Abstract
Objectives
Patients with coronavirus disease 2019 (COVID-19) are at increased risk of opportunistic fungal infections. This study aims to identify fungal pathogens among COVID positive and negative patients, assess their antifungal susceptibility and evaluate biofilm forming ability of Candida spp. A cross-sectional study was conducted among sputum samples from 135 COVID positive and 101 COVID negative cases. Fungal pathogens were identified by conventional culture methods. Antifungal susceptibility test of Candida isolates was done by disc diffusion method and biofilm production by microtiter plate method.
Results
The prevalence of fungal pathogens among COVID-positive and negative cases was 6.70% and 22.77% respectively. In COVID positive cases, Candida albicans (33.33%) was predominantly followed by Aspergillus flavus 2(22.22%) and Candida tropicalis, Mucor spp. and Aspergillus fumigatus. In COVID negative cases, Candida albicans (69.60%) prevailed followed by Trichosporon spp., Candida parapsilosis, Mucor and Alternaria. Age and gender were not associated with fungal infection. Most Candida spp. were susceptible to miconazole but resistant to ketoconazole. To the best of our knowledge, this study represents the first report from Nepal on critical and high priority fungal pathogens categorized by WHO. With fungal infections on the rise, enhanced clinical vigilanceand antifungal susceptibility testing are warranted.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13104-024-07010-4.
Keywords: COVID-19, Fungal infection, Antifungal susceptibility testing, Critical and high priority pathogens
Introduction
The novel severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) pandemic, declared a global emergency by WHO on 11 March, 2020, has led to an alarming effect on global health and the economy [1, 2]. Nepal has also recorded episodes of SARS-CoV-2 with the first confirmed case on January 13, 2020, and peak in October 2020, May 2021, and January 2022. Subsequently, the number of cases decreased with a total of 1,003,382 confirmed cases of COVID-19 and 12,031 deaths from COVID as of 26th July 2023 [3]. Severe COVID-19 patients with overexpression of pro-inflammatory (IL-1, IL-2, IL-6, TNF-α) and anti-inflammatory (IL-4, IL-10) cytokines as well as decreased CD4 and CD8 T-cells, appear to be more vulnerable to invasive fungus co-infections (IFI) [4]. The commonly reported COVID associated fungal pathogens include Aspergillus spp., Mucor spp., Candida spp., Rhizopus spp., Coccidioides, Cryptococcus, Saccharomyces cerevisiae, Histoplasma spp., Pneumocystis etc. [5, 6].
The identification of 19 fungal priority pathogens categorized by WHO as having the greatest public health threat underscores the urgency of addressing fungal infections [7]. Early diagnosis, proper treatment and surveillance might be breakthrough needed to reduce the fatality rate of secondary infections in COVID-19 patients [8]. Furthermore, previously neglected fungal pathogens have emerged as opportunistic pathogens [9, 10]. While antibiotic resistant bacteria from various clinical specimens have been previously reported, there is limited data on antifungal resistance of fungal pathogens from Nepal [11–13]. Therefore, this study aims to determine the prevalence of fungal pathogens and identify the types of fungi among COVID positive and negative patients. This study will also assess the antifungal susceptibility pattern and biofilm forming ability of the isolated Candida spp. Identification of etiological agents of secondary infections is important to timely combat their complications and helps determine the appropriate treatment regimens. By presenting these findings, we aim to contribute valuable insights into the prevalence and characteristics of fungal pathogens in the Nepalese context. This study not only sheds light on the current scenario but also lays the foundation for future research and strategic interventions to enhance the understanding and management of fungal infections.
Methods
Study design
This hospital based cross sectional study was conducted from July 2022 to July 2023 at Bhaktapur Hospital, Bhaktapur, and Central Department of Microbiology, Tribhuvan University (TU), Kirtipur, Kathmandu Nepal. Bhaktapur hospital is a referral hospital located at Bhaktapur district to which COVID suspected cases were referred from other hospitals and health centres within Bhaktapur district. Sample collection and processing of samples for culture and identification of fungi was done at Bhaktapur hospital. Antifungal susceptibility testing and detection of biofilm formation was done at Central Department of Microbiology, TU.
Study population
Patients visiting Bhaktapur Hospital confirmed as COVID positive by laboratory through either rapid antigen test (SURE STATUS), or PCR (XABT) were included in the study.
Patients with COVID negative results as confirmed by laboratory and suspected of fungal infections by the attending doctors were also included. Considering the estimated prevalence of fungal infection in COVID cases (p) to be 4% with a 95% confidence interval (z), and a 5% maximum tolerable error, the minimum sample size calculated was 30 [14]. However, all the COVID positive cases (n = 131) and COVID negative cases (n = 101) visiting the hospital during the study period were included in the study.
Sample collection and transportation
Sputum samples were collected from patients requesting the culture; both from COVID-positive and negative cases. Each patient was instructed to cough vigorously and collect the sputum not saliva preferably in the morning in a clean, dry, sterile, and leak-proof wide-mouth container. The samples were immediately transported to the laboratory and processed for fungal culture and antifungal susceptibility testing.
Culture of specimen
Each sample was processed for fungal culture by following standard microbiological techniques. The collected specimen was immediately inoculated on two Sabouraud’s Dextrose Agar (SDA) plates. One SDA plate was used for central point inoculation for the identification of filamentous fungi and incubated aerobically at 28 °C for upto 2 weeks. The second SDA plate was streaked and incubated at 37 °C for 24–48 h aerobically for identification of yeast and dimorphic fungi [15].
Identification of fungal pathogens
Identification of fungal pathogens was done by observation of colony morphology on culture plate (SDA), simple staining, lactophenol cotton blue staining, Germ tube test and HiCrome™ Candida differentiation media for identification of Candida species [16].
Antifungal susceptibility testing
Candida spp. were further processed for antifungal susceptibility testing by disk diffusion methods in Mueller–Hinton Agar (MHA) supplemented with 2% glucose, providing a suitable growth for most of the yeasts and 0.5 mg/l methylene blue dye with pH 7.2 to 7.4 following CLSI guideline (CLSI 2018).
Detection of biofilm production by microtiter plate method
Biofilm formation was detected by microtiter plate method. A single colony of each isolate was picked and inoculated into tubes containing 2 ml of freshly prepared Brain Heart Infusion Broth (BHIB) and incubated aerobically at 37 °C for 24 h. After incubation, all the broth cultures were diluted at a ratio of 1: 20 using fresh BHIB. Then, 200 μl of each diluted broth was placed into microtiter plates and then incubated at 37 °C for 24 h. After complete incubation, the microtiter plates were drained and rinsed with distilled water three times, inverted to blot. Then each well was filled with 200 μl of 1% crystal violet and incubated for 15 min. Following incubation, the microplates were again rinsed three times with distilled water. Then, 200 μl of ethanol: acetone mixture (80:20 w/v) were added to each well and were read at 450 nm using an ELISA reader, and optical density (OD) was recorded for each well. Sterile BHIB without microorganisms was used as the negative control [17].
The experiment was performed in triplicate. The cut-off value was determined by arithmetically averaging the OD of the wells containing sterile BHIB and by adding a standard deviation of + 2. Samples with an OD higher than the cut-off value were considered positive, whereas those with lower optical density than the cut-off was considered to be negative [18].
Data analysis
The obtained data was entered into SPSS (Statistical Package for Social Sciences) software. To ascertain the significance between the study variables, the Chi-square test was performed and a p-value of < 0.05 was considered to be statistically significant.
Results
The fungal growth among COVID-positive cases was 6.67% (9/135) and that among COVID-negative cases was 22.77% (23/101).
Fungal growth in different age groups among COVID positive and negative cases
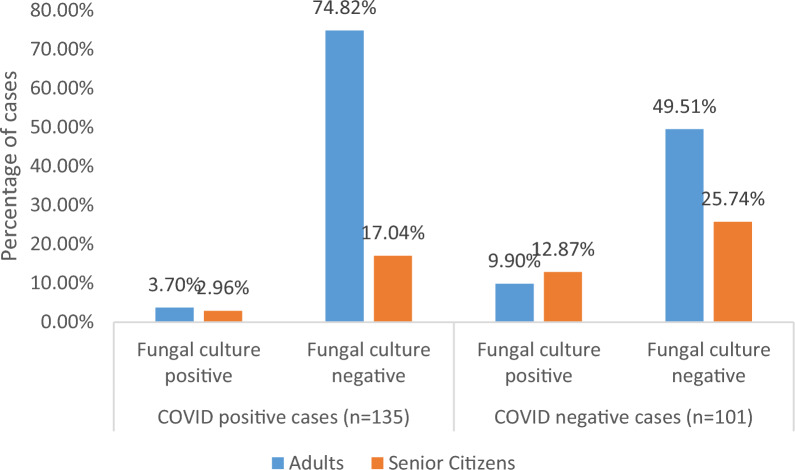
The fungi was not isolated from COVID positive children (n = 2) as well as COVID negative children (n = 2). The fungal infection was more among the senior citizens as compared to other age groups. However, the association between age-wise distribution of COVID-positive and COVID negative cases and presence of fungal growth was not statistically significant indicating all ages are equally vulnerable to fungal infection (Fig. 1).
Fig. 1.
Fungal growth pattern among different age groups in COVID-positive and negative cases
Fungal growth pattern among both gender in COVID-positive and negative cases
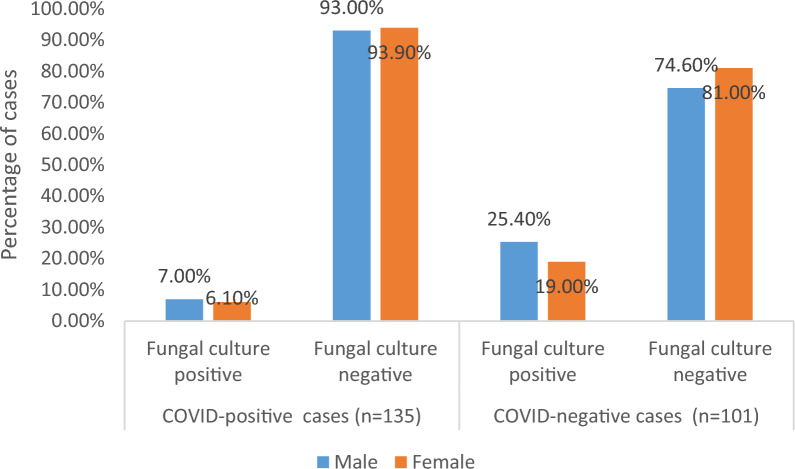
Fungal growth was slightly more among male patients (both COVID positive and negative) as compared to female patients. However, there was no significant association between the gender-wise distribution of COVID-positive (p = 0.848) and COVID-negative (p = 0.451) cases and fungal growth implying that fungal infection may affect both genders equally either COVID-positive or negative (Fig. 2).
Fig. 2.
Fungal growth pattern among both gender in COVID positive and negative cases
Distribution of fungal isolates in COVID-positive and COVID-negative patients
Candida albicans and Mucor were isolated from both COVID-positive and COVID-negative cases. The fungi categorized as critical priority –Aspergillus fumigatus and high priority- Candida tropicalis were isolated from only COVID positive cases. Similarly, Candida parapsilosis, Alternaria and Trichosporon were isolated only from COVID negative cases (Table 1).
Table 1.
Distribution of fungal isolates in COVID-positive and COVID-negative patients
| Organism isolated | COVID-positive No. (%) |
COVID-negative No. (%) |
Total No. (%) |
|---|---|---|---|
| Candida albicans | 3 (33.33%) | 16 (69.6%) | 19 (59.4%) |
| Candida tropicalis | 1 (11.11%) | 0 | 1 (3.1%) |
| Candida parapsilosis | 0 | 1 (4.3%) | 1 (3.1%) |
| Aspergillus flavus | 2 (22.22%) | 0 | 2 (6.3%) |
| Aspergillus fumigatus | 1 (11.11%) | 0 | 1 (3.1%) |
| Mucor | 1 (11.11%) | 1 (4.3%) | 2 (6.3%) |
| Unidentified budding yeast | 1 (11.11%) | 2 (8.7%) | 3 (9.4%) |
| Alternaria | 0 | 1 (4.3%) | 1 (3.1%) |
| Trichosporon spp. | 0 | 2 (8.7%) | 2 (6.3%) |
| Total | 9 (100%) | 23 (100%) | 32 (100%) |
Antifungal susceptibility pattern of Candida spp.
More than 60% of Candida albicans were susceptible to Miconazole and 38% were resistant to Ketoconazole. Similarly, both Candida parapsilosis and Candida tropicalis were susceptible to Miconazole, dose dependent susceptible to Ketoconazole and resistant to Nystatin (Additional file 1: Table S1).
Biofilm production
Out of 21 Candida spp, only one non-Candida albicans i.e. Candida tropicalis was weak biofilm producer (Additional file 2: Table S2).
Discussion
Neglected fungi are responsible for taking millions of lives globally and these trends are also being increased with COVID-19 complications [19]. Our study focuses on prevalence of fungal pathogens in both COVID-positive and COVID negative cases which showed slightly lower prevalence (6.67%) of fungal pathogens among COVID positive cases compared to the study conducted in the United States (13.4%) [20]. In a study conducted in Egypt, by Negm et al. (2023), secondary fungal infection was diagnosed in 32.8% of COVID-19 patients. Also, in a review study conducted by Seyedjavadi et al. (2022) from 1st January 2020 to 30th November 2021 on different continents, fungus co-infection occurred 49.7%, 23.2%, 19.8%, 6.6%, and 0.5% of COVID positive cases in Asia, America, Europe, Africa, and Australia, respectively [21]. Different factors including lifestyle, genetic, occupation, and other unknown factors may have contributed to the low fungal infection in our study [22]. Literatures suggest that mutations in nicotinamide adenine dinucleotide phosphate oxidase causes defects in phagocyte effector function which in turn predispose invasive infections by filamentous molds [23]. Similarly, IL-12/interferon γ signaling abnormalities predispose infections by dimorphic fungi [24]. Furthermore, impairement in IL-17 signaling has been related to increased susceptibility to Candida infections [25].The isolation of fungal pathogens from senior citizens as compared to other age groups in our study is in agreement with the study conducted by Seyedjavadi et al. (2022) in different continents (America, Europe, Australia, Asia, and Africa), where the maximum fungal coinfection was seen in COVID-related patients above 50 years of age [32]. Literatures suggest that various factors make the condition ideal for fungal infections in COVID positive cases which include low oxygen conditions due to patient’s hypoxemia, high glucose levels in case of diabetics, steroid induced hyperglycemia, and supressed immune response due to virus/ steroid treatment [26]. However, it is quite interesting that our study showed lower fungal growth in COVID-19 adults and senior citizens as compared to non-COVID-19 cases. Provided that COVID-19 further weakens the immune response increasing the chance of opportunistic fungal infection, it contradicts the general understanding. To better understand the reasons behind the lower fungal infections in COVID positive cases as compared to COVID negative cases, further investigations and analyses using a thorough review of patient characteristics, treatment regimens and immune responses may be necessary.
In contrast to a review study, showing 72.9% and 25.9% of fungal infection in males and females respectively, our study showed that the prevalence of fungal infection among COVID positive cases was not associated with gender. A study conducted by Bwire (2020) illustrated different factors responsible to robust immunity against COVID-19; Asian men express ACE2 in the lungs substantially more than Asian women do, according to single-cell RNA-sequencing investigations [27]. This genetic expression and cellular distribution pattern make males more vulnerable to SARS-CoV-2 infection than women. The expression of key immune components is increased in women due to the fact that they have two X chromosomes as opposed to one in males, and oestrogen and progesterone, which are found in female sex hormones, are also significant in initiating immune signalling and lowering inflammation. Also, lifestyle factors that include higher smoking and alcoholism in males were observed globally than in females. Besides, occupational factors and social factors like men are significantly higher in outdoor activities involving income generation that may expose them to crowded conditions making more airborne exposure [27].
The study conducted by Negm et al. (2023) reported the predominant fungi in critically ill I.C.U. admitted COVID-positive patient to be Candida followed by Aspergillus spp. and mucormycosis [19]. Similar to the present study, the study conducted by Peman et al. (2020) also reported different species of Candida and Aspergillus infection in COVID positive patients [4]. Similar to our study, a study conducted by Rafat et al. (2020) in Iran also confirmed fungal respiratory infection in 35.67% of the patients with the most predominant fungi being Candida albicans (37.22%) followed by Candida tropicalis (21.89%), Candida glabrata (12.4%), Candida krusei (5.83%), Candida parapsilosis (5.1%), Trichosporon asahii (2.18%), Geothricum candidum (2.18%), Aspergillus flavus (2.18%), Rhizopus orizae (0.72%), Aspergillus niger (0.72), Aspergillus fumigatus (0.72%) and Alternaria alternata (0.72%) [28].
Antifungal susceptibility testing of one Candida albicans out of 19 isolates in this study was not done as it couldn’t be revived. In contrast to the our study showing higher percentage of Candida albicans to be susceptible to Miconazole, the study conducted by Njunda et al. (2012) showed that the highest susceptibility of C. albicans with ketoconazole [29]. Similar to our study, the study conducted by Khadka et al. (2017) also illustrated that Candida isolates (C. albicans, C. tropicalis, C. krusei, C. glabrata) were highly resistant (86%) to ketoconazole whereas miconazole was mostly susceptible (44%) [30]. The study conducted by Tamai et al. (2020) reported that ketoconazole was susceptible to 31 isolates (62%), S-DD or intermediate to 5 isolates (10%) and 14 isolates (28%) were resistant [31]. In the present study among one each non-germ tube forming Candida species isolated (Candida parapsilosis and Candida tropicalis), both showed resistance to nystatin (50mcg), susceptible-dose dependent to ketoconazole (30mcg) and susceptibility to miconazole. Fungal growth is supported by the additional glucose supplementation in the MHA medium, and the zone edge is improved by the methylene blue dye [32].
Candida biofilms encompass the complex network of yeast, pseudo-hyphae, and hyphal cells protecting host immune defence and antifungal drugs [33]. Numerous studies emphasize the significance of biofilm as the possible virulence factor influencing microbial invasiveness and persistence, hence determining the severity of infection [34]. In our study out of 21 Candida species (both COVID and non-COVID patients) only one (4.76%) was a weak biofilm producer while the rest were biofilm non-producers.
Opportunistic fungal infections have been established in patients due to immune-compromised conditions brought on by COVID-19 infection, the existence of other co-morbidities and age-related variables. With the advent of drug-resistant pathogenic fungi, there are also few trustworthy, inexpensive diagnostic tools and few therapeutic options, posing a major danger to both the economy and public health. Therefore, it is essential that the fungal infection be correctly diagnosed and that inexpensive, efficient treatments and strategies be put in place in order to lower the morbidity. To the best of our knowledge, this is the first report from Nepal on fungal pathogens categorized as critical and high priority by WHO. With fungal infections on the rise, enhanced clinical vigilance and antifungal susceptibility testing are warranted.
Limitations
The major limitation of our study is the small sample size, making it difficult to generalize the data. Antifungal susceptibility tests for the molds were not done in the study. While SDA plates were employed for fungal culture, certain fungi may demand special techniques. In future studies, we recommend the use of alternative methods such as addition of selective media for culture or PCR to enhance the sensitivity and confirm the results.
Supplementary Information
Additional file 1: Table S1: Antifungal susceptibility pattern of Candida albicans (n=18).
Additional file 2: Table S2: Biofilm production among Candida isolates (n=21).
Acknowledgements
The author would like to acknowledge Central Department of Microbiology, Tribhuvan University, Nepal and Bhaktapur Hospital, Nepal for providing the laboratory facility and all the patients for providing samples.
Author contributions
Asmita Lamichhane, Sushma Regmi, Krishma Pandit, Sweety Upadhyay, Jyoti Acharya and Suprina Sharma reviewed the literature and performed the sample collection and laboratory analysis. Asmita Lamichhane drafted the manuscript. Srijana Koirala, Shreedhar Aryal, Krishna Gurung, Jiwan Thapa, Sanjib Adhikari, Pramod Poudel and Supriya Sharma designed the study, supervised the sample collection and laboratory analysis and critically reviewed the manuscript. All authors read and approved the final manuscript.
Funding
This work was partially supported by University Grants Commission (UGC), Nepal under UGC Collaborative research grant program (Grant no. CRG-79/79-S&T-02, Principal investigator- Dr. Supriya Sharma). The funder did not play any role in study design, data collection, analysis or interpretation of the data for the study or preparation of the manuscript and decision to publish.
Availability of data and materials
All data generated or analysed during this study are included in this published article and its supplementary information files.
Declarations
Ethics approval and consent to participate
This study was performed in accordance with the Declaration of Helsiki. Ethical approval of this study was obtained from the Institutional Review Committee (IRC) of the Institute of Science and Technology, Tribhuvan University (Ref. No.: IRC-IOST-22–0052). At the time of enrolment, written informed consent was taken from the patients or their legal guardians on behalf of the patients in case of participants under 16 years of age. Participants, parents or guardians were assured about the non-disclosure of information collected from them and were also informed about the use of data for analysis and using the results for improving patient care activities as well as publication without disclosing the name or identity of cases.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lai CC, Shih TP, Ko WC, Tang HJ, Hsueh PR. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrob Agents. 2020;55(3): 105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dawood FS, Ricks P, Njie GJ, Daugherty M, Davis W, Fuller JA, et al. Observations of the global epidemiology of COVID-19 from the prepandemic period using web-based surveillance: a cross-sectional analysis. Lancet Infect Dis. 2020;20(11):1255–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nepal: WHO Coronavirus Disease (COVID-19) Dashboard with vaccination data. https://covid19.who.int. Accessed 29 Jul 2023.
- 4.Pemán J, Ruiz-Gaitán A, García-Vidal C, Salavert M, Ramírez P, Puchades F, et al. Fungal co-infection in COVID-19 patients: Should we be concerned? Rev Iberoam Micol. 2020;37(2):41–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kundu R, Singla N. COVID-19 and plethora of fungal infections. Curr Fungal Infect Rep. 2022;16(2):47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silva LN, De Mello TP, De Souza RL, Branquinha MH, Roudbary M, Dos Santos ALS. Fungal infections in COVID-19-positive patients: a lack of optimal treatment options. CTMC. 2020;20(22):1951–7. [DOI] [PubMed] [Google Scholar]
- 7.Burki T. WHO publish fungal priority pathogens list. Lancet Microbe. 2023;4(2): e74. [DOI] [PubMed] [Google Scholar]
- 8.Meng Z, Guo S, Zhou Y, Li M, Wang M, Ying B. Applications of laboratory findings in the prevention, diagnosis, treatment, and monitoring of COVID-19. Sig Transduct Target Ther. 2021;6(1):316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma S, Acharya J, Rijal N, Chalise BS, Bhandari P, Banjara MR, et al. Cryptococcal meningitis in people living with human immunodeficiency virus in Nepal: Perspectives from resource limited setting. Mycoses. 2023;66(1):47–51. [DOI] [PubMed] [Google Scholar]
- 10.Rodrigues ML, Nosanchuk JD. Fungal diseases as neglected pathogens: a wake-up call to public health officials. PLoS Negl Trop Dis. 2020;14(2): e0007964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Acharya M, Joshi PR, Thapa K, Aryal R, Kakshapati T, Sharma S. Detection of metallo-β-lactamases-encoding genes among clinical isolates of Pseudomonas aeruginosa in a tertiary care hospital, Kathmandu. Nepal BMC Res Notes. 2017;10(1):718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shakya G, Kim DW, Clemens JD, Malla S, Upadhyaya BP, Dumre SP, et al. Phenotypic and genetic characterization of Vibrio cholerae O1 clinical isolates collected through national antimicrobial resistance surveillance network in Nepal. World J Microbiol Biotechnol. 2012;28(8):2671–8. [DOI] [PubMed] [Google Scholar]
- 13.Devkota SP, Sharma S, Bhatta DR, Paudel A, Sah AK, Kandel BP. Prevalence of the blaNDM gene among metallo-β-lactamase-producing gram-negative isolates from western Nepal. J Glob Antimicrob Resist. 2018;12:3–4. [DOI] [PubMed] [Google Scholar]
- 14.Soltani S, Zandi M, Faramarzi S, Shahbahrami R, Vali M, Rezayat SA, et al. Worldwide prevalence of fungal coinfections among COVID-19 patients: a comprehensive systematic review and meta-analysis. PHRP. 2022;13(1):15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong G, Miller HB, Allgood S, Lee R, Lechtzin N, Zhang SX. Use of selective fungal culture media increases rates of detection of fungi in the respiratory tract of cystic fibrosis patients. J Clin Microbiol. 2017;55(4):1122–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nadeem SG, Hakim ST, Kazmi SU. Use of CHROMagar Candida for the presumptive identification of Candida species directly from clinical specimens in resource-limited settings. Libyan J Med. 2010;5(1):2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marak MB, Dhanashree B. Antifungal susceptibility and biofilm production of Candida spp. isolated from clinical samples. Int J Microbiol. 2018;2018:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stepanovic S, Vukovic D, Dakic I, Savic B, Svabic-Vlahovic M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J Microbiol Methods. 2000;40(2):175–9. [DOI] [PubMed] [Google Scholar]
- 19.Negm EM, Mohamed MS, Rabie RA, Fouad WS, Beniamen A, Mosallem A, et al. Fungal infection profile in critically ill COVID-19 patients: a prospective study at a large teaching hospital in a middle-income country. BMC Infect Dis. 2023;23(1):246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanne JH. Fungal infections are especially dangerous for covid-19 patients. CDC study warns BMJ. 2023;15: p1378. [DOI] [PubMed] [Google Scholar]
- 21.Seyedjavadi SS, Bagheri P, Nasiri MJ, Razzaghi-Abyaneh M, Goudarzi M. Fungal infection in co-infected patients with COVID-19: an overview of case reports/case series and systematic review. Front Microbiol. 2022;6(13): 888452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lionakis MS. Genetic susceptibility to fungal infections in humans. Curr Fungal Infect Rep. 2012;6(1):11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Segal BH, Leto TL, Gallin JI, Malech HL, Holland SM. Genetic, biochemical, and clinical features of chronic granulomatous disease. Medicine. 2000;79(3):170–200. [DOI] [PubMed] [Google Scholar]
- 24.Rosenzweig SD, Holland SM. Defects in the interferon-gamma and interleukin-12 pathways. Immunol Rev. 2005;203:38–47. [DOI] [PubMed] [Google Scholar]
- 25.Mathis D, Benoist C. Aire. Annu Rev Immunol. 2009;27:287–312. [DOI] [PubMed] [Google Scholar]
- 26.Amin A, Vartanian A, Poladian N, Voloshko A, Yegiazaryan A, Al-Kassir AL, et al. Root causes of fungal coinfections in COVID-19 infected patients. Infect Dis Rep. 2021;13(4):1018–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bwire GM. Coronavirus: why men are more vulnerable to Covid-19 than women? SN Compr Clin Med. 2020;2(7):874–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rafat Z, Hashemi SJ, Ashrafi K, Nikokar I, Jafari A, Rahimi Foroushani A, et al. Fungal isolates of the respiratory tract in symptomatic patients hospitalized in pulmonary units: a mycological and molecular epidemiologic study. J Multidiscip Healthc. 2020;13:661–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Njunda AL, Nsagha DS, Assob JCN, Kamga HL, Teyim P. In vitro antifungal susceptibility patterns of Candida albicans from HIV and AIDS patients attending the Nylon health district hospital in Douala, Cameroon. J Public Health Africa. 2012;3(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khadka S, Sherchand JB, Pokhrel BM, Parajuli K, Mishra SK, Sharma S, et al. Isolation, speciation and antifungal susceptibility testing of Candida isolates from various clinical specimens at a tertiary care hospital. Nepal BMC Res Notes. 2017;10(1):218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamai IA, Pakbin B, Fasaei BN. Genetic diversity and antifungal susceptibility of Candida albicans isolates from Iranian HIV-infected patients with oral candidiasis. BMC Res Notes. 2021;14(1):93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alastruey-Izquierdo A, Melhem MSC, Bonfietti LX, Rodriguez-Tudela JL. Susceptibility test for fungi: clinical and laboratorial correlations in medical mycology. Rev Inst Med Trop Sao Paulo. 2015;57(Suppl 19):57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gulati M, Nobile CJ. Candida albicans biofilms: development, regulation, and molecular mechanisms. Microbes Infect. 2016;18(5):310–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Di Domenico E, Farulla I, Prignano G, Gallo M, Vespaziani M, Cavallo I, et al. Biofilm is a major virulence determinant in bacterial colonization of chronic skin ulcers independently from the multidrug resistant phenotype. IJMS. 2017;18(5):1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1: Antifungal susceptibility pattern of Candida albicans (n=18).
Additional file 2: Table S2: Biofilm production among Candida isolates (n=21).
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary information files.