ABSTRACT
Recurrent respiratory papillomatosis (RRP) is caused by human papillomaviruses (HPV) 6 and 11, but the role of their genomic variants in the disease's clinical course is unclear. This study investigated whether long‐term persistence of a particular HPV genotype, subtype or genomic variant influences the RRP clinical course. HPV genotyping was performed in paired baseline and follow‐up RRP laryngeal tissue specimens of 59 patients. HPV6 and HPV11 genomic variants were determined in paired tissue specimens taken at least 10 years apart in 20 selected patients. HPV was identified in 58/59 patients, most commonly HPV6 (40/58), followed by HPV11 (17/58). The most prevalent HPV genomic variant was HPV11 A2. HPV6 A and HPV6 B1 were most frequent in aggressive RRP. In all patients, identical HPV genomic variants were identified in both paired specimens. RRP results from a long‐term infection with the same HPV genomic variant that can be identified decades after disease onset. We report the longest duration of genetically confirmed persistent HPV infection in peer‐reviewed literature, during a 44‐year interval in a patient with HPB6 B1. This study suggests that infection with a particular HPV genotype, subtype, or genomic variant does not significantly influence the clinical course of RRP.
Keywords: genomic variant, HPV, human papillomaviruses, laryngeal papilloma, persistence, recurrent respiratory papillomatosis
Abbreviations
- CI
confidence interval
- HPV
human papillomaviruses
- IQR
interquartile range
- RRP
recurrent respiratory papillomatosis
- SD
standard deviation
1. Introduction
Recurrent respiratory papillomatosis (RRP) is a rare respiratory tract disease that mainly affects the larynx and clinically presents as benign squamous cell papillomas, which are caused by low‐risk human papillomaviruses (HPV), mainly HPV6 and HPV11 [1]. Based on the patient's age at disease presentation, juvenile‐ and adult‐onset RRP are distinguished, the first having a more aggressive course [2, 3]. The distinctive features of RRP are the unpredictable clinical course and the potential to spread to the distal respiratory tract mucosa, both defining RRP's aggressiveness [2, 4]. In addition, mucosal papillomas can reappear regularly at short intervals or following long remissions of up to a decade or even longer [5, 6]. According to immunological studies, RRP usually appears in individuals with a permissive and insufficient immune response to the viral infection, suggesting that recurrences of RRP could be immunologically mediated [7].
HPV DNA is identified in virtually all RRPs: most commonly HPV6, followed by HPV11, and with other low‐risk and high‐risk HPV genotypes being only rarely detected [1, 5, 6, 7, 8]. L1 gene nucleotide sequences of different HPV genotypes differ by more than 10%, while identification of major genomic variant lineages and sublineages is based on differences of ~1%– < 10% and 0.5%–1.0%, across the complete viral genome sequence, respectively [9, 10]. HPV6 genomic variants are classified into lineages A and B and are further divided into sublineages B1, B2, B3, B4, and B5 [10, 11], and HPV11 genomic variants are classified into lineage A, formed by sublineages A1, A2, A3, and A4, and lineage B [10, 12]. The most commonly identified genomic variants in RRP are HPV6 B and HPV11 A2 [10, 11, 12, 13, 14].
Specific gene mutations in HPV genomic variants can change the amino acid sequence and gene expression, they could also influence the viral biological and pathogenic effects on the host cells [10, 13, 15, 16]. Moreover, even though it has not been proven yet, a relationship between specific HPV genomic variants and RRP aggressiveness could potentially exist [16, 17, 18, 19, 20, 21, 22].
Surgical removal of papillomas to keep the airway patent is currently the primary treatment for RRP [7, 23]. However, such a therapeutic approach is not curative because the surrounding respiratory mucosa serves as an HPV reservoir that allows the (frequent) recurrence of papillomas [24, 25]. In line with this, adjuvant therapy, including HPV vaccination, has frequently been used in patients with an aggressive clinical course, showing promising results in reducing RRP recurrences [5, 7, 26]. Nonetheless, definitive treatment with complete HPV elimination from the respiratory mucosa has not been achieved yet, making RRP a practically incurable disease [5, 7, 27].
The main aim of this study was to investigate whether, in RRP patients, the long‐term persistence of a particular HPV genotype, HPV subtype or HPV genomic variant influences the clinical course of RRP, especially if it is linked with a more aggressive clinical course. HPV detection and genotyping were performed in paired baseline and follow‐up archival RRP laryngeal tissue specimens of 59 retrospectively selected eligible patients, and, in addition, determination of HPV genomic variants in paired tissue specimens taken at least 10 years apart was carried out in a subset of 20 eligible patients. The median clinical follow‐up of patients included in the main study was 18 years, the longest follow‐up time recorded in similar studies.
2. Methods
2.1. Patient Population
This retrospective study was performed on a cohort of 59 patients (36 males and 23 females) followed and treated for laryngeal RRP at the Department of Otorhinolaryngology and Cervicofacial Surgery of the University Medical Centre Ljubljana, Slovenia.
Three inclusion criteria were used for the main study: (i) duration of clinical follow‐up (calculated from the date of the first histologically confirmed RRP to the date of last clinical follow‐up) of at least 10 years; (ii) availability of baseline and at least one follow‐up RRP tissue specimen (laryngeal papilloma biopsies) to confirm the recurrent natural history of the disease; and (iii) absence of laryngeal papilloma malignant transformation during the follow‐up period. Thus, after reviewing the medical records and availability of baseline and follow‐up archival RRP tissue specimens, out of 217 RRP patients with histologically confirmed laryngeal papillomas treated between 1974 and 2018 at our institution, 158 patients were excluded: 151 patients with a clinical follow‐up shorter than 10 years, five patients due to lack of available baseline and follow‐up RRP tissue specimens, and two patients because of the malignant transformation of laryngeal papillomas, finally rendering 59 patients eligible for the main study.
All patients with available baseline and follow‐up RRP tissue specimens taken at least 10 years apart were selected for additional substudy (total of 20 patients), in which their HPV6 and HPV11 isolates were further characterized in detail using sequencing and HPV genomic variants determined.
2.2. Data Collection
The clinical data extracted from patients' medical records included sex, age at onset of RRP, the aggressiveness of the disease (10 or more laryngeal papilloma surgeries performed in total or three or more surgeries performed per year, the spread of laryngeal papilloma to the subglottis in the first 6 months after the diagnosis or spread to the distal respiratory tract at any point during follow‐up) [4], use of adjuvant treatment (acyclovir, valacyclovir, indol‐3‐carbinol, cidofovir), HPV vaccination status (quadrivalent HPV vaccine between 2006 and 2016 and nonavalent HPV vaccine since 2016), and follow‐up in years. Based on the age at RRP onset, patients 15 years or older were classified into adult‐onset RRP (39 patients) and those younger than 15 years into juvenile‐onset RRP (20 patients).
2.3. DNA Extraction and Human Papillomavirus Typing
Total DNA was extracted from formalin‐fixed paraffin‐embedded laryngeal papilloma tissue specimens using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany), as described previously [8, 28], and quantified using the NanoDrop ND‐2000c spectrophotometer (NanoDrop Technologies, Oxfordshire, UK). The quality of extracted DNA was assessed by amplifying a 150 bp fragment of the human beta‐globin gene [29], and the presence of HPV6 and HPV11 DNA was identified by the HPV6/11 FRET real‐time PCR (RT‐PCR), which allows the simultaneous, highly sensitive, and specific detection of HPV genotypes 6 and 11 and the differentiation between prototypic (HPV6b) and non‐prototypic (HPV6a) viral subtypes [30]. HPV6 and HPV11 DNA amplification and RT‐PCR product analysis were performed on the LightCycler v2.0 instrument (Roche Diagnostics, Mannheim, Germany), using the commercially available Quanti Tect Probe PCR Kit (Qiagen), as described previously [30].
2.4. Determination of Human Papillomavirus Genomic Variants
HPV6 and HPV11 genomic variants were determined based on the representative regions of 961 bp within the L2 gene and 208 bp at the 3′‐end of the E2 gene and 5′‐end of noncoding region 2, respectively, as previously proposed for phylogenetic clustering [11, 12].
The detailed procedures for amplifying and recording the targeted genomic sequence regions of HPV6 and HPV11 and their phylogenetic placement are provided in the supplementary text titled “Target DNA Sequence Amplification, Sequencing and Phylogenetic placement.”
2.5. Statistical Analysis
The normality of data distribution was assessed with the Shapiro–Wilk test. Normally distributed data were represented with mean, standard deviation (SD), and 95% confidence interval (CI), and non‐normally distributed data with median and interquartile range (IQR). The role of a particular HPV genomic variant as a risk factor for a more aggressive clinical course of RRP was tested with two‐tailed Fisher's exact test using R software version 4.4.0. A Type I error p‐value of ≤ 0.05 was considered statistically significant.
2.6. Ethical Approval
All tissue specimens used in the study were collected during routine surgical treatment of RRP, and all patients provided written informed consent. Institutional Review Boards of the Department of Otorhinolaryngology and Cervicofacial Surgery of the University Medical Center in Ljubljana, Slovenia, the Institute of Microbiology and Immunology, and the Institute of Pathology, Faculty of Medicine, at the University of Ljubljana approved the study protocol following Helsinki Declaration requirements, as well as the use of previously collected samples and data for this study. In addition, the National Medical Ethics Committee at the Ministry of Health of the Republic of Slovenia approved the study protocol (consent number 79/07/15).
3. Results
The mean age of 59 patients at RRP onset was 27.3 years (SD 19.7; range: 1–70), and the median clinical follow‐up time was 18 years (IQR: 14–27; range: 10–61).
HPV detection and HPV genotype determination were performed in both baseline and follow‐up archival RRP laryngeal tissue specimens of all 59 enrolled patients. The median time from baseline to follow‐up tissue specimens used for HPV detection for 59 patients was 5 years (IQR: 2–16, range: 1–44). The 150 bp fragment of the human beta‐globin gene was successfully amplified from all 118 paired RRP tissue specimens of 59 patients, indicating adequate sample quality for subsequent HPV testing/genotyping. HPV DNA was detected in 58/59 (98.3%) patients, with HPV6 (40/58; 68.9%) being the most common HPV genotype: HPV6a was detected in 27/40 (67.5%) and HPV6b in 12/40 (30.0%) patients, and one patient (1/40; 2.5%) had a coinfection with HPV6a and HPV6b in the baseline RRP tissue specimen. HPV11 was identified in 17/58 (29.3%) patients, and one patient (1/58; 1.7%) had coinfection with HPV6a and HPV11 in the baseline RRP tissue specimen. When comparing detected HPV (sub)types in paired baseline and follow‐up specimens of individual patients, the identical HPV (sub)type was identified in both samples in 56/58 (96.5%) patients. In the two patients with coinfection, at least one of the HPV (sub)types detected in baseline specimens was also present in the follow‐up specimen. In a single patient, both paired RRP tissue specimens tested negative for HPV.
HPV genomic variants were determined in 20 patients having available paired RRP tissue specimens taken at least 10 years apart. As shown in Table 1, HPV6 or HPV11 genomic variants were successfully detected in both paired RRP tissue specimens of 15/20 (75.0%) patients. In the remaining five patients, the genomic variant determination was successful from only one of the paired RRP tissue specimens (patients 1, 3, 9, 11 and 14; Table 1). In all 15/15 (100.0%) patients with genomic variant status determined in both paired specimens, the identical genomic variant was identified in both specimens, although tissue specimens were taken in a median 17.5 years apart (IQR: 12.5–26.5; range: 10–44) and regardless of the patients' adjuvant therapy history and/or HPV vaccination status (Table 1).
Table 1.
HPV6/HPV11 genomic variants in paired RRP laryngeal tissue specimens taken at least 10 years apart.
| Patient number | Sequence number | Age at RRP diagnosis | Aggressive RRP | Adjuvant therapy/HPV vaccine | Time between baseline and follow‐up surgical procedures (in years) | HPV genomic variant in baseline sample | HPV genomic variant in follow‐up sample |
|---|---|---|---|---|---|---|---|
| 1 | P‐1 | 5 | Yes | No/Yes | 25 | HPV6 B1 | NS |
| 2 | P‐6 | 4 | Yes | No/No | 44 | HPV6 B1 | HPV6 B1 |
| 3 | P‐10 | 41 | No | No/No | 10 | NS | HPV6 B1 |
| 4 | P‐12 | 1 | Yes | Yes/Yes | 13 | HPV6 B1 | HPV6 B1 |
| 5 | P‐14 | 1 | No | Yes/No | 34 | HPV6 B1 | HPV6 B1 |
| 6 | P‐8 | 58 | No | No/No | 16 | HPV6 B4 | HPV6 B4 |
| 7 | P‐2 | 23 | Yes | Yes/Yes | 11 | HPV6 A | HPV6 A |
| 8 | P‐3 | 11 | Yes | No/No | 24 | HPV6 A | HPV6 A |
| 9 | P‐4 | 33 | Yes | Yes/Yes | 20 | NS | HPV6 A |
| 10 | P‐13 | 38 | Yes | Yes/No | 17 | HPV6 A | HPV6 A |
| 11 | P‐16 | 21 | No | No/No | 34 | NS | HPV6 A |
| 12 | P‐19 | 70 | No | No/No | 17 | HPV6 A | HPV6 A |
| 13 | P‐5 | 30 | No | No/No | 30 | HPV11 A2 | HPV11 A2 |
| 14 | P‐7 | 31 | Yes | Yes/Yes | 16 | NS | HPV11 A2 |
| 15 | P‐9 | 13 | No | No/No | 12 | HPV11 A2 | HPV11 A2 |
| 16 | P‐11 | 30 | No | No/No | 11 | HPV11 A2 | HPV11 A2 |
| 17 | P‐15 | 39 | Yes | Yes/No | 22 | HPV11 A2 | HPV11 A2 |
| 18 | P‐17 | 49 | No | No/No | 18 | HPV11 A2 | HPV11 A2 |
| 19 | P‐18 | 4 | Yes | Yes/Yes | 28 | HPV11 A2 | HPV11 A2 |
| 20 | P‐20 | 56 | No | No/No | 10 | HPV11 A1 | HPV11 A1 |
As presented in Table 1 and Figures 1 and 2, the most common HPV6 genomic variants were HPV6 A (6/12; 50.0%) and HPV6 B1 (5/12; 41.6%), whereas HPV6 B4 was identified in a single patient. In the case of HPV11, the most common genomic variant was HPV11 A2, determined in 7/8 (87.5%) patients, whereas a single patient had an HPV11 A1 genomic variant.
Figure 1.
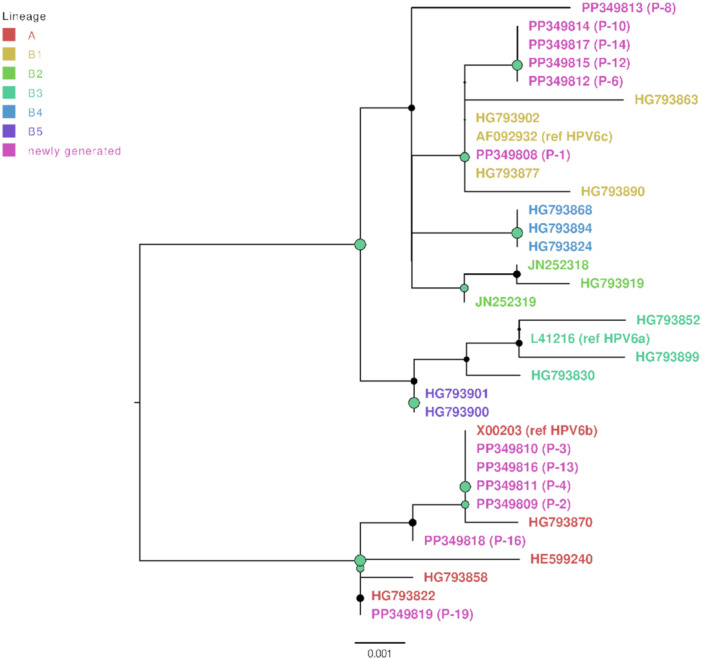
HPV6 genomic variants maximum likelihood phylogenetic tree. Represented are the HPV6 nucleotide sequences newly generated in this study (n = 12; marked with pink color) and context reference nucleotide sequences (n = 22). HPV6 genomic variant lineage A is colored red, sublineage B1 yellow, B2 bright green, B3 dark green, B4 bright blue, and B5 purple. The tree was rooted at the branch with lineage A. Node sizes are proportional to phylogenetic node UFBoot support (0–100), nodes receiving UFBoot scores > 70 are shown in green.
Figure 2.
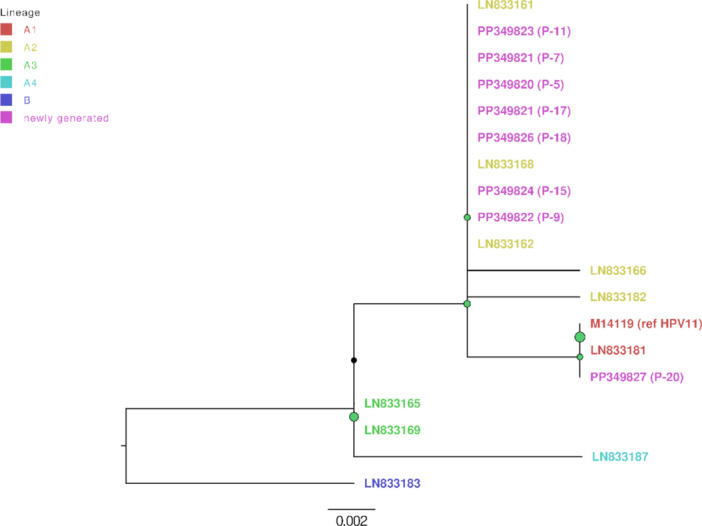
HPV11 genomic variants maximum likelihood phylogenetic tree. Represented are the HPV11 nucleotide sequences newly generated in this study (n = 8; marked in pink color) and context reference nucleotide sequences (n = 11). HPV11 genomic variant lineage B is colored dark blue, sublineage A1 red, A2 yellow, A3 green and A4 bright blue. The tree was rooted at the branch with lineage B. Node sizes are proportional to phylogenetic node UFBoot support (0–100), nodes receiving UFBoot scores > 70 are shown in green.
As presented in Table 2, the most common HPV genomic variants in patients with aggressive RRP (10/20, 50.0%) were HPV6 A (4/10, 40.0%) and HPV6 B1 (4/10, 30.0%). HPV11 A2 (4/10, 40.0%) was the most prevalent genomic variant identified in patients with nonaggressive RRP (10/20, 50.0%; Table 2). As shown in Table 2, no statistically significant differences were observed for these variables.
Table 2.
HPV genomic variants as potential risk factors for the aggressive clinical course of RRP.
| HPV genomic variant | Patients with nonaggressive RRP number of patients = 10 (%) | Patients with aggressive RRP number of patients = 10 (%) | Odds ratio (95% CI) | p value |
|---|---|---|---|---|
| HPV6 A | 2 (10.0%) | 4 (40.0%) | 2.54 (0.25–37.18) | 0.626 |
| HPV6 B1 | 2 (10.0%) | 3 (30.0%) | 1.67 (0.14–25.60) | 1 |
| HPV6 B4 | 1 (10.0%) | 0 (0%) | 0.00 (0.00–39.00) | 1 |
| HPV11 A1 | 1 (10.0%) | 0 (0%) | 0.00 (0.00–39.00) | 1 |
| HPV11 A2 | 4 (40.0%) | 3 (30.0%) | 0.66 (0.07–5.78) | 1 |
Abbreviations: CI, confidence interval; HPV, human papillomavirus; RRP, recurrent respiratory papillomatosis.
Table 3 shows the results of a similar analysis but using the whole patients' cohort (59 patients) and performed on HPV genotype/subtype level. No statistically significant differences were observed between the HPV genotype/subtype and the aggressive clinical course of RRP.
Table 3.
HPV genotype/subtype as potential risk factors for the aggressive clinical course of RRP.
| HPV genotype | Patients with nonaggressive RRP number of patients = 33 (%) | Patients with aggressive RRP number of patients = 26 (%) | Odds ratio (95% CI) | p value |
|---|---|---|---|---|
| HPV6a | 16 (48.5%) | 12 (46.2%) | 0.91 (0.29–2.88) | 1 |
| HPV6b | 8 (24.2%) | 5 (19.2%) | 0.75 (0.17–3.07) | 0.757 |
| HPV11 | 9 (27.3%) | 9 (34.6%) | 1.40 (0.40–4.97) | 0.580 |
| HPV neg | 1 (3.0%) | 0 (0%) | 0.00 (0.00–49.5) | 1 |
Abbreviations: CI, confidence interval; HPV, human papillomavirus; RRP, recurrent respiratory papillomatosis.
4. Discussion
Approximately 40 HPV genotypes from the Alpha genus are subdivided into two groups: high‐risk and low‐risk. High‐risk HPV genotypes, considered as group 1 carcinogens, are etiologically associated with 4.5% of all cancers worldwide and cause virtually all cervical and anal cancers, a substantial proportion of vaginal cancers, and a component of oropharyngeal, penile, and vulvar cancer. Cervical cancer, with 600 000 new cases per year and more than 340 000 deaths annually, is the high‐risk HPV‐driven cancer associated with the greatest morbidity and mortality. This cancer and its precursors are susceptible to highly effective treatment, especially when detected early through prevention strategies [31].
In this retrospective longitudinal study, we focused on RRP, relatively rare human benign tumors that are like more frequent anogenital warts etiologically linked with low‐risk HPV. Paired baseline and follow‐up RRP laryngeal tissue specimens obtained from 59 RRP patients with a median clinical follow‐up of 18 years were tested for the presence of HPV, which is the longest median clinical follow‐up time recorded in similar studies so far [16, 18, 19, 20, 21, 22, 32, 33, 34, 35, 36, 37]. This study showed that persistent infection with the same HPV genotype/subtype and a completely identical HPV6 and HPV11 genomic variant is responsible for the recurrent nature of RRP that could last at least a decade, if not for a lifetime.
HPV DNA was detected in tissue specimens of all but one of the 59 RRP patients, similar to previously reported HPV detection rates in RRP tissues, ranging from 80% to 100% [3, 6, 8, 32, 33, 34, 35, 38, 39]. In accordance with previous studies [3, 6, 32, 33, 34, 35, 39], HPV6 was detected in almost two‐thirds of our patients, with HPV6a being the most prevalent HPV subtype, and HPV11 being identified in one‐third of patients. No single high‐risk HPV genotype was detected in the RRP tissue specimens tested.
The same low‐risk HPV (sub)type was detected in paired RRP tissue specimens of 96.5% of patients. The remaining two HPV‐positive patients had HPV6a/HPV6b and HPV6a/HPV11 coinfections, which is in line with previously reported studies suggesting that HPV coinfections in RRP tissue specimens are rare, previously described in up to 7.2% of cases [3, 6, 32, 33, 35].
To explore long‐term HPV persistence in RRP, we have determined the HPV6 and HPV11 genomic variants in paired RRP tissue specimens taken at least 10 years apart in 20 RRP patients. The most commonly identified HPV6 genomic variants in our study clustered to the HPV6 lineage A and sublineage B1 and sublineage B4 was identified in a single patient. Our results are in agreement with those obtained in the largest HPV6 genomic variant study, performed on 176 RRP tissues collected worldwide, in which sublineage HPV6 B1 (47.2%) was most frequently detected, followed by lineage HPV6 A (21.6%) and sublineages HPV6 B3 (17.0%), HPV6 B2 (9.7%), HPV6 B5 (4.0%), and HPV6 B4 (0.6%) [11]. In addition, our study identified the HPV6 B4 genomic variant in Slovenia, which has previously been described in only 0.6% of HPV6 isolates collected worldwide, with only a single HPV6 isolate originating from RRP [11].
In a similar large international HPV11 genomic variant study on 433 HPV11 isolates collected worldwide, the most common HPV11 genomic variant clustered to sublineage HPV11 A2 (86.1%), followed by HPV11 A1 (12.5%), HPV11 A3 (0.7%), HPV11 A4 (0.5%), and lineage HPV11 B (0.2%), detected in a single African isolate [12]. Similarly, sublineage HPV11 A2 was the most common in our patients, followed by HPV11 A1 detected in a single patient.
In this study, HPV genomic variants were successfully determined in paired baseline and follow‐up RRP tissue specimens taken 10 years apart in 15 out of 20 patients. In all 15 patients, a completely identical HPV genomic variant was identified in both baseline and follow‐up RRP tissue specimens of individual patients with a median interval time between surgical biopsies of 17.5 years, confirming that RRP recurrences are a result of long‐term persistent laryngeal HPV infection with the same HPV genomic variant, which is in agreement with the results of our pilot study from 2013, which, based on the HPV E5a gene or whole‐genome sequence analysis, identified the identical HPV genomic variants in paired samples in 95.7% of 70 RRP patients studied [40]. However, the patients included in our pilot study in 2013 had a median time between baseline and follow‐up RRP tissue specimens of only 4 years [40], substantially shorter than in the present study. In addition, this study's 44‐year interval between baseline and follow‐up biopsy in a patient with aggressive RRP and HPV6 B1 infection represents the longest duration of genetically confirmed persistent HPV infection with the identical HPV genomic variant for any HPV genotype recorded in the currently published peer‐reviewed literature.
The potential influence of HPV genomic variants on the severity of the RRP clinical course has been previously studied, but with inconclusive results, mainly due to the small number of patients included (ranging from 1 to 60 patients), studies having a cross‐sectional design and short follow‐up [16, 17, 18, 19, 20, 21, 22, 36, 37, 41]. Gabbott et al. described a case report of RRP spreading into the distal respiratory tract, in which HPV11 causing these papillomas had specific mutations in the LCR region that could potentially cause the aggressive clinical course and even malignant transformation of papillomas in this patient [41]. Studies on Brazilian RRP patients have identified sublineage HPV6 B1 to be more common in juvenile‐compared to adult‐onset RRP [19, 22]. Whole genome sequence analysis of HPV6 and HPV11 in patients with aggressive RRP has identified mutations that could, at least theoretically, influence viral gene expression and cause a more severe disease course [18, 20, 37, 40], like the mutations in the LCR region and the E6 gene of HPV6 detected in 4/12 (33.3%) patients with an aggressive form of RRP [42]. In addition, in cross‐sectional studies comprising 13 and 36 RRP patients, Measso do Bonfim et al. and Sichero et al. respectively, failed to identify a link between infection with a particular HPV6 genomic variant and aggressive course of disease scored using the Derkay score [19, 22]. Similarly, the influence of HPV11 sublineages A2 and A1 on the RRP disease course was not established in two distinct studies, Xiao et al. and Sichero et al. in 18 and 24 RRP patients, respectively [21, 22].
In our 20 RRP patients with HPV genomic variants successfully determined in at least one RRP tissue specimen, HPV6 lineage A (36.4%) and sublineage B1 (36.4%) were the most prevalent genomic variants in patients with the aggressive course of the disease, but, compared to those with a nonaggressive disease, a statistically significant difference was not identified. This and previous studies seem to show that the mutation characteristics for any major HPV6 and HPV11 genomic variant most probably do not significantly impact the clinical course of RRP.
Because surgery is not a curative procedure for RRP, various adjuvant therapies are used in 25% to 48% of RRP patients with frequent disease recurrences [5, 34, 43, 44, 45, 46]. In addition, vaccination against HPV using prophylactic HPV vaccines containing HPV6 and HPV11 viral‐like particles has been recommended in RRP patients with an aggressive form of the disease based on studies showing a favorable influence of vaccination on the clinical course of the disease [26, 47, 48, 49]. The use of vaccination against HPV as an adjuvant therapy has also been recommended after primary surgical treatment of cervical precancerous lesions. Posttreatment HPV vaccination can reduce recurrences of cervical lesions related to new HPV infections and prevent appearance of HPV‐related lesions in the surrounding area (vulva, vagina, anus) [50, 51]. Our results show that adjuvant therapy (6/15, 40.0%) and vaccination against HPV (3/15, 20.0%) did not influence HPV elimination because the identical HPV genomic variant persisted in individual patients for years regardless of the aforementioned measures taken. Similarly, Gall et al. demonstrated that therapy with IFN‐alfa and cidofovir did not induce any changes in the HPV11 genome in three patients with aggressive juvenile‐onset RRP [17, 18]. Long‐term persistent infection with the same HPV genomic variant in treated patients suggests the inability of adjuvant therapy and/or HPV vaccination to clear already acquired HPV infections of the larynx. Whether some HPV genomic variants could be more susceptible to current or future specific antiviral adjuvant therapy remains elusive.
The main strength of this study is the close follow‐up of enrolled patients as well as the availability of the complete medical documentation since 1974, which enabled a longitudinal study with an exceptionally long clinical follow‐up, spanning up to 61 years, differentiating our study from previously published studies with substantially shorter follow‐ups. A long and close clinical follow‐up of RRP patients allows for capturing the great majority of the disease's clinical ups and downs occurring during the course of RRP over time, which would not be possible with the inclusion of individuals with short follow‐ups. Another strength of the study was the availability of baseline and follow‐up RRP tissue specimens taken at least 10 years apart and of sufficient quantity and quality for reliable HPV testing, allowing an undoubtful genetic confirmation of long‐lasting persistent HPV infection of the larynx.
The main limitations of this study are the low number of enrolled patients overall as well as the low number of paired RRP tissue specimens with successfully determined HPV genomic variants, which is a result of very strict inclusion criteria (duration of clinical follow‐up of at least 10 years and availability of baseline and follow‐up RRP tissue specimens taken at least 10 years apart, respectively) as well as technical issues connected with sequencing of the large fragments of HPV genome from archival tissues with partially degraded DNA. In addition, the HPV testing method(s) used in this study may have missed some coinfection(s) with other nontargeted HPV genotypes, which could also potentially influence the RRP course [52]. On the other hand, it is well established that HPV6 and HPV11 are the main causative HPV genotypes of RRP and that other HPV genotypes potentially identified with broader diagnostic approaches in RRP tissue usually represent innocent HPV bystanders with no clear active biological role [53]. The limited number of enrolled patients prevented in‐depth statistical analysis, including important parameters potentially influencing aggressive clinical course like the age at diagnosis, length of follow‐up, gender, juvenile vs adult presentation, smoking, adjuvant therapy used, histological specificities and others. Adding more parameters into statistical analysis would generate smaller and smaller patient subgroups, making any statistical analysis non‐reliable. Due to the low number of enrolled patients the statistical power of our study remains low, or rather false negative probability (type II error probability = 1 – statistical power) remains high. Based on the reported odds ratios and sample sizes the chances of incorrectly missing the effect of (i) HPV genotypes or (ii) HPV variants at threshold false positive rate 0.05 were between 92.5% and 96.5%, and 99.8% and 93.5%, respectively. To reduce this chance below 20%, for 80% statistical power we would need between 254 (for odds of HPV negatives) and 7200 (for odds of HPV6a) patients with aggressive and nonaggressive RRP in the case of HPV genotypes, and between 74 (for odds of HPV6 B4 HPV11 A1) and 360 (for odds of HPV11 A2) samples per group in case of HPV variants. This power calculation analysis for “ideal” sample sizes could be used in potential future studies on the subject. However, given the rarity of RRP and the recent further substantial reduction of their incidence as a result of worldwide implementation of gender‐neutral HPV vaccination, studies with statistical power as calculated above will most probably never be possible to conduct.
5. Conclusions
This study performed on paired baseline and follow‐up RRP laryngeal tissue specimens of 59 patients adds to the mounting evidence that the recurrent nature of RRP is a result of a long‐term, most likely lifelong, persistent infection of the laryngeal mucosa with the same HPV genotype, HPV subtype and HPV genomic variant, which can be identified in RRP patients many decades after disease onset and following several rounds of surgical treatment accompanied by adjuvant therapy and/or vaccination against HPV.
Despite long‐term HPV persistence, it seems that infection with a particular HPV genotype, HPV subtype, or HPV genomic variant does not significantly influence the clinical course of RRP, including disease aggressiveness, but further confirmatory studies with much higher number of enrolled patients are warranted to confirm this assumption. Most probably, other factors such as age at onset, host susceptibility, and patient immune status dictate clinical disease presentation, implying that future research should focus more on the host factors and immune response to HPV infection and away from the HPV itself.
Author Contributions
N.G., I.H.B., and M.P. conceptualized the study; D.G., L.H. and T.M.Z. performed the analyses and interpreted the data; D.G., L.H., and M.P. drafted the work; all Authors (D.G., L.H., T.M.Z., N.G., I.H.B., and M.P.) approved the submitted version and agreed both to be personally accountable for their own contributions and to ensure that questions related to the accuracy or integrity of any part of the work, even ones in which the Author was not personally involved, are appropriately investigated, resolved, and the resolution documented in the literature.
Disclosure
All tissue specimens used in the study were collected during routine surgical treatment of RRP, and all patients provided written informed consent. Institutional Review Boards of the Department of Otorhinolaryngology and Cervicofacial Surgery of the University Medical Center in Ljubljana, Slovenia, the Institute of Microbiology and Immunology, and the Institute of Pathology, Faculty of Medicine, at the University of Ljubljana approved the study protocol following Helsinki Declaration requirements, as well as the use of previously collected samples and data for this study. In addition, the National Medical Ethics Committee at the Ministry of Health of the Republic of Slovenia approved the study protocol (consent number 79/07/15).
Conflicts of Interest
The authors declare no conflicts of interest.
Supporting information
Supporting information.
Acknowledgments
The Authors would like to thank Tina Bukovec for sectioning of the archival tissue specimens. This research was funded by the Institute of Microbiology and Immunology, Faculty of Medicine, University of Ljubljana (internal funding), and the Slovenian Research and Innovation Agency ARIS (funding grant number: P3‐00083). M.P. is supported by the Horizon 2020 Framework Program for Research and Innovation of the European Commission, through the RISCC Network (funding grant number 847845).
Data Availability Statement
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/nuccore, reference number PP349808‐PP349827.
The data set supporting the conclusions of this article is available in the GenBank repository, PP349808‐PP349827.
References
- 1. Gale N., Ferluga D., Poljak M., Kambič V., and Fischinger J., “Laryngeal Papillomatosis: Molecular, Histopathological, and Clinical Evaluation,” Virchows Archiv 425 (1994): 291–295. [DOI] [PubMed] [Google Scholar]
- 2. Lindeberc H., Øster S., Oxlund I., and Elbrbnd O., “Laryngeal Papillomas: Classification and Course,” Clinical Otolaryngology 11 (1986): 423–429. [PubMed] [Google Scholar]
- 3. Buchinsky F. J., Valentino W. L., Ruszkay N., et al., “Age at Diagnosis, but not HPV Type, Is Strongly Associated With Clinical Course in Recurrent Respiratory Papillomatosis,” PLoS One 14 (2019): e0216697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Doyle D. J., Gianoli G. J., Espinola T., and Miller R. H., “Recurrent Respiratory Papillomatosis: Juvenile Versus Adult Forms,” The Laryngoscope 104 (1994): 523–527. [DOI] [PubMed] [Google Scholar]
- 5. Wiatrak B. J., Wiatrak D. W., Broker T. R., and Lewis L., “Recurrent Respiratory Papillomatosis: A Longitudinal Study Comparing Severity Associated With Human Papilloma Viral Types 6 and 11 and Other Risk Factors in a Large Pediatric Population,” The Laryngoscope 114 (2004): 1–23. [DOI] [PubMed] [Google Scholar]
- 6. Tjon Pian Gi R. E. A., San Giorgi M. R. M., Slagter‐Menkema L., et al., “Clinical Course of Recurrent Respiratory Papillomatosis: Comparison Between Aggressiveness of Human Papillomavirus‐6 and Human Papillomavirus‐11,” Head & Neck 37 (2015): 1625–1632. [DOI] [PubMed] [Google Scholar]
- 7. Aaltonen L. M., Rihkanen H., and Vaheri A., “Human Papillomavirus in Larynx,” The Laryngoscope 112 (2002): 700–707. [DOI] [PubMed] [Google Scholar]
- 8. Komloš K. F., Kocjan B. J., Košorok P., et al., “Tumor‐Specific and Gender‐Specific Pre‐Vaccination Distribution of Human Papillomavirus Types 6 and 11 in Anogenital Warts and Laryngeal Papillomas: A Study on 574 Tissue Specimens,” Journal of Medical Virology 84 (2012): 1233–1241. [DOI] [PubMed] [Google Scholar]
- 9. Bernard H. U., Burk R. D., Chen Z., van Doorslaer K., Hausen H., and de Villiers E. M., “Classification of Papillomaviruses (PVs) Based on 189 PV Types and Proposal of Taxonomic Amendments,” Virology 401 (2010): 70–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Burk R. D., Chen Z., Harari A., et al., “Classification and Nomenclature System for Human Alphapapillomavirus Variants: General Features, Nucleotide Landmarks and Assignment of HPV6 and HPV11 Isolates to Variant Lineages,” Acta dermatovenerologica Alpina, Pannonica, et Adriatica 20 (2011): 113–123. [PMC free article] [PubMed] [Google Scholar]
- 11. Jelen M. M., Chen Z., Kocjan B. J., et al., “Global Genomic Diversity of Human Papillomavirus 6 Based on 724 Isolates and 190 Complete Genome Sequences,” Journal of Virology 88 (2014): 7307–7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jelen M. M., Chen Z., Kocjan B. J., et al., “Global Genomic Diversity of Human Papillomavirus 11 Based on 433 Isolates and 78 Complete Genome Sequences,” Journal of Virology 90 (2016): 5503–5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kocjan B. J., Jelen M. M., Maver P. J., Seme K., and Poljak M., “Pre‐Vaccination Genomic Diversity of Human Papillomavirus Genotype 6 (HPV 6): A Comparative Analysis of 21 Full‐Length Genome Sequences,” Infection, Genetics and Evolution 11 (2011): 1805–1810. [DOI] [PubMed] [Google Scholar]
- 14. Maver P. J., Kocjan B. J., Seme K., Potočnik M., Gale N., and Poljak M., “Prevaccination Genomic Diversity of Human Papillomavirus Genotype 11: A Study on 63 Clinical Isolates and 10 Full‐Length Genome Sequences,” Journal of Medical Virology 83 (2011): 461–470. [DOI] [PubMed] [Google Scholar]
- 15. Kocjan B. J., Poljak M., Cimerman M., et al., “Prevaccination Genomic Diversity of Human Papillomavirus Genotype 6 (HPV 6),” Virology 391 (2009): 274–283. [DOI] [PubMed] [Google Scholar]
- 16. Szinai M., Nagy Z., Máté P., et al., “Comparative Analysis of Human Papillomavirus Type 6 Complete Genomes Originated From Head and Neck and Anogenital Disorders,” Infection, Genetics and Evolution 71 (2019): 140–150. [DOI] [PubMed] [Google Scholar]
- 17. Gáll T., Kis A., Fehér E., Gergely L., and Szarka K., “Virological Failure of Intralesional Cidofovir Therapy in Recurrent Respiratory Papillomatosis Is not Associated With Genetic or Epigenetic Changes of HPV11: Complete Genome Comparison of Sequential Isolates,” Antiviral Research 92 (2011): 356–358. [DOI] [PubMed] [Google Scholar]
- 18. Gáll T., Kis A., Tatár T. Z., Kardos G., Gergely L., and Szarka K., “Genomic Differences in the Background of Different Severity in Juvenile‐Onset Respiratory Papillomatoses Associated With Human Papillomavirus Type 11,” Medical Microbiology and Immunology 202 (2013): 353–363. [DOI] [PubMed] [Google Scholar]
- 19. Measso do Bonfim C., Simão Sobrinho J., Lacerda Nogueira R., et al., “Differences in Transcriptional Activity of Human Papillomavirus Type 6 Molecular Variants in Recurrent Respiratory Papillomatosis,” PLoS One 10 (2015): e0132325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Seedat R. Y., Combrinck C. E., Bester P. A., Lee J. Y., and Burt F. J., “Determination of the Complete Genome and Functional Analysis of HPV6 Isolate VBD19/10 From a Patient With Aggressive Recurrent Respiratory Papillomatosis,” Epidemiology and Infection 144 (2016): 2128–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xiao Y., Wang J., Ma L., Ren J., and Yang M., “Nucleotide and Phylogenetic Analysis of Human Papillomavirus Type 11 Isolated From Juvenile‐Onset Recurrent Respiratory Papillomatosis in China,” Journal of Medical Virology 88 (2016): 686–694. [DOI] [PubMed] [Google Scholar]
- 22. Sichero L., Ferreira S., López R. V. M., et al., “Prevalence of Human Papillomavirus 6 and 11 Variants in Recurrent Respiratory Papillomatosis,” Journal of Medical Virology 93 (2021): 3835–3840. [DOI] [PubMed] [Google Scholar]
- 23. Derkay C. S. and Bluher A. E., “Update on Recurrent Respiratory Papillomatosis,” Otolaryngologic Clinics of North America 52 (2019): 669–679. [DOI] [PubMed] [Google Scholar]
- 24. Glynn M., Sanford T., Hoover L., Kinsey W., Dobbs L., and Bruegger D., “Characterization of Human Papillomavirus in Airway Papillomas by Histologic and Biochemical Analysis,” Annals of Otology, Rhinology & Laryngology 108 (1999): 1073–1077. [DOI] [PubMed] [Google Scholar]
- 25. Forslund O., Schwartz S., Olofsson K., and Rydell R., “Viral Load and mRNA Expression of HPV Type 6 Among Cases With Recurrent Respiratory Papillomatosis,” The Laryngoscope 126 (2016): 122–127. [DOI] [PubMed] [Google Scholar]
- 26. Hočevar‐Boltežar I., Matičič M., Šereg‐Bahar M., et al., “Human Papilloma Virus Vaccination in Patients With an Aggressive Course of Recurrent Respiratory Papillomatosis,” European Archives of Oto‐Rhino‐Laryngology 271 (2014): 3255–3262. [DOI] [PubMed] [Google Scholar]
- 27. Ivancic R., Iqbal H., deSilva B., Pan Q., and Matrka L., “Current and Future Management of Recurrent Respiratory Papillomatosis,” Laryngoscope Investigative Otolaryngology 3 (2018): 22–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kocjan B. J., Hošnjak L., and Poljak M., “Detection of Alpha Human Papillomaviruses in Archival Formalin‐Fixed, Paraffin‐Embedded (FFPE) Tissue Specimens,” Journal of Clinical Virology 76, no. Suppl 1 (2016): S88–S97. [DOI] [PubMed] [Google Scholar]
- 29. Hošnjak L., Kocjan B. J., Pirš B., Seme K., and Poljak M., “Characterization of Two Novel Gammapapillomaviruses, HPV179 and HPV184, Isolated From Common Warts of a Renal‐Transplant Recipient,” PLoS One 10 (2015): e0119154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kocjan B. J., Seme K., and Poljak M., “Detection and Differentiation of Human Papillomavirus Genotypes HPV‐6 and HPV‐11 by Fret‐Based Real‐Time PCR,” Journal of Virological Methods 153 (2008): 245–249. [DOI] [PubMed] [Google Scholar]
- 31. Golia D'Augè T., Cuccu I., Etrusco A., et al., “State of the Art on HPV‐Related Cervical Lesions,” Italian Journal of Gynaecology and Obstetrics 36 (2024): 135–137. [Google Scholar]
- 32. Sanchez G. I., Jaramillo R., Cuello G., et al., “Human Papillomavirus Genotype Detection in Recurrent Respiratory Papillomatosis (RRP) in Colombia,” Head & Neck 35 (2013): 229–234. [DOI] [PubMed] [Google Scholar]
- 33. Omland T., Akre H., Lie K. A., Jebsen P., Sandvik L., and Brøndbo K., “Risk Factors for Aggressive Recurrent Respiratory Papillomatosis in Adults and Juveniles,” PLoS One 9 (2014): e113584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Amiling R., Meites E., Querec T. D., et al., “Juvenile‐Onset Recurrent Respiratory Papillomatosis in the United States, Epidemiology and HPV Types‐2015–2020,” Journal of the Pediatric Infectious Diseases Society 10 (2021): 774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nogueira R. L., Küpper D. S., do Bonfim C. M., et al., “HPV Genotype Is a Prognosticator for Recurrence of Respiratory Papillomatosis in Children,” Clinical Otolaryngology 46 (2021): 181–188. [DOI] [PubMed] [Google Scholar]
- 36. de Matos R. P. A., Sichero L., Mansur I. M., et al., “Nucleotide and Phylogenetic Analysis of Human Papillomavirus Types 6 and 11 Isolated From Recurrent Respiratory Papillomatosis in Brazil,” Infection, Genetics and Evolution 16 (2013): 282–289. [DOI] [PubMed] [Google Scholar]
- 37. Nagy Z., Pethő Z., Kardos G., Major T., Szűcs A., and Szarka K., “Effect of E2 and Long Control Region Polymorphisms on Disease Severity in Human Papillomavirus Type 11 Mediated Mucosal Disease: Protein Modelling and Functional Analysis,” Infection, Genetics and Evolution 93 (2021): 104948. [DOI] [PubMed] [Google Scholar]
- 38. Buchinsky F. J., Donfack J., Derkay C. S., et al., “Age of Child, More Than HPV Type, Is Associated With Clinical Course in Recurrent Respiratory Papillomatosis,” PLoS One 3 (2008): e2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cuello G., I Sánchez G., Jaramillo R., et al., “Clinical Characteristics and HPV Type in Recurrent Respiratory Papillomatosis in Colombia,” Salud Pública de México 55 (2013): 416–420. [DOI] [PubMed] [Google Scholar]
- 40. Kocjan B. J., Gale N., Hočevar Boltežar I., et al., “Identical Human Papillomavirus (HPV) Genomic Variants Persist in Recurrent Respiratory Papillomatosis for up to 22 Years,” The Journal of Infectious Diseases 207 (2013): 583–587. [DOI] [PubMed] [Google Scholar]
- 41. Gabbott M., Cossart Y. E., Kan A., Konopka M., Chan R., and Rose B. R., “Human Papillomavirus and Host Variables as Predictors of Clinical Course in Patients With Juvenile‐Onset Recurrent Respiratory Papillomatosis,” Journal of Clinical Microbiology 35 (1997): 3098–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Combrinck C. E., Seedat R. Y., Randall C., Roodt Y., and Burt F. J., “Novel HPV‐6 Variants of Human Papillomavirus Causing Recurrent Respiratory Papillomatosis in Southern Africa,” Epidemiology and Infection 140 (2012): 1095–1101. [DOI] [PubMed] [Google Scholar]
- 43. Reeves W. C., Ruparelia S. S., Swanson K. I., Derkay C. S., Marcus A., and Unger E. R., “National Registry for Juvenile‐Onset Recurrent Respiratory Papillomatosis,” Archives of Otolaryngology–Head & Neck Surgery 129 (2003): 976–982. [DOI] [PubMed] [Google Scholar]
- 44. Schraff S., Derkay C. S., Burke B., and Lawson L., “American Society of Pediatric Otolaryngology Members' Experience With Recurrent Respiratory Papillomatosis and the Use of Adjuvant Therapy,” Archives of Otolaryngology–Head & Neck Surgery 130 (2004): 1039–1042. [DOI] [PubMed] [Google Scholar]
- 45. Campisi P., Hawkes M., and Rn K. S., “The Epidemiology of Juvenile Onset Recurrent Respiratory Papillomatosis Derived From a Population Level National Database,” The Laryngoscope 120 (2010): 1233–1245. [DOI] [PubMed] [Google Scholar]
- 46. Tjon Pian Gi R. E. A., Ilmarinen T., van den Heuvel E. R., et al., “Safety of Intralesional Cidofovir in Patients With Recurrent Respiratory Papillomatosis: An International Retrospective Study on 635 RRP Patients,” European Archives of Oto‐Rhino‐Laryngology 270 (2013): 1679–1687. [DOI] [PubMed] [Google Scholar]
- 47. Young D. L., Moore M. M., and Halstead L. A., “The Use of the Quadrivalent Human Papillomavirus Vaccine (gardasil) as Adjuvant Therapy in the Treatment of Recurrent Respiratory Papilloma,” Journal of Voice 29 (2015): 223–229. [DOI] [PubMed] [Google Scholar]
- 48. Tjon Pian Gi R. E. A., San Giorgi M. R. M., Pawlita M., et al., “Immunological Response to Quadrivalent HPV Vaccine in Treatment of Recurrent Respiratory Papillomatosis,” European Archives of Oto‐Rhino‐Laryngology 273 (2016): 3231–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Matsuzaki H., Makiyama K., Hirai R., Suzuki H., Asai R., and Oshima T., “Multi‐Year Effect of Human Papillomavirus Vaccination on Recurrent Respiratory Papillomatosis,” The Laryngoscope 130 (2020): 442–447. [DOI] [PubMed] [Google Scholar]
- 50. Bogani G., Sopracordevole F., Ciavattini A., et al., “HPV‐Related Lesions After Hysterectomy for High‐Grade Cervical Intraepithelial Neoplasia and Early‐Stage Cervical Cancer: A Focus on the Potential Role of Vaccination,” Tumori Journal 110 (2024): 139–145. [DOI] [PubMed] [Google Scholar]
- 51. Joshi S., Anantharaman D., Muwonge R., et al., “Evaluation of Immune Response to Single Dose of Quadrivalent HPV Vaccine at 10‐Year Post‐Vaccination,” Vaccine 41 (2023): 236–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bertinazzi M., Gheit T., Polesel J., et al., “Clinical Implications of Alpha, Beta, and Gamma HPV Infection in Juvenile Onset Recurrent Respiratory Papillomatosis,” European Archives of Oto‐Rhino‐Laryngology 279 (2022): 285–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Odar K., Kocjan B. J., Hošnjak L., Gale N., Poljak M., and Zidar N., “Verrucous Carcinoma of the Head and Neck–Not a Human Papillomavirus‐Related Tumour?,” Journal of Cellular and Molecular Medicine 18 (2014): 635–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Data Availability Statement
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/nuccore, reference number PP349808‐PP349827.
The data set supporting the conclusions of this article is available in the GenBank repository, PP349808‐PP349827.


