Abstract
Coronaviruses (CoVs) pose a threat to human health globally, as highlighted by severe acute respiratory syndrome (SARS), Middle East respiratory syndrome (MERS) and the COVID-19 pandemic. Bats from the Greater Mekong Subregion (GMS) are an important natural reservoir for CoVs. Here we report the differential prevalence of CoVs in bats within Yunnan Province across biological and ecological variables. We also show the coexistence of CoVs in individual bats and identify an additional putative host for SARS-related CoV, with higher dispersal capacity than other known hosts. Notably, 11 SARS-related coronaviruses (SARSr-CoVs) were discovered in horseshoe bats (family Rhinolophidae) and a Chinese water myotis bat (Myotis laniger) by pan-CoV detection and Illumina sequencing. Our findings facilitate an understanding of the fundamental features of the distribution and circulation of CoVs in nature as well as zoonotic spillover risk in the One health framework.
Keywords: Bats, Coronaviruses, Prevalence, Spillover risk, Coexistence
1. Introduction
Coronaviruses (CoVs) have gained global attention with the emergence of severe acute respiratory syndrome (SARS), Middle East respiratory syndrome (MERS) and the coronavirus 2019 (COVID-19) pandemic over the last two decades. These viruses are speculated to have zoonotic origins [1,2], that spilled over into the human population, with the possible involvement of intermediate amplifying hosts [[3], [4], [5], [6]]. In assessing the risk and possible extent of such spillovers, it is necessary to delineate the range of hosts, within which these coronaviruses and related viruses within the family may replicate [7]. Such knowledge would provide a crucial foundation for future work to prevent the spillover of novel CoVs from either their natural hosts or susceptible intermediate hosts to humans.
Coronaviruses are classified into four genera: Alphacoronavirus (α-CoV), Betacoronavirus (β-CoV), Gammacoronavirus (γ-CoV) and Deltacoronavirus (δ-CoV), as defined by the International Committee on Taxonomy of Viruses (ICTV) in 2020 [8]. Some members of the β-CoV subgenus Sarbecovirus and Merbecovirus are known to cause severe disease in humans, including severe acute respiratory syndrome coronavirus (SARS-CoV) [[9], [10], [11]], SARS-CoV-2 [12] and Middle East respiratory syndrome coronavirus (MERS-CoV) [2], whereas severe acute respiratory syndrome-related coronaviruses (SARSr-CoVs) also pose potential risks [[13], [14], [15], [16]]. Over the past two decades, SARS-CoV, MERS-CoV and SARS-CoV-2 have posed significant threats to public health [17,18] with negative impacts on all elements of society [19,20]. Mitigating the adverse impacts of disease outbreaks caused by sarbecoviruses and merbecoviruses is therefore of vital importance to the human population.
Coronaviruses infect a wide range of hosts: α-CoV and β-CoV can infect humans [21] and other mammals [[22], [23], [24], [25]]. In addition, γ-CoV and δ-CoV have also been detected in wild birds [26] and poultry [27]. With notably different immune systems from human [28], bats are implicated as important reservoir hosts for CoVs, especially as the hosts of the ancestors of five of the seven human CoVs, including human coronavirus 229E (HCoV-229E) and human coronavirus NL63 (HCoV-NL63) belonging to α-CoV, and MERS-CoV, SARS-CoV and SARS-CoV-2 belonging to β-CoV [16,[29], [30], [31], [32]].
Understanding hotspots of potential vectors is crucial to understand the potential risk of spillover of novel CoVs [33]. Rhinolophids, which are distributed across the entire Old World, are a major reservoir of CoVs [31,34]. Rhinolophids show the highest levels of richness in the Greater Mekong Subregion (GMS) [35] of Southeast Asia, in which potentially over half of the rhinolophid species in the region are yet to be scientifically described [36]. The GMS includes territories from six countries: Myanmar, Thailand, Lao People's Democratic Republic (Lao PDR), Vietnam, Cambodia, as well as Yunnan and Guangxi Provinces of China [37,38]. Coronaviruses of note, in the context of SARS-CoV and SARS-CoV-2, include two SARS-CoV-2 related coronaviruses (RshSTT182 and RshSTT200), which were found in Shamel's horseshoe bats (Rhinolophus shameli) collected from Cambodia in 2010 [39]. One SARS-related coronavirus (CoV) was identified in a Horsfield's leaf-nosed bat (Hipposideros larvatus) collected from Eastern Thailand in 2013 [40]. RaTG13 was identified from an intermediate horseshoe bat (Rhinolophus affinis) sampled from Yunnan Province in 2013 [41], and is understood to be among the closest relatives to the initial SARS-CoV-2 isolate responsible for the COVID-19 pandemic [42,43]. Three new β-CoVs were detected in leaf-nosed bats (Hipposideros) sampled in Myanmar during 2016–2018 [44]. There is molecular and serological evidence indicating the presence of SARS-CoV-2 related coronaviruses (RmYN02 and RacCS203) from horseshoe bats (Rhinolophus) circulating in Southeast Asia in 2019 and 2020, respectively [45]. A SARS-CoV-2 related bat coronavirus (BANAL-236) was isolated in a Marshall's horseshoe bat (Rhinolophus marshalli) captured in northern Laos in 2020, in which the viral spike protein was shown to mediate viral entry into hACE2-expressing human cells [46]. Previous studies have shown that the GMS is a hotspot for a variety of emerging CoVs of bat-origin [[39], [40], [41], [42], [43], [44], [45], [46]].
While these studies show the presence and diversity of SARS-CoV-2 like viruses among bat species endemic to the GMS, other β-CoVs are also present in the region [47]. The scope of animal hosts susceptible to infection by these viruses, and their prevalence within the GMS, are not well characterized. To fill in these knowledge gaps, we identified CoVs in bats and determined their prevalence by host species and sampling locations across Yunnan Province, China. We then investigated the CoV diversity with a focus on SARS-rCoVs in various bat species, with the view of further understanding the potential drivers behind the emergence of novel CoVs, their spread within animal populations and spillover to humans.
2. Results
2.1. Sample collection from various bat species from multiple trapping locations
A total of 716 bat rectal swabs (from 376 females/287 males, 53 no record), belonging to 17 bat species, were collected in 10 different caves (site A-J) at three locations (KM; YX; and XSBN) (Fig. 1-A-C, Table S1 and Table S2). The trapped bats belonged to six bat families including Pteropodidae (3 in total, 1 female/2 male), Hipposideridae (123, 75/46/2 unidentified sex), Vespertilionidae (228, 109/114/5), Miniopteridae (60, 31/20/9), Megadermatidae (2, both male), and Rhinolophidae (300, 160/103/37). Further details on sampled bats are shown in Table S1 and Table S2.
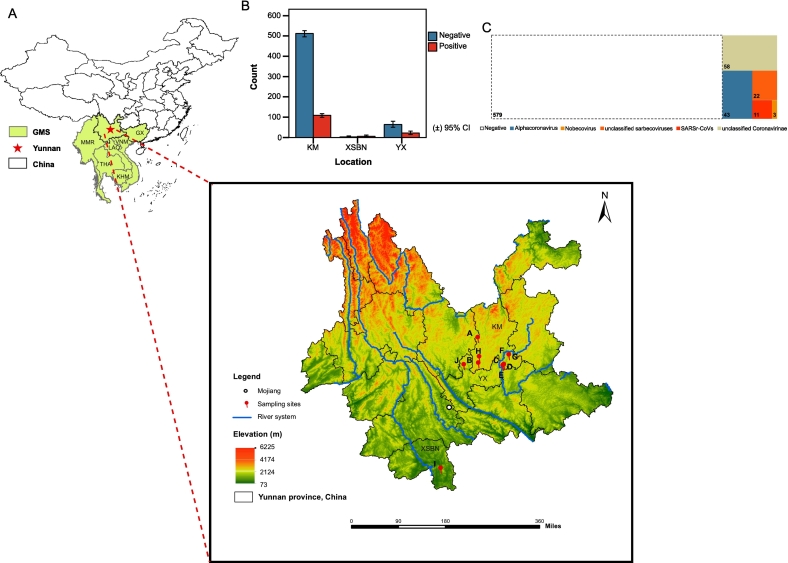
Fig. 1.
Geographical distribution of bat sampling sites and prevalence of coronavirus in the investigation. (A) The location of Yunnan province in relation to China and the Greater Mekong Subregion (GMS) and sampling sites. The white dot mark Mojiang county. Numbers and corresponding colors denote the different elevations (in Meters) in each place. A-J denotes ten different sampling sites from around Kunming (KM) City (sites: A-H), Yuxi (YX) City (site J) and Xishuangbanna (XSBN) Prefecture (site I) of Yunnan Province. The map shows countries and provinces comprising the GMS, including: Myanmar (MMR), Thailand (THA), Lao People's Democratic Republic (LAO), Vietnam (VNM), Cambodia (KHM), Guangxi Provinces of China (GX) and Yunnan Province. (B) Coronavirus prevalence in three sampling locations (KM, XSBN and YX). (C) Treemap showing individual counts of bats found positive, or negative for coronaviruses in the investigation. Related to Table S1 and Table S3.
2.2. Coronavirus prevalence
Based on pan-CoV semi-nested PCR detection and Sanger sequencing, the overall CoV prevalence was 19.1 % (95 % confidence interval (CI): 16.2–22.0), which corresponded to 137 CoV-positive individuals (see Table S3 for details of BLASTn confirmation) out of 716 total (Fig. 1-C), and the negative rate is 80.9 % (95 % CI: 77.75–83.65, 579 negative samples) (Table S1). After PCR, the samples are run by agarose gel electrophoresis, in which the target DNA band (236 bp) will be visualized in positive samples, as shown by a representative gel in Fig. S1. The percentages and counts in each category are given for incidence by genera, sites, species, and sex (Fig. 2-A, Fig. 2-B, Table S1, Table S4 and Table S5), as discussed below.
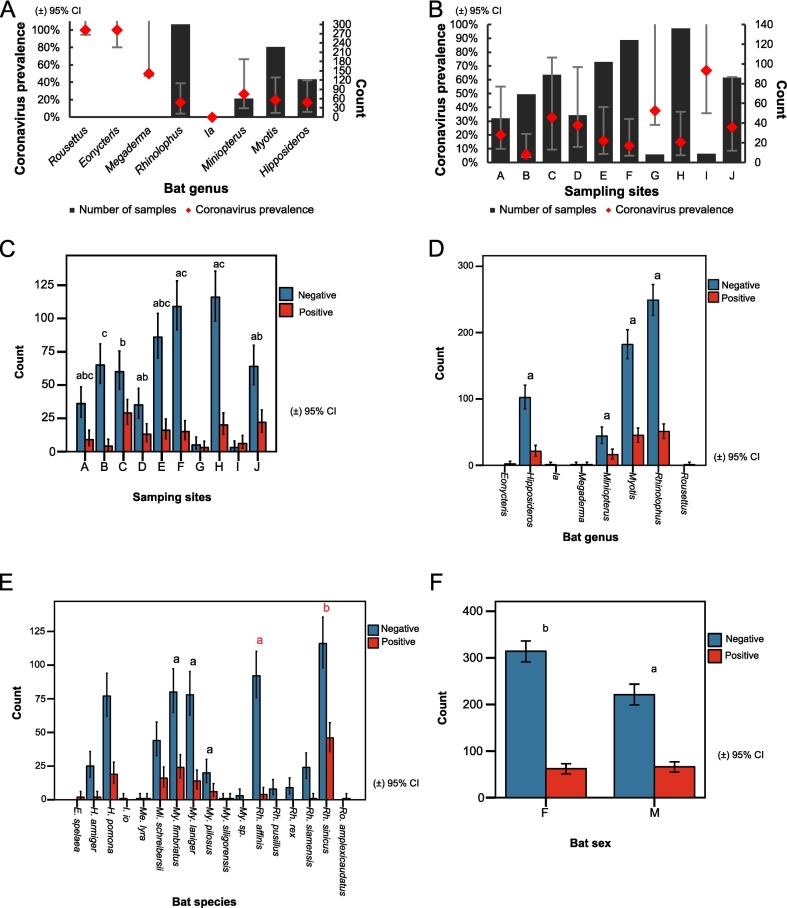
Fig. 2.
Coronavirus prevalence and analysis of significant association across different factors. (A) Prevalence of coronavirus and number of samples in each bat genus (B) Prevalence of coronavirus and number of samples in each sampling site. The x-axis indicates bat genera (A) and sampling sites (B). The y-axis indicates coronavirus prevalence (left) and the number of samples (right). The black column indicates the number of samples collected. The red line indicates coronavirus prevalence. (C) Coronavirus prevalence and significance of association with sampling sites. D—F show prevalence and significance of association for (D) genera. (E) species. (F) sex. Error bars represent 95 % confidence interval (CI). Related to Table S1, Table S2 and Table S6. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.3. Analysis of CoV prevalence based on sampling site
CoV prevalence was found to be weakly associated with sampling site (Pearson χ2 (7, N = 699) = 29.791, p < 0.001) (Cramer's V = 0.206, p < 0.001, and Goodman-Kruskal tau's coefficient is 0.043, p < 0.001) (Table S6). The prevalence of bat-borne CoVs in our study ranged between 5.8 %–66.7 % (5.8 %, 95 % CI: 1.87–14.93; 66.7 %, 30.92–90.96) for all ten sampling sites (A-J) (Fig. 2-B and Table S1) and between 17.6 %–66.7 % (17.6 %, 95 % CI: 14.69–20.82) for the three categories of sites (KM, YX and XSBN) (Fig. 1-B and Table S1). Prevalence ranged between 5.8 % (site B) and 32.6 % (95 % CI: 23.24–43.44, site C) for all sites collectively (there are at least 45 samples per site in the comparison), as shown in Fig. 2-B, Table S1, Table S7, and highest in site C in Kunming (Fig. 2-B and Table S1), with significant differences (p < 0.05) with other sites (Fig. 2-C).
2.4. Analysis of CoV prevalence based on bat genera and species
Thirteen out of 17 species were positive for CoVs (Table S1, Table S4). CoV prevalence was significantly associated with bat species within the genus Rhinolophus (Pearson χ2 (1, N = 258) = 22.648, p < 0.001) (Table S6). Within Rhinolophus, Rh. sinicus were found to have a higher CoV prevalence than Rh. affinis (p < 0.05), but there were not significant differences between My. laniger, My. fimbriatus and My. pilosus (Fig. 2-E, Table S1). CoV prevalence was between 17 % (95 % CI: 13.02–21.84, Rhinolophus bats) and 26.7 % (95 % CI: 16.45–39.89, Miniopterus bats) (there are at least 60 samples per genus in the comparison) (Fig. 2-A and Table S1). CoV prevalence varies across different bat genera, and species are shown in Fig. 2-A, Fig. 2-D, Fig. 2-E, Table S1, Table S5 and Table S7.
2.5. Analysis of CoV prevalence based on bat sex
CoV prevalence was 16.5 % (95 % CI: 12.7–20.3, 62 positives/376 samples) for female and 23.0 % (95 % CI: 18.1–27.9, 66/287) for male bats (Table S4 and Table S5). CoV prevalence was found to be associated with bat sex (Pearson χ2 (1, N = 663) = 4.424, p < 0.05) (Cramer's V = 0.082, p < 0.05, and Goodman-Kruskal tau's coefficient = 0.007, p < 0.05) (Table S6), with male bats having a higher prevalence. Rh. sinicus (28.7 %, 95 % CI: 19.79–39.59, females; 28.6 %, 95 % CI: 18.24–41.53, males) and Rh. affinis (2.1 %, 95 % CI: 0.11–12.73, females; 11.5 %, 95 % CI: 3.03–31.28, males) were found to correspond to the highest and lowest CoV prevalence, respectively, in both sexes (there are at least 23 samples per species in the comparison) (Table S4). Higher CoV prevalence was observed in males: Hipposideros (14.7 %, 95 % CI: 7.9–25.16, females; 21.7 %, 95 % CI: 11.45–36.76, males), Miniopterus (19.4 %, 8.12–38.05; 20.0 %, 6.61–44.27), Myotis (15.6 %, 9.61–24.09; 24.6 %, 17.19–33.67), Rhinolophus (16.9 %, 11.60–23.79; 20.4 %, 13.34–29.70) (Table S5) (there are at least 51 samples per genus in the comparison).
2.6. Differential prevalence of α-, β-, and unclassified-CoVs across sites and taxa
The compositions of CoVs (either α, β or unclassified CoVs) varied across all samples (Fig. 1-C, Fig. 3, Fig. S2 and Table S7). α-CoV accounted for 31.4 % (95 % CI: 23.5–39.3, 43/137) of CoV positive samples, whereas β-CoV accounted for 26.3 % (95 % CI: 18.8–33.7, 36/137) of CoV positive samples (Table S3 and Table S7). The rest of the CoV positive samples (42.3 %, 95 % CI: 34.0–50.7, 58/137) belonged to unclassified CoVs (Table S3 and Table S7). Table S3 shows the best NCBI BLASTn hit for each of the 137 CoV positive samples. Phylogenetic analysis for all 137 coronaviruses based on CoV RdRp sequences were visualized in a polar tree (Fig. 4-A and Fig. S3). Deep sequencing by MGI confirmed the presence of α-CoV (and coinfection with multiple α-CoV) and unclassified-CoV in 9 CoV positive samples as shown in Table S8.
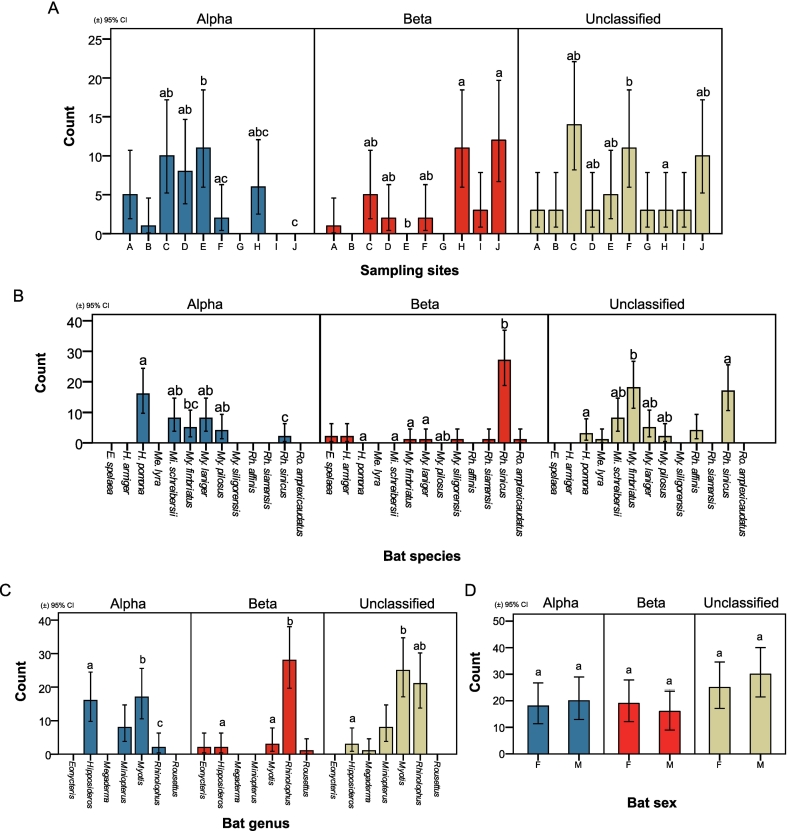
Fig. 3.
Differential prevalence of α-, β-, and unclassified-CoV and analysis of significant association across different factors. (A) Differential prevalence of CoV in different genera and significance of association with sampling site. B-D: Differential prevalence of CoV in different genera and significance of association with (B) species. (C) genera. (D) sex. Related to Fig. S2, Table S1, Table S4, Table S5, Table S7 and Table S9.
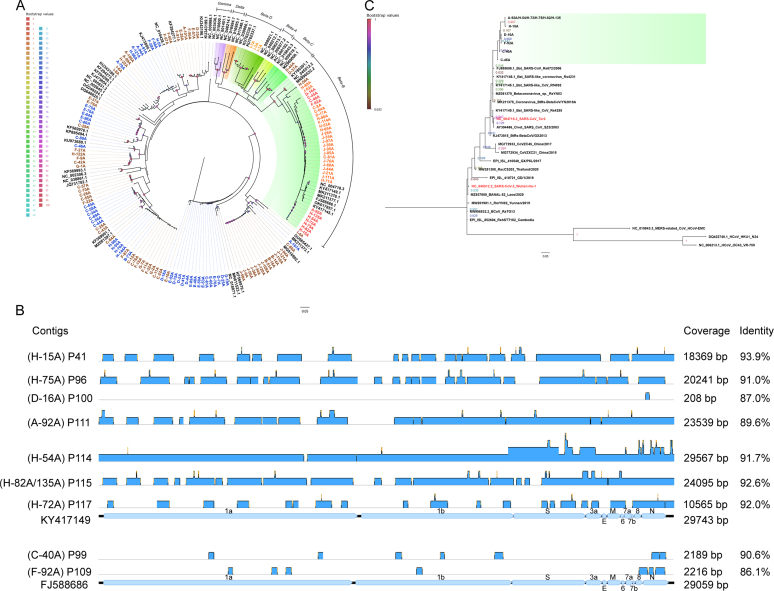
Fig. 4.
Phylogenetic analysis of targeted RdRp sequences of coronaviruses and identification of contigs in 11 SARSr-CoVs positive related libraries from NGS data. (A) Phylogenetic tree for all 137 CoV positives. (B) The blue blocks represent contigs in NGS libraries corresponding to 11 SARSr-CoV positive individuals while mapped onto their best GenBank hits (KY417149 and FJ588686, related to Table S11). (C) Phylogenetic tree for the 11 SARSr-CoVs. The green block shows RdRp sequences for the SARSr-CoVs identified in this study while the entries in red are SARS-CoV and SARS-CoV-2. (C) and (A) are both based on 195 bp targeted RdRp from Pan-CoV PCR. Seven sequences are less than 195 bp, related to Fig. S3. Phylogenetic analysis was conducted in MEGA11 using the Maximum-likelihood (ML) method, Tamura 3-parameter (TN92) model with Gamma distribution (+G) for Fig. 4-C. General Time Reversible (GTR) model with Gamma distribution (+G) and evolutionarily invariable (+I) for Fig. 4-A, which best model was selected in MEGA11 and the tree was established in QIAGEN CLC Genomics Workbench 22.0.2. The scale bar denotes the substitutions per site (0.05). Each letter number combination presents the targeted RdRp from bat rectal samples: alphacoronavirus is marked in blue, nobecoviruses are marked in light orange, sarbecoviruses are marked in red and unclassified sarbecoviruses are marked in orange, and unclassified coronavirus is marked in brown (based on the best hit from BLASTn). Others show reference sequences of coronaviruses. The different numbers below the branches indicate the support values of branch points. Node support was estimated from 1000 bootstrap replicates. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.7. Analysis of differential prevalence based on sampling site
The differential prevalences of either α-, β-, or unclassified-CoV in bats was most significantly associated with sampling site (Pearson χ2 (10, N = 115) = 46.564, p < 0.001) (Cramer's V = 0.450, p < 0.001, and Goodman-Kruskal tau's coefficient = 0.197, p < 0.001) and location (Pearson χ2 (4, N = 137) = 21.222, p < 0.001) (Cramer's V = 0.278, p < 0.001, and Goodman-Kruskal tau's coefficient = 0.069, p < 0.001) (Table S9). The northern region (KM) (6.9 %, 95 % CI: 5.11–9.28) has a higher prevalence in α-CoV than the southern region (XSBN) (0 %, 95 % CI: 0–5.21), with lower prevalence of β-CoV in KM (3.4 %, 95 % CI: 2.16–5.21) than XSBN (14.0 %, 95 % CI: 7.73–23.49) (Table S7). The highest prevalence of α-CoV in Site D (16.7 %, 95 % CI: 7.97–30.77), of β-CoV in site J (14.0 %, 7.73–23.49) and of unclassified-CoV in site C (15.7 %, 9.17–25.33) (sites with at least 45 samples are included in the comparison) (Fig. 3-A and Table S7). We also show pair-wise comparisons for differential CoV prevalence by site for pairs of sites (Fig. 3-A and Table S7).
2.8. Analysis of differential prevalence based on bat genera and species
Differential CoV prevalence was also most significantly associated with bat genus (Pearson χ2 (4, N = 117) = 56.203, p < 0.001) (Cramer's V = 0.490, p < 0.001, and Goodman-Kruskal tau's coefficient = 0.223, p < 0.001) and bat species (Pearson χ2 (10, N = 125) = 81.907, p < 0.001) (Cramer's V = 0.572, p < 0.001, and Goodman-Kruskal tau's coefficient = 0.303) (Table S9). For α-CoVs, H. pomona (16.7 %, 95 % CI: 10.12–25.96) has the highest prevalence and Rh. sinicus (1.2 %, 0.21–4.85) has the lowest prevalence (Fig. 3-B and Table S7). For β-CoVs, Rh. sinicus (16.7 %, 11.46–23.51) is significantly higher (p < 0.05), with most other species showing a prevalence of under 2 % (Fig. 3-B and Table S7).
Rhinolophus (0.7 %, 95 % CI: 0.12–2.66) has significantly lower (p < 0.05) α-CoV prevalence than Hipposideros (13.0 %, 7.85–20.56). Conversely, Rhinolophus (9.3 %, 6.39–13.34) have significantly higher (p < 0.05) β-CoV prevalence than both Myotis (1.3 %, 0.34–4.13) and Hipposideros (1.6 %, 0.28–6.34) (Fig. 3-C and Table S7).
2.9. Analysis of differential prevalence based on bat sex
Although CoVs showed a higher prevalence in male (23.0 %, 95 % CI: 18.35–28.39) than female (16.5 %, 95 % CI: 12.96–20.72) bats overall (Fig. 2-F, Table S4 and Table S5), we observed no significant difference (p < 0.05) in differential prevalence of α-, β-, and unclassified-CoV between male and female bats (Fig. 3-D and Table S9). However, it was noted, some male bats had a lower prevalence than females in Mi. schreibersii (female: 9.7 %, 95 % CI: 2.53–26.90/male: 5.0 %, 0.26–26.94) and My. laniger (10.0 %, 3.74–22.59/7.1 %, 1.86–20.55) bats of α-CoVs, as well as in H armiger (10.5 %, 1.85–34.54/0 %, 0.00–40.23) and Rh. siamensis (6.7 %, 0.35–33.97/0 %, 0.00–40.23) of β-CoV (Table S4). Conversely, male H. pomona (female: 14.3 %, 95 % CI: 6.80–26.78/male: 21.05 %, 10.1–37.78), My. pilosus (0 %, 0.00–60.42/18.2 %, 5.99–41.01) and My. fimbriatus (1.9 %, 0.10–11.18/8.2 %, 2.65–20.48) bats had a higher prevalence of α-CoV, whereas male Rh. sinicus (16.1 %, 9.39–25.87/19.0 %, 10.64–31.29) and My. laniger (0 %, 0.00–8.89/2.4 %, 0.12–14.09) had a higher prevalence of β-CoV (Table S4) (there are at least 23 samples per species in the comparison). Among female bats, H. armiger and Rh. sinicus had the highest prevalence for β-CoV (Table S4).
2.10. Beta-coronaviruses and SARSr-CoVs prevalence
Of 36 β-CoV positive samples, 3 were Nobecovirus (“Beta-D" in Fig. 4-A) and 33 were Sarbecovirus (“Beta-B" in Fig. 4-A) positive, including 22 unclassified sarbecoviruses and 11 SARSr-CoVs (Fig. 1-C and Table S3). The β-CoVs were detected in bats of the genera Eonycteris (2 samples), Hipposideros (2), Myotis (2), Rhinolophus (29) and Rousettus (1) (Table S5). The distribution according to bat species is shown in Table S4 (across sex) and Table S7 (across sites), and further genomic barcoding for bats found positive for SARSr-CoVs is shown in Table S10.
Following read assembly in NGS libraries sequenced from the 11 SARSr-CoVs positive individuals, contigs mapping to 35.5 %–99.4 % of the reference genomes (Pairwise identity: 89.6 %–93.9 %) for the corresponding SARSr-CoV references (GenBank references: KY417149.1, Bat SARS-like coronavirus Rs4255; FJ588686.1, SARS coronavirus Rs_672/2006) were found in 9 corresponding NGS libraries (Fig. 4-B and Table S11). This was with the exception of library P100 (Individual ID: D-16 A) from which a 208 bp contig (Coverage: 0.7 %, Pairwise identity: 87.0 %) was obtained (Fig. 4-B and Table S11). For libraries P111 (A-92 A), P114 (H-54 A) and P115 (H-82 A/135 A), the contigs covered 11 major open reading frames encoding ORF1a, ORF1b, S, 3a, E, M, 6, 7a, 7b, 8 and N on the two references. Notably, over 99 % of the whole genome was covered by the contigs assembled from library P114 (H-54 A) (Fig. 4-B and Table S11). Phylogenetic analysis showed that the 11 SARSr-CoVs may share a common ancestor with Bat SARS-CoV Rs672, Bat SARS-like CoV Rs 4231, Betacoronavirus RsYN03, Bat SARS-like CoV Rf4092 and BtRs-BetaCoV/YN2018 (Fig. 4-C).
2.11. Other vertebrate-infecting viruses in SARSr-CoVs positive libraries
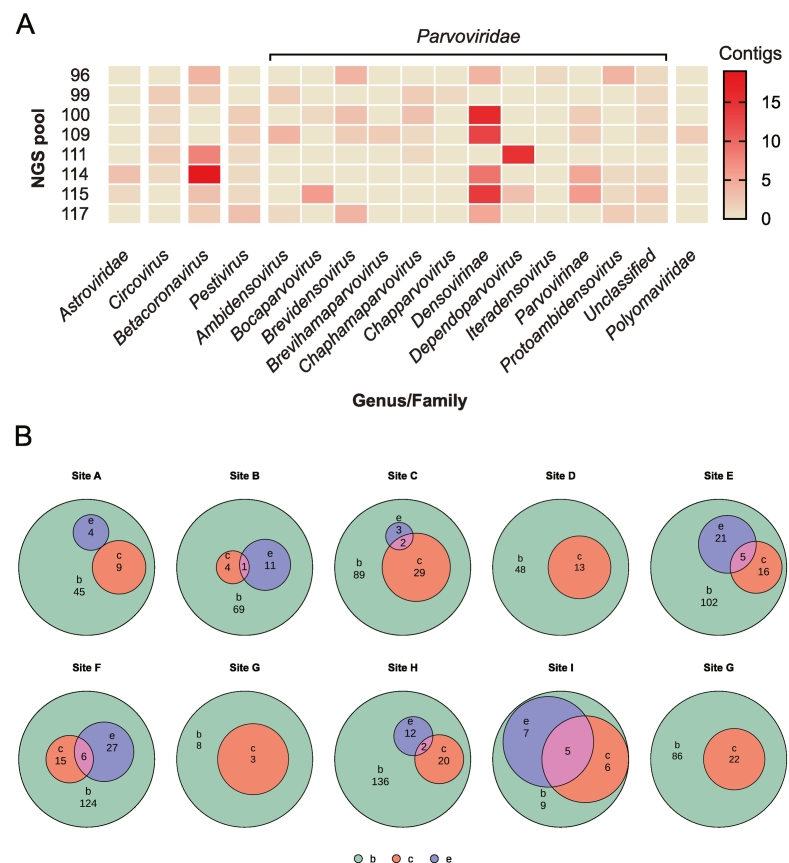
Six families of vertebrate-infecting viruses, including Astroviridae, Circoviridae, Coronaviridae, Flaviviridae, Parvoviridae, and Polyomaviridae, were detected in samples from SARSr-CoV positive individuals in horseshoe bats. In the ten libraries, most were rich in densoviruses (Family: Parvoviridae) (Fig. 5-A).
Fig. 5.
Vertebrate-infecting viruses in SARSr-CoVs positive libraries and intersection among bats with or without CoVs and bats with ectoparasites.
(A) Heat map of contigs of vertebrate-infecting viruses in the Illumina sequencing libraries including the 11 SARSr-CoV positive individuals. The virome is shown by genus, if it is unclassified, shown by the family. (B) The Euler diagram of bats with or without CoVs and bats with ectoparasites in different locations. Violet indicates the number of bats with ectoparasites (e). Red indicates the number of bats with CoVs positive (c). Green indicates the number of bats collected in each site (b). The circle was scaled by the counts. The sizes of intersection areas (pink) positively correlated with number of intersection elements. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.12. Interaction between parasitism and infection
In total, 11.6 % (83/716, 95 % CI: 9.38–14.22) of bats were parasitized (Fig. 5-B). No obvious trend was observed between parasitized bats and CoV prevalence overall (Fig. 5-B). However, CoV-positive bats with ectoparasites were distributed across sites B, C, E, H, and I. A higher proportion of bats were parasitized in the northern regions (78/707, 11.1 %, 95 % CI: 8.86–13.63) (i.e., the Kunming area) than the southern regions (5/9, 55.6 %, 22.66–84.66) (i.e., the Xishuangbanna area) of our study region (Table S12). Among parasitized bats, α-CoVs were detected in three My. pilosus and three Mi. schreibersii, and two β-CoV positive bats (E. spelaea and Me. lyra) were parasitized, whereas the rest of the parasitized bats contained unclassified CoVs(Table S12).
2.13. Understanding patterns of infection
After multinominal logistic regression modelling, multiple factors (sampling sites, species, genus and sex) appeared to have evident interaction. When the interaction effects were introduced in the model, a more fitting model was obtained (Pseudo R2: larger McFadden value >0.2, p < 0.001) than just the main effects to be considered (Table S13 and Table S14). With the interaction incorporated, the model of “Site * Sex * Species” both in CoV prevalence (McFadden value = 0.286, p < 0.001) and CoV differential distribution (in α-CoV, β-CoV and unclassified CoV) (McFadden value = 0.598, p < 0.001) produced the best model fitting.
When only considering main effects, species have a greater influence on CoV prevalence (AIC: 251.110 < 256.875; p < 0.001) than sites, whereas sites have the greatest influence on CoV differential distribution (AIC: 138.189 < 171.348, p < 0.05; 137.631 < 157.881, p < 0.01) compared to species or genus.
3. Discussion
In determining the risk of CoV spillover, the surveillance of CoV prevalence in possible reservoir hosts is essential. Bats are an important order to study due to their demonstrated ability to host CoVs [12], as well as other diverse viruses. To investigate the occurrence of CoVs in bat populations, we determined CoV prevalence by pan-CoV semi-nested PCR and high-throughput sequencing, then proceeded to evaluate its interaction with risk factors across multiple sampling sites, different host genera, species, and sex in Yunnan Province, China. This region is of particular importance due to previous identification and/or isolation of human ACE2 utilizing relatives of human infecting β-CoVs [12,13,31].
3.1. The impact of site and habitat health on pathogen prevalence
Zoonotic spillover and transmission of emergent pathogens are influenced by both biological and ecological factors [33,[48], [49], [50], [51], [52]]. The impact of these drivers is poorly characterized but may contribute to varying ecophysiological stress among different bat hosts. They may include bat species, bat sex, life history traits, or ecological factors related to their habitats. While the biological factors e.g., sex, and species are immutable, habitat quality may be disturbed. This further exacerbates physiological stress in these hosts, a factor that has been shown to increase pathogen spillover risk [53,54]. For example, Hendra virus spillover risk increases exponentially following periods of drought and habitat loss, as in conjunction these conditions stress bats, and force them into the vicinity of horses (foraging in shade trees) leading to exposure in horses [52]. Consequently, species, sex, hibernation, pregnancy, lactation, and so on as well as environmental stressors (like agricultural intensification, deforestation and climate change) are important and fundamental considerations in identifying risk factors that contribute to the initial spillover and spread of bat-borne CoVs to new hosts [48,51,[53], [54], [55]]. Greater sampling in these regions would illuminate on whether this is merely the result of a small sample size in XSBN, considering that the highest proportion of β-CoV positive bats were found in Yuxi. In addition, in phylogenetics analysis, we have noted an identical sequence in 11 SARSr-CoVs appears in site A (one Rh. cf. thomasi) and site H (four Rh. cf. thomasi and one Rh. siamensis). It indicates migration may occur in some of these bat species [31,56], and may be a factor leading to the acquisition of SARSr-CoVs within bats from site H to site A leading to a wider distribution of SARSr-CoVs.
3.2. β-CoVs hosts
SARSr-CoVs was detected in Chinese water myotis (My. laniger), Thomas's horseshoe bat (Rh. cf. thomasi) and the Thai horseshoe bat (Rh. siamensis) from Kunming city (Table S10). While SARSr-CoVs have been mainly identified in bats of the genus Rhinolophus and Hipposideros [47,[57], [58], [59]], also in Aselliscus and one member of Vespertilionidae [54,60] before, to our knowledge, it is the first identification of SARSr-CoVs in Myotis, despite previous analysis of the group [[61], [62], [63]]. This is also important as Myotis is more capable of dispersal, including over open environments than Rhinolophus, thus it has a disproportionate ability to enable the spread of pathogens across greater distances, or new regions than other known hosts [64,65]. Rf4092, which was characterized from Rh. ferrumequinum in 2012 in Kunming city of Yunnan Province, shares a more recent common ancestor with the SARSr-CoVs identified in this study than with SARS-CoV and SARS-CoV-2. It does, however, possess considerable sequence identity to SARS-CoV (similarity in the variable region is ORF8 (97.5 %), ORF3b (95.6 %) and S1 (63.3 %) and S2 (95.3 %)) [16]. Previous studies [7,66] suggested host-switching of CoVs between bats and showed ACE2 of mouse-eared bats to enable SARS-CoV and SARS-CoV-2 pseudovirus entry. There was, however, no evidence of β-CoV host switching between mouse-eared bats and horseshoe bats [7,66]. Due to the close phylogenetic proximity of Rf4092 to the SARSr-CoVs identified here, it is possible that host switching of β-CoV between Myotis and Rhinolophus bats does occur.
3.3. Co-existing CoVs
We observed the coexistence of α-CoV in individual bats (Table S8), which has also been reported previously in Rh. sinicus, Rh. affinis, Rh. thomasi and Mi. pusillus [42,60,67]. It is, however, unclear whether and how co-infection impacts specifically on spillover, evolution and recombination of CoVs [68,69]. Coexistence of different CoV species in individuals may enable recombination, which when coupled with random mutation enables the emergence of novel lineages of CoVs, further contributing to the diversity of CoVs circulating in bats [70]. Although it is difficult to determine the specific relationships between other vertebrate viruses and CoVs, densoviruses were found in most of the SARSr-CoVs positive NGS libraries (Fig. 5-A). Densoviruses are common mosquito-borne pathogens [71], and may be related to insectivorous bats as they are arthropod predators [72].
3.4. Understanding the role of parasites
Parasitism in mammals is associated with other physical symptoms of disease. While they are understudied, bat ectoparasites are often shown to contain viruses which they may transmit either actively or passively by mechanical transfer [73], and may also be an indicator of poorer health [74,75]. However, we did not find any definite overall trends between bats with ectoparasites and CoV prevalence (Fig. 5-B and Table S12) [76]. The types and incidence of ectoparasites within different species may be influential and should be considered in the future, both in terms of understanding their roles in bat health status in general, and their roles in transmitting viruses between potential hosts [77], although not as the main source of bat-borne viruses [78].
3.5. Next steps
Phylogenetic analysis of the 11 SARSr-CoV identified here based on the targeted RdRp showed them to be more closely related to SARS-CoV than to SARS-CoV-2 (Fig. 4-C). We however showed from analysis of NGS data that contigs in 10/11 SARSr-CoV samples covered from 35.5 % to 99.4 % of the best-hit reference. Isolation of these viruses would enable assessment of their capacity to infect humans, or other mammals through ACE2 receptor binding assays [13,69]. It will also be important to derive the full-length genome of the SARSr-CoV identified in the My. laniger, as it will shed light on the evolution, mutation and recombination of CoVs in nature, which is key to understanding its incidence in the new host.
Dynamics of pathogenic infection in hosts may modulate spillover risk and surveillance of other specimen types e.g., plasma, bat guano or urine may offer a supplementary method to assess the risk of spillover. Seasonal trends exist in coronavirus prevalence when studies have been conducted longitudinally [79,80], and there is extreme variation (0 %–80 %) in the shedding of bat-borne CoVs through various phases of the breeding season [81,82]. Surveillance of CoV prevalence in bats with seasonal changes and longitudinally through successive life cycle stages should be prioritized in future research, especially with different patterns of roosting, ecophysiological stressors (such as breeding) and hibernation. While bat age or maturity were not extensively analyzed in this study, these are important considerations for future studies. A longer period of sampling through successive seasons, capturing bats from different age groups over a greater geographical area than Yunnan province, would be beneficial to assessment of spillover risk within bat communities concerning diversity [83]. Multiple ecological, biological and social factors simultaneously influence coronavirus spillover risk in nature [33,[84], [85], [86], [87]]. Therefore, future analysis and modelling of coronavirus prevalence and spillover risk should incorporate these internal and external factors on the background of a much larger sample size.
4. Conclusion
This research takes us a step closer to understanding the dynamics of CoVs in Asian bats. Not only did we find a broader range of bat hosts of SARSr-CoVs, with the identification in Myotis laniger, but we confirmed previous findings by others on the ability of bats to host more than a single CoV concurrently [42,60,67], providing the potential for recombination and the generation of further novel CoVs. Furthermore, we found that within and between species and sites as well as sex, there was a considerable ability for bats to host different CoVs, highlighting the need for more work to understand the mechanisms behind CoV transmission and evolution between bats, and how this varies across space and time. Although our samples come only from Yunnan province and cover a limited period, we find a diversity of CoVs and the prevalence at this specific location and time, reinforcing the need for further sustained work across the region. This work also reiterates the potential for the emergence of novel CoVs from bats, due to the interface between bats and animals with the potential to encounter humans.
5. Materials and methods
5.1. Ethical approval and sample collection
The ethical approval for bat sample collection in Yunnan Province was provided by the Ethics Committee of Life Sciences, Kunming Institute of Zoology (KIZ), Chinese Academy of Sciences (CAS). The ethical approval number is SMKX-20200210-01. Sampling approval was provided from the provincial office in Jinghong, Xishuangbanna Nature Reserve office as well as from XTBG.
Rectal swabs were collected from bats in 10 different caves in Kunming (KM) City (sites: A-H), Yuxi (YX) City (site: J) and Xishuangbanna (XSBN) Prefecture (site: I) in Yunnan Province, China, from September to November 2020. Further details of samples collection are provided in Supplementary Method 1.
5.2. Nucleic acid extraction from bat rectal swabs
The samples were retrieved from −80 °C, thawed at 4 °C, vortexed for 3–5 min and centrifuged at 17000 ×g, 4 °C for 3 min. The supernatant was transferred into a 0.45 μM filter microtube (Corning, NY, USA), then centrifuged at 15000 ×g, 4 °C for 1 min. The flow-through was used for nucleic acid purification. Nucleic acids were extracted using the GeneJet Viral DNA and RNA Purification Kit (Thermo Fisher Scientific, MA, USA) and host depletion done using enzyme cocktail digestion [88,89]. Further details on extraction are in Supplementary Method 2.
5.3. cDNA synthesis and pan-CoV semi-nested PCR detection
cDNA synthesis was performed with TAKARA PrimerScript™ 1st Strand cDNA Synthesis Kit (TAKARA, Dalian, China), following manufacturer instructions. The pan-CoV semi-nested PCR detection consisted of two sets of PCR, the pan-CoV outer PCR and the pan-CoV inner PCR. The primers (synthesized by Tsingke, Beijing, China) are described in Supplementary Method 3. Further details of cDNA synthesis and PCR are in Supplementary Method 4. After PCR, the results were observed by DNA agarose gel electrophoresis and checked by Sanger sequencing, for which details are given in Supplementary Method 5. Further details of identity verification of bats with SARSr-CoVs after detection are described in Supplementary Method 6.
5.4. BLAST and analysis of coronavirus prevalence and diversity
The SnapGene software was used to analyze the Sanger sequencing results, by aligning the forward and reverse sequences and electropherogram analysis. After removing the primer sequences, BLASTn was performed on NCBI for all sequences successively. CoVs-positive individuals were identified based on query of targeted RdRp sequences using BLASTn and the best hit virus species were retrieved for subsequent analyses. TaxonKit was used to obtain lineage information for virus species. The classified coronaviruses (α-CoV, β-CoV and unclassified CoV) by the names corresponding to their NCBI Taxonomy IDs. CoV positive and negative individuals were sorted by different bat species and genera as well as sampling sites in Microsoft Excel (2016). The overlap between the number of bats with ectoparasites and the number of bats with or without CoVs were analyzed and displayed in Euler diagrams by Evenn [90].
5.5. Statistical analysis
Statistical analysis was performed in SPSS (version 24.0). The count for each category as a frequency variate was weighted before descriptive statistics. Crosstabs analysis was used for Chi-square tests. Goodman-Kruskal's lambda and Cramer's V coefficient were measured for association analysis. Bonferroni method was chosen to adjust p-values. Different letters (a, b and c in Fig. 2 and Fig. 3) indicate statistically significant differences below the p-value = 0.05 threshold. Multinominal (Polytomous) logistic regression analysis is used to depict multi-effects of bats in CoV prevalence status (main effects and full factorial modelling). Confidence intervals (95 % CI) were calculated by SPSS and VassarStats online.
5.6. Library preparation for Next generation sequencing (NGS)
Illumina sequencing: Whole transcriptome amplification was performed before library preparation. The WTA2 whole transcriptome amplification (WTA) kit (Sigma-Aldrich, MA, USA) was used for reverse transcription and cDNA synthesis based on the protocol supplied. NGS libraries were prepared by NEBNext® Ultra™ II DNA Library Prep Kit for Illumina (NEB, MA, USA) following manufacturer instructions and sequencing was conducted by NovaSeq 6000 (PE150) (Novogene, Shanghai, China).
MGI sequencing: The amplification product of pan-CoV semi-nested PCR was purified and concentrated by the Monarch PCR & DNA Cleanup Kit (NEB), according to the protocol supplied. The libraries were prepared using the MGIEasy FS DNA Library Prep kit (BGI, Shenzhen, China) following manufacturer instructions. The library was sent to BGI for library cyclization and sequencing, and the MGISEQ-2000 > PE150 was requested.
5.7. Bioinformatic analysis on NGS data
All NGS data were deposited to the open access database NMDC, China National Microbiology Data Center (https://nmdc.cn/en) (Table S15). Illumina libraries were prepared from samples found positives in the SARSr-CoVs detection. All reads were trimmed by Trimgalore and filtered based on the SILVA database. Contigs were assembled from the reads using SPAdes. Local blastn (Blast+ suite) was utilized to obtain matches in a local blast database consisting of all 2865 CoV sequences initially employed for Pan-CoV PCR primers design using the assembled contigs as queries (contigs in each library). The detailed workflow for this section of the analysis is compiled in Snakemake [91] and may be found at https://github.com/aatendu/Covs_differential_prevalence_study_yunnan/blob/main/Snakefile. Contig mapping was performed in Geneious R9.
NGS data from Illumina was also processed for the detection of vertebrate-infecting viruses [92]: After read trimming by Trimmomatic, clean reads were aligned to a local database of genomic and transcriptional sequences of bats with Bowtie2. The reads that did not align against the bat sequences were aligned to the human genome. After removing bat and human sequences, de novo assembly was performed with the remaining reads using the MEGAHIT software. The contigs were clustered with CD-HIT-EST software requiring 99 % identity and 100 % coverage for the shortest sequence. The representative sequences of each cluster were aligned with BLASTN against the Reference Viral Database (RVDB) version 22. The best hit was selected according to e-value, identity, and coverage, in this order of importance. The taxonomic classification of the contigs was assigned according to the taxonomy of the best hit. In parallel, the MEGAN blast2lca tool determined the taxonomy of the contigs according to the lowest common ancestor (LCA) criteria. This information was combined by manual curation and the taxonomies for vertebrate-infecting viruses obtained. The contigs were re-grouped according to the target sequence of the best hit and realigned with the LASTZ tool. From this realignment, the coverage was estimated with Samtools. The contigs were also aligned with Diamond software (The pipeline was implemented by Dr. Alix Armero). The heatmap for vertebrate-host virus was drawn by Graphpad.
MGI sequencing data was utilized for verification of CoV co-infection. Analysis of the amplicon obtained after performing a nested PCR of targeted RdRp region of the CoV genomes consisted of two steps: 1) to estimate the diversity of existing CoVs sequences by the algorithm Emu [93] in conjunction with a fresh homemade genomic CoVs database (accession numbers for the sequences that comprise the genomic CoVs database are provided in an accompanying .txt file as supporting Information). The local CoVs database consists of CoV genome sequences downloaded from GenBank [94] and Virus Pathogen Resource (ViPR) [95] and duplicated sequences were removed; 2) generate the amplification sequence of RdRp regions of CoVs in each sample using an in-house script relying on Minimap2 [96] and alignment produced by Emu in the first step.
5.8. Phylogenetic analysis
The phylogenetic analysis was performed based on the 195 bp Sanger sequences obtained from the amplicons from Pan-CoV semi-nested PCR targeting the RdRp sequences of SARSr-CoVs. The same 195 bp RdRp region of reference sequences was also used in the phylogenetic tree. α-CoVs were designated as the outgroups of branches. All of these DNA sequences were aligned by ClustalW algorithm [97]. Models with the lowest BIC scores (Bayesian Information Criterion) [98] are considered to best describe the substitution pattern. The alignment and the best model selection was conducted in MEGA11 [99]. The phylogenetic tree was performed by MEGA11 and QIAGEN CLC Genomics Workbench 22.0.2. The node support was estimated from 1000 bootstrap replicates [100].
CRediT authorship contribution statement
Ruiya Li: Writing – review & editing, Writing – original draft, Visualization, Validation, Project administration, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Alexander Tendu: Writing – review & editing, Methodology, Formal analysis, Data curation. Yakhouba Kane: Writing – review & editing, Methodology, Data curation. Victor Omondi: Data curation. Jiaxu Ying: Data curation. Lingjing Mao: Data curation. Shiman Xu: Data curation. Rong Xu: Methodology, Data curation. Xing Chen: Resources, Methodology, Data curation. Yanhua Chen: Resources, Methodology, Data curation. Stéphane Descorps-Declère: Visualization, Methodology, Formal analysis, Data curation. Kathrina Mae Bienes: Methodology, Data curation. Meriem Fassatoui: Methodology, Data curation. Alice C. Hughes: Writing – review & editing, Supervision, Resources, Methodology, Investigation, Data curation, Conceptualization. Nicolas Berthet: Writing – review & editing, Supervision, Project administration, Methodology, Funding acquisition, Formal analysis, Data curation, Conceptualization. Gary Wong: Writing – review & editing, Validation, Supervision, Resources, Methodology, Funding acquisition, Formal analysis, Conceptualization.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgments
We thank Canping Huang for participating in field sampling, and design of pan-CoV primers. We thank Alix Armero for preliminary analysis of the NGS data. We also thank Shengjie Shao, Jialei Chen, Jihao Li for supporting sample collection. We gratefully acknowledge all researchers for sharing their genome data on GISAID. This project was supported by the Ministry of Science and Technology of China (grant no. 2021YFC0863400, 2022YFE0114700), the Alliance of International Scientific Organizations (grant no. ANSO-CR-SP-2020-02), the Shanghai Municipal Science and Technology Major Project (grant no. 2019SHZDZX02), G4 funding from Institut Pasteur, Fondation Merieux and Chinese Academy of Sciences to G.W., and the International Affairs Department of the Institut Pasteur of Paris. A.T. was supported by the ANSO Scholarship for Young Talents. Y.K. was supported by the CAS-TWAS Fellowship for International Doctoral Students.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.onehlt.2024.100923.
Contributor Information
Nicolas Berthet, Email: nicolas.berthet@pasteur.fr.
Gary Wong, Email: gckwong@pasteur-kh.org.
Appendix A. Supplementary data
Supplementary material 1
Supplementary material 2
Supplementary material 3
Supplementary material 4
Supplementary material 5
Data availability
I have shared the link to my data.
References
- 1.Boni M.F., Lemey P., Jiang X. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat. Microbiol. 2020;5(11):1408–1417. doi: 10.1038/s41564-020-0771-4. [DOI] [PubMed] [Google Scholar]
- 2.Memish Z.A., Perlman S., Van Kerkhove M.D. Middle East respiratory syndrome. Lancet. 2020;395(10229):1063–1077. doi: 10.1016/s0140-6736(19)33221-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guan Y., Zheng B.J., He Y.Q. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302(5643):276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 4.Wang Q., Chen H., Shi Y. Tracing the origins of SARS-CoV-2: lessons learned from the past. Cell Res. 2021;31(11):1139–1141. doi: 10.1038/s41422-021-00575-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holmes E.C., Goldstein S.A., Rasmussen A.L. The origins of SARS-CoV-2: A critical review. Cell. 2021;184(19):4848–4856. doi: 10.1016/j.cell.2021.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Worobey M., Levy J.I., Malpica Serrano L. The Huanan seafood wholesale market in Wuhan was the early epicenter of the COVID-19 pandemic. Science. 2022;377(6609):951–959. doi: 10.1126/science.abp8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan H., Jiao H., Liu Q. ACE2 receptor usage reveals variation in susceptibility to SARS-CoV and SARS-CoV-2 infection among bat species. Nat Ecol Evol. 2021;5(5):600–608. doi: 10.1038/s41559-021-01407-1. [DOI] [PubMed] [Google Scholar]
- 8.Walker P.J., Siddell S.G., Lefkowitz E.J. Changes to virus taxonomy and to the international code of virus classification and nomenclature ratified by the international committee on taxonomy of viruses (2021) Arch. Virol. 2021;166(9):2633–2648. doi: 10.1007/s00705-021-05156-1. [DOI] [PubMed] [Google Scholar]
- 9.Drosten C., Günther S., Preiser W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 10.Booth C.M., Matukas L.M., Tomlinson G.A. Clinical features and short-term outcomes of 144 patients with SARS in the greater Toronto area. Jama. 2003;289(21):2801–2809. doi: 10.1001/jama.289.21.JOC30885. [DOI] [PubMed] [Google Scholar]
- 11.Zhong N.S., Zheng B.J., Li Y.M. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet. 2003;362(9393):1353–1358. doi: 10.1016/s0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou P., Yang X.L., Wang X.G. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ge X.Y., Li J.L., Yang X.L. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503(7477):535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Menachery V.D., Yount B.L., Jr., Debbink K. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med. 2015;21(12):1508–1513. doi: 10.1038/nm.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang X.-L., Hu B., Wang B. Isolation and characterization of a novel bat coronavirus closely related to the direct progenitor of severe acute respiratory syndrome coronavirus. J. Virol. 2015;90(6):3253–3256. doi: 10.1128/JVI.02582-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu B., Zeng L.P., Yang X.L. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog. 2017;13(11) doi: 10.1371/journal.ppat.1006698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.K.K. To, Sridhar S., Chiu K.H. Lessons learned 1 year after SARS-CoV-2 emergence leading to COVID-19 pandemic. Emerg Microbes Infect. 2021;10(1):507–535. doi: 10.1080/22221751.2021.1898291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abdelghany T.M., Ganash M., Bakri M.M. SARS-CoV-2, the other face to SARS-CoV and MERS-CoV: future predictions. Biom. J. 2021;44(1):86–93. doi: 10.1016/j.bj.2020.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.AllenM M.B. Mirsaeidi health and economy in COVID-19 era: A plan for reconstituting long-term economic security. Front. Public Health. 2020;8:235. doi: 10.3389/fpubh.2020.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akter R., Rahman M.H., Bhattacharya T. Novel coronavirus pathogen in humans and animals: an overview on its social impact, economic impact, and potential treatments. Environ. Sci. Pollut. Res. Int. 2021;28(48):68071–68089. doi: 10.1007/s11356-021-16809-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirtipal N., S. BharadwajS.G. Kang from SARS to SARS-CoV-2, insights on structure, pathogenicity and immunity aspects of pandemic human coronaviruses. Infect. Genet. Evol. 2020;85 doi: 10.1016/j.meegid.2020.104502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang X.C., Zhang J.X., Zhang S.Y. Prevalence and genetic diversity of coronaviruses in bats from China. J. Virol. 2006;80(15):7481–7490. doi: 10.1128/jvi.00697-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang W., Lin X.D., Liao Y. Discovery of a highly divergent coronavirus in the Asian house shrew from China illuminates the origin of the Alphacoronaviruses. J. Virol. 2017;91(17) doi: 10.1128/jvi.00764-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oreshkova N., Molenaar R.J., Vreman S. SARS-CoV-2 infection in farmed minks, the Netherlands, April and may 2020. Euro Surveill. 2020;25(23) doi: 10.2807/1560-7917.Es.2020.25.23.2001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X., Wang L., Liu P. A novel potentially recombinant rodent coronavirus with a polybasic cleavage site in the spike protein. J. Virol. 2021;95(22) doi: 10.1128/jvi.01173-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.SnoeckS C.J. Zohari detection and discovery of coronaviruses in wild bird populations. Methods Mol. Biol. 2020;2203:41–53. doi: 10.1007/978-1-0716-0900-2_3. [DOI] [PubMed] [Google Scholar]
- 27.C.H. CunninghamH.O. Stuart cultivation of the virus of infectious bronchitis of chickens in embryonated chicken eggs. Am. J. Vet. Res. 1947;8(27):209–212. [PubMed] [Google Scholar]
- 28.Irving A.T., Ahn M., Goh G. Lessons from the host defences of bats, a unique viral reservoir. Nature. 2021;589(7842):363–370. doi: 10.1038/s41586-020-03128-0. [DOI] [PubMed] [Google Scholar]
- 29.Li W., Shi Z., Yu M. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310(5748):676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 30.Lau S.K., Woo P.C., Li K.S. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA. 2005;102(39):14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou H., Ji J., Chen X. Identification of novel bat coronaviruses sheds light on the evolutionary origins of SARS-CoV-2 and related viruses. Cell. 2021;184(17):4380–4391.e14. doi: 10.1016/j.cell.2021.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gozalo A.S., T.S. ClarkD.M. Kurtz coronaviruses: troubling crown of the animal kingdom. Comp. Med. 2023;73(1):6–44. doi: 10.30802/aalas-cm-21-000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muylaert R.L., Wilkinson D.A., Kingston T. Using drivers and transmission pathways to identify SARS-like coronavirus spillover risk hotspots. Nat. Commun. 2023;14(1):6854. doi: 10.1038/s41467-023-42627-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Z., Huang G., Huang M. Global patterns of phylogenetic diversity and transmission of bat coronavirus. Sci. China Life Sci. 2023;66(4):861–874. doi: 10.1007/s11427-022-2221-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Brakel M., HambreyS J. 2011. Bunting, Regional Case Study: R6 Mekong – Inland Fisheries and Aquaculture. [Google Scholar]
- 36.Chornelia A., J. LuA.C. Hughes how to accurately delineate morphologically conserved taxa and diagnose their phenotypic disparities: species delimitation in cryptic Rhinolophidae (Chiroptera) Front. Ecol. Evol. 2022:10. doi: 10.3389/fevo.2022.854509. [DOI] [Google Scholar]
- 37.Richter C.H., Custer B., Steele J.A. Intensified food production and correlated risks to human health in the greater Mekong subregion: a systematic review. Environ. Health. 2015;14:43. doi: 10.1186/s12940-015-0033-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Namkhan M., Gale G.A., Savini T. Loss and vulnerability of lowland forests in mainland Southeast Asia. Conserv. Biol. 2021;35(1):206–215. doi: 10.1111/cobi.13538. [DOI] [PubMed] [Google Scholar]
- 39.Delaune D., Hul V., Karlsson E.A. A novel SARS-CoV-2 related coronavirus in bats from Cambodia. Nat. Commun. 2021;12(1):6563. doi: 10.1038/s41467-021-26809-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wacharapluesadee S., Duengkae P., Rodpan A. Diversity of coronavirus in bats from eastern Thailand. Virol. J. 2015;12:57. doi: 10.1186/s12985-015-0289-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou H., Chen X., Hu T. A novel bat coronavirus closely related to SARS-CoV-2 contains natural insertions at the S1/S2 cleavage site of the spike protein. Curr. Biol. 2020;30(11):2196–2203.e3. doi: 10.1016/j.cub.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ge X.Y., Wang N., Zhang W. Coexistence of multiple coronaviruses in several bat colonies in an abandoned mineshaft. Virol. Sin. 2016;31(1):31–40. doi: 10.1007/s12250-016-3713-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li L.L., Wang J.L., Ma X.H. A novel SARS-CoV-2 related coronavirus with complex recombination isolated from bats in Yunnan province, China. Emerg Microbes Infect. 2021;10(1):1683–1690. doi: 10.1080/22221751.2021.1964925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valitutto M.T., Aung O., Tun K.Y.N. Detection of novel coronaviruses in bats in Myanmar. PLoS One. 2020;15(4) doi: 10.1371/journal.pone.0230802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wacharapluesadee S., Tan C.W., Maneeorn P. Evidence for SARS-CoV-2 related coronaviruses circulating in bats and pangolins in Southeast Asia. Nat. Commun. 2021;12(1):972. doi: 10.1038/s41467-021-21240-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Temmam S., Vongphayloth K., Baquero E. Bat coronaviruses related to SARS-CoV-2 and infectious for human cells. Nature. 2022;604(7905):330–336. doi: 10.1038/s41586-022-04532-4. [DOI] [PubMed] [Google Scholar]
- 47.Fan Y., Zhao K., Shi Z.L. Bat Coronaviruses in China. Viruses. 2019;11:3. doi: 10.3390/v11030210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Plowright R.K., Sokolow S.H., Gorman M.E. Causal inference in disease ecology: investigating ecological drivers of disease emergence. Front. Ecol. Environ. 2008;6(8):420–429. doi: 10.1890/070086. [DOI] [Google Scholar]
- 49.Altizer S., R. BartelB.A. Han animal migration and infectious disease risk. Science. 2011;331(6015):296–302. doi: 10.1126/science.1194694. [DOI] [PubMed] [Google Scholar]
- 50.Ruiz-Aravena M., McKee C., Gamble A. Ecology, evolution and spillover of coronaviruses from bats. Nat. Rev. Microbiol. 2022;20(5):299–314. doi: 10.1038/s41579-021-00652-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carlson C.J., Albery G.F., Merow C. Climate change increases cross-species viral transmission risk. Nature. 2022;607(7919):555–562. doi: 10.1038/s41586-022-04788-w. [DOI] [PubMed] [Google Scholar]
- 52.Eby P., Peel A.J., Hoegh A. Pathogen spillover driven by rapid changes in bat ecology. Nature. 2023;613(7943):340–344. doi: 10.1038/s41586-022-05506-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Afelt A., Lacroix A., Zawadzka-Pawlewska U. Distribution of bat-borne viruses and environment patterns. Infect. Genet. Evol. 2018;58:181–191. doi: 10.1016/j.meegid.2017.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sánchez C.A., Li H., Phelps K.L. A strategy to assess spillover risk of bat SARS-related coronaviruses in Southeast Asia. Nat. Commun. 2022;13(1):4380. doi: 10.1038/s41467-022-31860-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grange Z.L., Goldstein T., Johnson C.K. Ranking the risk of animal-to-human spillover for newly discovered viruses. Proc. Natl. Acad. Sci. USA. 2021;118(15) doi: 10.1073/pnas.2002324118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Y.Y. YouJ.F. Du [A long distance colonization event of Chinese endemic bat Myotis davidii] Ying Yong Sheng Tai Xue Bao. 2011;22(3):773–778. [PubMed] [Google Scholar]
- 57.Wang L.F., Shi Z., Zhang S. Review of bats and SARS. Emerg. Infect. Dis. 2006;12(12):1834–1840. doi: 10.3201/eid1212.060401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.L.F. WangB.T. Eaton bats, civets and the emergence of SARS. Curr. Top. Microbiol. Immunol. 2007;315:325–344. doi: 10.1007/978-3-540-70962-6_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yuan J., Hon C.C., Li Y. Intraspecies diversity of SARS-like coronaviruses in Rhinolophus sinicus and its implications for the origin of SARS coronaviruses in humans. J. Gen. Virol. 2010;91(Pt 4):1058–1062. doi: 10.1099/vir.0.016378-0. [DOI] [PubMed] [Google Scholar]
- 60.Si H.R., Wu K., Su J. Individual virome analysis reveals the general co-infection of mammal-associated viruses with SARS-related coronaviruses in bats. Virol. Sin. 2024;39(4):565–573. doi: 10.1016/j.virs.2024.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lacroix A., Duong V., Hul V. Genetic diversity of coronaviruses in bats in Lao PDR and Cambodia. Infect. Genet. Evol. 2017;48:10–18. doi: 10.1016/j.meegid.2016.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lo V.T., Yoon S.W., Noh J.Y. Long-term surveillance of bat coronaviruses in Korea: diversity and distribution pattern. Transbound. Emerg. Dis. 2020;67(6):2839–2848. doi: 10.1111/tbed.13653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Caraballo D.A., Sabio M.S., Colombo V.C. The role of Molossidae and Vespertilionidae in shaping the diversity of Alphacoronaviruses in the Americas. Microbiol Spectr. 2022;10(6) doi: 10.1128/spectrum.03143-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moussy C., Hosken D.J., Mathews F. Migration and dispersal patterns of bats and their influence on genetic structure. Mammal Rev. 2013;43(3):183–195. doi: 10.1111/j.1365-2907.2012.00218.x. [DOI] [Google Scholar]
- 65.Liu T., Jia J., Liu L. New insights into the taxonomy of Myotis bats in China based on morphology and multilocus phylogeny. Diversity. 2023;15(7):805. [Google Scholar]
- 66.Latinne A., Hu B., Olival K.J. Origin and cross-species transmission of bat coronaviruses in China. Nat. Commun. 2020;11(1):4235. doi: 10.1038/s41467-020-17687-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 67.Chu D.K.W., Peiris J.S.M., Chen H. Genomic characterizations of bat coronaviruses (1A, 1B and HKU8) and evidence for co-infections in Miniopterus bats. J. Gen. Virol. 2008;89(Pt 5):1282–1287. doi: 10.1099/vir.0.83605-0. [DOI] [PubMed] [Google Scholar]
- 68.Jones B.D., E.J. KaufmanA.J. Peel viral co-infection in bats: A systematic review. Viruses. 2023;15(9) doi: 10.3390/v15091860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang J., Pan Y.F., Yang L.F. Individual bat virome analysis reveals co-infection and spillover among bats and virus zoonotic potential. Nat. Commun. 2023;14(1):4079. doi: 10.1038/s41467-023-39835-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Su S., Wong G., Shi W. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24(6):490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.R.M. JohnsonJ.L. Rasgon Densonucleosis viruses ('densoviruses') for mosquito and pathogen control. Curr Opin. Insect Sci. 2018;28:90–97. doi: 10.1016/j.cois.2018.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang Q., Feng J., Wu H. Insectivorous bats provide more pest suppression services than disservices - a case study in China. Biol. Control. 2024;188 doi: 10.1016/j.biocontrol.2023.105435. [DOI] [Google Scholar]
- 73.Tendu A., Hughes A.C., Berthet N. Viral Hyperparasitism in bat Ectoparasites: implications for pathogen maintenance and transmission. Microorganisms. 2022;10(6) doi: 10.3390/microorganisms10061230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Giorgi M.S., Arlettaz R., Christe P. The energetic grooming costs imposed by a parasitic mite (Spinturnix myoti) upon its bat host (Myotis myotis) Proc. Biol. Sci. 2001;268(1480):2071–2075. doi: 10.1098/rspb.2001.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.PostawaZ T. Nagy variation of parasitism patterns in bats during hibernation: the effect of host species, resources, health status, and hibernation period. Parasitol. Res. 2016;115(10):3767–3778. doi: 10.1007/s00436-016-5138-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Szentiványi T., Markotter W., Dietrich M. Host conservation through their parasites: molecular surveillance of vector-borne microorganisms in bats using ectoparasitic bat flies. Parasite. 2020;27:72. doi: 10.1051/parasite/2020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gay N., Olival K.J., Bumrungsri S. Parasite and viral species richness of southeast Asian bats: fragmentation of area distribution matters. Int J Parasitol Parasites Wildl. 2014;3(2):161–170. doi: 10.1016/j.ijppaw.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tendu A., Li R., Kane Y. Viromes of arthropod parasites and their hosts: the case of bats and bat ectoparasites. Acta Trop. 2024;259 doi: 10.1016/j.actatropica.2024.107375. [DOI] [PubMed] [Google Scholar]
- 79.Cappelle J., Furey N., Hoem T. Longitudinal monitoring in Cambodia suggests higher circulation of alpha and betacoronaviruses in juvenile and immature bats of three species. Sci. Rep. 2021;11(1):24145. doi: 10.1038/s41598-021-03169-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wacharapluesadee S., Duengkae P., Chaiyes A. Longitudinal study of age-specific pattern of coronavirus infection in Lyle's flying fox (Pteropus lylei) in Thailand. Virol. J. 2018;15(1):38. doi: 10.1186/s12985-018-0950-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Joffrin L., Hoarau A.O.G., Lagadec E. Seasonality of coronavirus shedding in tropical bats. R. Soc. Open Sci. 2022;9(2) doi: 10.1098/rsos.211600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Montecino-Latorre D., Goldstein T., Kelly T.R. Seasonal shedding of coronavirus by straw-colored fruit bats at urban roosts in Africa. PLoS One. 2022;17(9) doi: 10.1371/journal.pone.0274490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Meyer M., Melville D.W., Baldwin H.J. Bat species assemblage predicts coronavirus prevalence. Nat. Commun. 2024;15(1):2887. doi: 10.1038/s41467-024-46979-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Plowright R.K., Parrish C.R., McCallum H. Pathways to zoonotic spillover. Nat. Rev. Microbiol. 2017;15(8):502–510. doi: 10.1038/nrmicro.2017.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wood J.L., Leach M., Waldman L. A framework for the study of zoonotic disease emergence and its drivers: spillover of bat pathogens as a case study. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2012;367(1604):2881–2892. doi: 10.1098/rstb.2012.0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Clifford Astbury C., Lee K.M., McLeod R. Policies to prevent zoonotic spillover: a systematic scoping review of evaluative evidence. Glob. Health. 2023;19(1):82. doi: 10.1186/s12992-023-00986-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lau S.K., Li K.S., Huang Y. Ecoepidemiology and complete genome comparison of different strains of severe acute respiratory syndrome-related Rhinolophus bat coronavirus in China reveal bats as a reservoir for acute, self-limiting infection that allows recombination events. J. Virol. 2010;84(6):2808–2819. doi: 10.1128/jvi.02219-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wu Z., Ren X., Yang L. Virome analysis for identification of novel mammalian viruses in bat species from Chinese provinces. J. Virol. 2012;86(20):10999–11012. doi: 10.1128/jvi.01394-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang J., Yang F., Ren L. Unbiased parallel detection of viral pathogens in clinical samples by use of a metagenomic approach. J. Clin. Microbiol. 2011;49(10):3463–3469. doi: 10.1128/JCM.00273-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang M., Chen T., Liu Y.X. Visualizing set relationships: EVenn's comprehensive approach to Venn diagrams. Imeta. 2024;3(3) doi: 10.1002/imt2.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mölder F., Jablonski K.P., Letcher B. Sustainable data analysis with Snakemake. F1000Res. 2021;10:33. doi: 10.12688/f1000research.29032.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Armero A., Li R., Bienes K.M. Myotis fimbriatus Virome, a window to virus diversity and evolution in the genus Myotis. Viruses. 2022;14(9) doi: 10.3390/v14091899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Curry K., Wang Q., Nute M. 2021. Emu: Species-Level Microbial Community Profiling for Full-Length Nanopore 16S Reads. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Benson D.A., Cavanaugh M., Clark K. GenBank. Nucleic Acids Res. 2013;41(Database issue):D36–D42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pickett B.E., Sadat E.L., Zhang Y. ViPR: an open bioinformatics database and analysis resource for virology research. Nucleic Acids Res. 2012;40(Database issue):D593–D598. doi: 10.1093/nar/gkr859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.H. Li Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34(18):3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Thompson J.D., Gibson T.J., Higgins D.G. 2002. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics. Chapter 2: p. Unit 2.3. [DOI] [PubMed] [Google Scholar]
- 98.G. Schwarz estimating the dimension of a model. Ann. Stat. 1978;6(2):461–464. [Google Scholar]
- 99.Tamura K., StecherS G. Kumar MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021;38(7):3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.J. Felsenstein confidence Limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39(4):783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2
Supplementary material 3
Supplementary material 4
Supplementary material 5
Data Availability Statement
I have shared the link to my data.