S., et al. Occurrence and distribution of antibiotics and antibiotic resistance genes in Ba River, China. Sci. Tot. Environ. 2018;642:1136–1144. doi: 10.1016/j.scitotenv.2018.06.149. [DOI] [PubMed] [Google Scholar]
- 93.Stevenson E.M., Buckling A., Cole M., Lindeque P.K., Murray A.K. Selection for antimicrobial resistance in the plastisphere. Sci. Tot. Environ. 2024;908 doi: 10.1016/j.scitotenv.2023.168234. [DOI] [PubMed] [Google Scholar]
- 94.Murray A.K., Zhang L., Yin X., Zhang T., Buckling A., Snape J., et al. Novel insights into selection for antibiotic resistance in complex microbial communities. mBio. 2018;9 doi: 10.1128/mBio.00969-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Guan Y., Jia J., Fan X., Li K., Wang Z. Anthropogenic impacts on antibiotic resistance genes and their hosts from pristine to urban river using metagenomic and binning approaches. Aquat. Toxicol. 2022;249 doi: 10.1016/j.aquatox.2022.106221. [DOI] [PubMed] [Google Scholar]
- 96.Imran M., Das K.R., Naik M.M. Co-selection of multi-antibiotic resistance in bacterial pathogens in metal and microplastic contaminated environments: an emerging health threat. Chemosphere. 2019;215:846–857. doi: 10.1016/j.chemosphere.2018.10.114. [DOI] [PubMed] [Google Scholar]
- 97.Baker-Austin C., Wright M.S., Stepanauskas R., McArthur J.V. Co-selection of antibiotic and metal resistance. Trends Microbiol. 2006;14:176–182. doi: 10.1016/j.tim.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 98.Zhang Y., Gu A.Z., He M., Li D., Chen J. Subinhibitory concentrations of disinfectants promote the horizontal transfer of multidrug resistance genes within and across genera. Environ. Sci. Technol. 2017;51:570–580. doi: 10.1021/acs.est.6b03132. [DOI] [PubMed] [Google Scholar]
- 99.Guo M.T., Yuan Q.B., Yang J. Distinguishing effects of ultraviolet exposure and chlorination on the horizontal transfer of antibiotic resistance genes in municipal wastewater. Environ. Sci. Technol. 2015;49:5771–5778. doi: 10.1021/acs.est.5b00644. [DOI] [PubMed] [Google Scholar]
- 100.Toduka Y., Toyooka T., Ibuki Y. Flow cytometric evaluation of nanoparticles using side-scattered light and reactive oxygen species-mediated fluorescence-correlation with genotoxicity. Environ. Sci. Technol. 2012;46:7629–7636. doi: 10.1021/es300433x. [DOI] [PubMed] [Google Scholar]
- 101.Ding P., Lu J., Wang Y., Schembri M.A., Guo J. Antidepressants promote the spread of antibiotic resistance via horizontally conjugative gene transfer. Environ. Microbiol. 2022;24:5261–5276. doi: 10.1111/1462-2920.16165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lu J., Ding P., Wang Y., Guo J. Antidepressants promote the spread of extracellular antibiotic resistance genes via transformation. ISME Commun. 2022;2:63. doi: 10.1038/s43705-022-00147-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang J., Wang J., Zhao Z., Chen J., Lu H., Liu G., et al. PAHs accelerate the propagation of antibiotic resistance genes in coastal water microbial community. Environ. Pollut. 2017;231:1145–1152. doi: 10.1016/j.envpol.2017.07.067. [DOI] [PubMed] [Google Scholar]
- 104.Li S., Duan G., Xi Y., Chu Y., Li F., Ho S.H. Insights into the role of extracellular polymeric substances (EPS) in the spread of antibiotic resistance genes. Environ. Pollut. 2023;343 doi: 10.1016/j.envpol.2023.123285. [DOI] [PubMed] [Google Scholar]
- 105.Li Y.P., You L.X., Yang X.J., Yu Y.S., Zhang H.T., Yang B., et al. Extrapolymeric substances (EPS) in Mucilaginibacter rubeus P2 displayed efficient metal(loid) bio-adsorption and production was induced by copper and zinc. Chemosphere. 2022;291 doi: 10.1016/j.chemosphere.2021.132712. [DOI] [PubMed] [Google Scholar]
- 106.Lin Z., Yuan T., Zhou L., Cheng S., Qu X., Lu P., et al. Impact factors of the accumulation, migration and spread of antibiotic resistance in the environment. Environ. Geochem. Health. 2021;43:1741–1758. doi: 10.1007/s10653-020-00759-0. [DOI] [PubMed] [Google Scholar]
- 107.Haffiez N., Zakaria B.S., Azizi S.M.M., Dhar B.R. Fate of intracellular, extracellular polymeric substances-associated, and cell-free antibiotic resistance genes in anaerobic digestion of thermally hydrolyzed sludge. Sci. Tot. Environ. 2023;855 doi: 10.1016/j.scitotenv.2022.158847. [DOI] [PubMed] [Google Scholar]
- 108.He P., Zhou Y., Shao L., Huang J., Yang Z., Lü F. The discrepant mobility of antibiotic resistant genes: evidence from their spatial distribution in sewage sludge flocs. Sci. Tot. Environ. 2019;697 doi: 10.1016/j.scitotenv.2019.134176. [DOI] [PubMed] [Google Scholar]
- 109.Guo T., Lou C., Zhai W., Tang X., Hashmi M.Z., Murtaza R., et al. Increased occurrence of heavy metals, antibiotics and resistance genes in surface soil after long-term application of manure. Sci. Tot. Environ. 2018;635:995–1003. doi: 10.1016/j.scitotenv.2018.04.194. [DOI] [PubMed] [Google Scholar]
- 110.Zheng Z., Huang Y., Liu L., Wang L., Tang J. Interaction between microplastic biofilm formation and antibiotics: effect of microplastic biofilm and its driving mechanisms on antibiotic resistance gene. J. Hazard. Mater. 2023;459 doi: 10.1016/j.jhazmat.2023.132099. [DOI] [PubMed] [Google Scholar]
- 111.Perveen S., Pablos C., Reynolds K., Stanley S., Marugán J. Growth and prevalence of antibiotic-resistant bacteria in microplastic biofilm from wastewater treatment plant effluents. Sci. Tot. Environ. 2023;856 doi: 10.1016/j.scitotenv.2022.159024. [DOI] [PubMed] [Google Scholar]
- 112.Yu X., Zhou Z.C., Shuai X.Y., Lin Z.J., Liu Z., Zhou J.Y., et al. Microplastics exacerbate co-occurrence and horizontal transfer of antibiotic resistance genes. J. Hazard. Mater. 2023;451 doi: 10.1016/j.jhazmat.2023.131130. [DOI] [PubMed] [Google Scholar]
- 113.Cao M., Wang F., Zhou B., Chen H., Yuan R., Ma S., et al. Nanoparticles and antibiotics stress proliferated antibiotic resistance genes in microalgae-bacteria symbiotic systems. J. Hazard. Mater. 2023;443 doi: 10.1016/j.jhazmat.2022.130201. [DOI] [PubMed] [Google Scholar]
- 114.Jia J., Liu Q., Wu C. Microplastic and antibiotic proliferated the colonization of specific bacteria and antibiotic resistance genes in the phycosphere of Chlorella pyrenoidosa. J. Hazard. Mater. 2023;455 doi: 10.1016/j.jhazmat.2023.131618. [DOI] [PubMed] [Google Scholar]
- 115.Liu Q., Jia J., Hu H., Li X., Zhao Y., Wu C. Nitrogen and phosphorus limitations promoted bacterial nitrate metabolism and propagation of antibiotic resistome in the phycosphere of Auxenochlorella pyrenoidosa. J. Hazard. Mater. 2024;468 doi: 10.1016/j.jhazmat.2024.133786. [DOI] [PubMed] [Google Scholar]
- 116.Ashraf N., Ahmad F., Lu Y. Synergy between microalgae and microbiome in polluted waters. Trends Microbiol. 2023;31:9–21. doi: 10.1016/j.tim.2022.06.004. [DOI] [PubMed] [Google Scholar]
- 117.Seymour J.R., Amin S.A., Raina J.B., Stocker R. Zooming in on the phycosphere: the ecological interface for phytoplankton-bacteria relationships. Nat. Microbiol. 2017;2 doi: 10.1038/nmicrobiol.2017.65. [DOI] [PubMed] [Google Scholar]
- 118.Junaid M., Liu X., Wu Y., Wang J. Selective enrichment of antibiotic resistome and bacterial pathogens by aquatic microplastics. J. Hazard. Mater. Adv. 2022;7 [Google Scholar]
- 119.Li R., Zhu L., Cui L., Zhu Y.G. Viral diversity and potential environmental risk in microplastic at watershed scale: evidence from metagenomic analysis of plastisphere. Environ. Int. 2022;161 doi: 10.1016/j.envint.2022.107146. [DOI] [PubMed] [Google Scholar]
- 120.Metcalf R., Messer L.F., White H.L., Ormsby M.J., Matallana-Surget S., Quilliam R.S. Evidence of interspecific plasmid uptake by pathogenic strains of Klebsiella isolated from microplastic pollution on public beaches. J. Hazard. Mater. 2024;461 doi: 10.1016/j.jhazmat.2023.132567. [DOI] [PubMed] [Google Scholar]
- 121.Oberbeckmann S., Kreikemeyer B., Labrenz M. Environmental factors support the formation of specific bacterial assemblages on microplastics. Front. Microbiol. 2017;8:2709. doi: 10.3389/fmicb.2017.02709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lai K.P., Tsang C.F., Li L., Yu R.M.K., Kong R.Y.C. Microplastics act as a carrier for wastewater-borne pathogenic bacteria in sewage. Chemosphere. 2022;301 doi: 10.1016/j.chemosphere.2022.134692. [DOI] [PubMed] [Google Scholar]
- 123.Liang H., de Haan W.P., Cerdà-Domènech M., Méndez J., Lucena F., García-Aljaro C., et al. Detection of faecal bacteria and antibiotic resistance genes in biofilms attached to plastics from human-impacted coastal areas. Environ. Pollut. 2023;319 doi: 10.1016/j.envpol.2022.120983. [DOI] [PubMed] [Google Scholar]
- 124.Naudet J., d'Orbcastel E.R., Bouvier T., Godreuil S., Dyall S., Bouvy S., et al. Identifying macroplastic pathobiomes and antibiotic resistance in a subtropical fish farm. Mar. Pollut. Bull. 2023;194 doi: 10.1016/j.marpolbul.2023.115267. [DOI] [PubMed] [Google Scholar]
- 125.Lyons M.M., Ward J.E., Gaff H., Hicks R.E., Drake J.M., Dobbs F.C. Theory of island biogeography on a microscopic scale: organic aggregates as islands for aquatic pathogens. Aquat. Microb. Ecol. 2010;60:1–13. [Google Scholar]
- 126.Qiu C., Zhou Y., Wang H., Chu Y., Zheng L., Chen Y., et al. Microplastics enrichment characteristics of antibiotic resistance genes and pathogens in landfill leachate. Chemosphere. 2023;341 doi: 10.1016/j.chemosphere.2023.140100. [DOI] [PubMed] [Google Scholar]
- 127.Silva I., Rodrigues E.T., Tacão M., Henriques I. Microplastics accumulate priority antibiotic-resistant pathogens: evidence from the riverine plastisphere. Environ. Pollut. 2023;332 doi: 10.1016/j.envpol.2023.121995. [DOI] [PubMed] [Google Scholar]
- 128.Castro-Castellon A.T., Horton A.A., Hughes J.M.R., Rampley C., Jeffers E.S., Bussi G., et al. Ecotoxicity of microplastics to freshwater biota: considering exposure and hazard across trophic levels. Sci. Tot. Environ. 2022;816 doi: 10.1016/j.scitotenv.2021.151638. [DOI] [PubMed] [Google Scholar]
- 129.Lamb J.B., Willis B.L., Fiorenza E.A., Couch C.S., Howard R., Rader D.N., et al. Plastic waste associated with disease on coral reefs. Science. 2018;359:460–462. doi: 10.1126/science.aar3320. [DOI] [PubMed] [Google Scholar]
- 130.Fabra M., Williams L., Watts J.E.M., Hale M.S., Couceiro F., Preston J. The plastic Trojan horse: biofilms increase microplastic uptake in marine filter feeders impacting microbial transfer and organism health. Sci. Tot. Environ. 2021;797 doi: 10.1016/j.scitotenv.2021.149217. [DOI] [PubMed] [Google Scholar]
- 131.Chen X., Wang X., Huang Y., Zhu Z., Li T., Cai Z., et al. Combined effects of microplastics and antibiotic-resistant bacteria on Daphnia magna growth and expression of functional genes. Sci. Tot. Environ. 2023;905 doi: 10.1016/j.scitotenv.2023.166880. [DOI] [PubMed] [Google Scholar]
- 132.Thacharodi A., Meenatchi R., Hassan S., Hussain N., Bhat M.A., Arockiaraj J., et al. Microplastics in the environment: a critical overview on its fate, toxicity, implications, management, and bioremediation strategies. J. Environ. Manag. 2024;349 doi: 10.1016/j.jenvman.2023.119433. [DOI] [PubMed] [Google Scholar]
- 133.Zhang P., Lu G., Sun Y., Yan Z., Dang T., Liu J. Metagenomic analysis explores the interaction of aged microplastics and roxithromycin on gut microbiota and antibiotic resistance genes of Carassius auratus. J. Hazard. Mater. 2022;425 doi: 10.1016/j.jhazmat.2021.127773. [DOI] [PubMed] [Google Scholar]
- 134.Yu Z., Qiu D., Zhou T., Zeng L., Yan C. Biofilm enhances the interactive effects of microplastics and oxytetracycline on zebrafish intestine. Aquat. Toxicol. 2024;270 doi: 10.1016/j.aquatox.2024.106905. [DOI] [PubMed] [Google Scholar]
- 135.Zhang S., Wang H., Liu M., Yu H., Peng J., Cao X., et al. Press perturbations of microplastics and antibiotics on freshwater micro-ecosystem: case study for the ecological restoration of submerged plants. Water Res. 2022;226 doi: 10.1016/j.watres.2022.119248. [DOI] [PubMed] [Google Scholar]
- 136.Hong J., Huang X., Wang Z., Luo X., Huang S., Zheng Z. Combined toxic effects of enrofloxacin and microplastics on submerged plants and epiphytic biofilms in high nitrogen and phosphorus waters. Chemosphere. 2022;308 doi: 10.1016/j.chemosphere.2022.136099. [DOI] [PubMed] [Google Scholar]
- 137.Mao H., Yang H., Xu Z., Yang Y., Zhang X., Huang F., et al. Microplastics and co-pollutant with ciprofloxacin affect interactions between free-floating macrophytes. Environ. Pollut. 2023;316 doi: 10.1016/j.envpol.2022.120546. [DOI] [PubMed] [Google Scholar]
- 138.Zhang Y., Wang J.X., Liu Y., Zhang J.T., Wang J.H., Chi Z.Y. Effects of environmental microplastic exposure on Chlorella sp. biofilm characteristics and its interaction with nitric oxide signaling. Sci. Tot. Environ. 2024;912 doi: 10.1016/j.scitotenv.2023.169659. [DOI] [PubMed] [Google Scholar]
- 139.Ma L., Li B., Jiang X.T., Wang Y.L., Xia Y., Li A.D., et al. Catalogue of antibiotic resistome and host-tracking in drinking water deciphered by a large scale survey. Microbiome. 2017;5:154. doi: 10.1186/s40168-017-0369-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sabatino R., Zullo R., Di Cesare A., Piscia R., Musazzi S., Corno G., et al. Traditional and biodegradable plastics host distinct and potentially more hazardous microbes when compared to both natural materials and planktonic community. J. Hazard. Mater. 2023;465 doi: 10.1016/j.jhazmat.2023.133166. [DOI] [PubMed] [Google Scholar]
- 141.He Y., Bai M., He Y., Wang S., Zhang J., Jiang S., et al. Suspended particles are hotspots for pathogen-related bacteria and ARGs in coastal beach waters of northern China. Sci. Tot. Environ. 2022;817 doi: 10.1016/j.scitotenv.2022.153004. [DOI] [PubMed] [Google Scholar]
- 142.Pham D.N., Clark L., Li M. Microplastics as hubs enriching antibiotic-resistant bacteria and pathogens in municipal activated sludge. J. Hazard. Mater. Lett. 2021;2 [Google Scholar]
- 143.Metcalf R., White H.L., Ormsby M.J., Oliver D.M., Quilliam R.S. From wastewater discharge to the beach: survival of human pathogens bound to microplastics during transfer through the freshwater-marine continuum. Environ. Pollut. 2023;319 doi: 10.1016/j.envpol.2022.120955. [DOI] [PubMed] [Google Scholar]