Abstract
Candida albicans is an opportunist pathogen responsible for a broad spectrum of infections, from superficial mycosis to the systemic disease candidiasis. C. albicans has various morphological forms, including unicellular budding yeasts, filamentous pseudohyphae and true hyphae, and the ability to switch from yeast to hyphal forms is a key survival mechanism underlying the adaptation of the pathogen to the microenvironments encountered within the host. Filamentation is regulated by multiple signalling pathways and its induction in different growth media in vitro has often led to conflicting results. In this study, we performed quantitative proteomic analyses to compare the response of C. albicans yeast cells grown in YNB minimal medium to those of cells grown in four media widely used in the literature to induce the yeast-to-hyphae transition: YNB-Serum, YNB-N-acetylglucosamine (YNB-NAG), Lee medium and rich Spider medium. We show that each growth medium induces a unique pattern of response in C. albicans cells, and that some conditions trigger an original and specific adaptive metabolic response, showing significant differences in the intracellular content of the various filamentous forms. Moreover, this comparison of proteomic profiles indicates that the medium used can modify the thiol-dependent redox status of the cells, particularly in YNB-Serum and Lee medium and, to a lesser extent, in Spider medium, confirming the role of oxidative stress in the filamentation process. Overall, our data indicate that some of the media routinely used to induce hyphae cause significant changes in proteomic signature that should be taken account more carefully when exploring the hyphal transition in this pathogen.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12866-024-03627-4.
Keywords: Candida albicans, Oxidative stress, Metabolic adaptation, Filamentation, Proteomics
Introduction
Candida albicans is a major systemic fungal pathogen in humans and a commensal organism of the mucosal surfaces of the oral and vaginal cavities and the digestive tract [1–3]. C. albicans causes a broad range of infections, extending from superficial mycosis to life-threatening opportunistic bloodstream infections known as candidemia, which can develop into disseminated candidiasis, particularly in patients with compromised immunity. C. albicans can grow in various morphological forms, including unicellular budding yeasts, filamentous pseudohyphae and true hyphae, and some less common forms, such as chlamydospores and opaque cells [1, 4–8]. This morphological flexibility and the ability to utilize a variety of carbon sources is a key survival mechanism enabling the pathogen to adapt to the microenvironments encountered within the host [9–14]. Indeed, the hyphal form is invasive and can promote tissue penetration in the early stages of infection. Moreover, the yeast form can facilitate dissemination in the bloodstream [13, 15–17] but some studies have also shown that yeast-locked strains can efficiently penetrate tissues and structures [18–21].
The signalling pathways transducing environmental signals into morphological switching involve cAMP, MAPKinase, pH response and the Cph1-Efg1 transcription factors. These signals, which dynamically coordinate the transition from yeast to filamentous growth in response to diverse stimuli encountered in the host environment, are complex and have been extensively studied [5, 6, 9, 22–24]. In vitro, morphogenesis can be induced in various environmental conditions, such as culture at a temperature of 37 °C, in the presence of serum [25] and N-acetylglucosamine [26], an alkaline pH [27] or 5% CO2 [28], and the use of different conditions has often led to conflicting results (for a review see [5]). For example, we showed, in a precedent study, that the depletion in intracellular glutathione pools observed in the filamentous forms was associated with glutathione excretion into the culture medium and activation of glutathione-S-transferase Gst (fivefold increase) in Spider medium, but not in YNB-Serum [29]. In the same work, we observed a lower activation of the citrate synthase (2–threefold increase) in YNB-Serum than the one observed in the Spider medium (tenfold increase). Moreover, we also observed significant differences in the response to iron when the yeast-to-hyphae transition was induced in Spider or in the presence of serum [30].
YNB medium, which is known to promote the yeast form of the pathogen at 30 °C, is a minimal medium containing only essential nutrient. It can be supplemented with specific carbon sources such as glucose (YNB-glucose medium). Filamentation is enhanced by culture at a temperature of 37 °C in the presence of foetal bovine serum (FBS), which contains additional growth factors and signalling molecules [25]. The addition of N-acetylglucosamine, a derivative of glucose, to the minimal YNB medium, also promotes the yeast-to-hyphae transition [26]. Hyphal growth is also often induced in synthetic growth media, such as Lee's medium or the semi-synthetic Spider medium [31, 32]. Lee’s medium, widely used in published studies, provides specific nutrients such as amino-acids, salts and glucose as the carbon source, and it induces robust filamentation. The rich Spider medium, with mannitol as the carbon source, is also routinely used to promote morphogenesis. Hence, each of these media provides a unique environment and nutrient conditions triggering filamentation.
We have shown that the adaptive response of the pathogen in terms of filamentation in both YNB-serum and Spider medium is associated with a redistribution of bioenergetic pathways and a strong decrease in intracellular glutathione levels [29, 33, 34]. The ability of C. albicans cells to respond to environmental conditions therefore probably depends on the metabolic state of the cells and the external availability of nutrients. It is, thus, crucial to understand the adaptive strategies of the pathogen in response to different growth media, and global proteomic studies of the different filamentation conditions are therefore required.
Global proteomic investigations of C. albicans, including some by our group, have been published [30, 33, 35–41]. However, to our knowledge, there have been no direct comparisons of the proteomes of C. albicans cultured in the different growth media routinely used. The study of the proteomes of yeast and filamentous forms, induced under different conditions, should make it possible to identify the proteins and specific metabolic pathways associated with the morphological transition and to develop a correlation between the various published results related to the yeast-to hyphae transition.
We performed quantitative global proteomic analyses to investigate the response of C. albicans cells to four media widely used to induce the yeast to hyphae transition: YNB-Serum, YNB-NAG, Lee and Spider medium. We ran a global proteomic analysis focusing on the overall differences in protein abundance between the yeast form and the four filamentous forms. We found that the different growth conditions triggered changes in the abundances of specific sets of proteins, indicating a specific metabolic adaptive response to each medium, with oxidative stress molecules implicated in these changes in at least two of the yeast-to-hyphae transition-inducing media. These data indicate significant differences in the intracellular content of each filamentous form, and suggest that, under the term filamentation, the details of the experimental procedures and the medium used to induce the transition should be more carefully considered in future explorations of the pathogen.
Results
We compared the proteomic profiles associated with the various filamentation conditions for C. albicans, by culturing cells in minimal YNB-glucose medium at 30 °C, conditions known to favour the yeast form, and in four growth media routinely used to induce the filamentation process at 37 °C: YNB-Serum (containing 10% bovine foetal serum), YNB-N-acetylglucosamine 2.5 mM (YNB-NAG), Lee medium and the rich Spider medium, with mannitol as the main carbon source. In YNB-glucose, YNB-Serum and Lee medium the carbon source was 0.1% glucose, and the conditions in these media therefore resembled those to be encountered by the pathogen within the host, in which glucose levels range from 0.06 to 0.1% (3–5 mM) in the bloodstream during systemic infections, reaching 0.5% in vaginal secretions [42, 43]. We controlled for variation due to differences in pH between the media by carefully checking the pH of each medium before inoculation, and when necessary, adjusting the pH to 7.0. The four culture media tested are representative of the conditions routinely used to induce the yeast-to-hyphae transition.
Phenotypic analysis of the influence of the various filamentation conditions
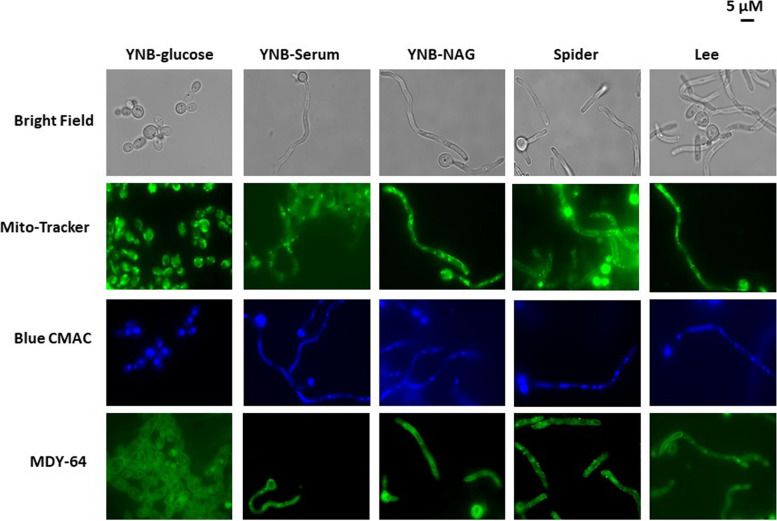
Before performing quantitative proteomic experiments, we used liquid culture assays to assess the ability of C. albicans cells to grow under the various filamentous conditions. For microscopy, cell cultures were established by inoculating fresh medium, with an overnight culture in YNB-glucose medium, to an OD of 0.1. The cells were cultured for six hours (corresponding to the time required for yeast cells to enter to exponential growth phase) and at this time point, microscopy studies were performed and the cells used for proteomic analyses were harvested. Microscopic analysis of C. albicans cultures in the minimal YNB-glucose medium revealed, as expected, a mixture of unicellular and budding yeast forms (Fig. 1). The presence of hyphae and/or pseudo-hyphae was observed when the cells were grown in the four hypha-inducing conditions. These findings were consistent with expectations, but it was important to verify the cell morphology associated with each set of growth conditions before performing quantitative global proteome analysis. Various fluorescent dyes were also used to further investigate the morphological variability induced by the filamentation-inducing media more specifically (Fig. 1).
Fig. 1.
Effect of various hypha-inducing media on the morphology of Candida albicans SC5314 cells. Cells from an overnight culture in YNB-glucose medium were used to inoculate the desired medium at an OD600nm of 0.1 and were then cultured until an OD600nm of 0.8 was reached. Images were then taken with a phase-contrast microscope (100X), as described in the experimental procedures. The scale bar represents 5 μM
The cell-permeant green MitoTracker probe diffuses passively across the plasma membrane and accumulate in active mitochondria. In YNB-glucose medium, a clear continuum of fluorescence was observed, typical of a mitochondrial tubular network and accumulation of mitochondria towards the cell wall (Fig. 1). During filamentation, as the cell elongates the cell organelle morphology gets changed. Moreover, in fungi, growth on different carbon sources can affect mitochondrial organization, and we expected mitochondria to be organized differently depending on the growth medium. Interestingly, in the YNB-NAG and Lee media, the mitochondrial network appeared to be partly fragmented and very different from the continuous network observed in the yeast forms. In the presence of serum, mitochondrial fragmentation increased, as indicated by the multiple green fluorescent dots in Fig. 1. Interestingly, when cells were cultured in the rich Spider medium, they appeared to have a lower mitochondrial density of the cells, which may be the result of the morphologic changes in the cells, but could also suggest a potentially different energetic profile. Taken together, the images of Fig. 1 show that the morphological changes associated to the transition to hyphae are associated with modifications of the mitochondrial network potentially correlated with changes in proteome profile.
The yeast vacuole marker green MDY-64, which stains the vacuole membrane and endosomes, and blue CMAC, which stains the vacuole lumen, were also used in the explorations shown in Fig. 1. In YNB-glucose medium, the blue florescence occupied most of the volume of the vacuole, as classically observed in the yeast Saccharomyces cerevisiae. In YNB-NAG, the vacuoles were smaller than those observed in yeast forms. Moreover, in the YNB-Serum and Lee media, vacuole size decreased from the cell body to the end of the hyphae (blue CMAC staining). Again, as observed with the green Mitotracker, the results obtained for Spider medium were different from those obtained in other conditions. Indeed, endocytosis vesicles were also observed when cells were grown in Spider medium (Fig. 1, MDY-64 staining), but not when they were grown in the other yeast-to-hyphae transition-inducing conditions. These observations suggest that endocytosis rates may be higher in Spider medium. Altogether, the microscopy analyses of Fig. 1 showed that the four media allow the production of filamentous forms, and the use of fluorescent dye suggested changes in the intracellular content. Overall, the morphology of the cells subjected to proteomic analyses was as anticipated and made it possible to explore the global proteome in these different growth conditions.
Proteomic analyses and principal component analysis
We performed a label-free quantitative proteomic analysis, to identify the principal proteins affected in the different filamentation growth media. Cells were cultured to exponential growth phase in YNB-glucose minimal medium at 30 °C as a control for yeast production. The filamentation media used were YNB-Serum, YNB-NAG, the synthetic Lee medium and the rich Spider medium. The cell extracts obtained under each set of conditions were subjected to mass spectrometry.
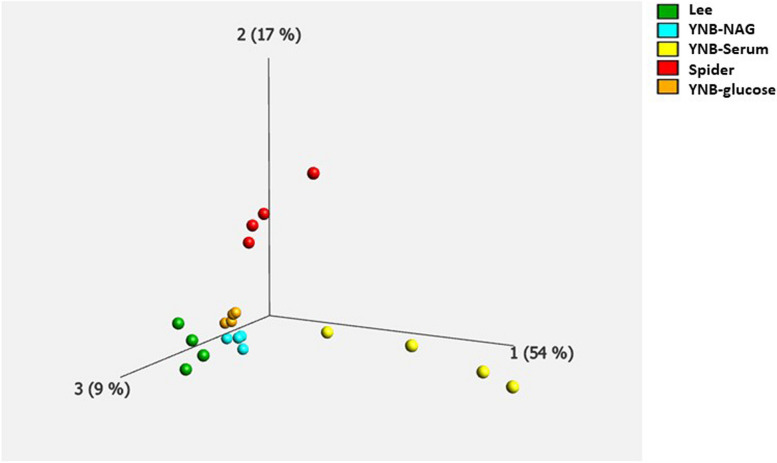
We performed a Principal Component Analysis (PCA) on the quantitative proteomic data for the 1454 identified proteins, to obtain an overview of the proteomic profile in each set of conditions. A plot of the three main eigenvectors is shown in Fig. 2, using a q value of 0.05. The data for each set of conditions are aligned along the same axis, indicating that the data for biological replicates are robust. Interestingly, the data obtained with the YNB-serum medium (yellow dots) were slightly less homogeneous within replicates, than those obtained with the other media, as also observed in previous published work of our laboratory [29, 33, 34]. In addition, the groups corresponding to each set of growth conditions are clearly separated, providing evidence for specific responses according to the culture medium used (Fig. 2). Moreover, the different sets of conditions contribute to proteome variability to different extents. For example, the YNB-Serum and Spider conditions accounted for 54% and 17%, respectively, of the differences in normalized abundances within the different conditions (Fig. 2, yellow and red dots). Separate groups were observed when cells were grown in Lee medium (green), YNB-Serum (yellow) or Spider medium (red), indicating that each set of growth conditions triggers specific variations and responses. The proteins abundance profiles in the presence of YNB-glucose and YNB-NAG media were quite similar (orange and light blue dots on Fig. 2), as expected given that the only difference between these two growth media is the carbon source. These findings suggest that the proteins identified in these two conditions are likely to be different from those identified in the other conditions. Taken together, the PCA analysis and the microscopy results presented in Fig. 1, show that the different media used in our study trigger different sets of specific responses and modifications of the global proteome.
Fig. 2.
Principal component analysis (PCA) on the levels of 1454 proteins differentially expressed between media. The significance of the observed differences was determined with a q-value threshold of 0.01 (values below this threshold indicate that the differences are statistically significant at a high level of confidence). The PCA revealed five distinct protein groups, each representing a different set of experimental conditions: yeast nitrogen base (YNB-glucose) in orange, YNB-Serum in yellow, Lee medium in green, YNB-NAG in blue, and Spider medium in red. The percentage of the variance associated with each principal component is indicated on the axis legend
Proteomic analyses and clustering
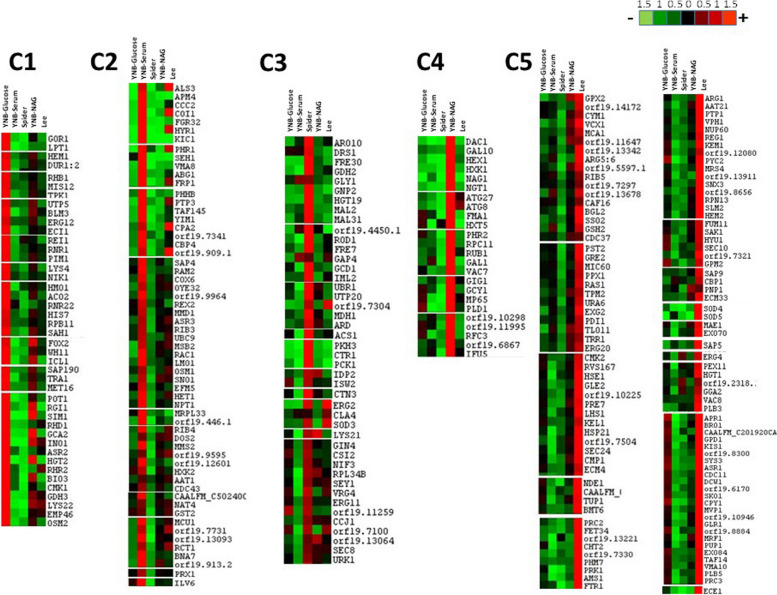
The differentially expressed proteins were then classified with the Autoclass@IJM Bayesian clustering algorithm [44], in a specific clustering analysis of the label-free data for direct comparisons of protein abundance between the five sets of conditions (Fig. 3). This classification identified groups of proteins that could not be directly deduced from a classical visual examination of protein abundance profiles, and the list of proteins identified in the five clusters of Fig. 3 is presented in Supplementary Table 1. The results of the clustering analysis reflected a differential response of C. albicans to the four sets of filamentation conditions, and confirmed the importance of comparing these growth media. Furthermore, approximately 50% of the proteins identified in the proteomics analysis were specifically more abundant in one particular set of growth conditions. For example, Cluster 1 contained proteins that were specifically more abundant in YNB-glucose medium (yeast forms), whereas Cluster 2 mostly contained proteins more abundant in the presence of serum. Similarly, Clusters 3, 4 and 5 identified sets of proteins specifically induced in Spider, YNB-NAG and Lee medium, respectively.
Fig. 3.
Overview of the JavaTreeView output for the AutoClass clustering of proteins with different normalised abundances (ANOVA p value < 0.05). The data used for this classification were obtained in four independent biological experiments and the Autoclass Bayesian classification was based on the mean of the means of the normalised abundances of the different proteins obtained in the proteomic analyses. The classified mean of means values are shown on a red-green colour scale, where red corresponds to higher abundances and green to lower abundances. Differences in protein levels are indicated on a linear scale. The list of proteins displaying significant changes in abundance is presented in Supplementary Table 1
Hyphal development in C. albicans requires several hyphae-specific genes [22, 24, 45]. The most highly expressed genes encode the cell wall protein hyphal wall protein 1 (Hwp1), the secreted aspartyl protease (SAP) family proteins Sap4, Sap5 and Sap6, cell surface adhesin 3 (Als3) and cell elongation protein 1 (Ece1). As expected, with the exception of the hyphal wall protein Hwp1, most of the hypha-specific proteins (Ece1, Als1, Als3, Als4, Hyr1, Rbt1, Sap 4, Sap5, Sap6…) were abundant and identified in the various inducing conditions, and the changes in abundance of the major hypha-specific proteins are shown in the box plots of supplementary Fig. 1. Hyphally regulated protein (Hyr1), cell surface adhesin 3 (Als3), cell surface adhesin 1 (Als1), aspartyl proteinase 4 (Sap4) and aspartyl proteinase 6 (Sap6) were particularly abundant in the presence of serum (Fig. 3 Cluster 2). Moreover, Als3, Hyr1 and Sap6 were also induced in Lee medium and, to a lesser extent, in YNB-NAG medium (Fig. 3 cluster 2 and Supplementary Fig. 1). Aspartyl proteinase-5 (Sap5) was greatly induced in Lee medium, and, to a much lesser extent, when the filamentation was induced in presence of serum or YNB-NAG. In addition, the extent of elongation protein Ece1 was identified in cluster 5, as being particularly induced when cells were grown in Lee medium (Fig. 3 Cluster 5 and Supplementary Fig. 1).
Tricarboxylic acid (TCA) cycle proteins, such as citrate synthase (Lys22), aconitase (Aco2), fumarate reductase (Osm2), or the glyoxylate cycle enzymes isocitrate lyase (Icl1) and glyoxylate reductase (Gor1) were more abundant in the yeast forms observed in YNB-glucose medium (Cluster 1). Both YNB-Serum and Spider media (Clusters 2 and 3) also stimulated the production of proteins involved in bioenergetics. Cluster 2 contained a set of proteins induced in YNB-serum, including fumarate reductase (Osm1), components of complexes I (NADH dehydrogenase) and IV (Cox6) of the mitochondrial respiratory chain, and ATP synthase subunits. In the presence of serum, hexokinase (Ilv6), which is involved in the glycolytic pathway, was also induced (Fig. 3 Cluster 2). ATP synthase subunits were also identified in Cluster 3 in Spider medium, altogether with malate dehydrogenase (Mdh1), homocitrate synthase (Lys21) isocitrate dehydrogenase (Idp2), acetyl-coA synthetase (Acs1) and PEP carboxykinase (Pck1) reflecting the strong stimulation of energetic and respiratory pathways in these conditions. The TCA cycle enzymes malate dehydrogenase (Mae1) and fumarate reductase (Fum11) were also induced in Lee medium (Fig. 3 cluster 5 and Supplementary Table 1). Interestingly, the abundance of proteins involved in glycolysis or the respiratory pathway was not significantly higher in Cluster 4, specific to YNB-NAG growth conditions. However, as expected, proteins involved in the GlcNac catabolic pathway were only identified in Cluster 4, including the GlcNAc-specific transporter (Ngt1), N-acetylglucosamine-6-phosphate deacetylase (Dac1) and the glucosamine-6-phosphate deaminase (Nag1). These data suggest that there is no significant difference in the proteomic profiles of C. albicans cells cultured in YNB-glucose and YNB-NAG conditions, consistent with the principal component analysis shown in Fig. 2.
The clustering analyses in Fig. 3 shows that sets of proteins involved in defences against oxidative stress were identified mainly in YNB-Serum and Lee medium (Fig. 3 clusters 2 and 5 and supplementary Table 1). In Cluster 5 (Lee medium), we detected the induction of a major set of glutathione-dependent proteins, including the main enzyme responsible for glutathione synthesis glutathione synthetase (Gsh2), together with glutathione reductase (Glr1), glutathione peroxidase (Gpx2) glutathione transferase (Ecm4) and thioredoxin reductase (Trr1), suggesting that thiol-dependent redox status may play a role in these conditions. Superoxide dismutases Sod3, Sod4 and Sod5 were also induced in Lee medium, together with the metacaspase Mca1. These data indicate that the primary defences against oxidative stress are activated when C. albicans cells are cultured in Lee medium. Moreover, several proteasome subunits were also identified in Cluster 5 (endopeptidase Pup1, proteasome subunits Rpn13 and Pre7). In the presence of serum, we also observed, albeit to a lesser extent, an induction of oxidative stress proteins, such as glutathione transferase (Gst2), thioredoxin peroxidase (Prx1) and glutathione peroxidase (Hyr1) (Fig. 3 Cluster 2). Interestingly, no strong induction of oxidative stress proteins was observed in YNB-glucose, YNB-NAG or spider medium, with the exception of the induction of Sod3 in Spider medium (Fig. 3 Clusters 1, 3 and 4).
Interestingly, as mentioned above for some of the hypha specific proteins, (As3, Hyr1 and Sap6), other clusters identified sets of proteins induced in more than one filamentation conditions, and the data are presented in the clusters of Supplementary Fig. 1. For example, Cluster 6 includes a set of proteins induced in both YNB-serum and YNB-NAG conditions, such as the acylcarrier proteins Acp1 and Acp12, the cytochrome oxidase subunit Cox19 or proteins of the oxidative stress defences such as glutaredoxin 3 (grx3) or superoxide dismutase (Sod1). Moreover, nine subunits of ATP synthase (ATT 14, ATP3, ATP2, ATP4, ATP20, ATP16, ATP7, ATP1, ATP5) were induced both in YNB-NAG and Lee media (Supplementary Fig. 1 Cluster 7), suggesting a strong need for energy in these two hypha-inducing conditions.
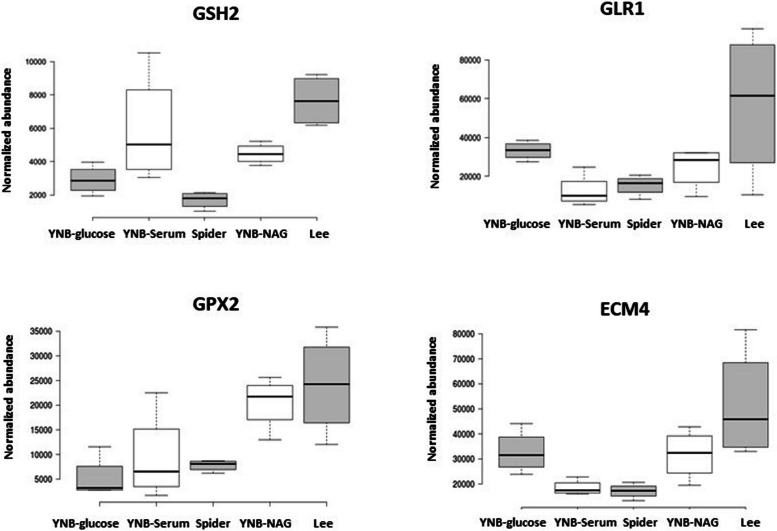
A closer analysis of the normalized abundances of various glutathione-dependent proteins confirmed the involvement of the thiol-dependent redox status in the filamentation process (Fig. 4). The glutathione synthetase Gsh2 was strongly induced in Lee medium and, to a lesser extent, in YNB-Serum and YNB-NAG, suggesting that the induction of hyphae is associated with an increase in the need for glutathione under these growth conditions. Moreover, this increase in glutathione synthesis was associated with an increase in the abundance of glutathione reductase Glr1 in Lee medium, suggesting the potential occurrence of a glutathione-dependent oxidative stress and the need to replenish the pools of reduced glutathione GSH. There was also an increase in glutathione peroxidase Gpx2 abundance in both Lee and YNB-NAG media, consistent with the potential occurrence of an oxidative stress and accumulation of the oxidised form, GSSG (Fig. 4). These findings reflect the induction of the cytoplasmic glutathione S-transferase Ecm4 in the Lee medium, which may be involved in the export of oxidised glutathione.
Fig. 4.
Comparison of the normalised abundances of various glutathione-dependent proteins. The data used for this representation were obtained in four independent biological experiments and the box plots were generated with the Chemgrid.org website. The lines in the middle indicate the medians obtained with the data from four biological replicates; the limits of the box indicate the 25th and 75th percentiles as determined by R software; the whiskers extend to 1.5 times the interquartile range from the 25th and 75th percentiles, outliers are represented by dots
Cluster 2 and 5 also include a set of proteins related to C. albicans virulence and/or responses to antifungal agents. In addition to the Sap family proteins mentioned above, the virulence factors vacuolar aspartic proteinase Apr1 and phospholipases B (Plb5 and Plb3) were induced in Lee medium (Cluster 5).
Comparison of the global proteomes of C. albicans cultured as the yeast forms and in various filamentation conditions
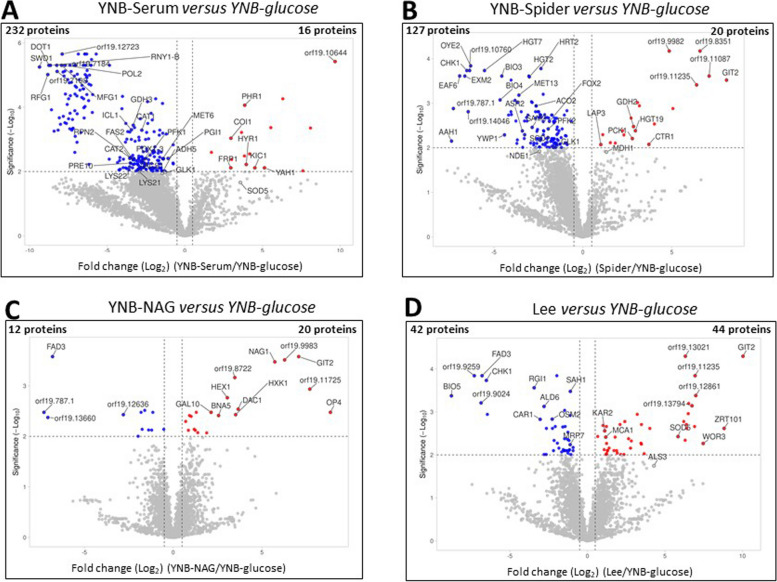
After comparing the global proteomes of C. albicans cultured in the five sets of growth conditions (Sects. 2.2 and 2.3), we performed pairwise comparisons of the data for the four sets of hypha-inducing conditions with the yeast form (YNB-glucose): i.e. YNB-NAG versus YNB-glucose, YNB-serum versus YNB-glucose, Spider versus YNB-glucose and Lee versus YNB-glucose. The quantitative data obtained were used to construct volcano plots showing the significant differences relative to YNB-glucose medium (q values) versus the magnitude of change (fold change, e.g., YNB-serum/YNB-glucose). The volcano plots facilitate the rapid identification of proteins with large fold-change differences that are statistically significant. In Fig. 5, points above the non-axial horizontal line represent proteins with abundances differing significantly (q < 0.01) between each of the filamentation-inducing media and YNB-glucose. Points to the left of the left-most non-axial vertical line correspond to fold-changes in protein levels of less than −1.5, whereas points to the right of the right-most non-axial vertical line correspond to fold-changes in proteins levels greater than 1.5 (the lists of proteins identified from the volcano plots are provided in Supplementary Table 2). The most striking feature of Fig. 5 is that the largest numbers of proteins with significant differences in abundance between media were obtained in comparisons of YNB-Serum and Spider media with the YNB-glucose condition. In these two media, more than 100 proteins were found to be present at levels significantly different from those in YNB-glucose. Moreover, the vast majority of these proteins, 232 proteins in YNB-serum and 127 in Spider, were downregulated compared to the YNB-glucose condition, whereas only about 20 proteins were upregulated in these two hypha-inducing conditions (Fig. 5A and B). The induction of hyphal growth in YNB-Serum and Spider conditions was, therefore, associated with changes in the abundance of a whole set of proteins relative to YNB-glucose medium, in response to the filamentation-inducing conditions. Proteins downregulated in these two hypha-inducing media included isocitrate lyase (Icl1) and glyoxylate reductase (Gor1), hexokinase 2 (Hxk2) and homoaconitase (Lys4) (Supplementary Table 2), consistent with the clustering analysis of Fig. 3. Moreover, the hypha-specific proteins Hyr1 was upregulated in YNB-serum when compared to YNB-glucose (Fig. 5). In addition, a number of proteins involved in sulfur metabolism was found to be downregulated in Spider medium. Comparison of the proteomes of the filamentous forms produced in Lee medium with the proteome of the yeast form grown in YNB-glucose medium yielded a very different pattern, with an almost equal numbers of proteins upregulated (44 proteins) and downregulated (42 proteins) (Fig. 5D). Isocitrate lyase (Icl1) and glyoxylate reductase (Gor1) were also differentially expressed, consistent with a lower level of involvement of the glyoxylate cycle in the filamentation process, as well as fumarate reductase (Osm2) (Supplementary Table 2). Consistent with our previous findings, the proteomic profiles of C. albicans cells cultured in YNB-NAG and YNB-glucose were largely similar (Fig. 5C), with only 20 and 12 proteins, respectively, showing significant changes in normalized abundances.
Fig. 5.
Comparison of the proteomes of the yeast form and the four filamentous forms. The -log10 q value was plotted against the log2 fold-change difference (filamentation/YNB-glucose) and the volcano plot highlights the proteins for which abundances differ significantly between the two sets of conditions. Points above the non-axial horizontal line represent proteins with significantly different abundances (q < 0.05). Points to the left of the left-most non-axial vertical line correspond to fold-changes in protein levels of than −1.5, whereas points to the right of the right-most non-axial vertical line correspond to fold-changes in protein levels greater than 1.5. The list of proteins displaying significant differences in abundance between conditions is presented in Supplementary Table 2
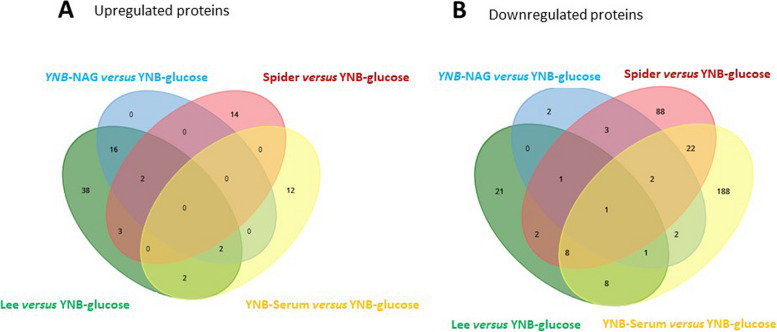
The lists of proteins identified as significantly up- or downregulated on the volcano plots in Fig. 5 are plotted on the Venn diagrams in Fig. 6 (the lists of proteins is presented in Supplementary Table 3). This graphic representation clearly shows that only a very small number of proteins are up- or downregulated in more than one set of filamentation conditions, consistent with the notion that each filamentation medium induces a unique differential response with respect to the yeast form. More than 80% of the 232 and 127 proteins downregulated in the filamentous forms obtained in YNB-Serum or Spider medium compared to the yeast form (YNB-glucose medium) were specific to only one set of growth conditions. There were 88 proteins specifically downregulated in the spider medium, 188 proteins in YNB-Serum and 21 proteins exclusively in Lee medium (Fig. 6B). These proteins represent the unique proteomic signature of the different yeast-to-hyphae inducing conditions (Supplementary Table 3). Interestingly, the number of proteins specifically upregulated (Fig. 6A) was significantly lower than the number of downregulated for each set of growth conditions. Moreover, the glyoxylate cycle proteins Icl1 and Gor1 were downregulated in these hypha-inducing media, in agreement with the results presented in Fig. 3. Only a few differentially expressed proteins were common to the Lee and Spider media. Furthermore, only two proteins were found to be downregulated exclusively in YNB-NAG growth conditions and not in YNB-glucose, consistent with the PCA findings shown in Fig. 2 and the clustering analyses (Fig. 3), suggesting that the global proteomes of the yeast forms and the filamentous forms induced in presence of N-acetylglucosamine are very similar.
Fig. 6.
Comparison of the total number of proteins in the yeast and filamentous forms and identification of the proteins specific to different forms. The total number of proteins and the number of proteins specific and common to different forms are shown in a Venn diagram generated with https://www.biotools.fr/misc/venny from the list of proteins generated from the volcano plots in Fig. 5A) Upregulated proteins, B) Downregulated proteins. YNB-Serum is shown in yellow, YNB-NAG in light blue, Spider medium in red and Lee medium in green. The list of proteins is presented in Supplementary Table 3
This is the first comparison of quantitative proteomic data for C. albicans cells grown in four classic hypha-inducing growth media. The various analyses indicate that each set of filamentation conditions is associated with a unique proteomic signature involving modifications to metabolic pathways. Moreover, our results highlight the importance of maintaining the thiol-dependent oxidative stress status of the cells during the filamentation process, especially in YNB-Serum and Spider media. These data shed new light on the intracellular status of C. albicans cells grown in four media widely used to induce hyphal growth, an aspect that should be considered in explorations of filamentation processes in this pathogen.
Discussion
Metabolic flexibility is very important in C. albicans, for proliferation, virulence and adaptation to the environments encountered within the host. In vitro, the production of hyphal forms can be induced by different growth media containing different nutrients and promoting the transition to hyphae via different signalling pathways; this use of different conditions to induce the production of hyphae has led to published results that are often complex and contradictory. Indeed, the composition of the medium is known to affect pathogen growth, but there are currently no robust data on the proteome of C. albicans cultured in the diverse widely used classical growth media. Proteomic data are progressively emerging for this pathogen [30, 33, 35–40]. A recent study compared the proteomic profiles of C. albicans cultured in solid and liquid media [46]. Its conclusions suggest that access to nutrients is restricted in solid media, forcing changes to metabolism and a search for alternative energy sources to replace the glucose readily available at earlier stages of culture in suspension in liquid medium. Moreover, we recently demonstrated that, in response to alternative carbon sources, C. albicans displays unique proteomic profiles, involving thiol-dependent defences against oxidative stress [33]. In addition to understanding the signalling pathways involved in the filamentation process, which have been extensively studied, we also need to decipher the metabolic changes associated with the various hypha-inducing media used in published studies. Here, we investigated the effect of hypha-inducing media, by exploring the concomitant changes to the proteome of C. albicans induced by culture as the yeast form and in four media routinely used to induce the filamentation process: YNB-Serum, YNB-NAG, Spider and Lee media.
Morphological analyses confirmed the production of unicellular budding yeasts in YNB-glucose and of filamentous forms in the four hypha-inducing growth media. It was important to perform this morphological control before the proteomic analysis, to establish associations between morphological changes and the proteomic profiles observed. In yeast, fragmentation of the mitochondrial network is an active process and a common response to stress, and the degree of mitochondrial fragmentation can provide information about the metabolic status of the cells [47, 48]. A typical dense and continuous mitochondrial network was observed in yeast cells, whereas fragmentation of the mitochondrial network was observed in the filamentous forms, consistent with the morphological changes of the cells.
The principal component analysis shown in Fig. 3 provides evidence for different responses in each set of conditions, as indicated by the distinct groups observed. The YNB-Serum and Spider media accounted for 54% and 17%, respectively, of the variability of the proteome. As both Spider and YNB-Serum growth media are widely used in the literature to induce the filamentation process, these data indicate that the experimental differences should be more considered when investigating the pathogen. The proteomes obtained in YNB-glucose and YNB-NAG conditions were very similar and appeared to differ from those observed when the cells were grown in other hypha-inducing media. However, the budding yeast form was observed in YNB-glucose, whereas hyphal forms were induced in YNB-NAG medium (Fig. 2). This suggests that morphological differences are not always associated with major changes in the global proteome. Conversely, it raises questions about whether similar morphologies may be associated with significantly different protein abundances.
Clustering analysis (Fig. 3) provided information on the principal metabolic pathways affected by the medium used to induce the filamentation process. Most of the hypha-specific proteins (Ece1, Als1, Als3, Hyr1, Sap4…) were identified in the filamentation conditions (Fig. 3 and Supplementary Fig. 1), confirming the induction of these proteins in our different hypha-inducing conditions. Interestingly, some of these proteins, i.e. Hyr1, Als3, Sap 4 or Sap6 were particularly abundant in YNB-serum and/or Lee medium, with a lesser increase of abundance in YNB-NAG or Spider medium, whereas the elongation protein Ece1 was highly induced when cells were grown in Lee medium.
Altogether, the clustering analysis shows that a very specific set of proteins was induced, depending on the medium, indicating a differential response. N-acetylglucosamine (NAG) is used as a nutrient by many micro-organisms and plays a crucial role in signalling in both bacterial and mammalian cells. It regulates several biological processes, including morphological transitions, virulence, and responses to oxidative stresses and antifungal agents, in several fungal species, including C. albicans [26, 49–55]. A comparison of clusters 1 and 4 suggested that, depending on the medium, the yeast-to-hyphae transition is not always associated with major changes in the proteome. Indeed, only a few differences in protein abundance were observed, as already suggested by the PCA shown in Fig. 2 (orange and light blue clusters). Moreover, the main proteins identified in Fig. 3 Cluster 4 were those involved in NAG transport and catabolism: Ngt1, Dac1 and Nag1 [51, 56].
Specific responses from the YNB-Serum and Spider media (Fig. 3 clusters 2 and 3) involve proteins of the bioenergetic pathways such as hexokinase, homocitrate synthase or malate dehydrogenase, complexes I and IV of the mitochondrial respiratory chain or ATP synthase subunits. In addition, the YNB-serum and Spider conditions were the growth conditions identified with the major significant changes in protein abundances compared to the YNB-glucose medium, as indicated by the PCA and volcano plots analyses. Volcano plots of Fig. 5 also identified isocitrate lyase and glyoxylate reductase to be less abundant in YNB-serum and Spider media than in YNB-glucose, and these two proteins were some of the few proteins identified in both growth conditions on the Venn diagram of Fig. 6. These data agree with some of our previous works showing a decrease of isocitrate lyase activity when the filamentation was induced in YNB-Serum or Spider medium, due to a glutathionylation process of the protein [29, 34]. Overall, our findings indicate that filamentation is associated with a medium-dependent adaptation of the energetic pathways.
Interestingly, the proteomic analyses also provided evidence for modifications to proteins involved in defence against oxidative stress. The levels of glutathione synthase (Gsh2), the key enzyme in glutathione biosynthesis, were significantly higher in both Lee medium and YNB-Serum than in the other media (Fig. 3 and Fig. 4) and, in Lee medium, this increase was coupled with an increase in the abundance of glutathione reductase, an essential enzyme for the maintenance of glutathione in its reduced state. Gsh2, Gst2 and Gpx1 were also overexpressed in the presence of serum. Together with the observed increases in the levels of glutathione peroxidase (Gpx2) and glutathione transferase (Ecm4), these data indicate a disturbance of glutathione-dependent redox status when filamentation is induced in the presence of serum or in Lee medium. They also suggest that an increase in glutathione biosynthesis is required, probably to compensate for the decrease in glutathione content and the accumulation of the oxidised form GSSG. These results are consistent with previous studies by our group showing a strong decrease in total glutathione levels and GSSG accumulation when filamentation is induced in YNB-Serum or Spider medium [29, 34]. The involvement of primary defences against oxidative stress in hypha induction in several media was confirmed by the induction of the superoxide dismutases Sod3, Sod4 and Sod 5 in Lee medium. Overall, our data suggest that filamentation of C. albicans in YNB-Serum or in Lee’s medium or, to a lesser extent, in Spider medium, is associated with intracellular oxidative stress and a disturbance of the thiol-dependent response of cells. Interestingly, there was no evidence for an increase in the abundance of proteins involved in oxidative stress in the presence of NAG. These findings raise questions about the relevance of comparisons between cells grown in different filamentation media, and confirm that morphologically similar forms are not always associated with similar intracellular content, depending on the growth medium, and that this methodological aspect should be more taken account in the studies of the pathogen.
Conclusions
In this study, we compared the proteomic profiles of C. albicans cells cultured in four different media routinely used to induce the yeast-to-hyphae transition: YNB-Serum, YNB-NAG, Lee medium and Spider medium. Overall, our data indicate that the different growth conditions trigger the different changes in the abundances of specific sets of proteins, indicating a unique pattern of specific metabolic adaptive response involving proteins of the energetic pathways in all the media tested and oxidative stress molecules in at least two of the media tested. These fundamental experimental differences should be taken account more carefully when exploring filamentation processes in this pathogen.
Methods
Yeast strains, media, and growth conditions
The virulent C. albicans wild-type strain SC5314 [57] was routinely cultured in complete synthetic YNB medium consisting of minimal YNB (Difco yeast nitrogen base) supplemented with 2% ammonium sulphate and a complete amino-acid mixture (CSM, Difco), in the presence of 0.1% glucose (YNB-glucose) and 10% bovine foetal serum when indicated (YNB-Serum). For the YNB-NAG medium, 2.5 mM N-acetylglucosamine was added as the carbon source. The Spider and Lee media were prepared as previously described [31, 32]. Differences in proteomic outputs due to differences in pH between the media were avoided by carefully checking the pH of each medium before inoculation and adjusting to pH 7.0 if necessary. All cultures were established from an overnight culture in liquid YNB containing 2% glucose, which was used to inoculate prewarmed medium to an OD of 0.1 in the morning, before vigorous shaking on a rotary shaker for six hours at 30 °C (yeast form) or 37 °C (filamentous forms) until exponential growth phase was reached.
Preparation of C. albicans cell extracts
C. albicans cultures were inoculated at an OD600nm of 0.1 with an overnight culture in YNB-glucose medium. Cells were cultured for six hours, corresponding to early exponential growth phase, and were then harvested by centrifugation. The pellets were whased three times in the appropriate medium containing 0.1% Triton X100. The pellets were then resuspended in 50 mM potassium phosphate buffer pH 7.8 (containing 0.1% Triton X100), washed three times, resuspended in the presence of protease inhibitors, and the cells were disrupted with glass beads and centrifuged at 5,000 × g for 30 min. The supernatant was used as the crude cell extract.
Microscopy
Cells were imaged at room temperature with a motorized Olympus BX-61 fluorescence microscope equipped with an Olympus PlanApo 100 × oil-immersion objective (1.40 NA), a QiClick cooled monochrome camera (QImaging, Surrey, BC, Canada), and MetaVue acquisition software (Molecular Devices, Sunnyvale, CA). Cells stained with MitoTracker and MDY64 were visualized with a GFP filter set (41,020 from Chroma Technology, Bellows Falls, VT; excitation HQ480/20 × , dichroic Q505LP, emission HQ535/50 m. The CMAC signal was visualized with a DAPI filter set (31013v2; Chroma Technology Corp.; excitation D365/10 × , dichroic 400dclp, emission D460/50 m). Images were then analyzed and processed in ImageJ (National Institutes of Health).
Quantitative analysis in label-free experiments
For global proteomic analyses, an overnight culture in YNB-glucose medium was used to inoculate the appropriate medium at an OD of 0.1, and the resulting culture was incubated until early exponential growth phase was reached (OD600nm = 0.8). Proteomic data were obtained from four independent samples. Cell extracts were then prepared as described above and the protein content of the samples was determined, to determine the amount of protein to be precipitated with acetone.
Materials
MS grade acetonitrile (ACN), MS grade H2O and MS grade formic acid (FA) were obtained from Thermo Fisher Scientific (Waltham, MA, USA). Sequencing-grade trypsin/Lys C mix was obtained from Promega (Madison, WI, USA). Trifluoroacetic acid (TFA), dithiothreitol (DTT), S-methyl methanethiosulphonate (MMTS) and ammonium bicarbonate (NH4HCO3) were obtained from Sigma-Aldrich (Saint-Louis, MO, USA).
Sample preparation for LC–MS/MS analysis
Six volumes of cold acetone (−20 °C) were added to a sample containing 10 µg of proteins. The tubes were vortexed tubes and incubated overnight at −20 °C, and were then centrifuged at 11,000 rpm, 4 °C, for 10 min. The protein pellets were dissolved in a buffer containing 8 M urea and 25 mM NH4HCO3. Samples were then reduced by incubation with 10 mM DTT and alkylated with 20 mM MMTS. After 16-fold dilution in NH4HCO3, samples were then digested overnight at 37 °C with a mixture of trypsin/Lys C (1/20 enzyme/substrate ratio). The digested peptides were loaded onto Evotips supplied by Evosep One (Odense, Denmark) and desalted according to the manufacturer’s procedure.
LC–MS/MS acquisition
Samples were analysed on a timsTOF Pro 2 mass spectrometer (Bruker Daltonics, Bremen, Germany) coupled to an Evosep One system (Evosep, Odense, Denmark) using the 30SPD method developed by the manufacturer. Briefly, this method is based on a 44-min gradient and a total cycle time of 48 min using a C18 analytical column (0.15 × 150 mm, 1.9 µm beads, ref EV-1106) equilibrated at 40 °C and operated at a flow rate of 500 nL/min. Solvent A was 0.1% FA in H2O and solvent B was 0.1% FA in ACN. The timsTOF Pro 2 was operated in PASEF mode1 with a cycle time of 1.3 s. Mass spectra for MS and MS/MS scans were recorded between 100 and 1700 m/z.
Data analysis
MS raw data files were processed using PEAKS Online X (build 1.8, Bioinformatics Solutions Inc.). Data were searched against the SwissProt Candida albicans database (strain SC5314/ATCC MYA-2876, downloaded January 2023, total entries 6035, YEAST). Parent mass tolerance was set to 20 ppm, and the fragment mass tolerance to 0.05 Da. Specific tryptic cleavage was selected and a maximum of two missed cleavages were allowed. For identification, the following post-translational modifications were included: acetylation (protein N-terminal), oxidation (M), deamidation (NQ) as variables and beta-methylthiolation (C) as fixed modifications. Identifications were filtered based on a 1% FDR (false discovery rate) threshold at both the peptide and protein group levels. Label-free quantification was performed with the PEAKS Online X quantification module, allowing a mass tolerance of 10 ppm, a CCS error tolerance of 0.02 and a 0.5-min retention time shift tolerance of 0.5 min for matching between runs. Protein abundances were inferred using the Top N peptide method and normalized with TIC.
Normalised abundances of proteins from trypsin digests with similar normalised abundance variations (ANOVA p < 0.05) were classified together by the AutoClass Bayesian clustering system [44] and visualised with Javatreeview (https://jtreeview.sourceforge.net/)
Data availability statement
The mass spectrometry proteomics data have been submitted to the ProteomeXchange Consortium via the PRIDE partner repository, with the dataset identifier PXD052475 [58].
The mass spectrometry proteomics data have been submitted to the ProteomeXchange Consortium via the PRIDE partner repository ProteomeXchange title: C. albicans cells exhibit specific proteomic profiles depending on the medium used to induce the filamentation process ProteomeXchange accession: PXD052475 Project Webpage: http://www.ebi.ac.uk/pride/archive/projects/PXD052475 FTP Download: https://ftp.pride.ebi.ac.uk/pride/data/archive/2024/10/PXD052475.
ProteomeXchange title: C. albicans cells exhibit specific proteomic profiles depending on the medium used to induce the filamentation process. ProteomeXchange accession: PXD052475.
Project Webpage: http://www.ebi.ac.uk/pride/archive/projects/PXD052475.
FTP Download: https://ftp.pride.ebi.ac.uk/pride/data/archive/2024/10/PXD052475.
Supplementary Information
Acknowledgments
We would like to thank Alex edelman associates for english scientific editing We would like to thank Yves Vesco for helping with data analyses
Authors’ contributions
FA designed experiments, curated data, supervised project and wrote and reviewed manuscript. VA runned the microscopy analyses. VL runned the mass spectrometry analyses and reviewed manuscript. GC runned PCA analysis. JMC reviewed manuscript.
Funding
No funding.
Data availability
The mass spectrometry proteomics data have been submitted to the ProteomeXchange Consortium via the PRIDE partner repository ProteomeXchange title: C. albicans cells exhibit specific proteomic profiles depending on the medium used to induce the filamentation process ProteomeXchange accession: PXD052475 Project Webpage: http://www.ebi.ac.uk/pride/archive/projects/PXD052475 FTP Download: https://ftp.pride.ebi.ac.uk/pride/data/archive/2024/10/PXD052475
Declarations
Ethics approval and consent to participate
Not applicable. No clinical trial involved in this study.
Consent for publication
Not applicable.
Competing interests
None
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Calderone RA. Candida and Candidiasis. Washington DC: American Society for Microbiology; 2002.
- 2.Mavor AL, Thewes S, Hube B. Systemic fungal infections caused by Candida species: epidemiology, infection process and virulence attributes. Curr Drug Targets. 2005;6(8):863–74. [DOI] [PubMed] [Google Scholar]
- 3.Odds FC: Candida and candidosis, 2nd edition edn. London; 1988.
- 4.Gow NA. Germ tube growth of Candida albicans. Curr Top Med Mycol. 1997;8(1–2):43–55. [PubMed] [Google Scholar]
- 5.Sudbery P, Gow N, Berman J. The distinct morphogenic states of Candida albicans. Trends Microbiol. 2004;12(7):317–24. [DOI] [PubMed] [Google Scholar]
- 6.Whiteway M, Bachewich C. Morphogenesis in Candida albicans. Annu Rev Microbiol. 2007;61:529–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown DH Jr, Giusani AD, Chen X, Kumamoto CA. Filamentous growth of Candida albicans in response to physical environmental cues and its regulation by the unique CZF1 gene. Mol Microbiol. 1999;34(4):651–62. [DOI] [PubMed] [Google Scholar]
- 8.Gow NA, Brown AJ, Odds FC. Fungal morphogenesis and host invasion. Curr Opin Microbiol. 2002;5(4):366–71. [DOI] [PubMed] [Google Scholar]
- 9.Brock M. Fungal metabolism in host niches. Curr Opin Microbiol. 2009;12(4):371–6. [DOI] [PubMed] [Google Scholar]
- 10.Brown AJ, Brown GD, Netea MG, Gow NA. Metabolism impacts upon Candida immunogenicity and pathogenicity at multiple levels. Trends Microbiol. 2014;22(11):614–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown AJ, Odds FC, Gow NA. Infection-related gene expression in Candida albicans. Curr Opin Microbiol. 2007;10(4):307–13. [DOI] [PubMed] [Google Scholar]
- 12.Miramon P, Lorenz MC. A feast for Candida: Metabolic plasticity confers an edge for virulence. PLoS Pathog. 2017;13(2):e1006144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitchell AP. Dimorphism and virulence in Candida albicans. Curr Opin Microbiol. 1998;1(6):687–92. [DOI] [PubMed] [Google Scholar]
- 14.Pellon A, Begum N, Sadeghi Nasab SD, Harzandi A, Shoaie S, Moyes DL: Role of Cellular Metabolism during Candida-Host Interactions. Pathogens. 2022;11(2):184. 10.3390/pathogens11020184. [DOI] [PMC free article] [PubMed]
- 15.Calderone RA, Fonzi WA. Virulence factors of Candida albicans. Trends Microbiol. 2001;9(7):327–35. [DOI] [PubMed] [Google Scholar]
- 16.Lo HJ, Kohler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90(5):939–49. [DOI] [PubMed] [Google Scholar]
- 17.Soll DR. Candida commensalism and virulence: the evolution of phenotypic plasticity. Acta Trop. 2002;81(2):101–10. [DOI] [PubMed] [Google Scholar]
- 18.Chen X, Kumamoto CA. A conserved G protein (Drg1p) plays a role in regulation of invasive filamentation in Candida albicans. Microbiology (Reading). 2006;152(Pt 12):3691–700. [DOI] [PubMed] [Google Scholar]
- 19.Grubb SE, Murdoch C, Sudbery PE, Saville SP, Lopez-Ribot JL, Thornhill MH. Adhesion of Candida albicans to endothelial cells under physiological conditions of flow. Infect Immun. 2009;77(9):3872–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saville SP, Lazzell AL, Monteagudo C, Lopez-Ribot JL. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot Cell. 2003;2(5):1053–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saville SP, Thomas DP, Lopez Ribot JL. A role for Efg1p in Candida albicans interactions with extracellular matrices. FEMS Microbiol Lett. 2006;256(1):151–8. [DOI] [PubMed] [Google Scholar]
- 22.Biswas S, Van Dijck P, Datta A. Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol Mol Biol Rev. 2007;71(2):348–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim J, Sudbery P. Candida albicans, a major human fungal pathogen. J Microbiol. 2011;49(2):171–7. [DOI] [PubMed] [Google Scholar]
- 24.Sudbery PE. Growth of Candida albicans hyphae. Nat Rev Microbiol. 2011;9(10):737–48. [DOI] [PubMed] [Google Scholar]
- 25.Taschdjian CL, Burchall JJ, Kozinn PJ. Rapid identification of Candida albicans by filamentation on serum and serum substitutes. AMA J Dis Child. 1960;99:212–5. [DOI] [PubMed] [Google Scholar]
- 26.Simonetti N, Strippoli V, Cassone A. Yeast-mycelial conversion induced by N-acetyl-D-glucosamine in Candida albicans. Nature. 1974;250(464):344–6. [DOI] [PubMed] [Google Scholar]
- 27.Buffo J, Herman MA, Soll DR. A characterization of pH-regulated dimorphism in Candida albicans. Mycopathologia. 1984;85(1–2):21–30. [DOI] [PubMed] [Google Scholar]
- 28.Mardon D, Balish E, Phillips AW. Control of dimorphism in a biochemical variant of Candida albicans. J Bacteriol. 1969;100(2):701–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guedouari H, Gergondey R, Bourdais A, Vanparis O, Bulteau AL, Camadro JM, Auchère F. Changes in glutathione-dependent redox status and mitochondrial energetic strategies are part of the adaptive response during the filamentation process in Candida albicans. Biochim Biophys Acta. 2014;1842(9):1855–69. [DOI] [PubMed] [Google Scholar]
- 30.Duval C, Macabiou C, Garcia C, Lesuisse E, Camadro JM, Auchère F. The adaptive response to iron involves changes in energetic strategies in the pathogen Candida albicans. Microbiologyopen. 2020;9(2):e970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee KL, Buckley HR, Campbell CC. An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida Albicans. Sabouraudia. 1975;13(2):148–53. [DOI] [PubMed] [Google Scholar]
- 32.Liu H, Kohler J, Fink GR. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266(5191):1723–6. [DOI] [PubMed] [Google Scholar]
- 33.Bayot J, Martin C, Chevreux G, Camadro JM, Auchère F. The adaptive response to alternative carbon sources in the pathogen Candida albicans involves a remodeling of thiol- and glutathione-dependent redox status. Biochem J. 2023;480(3):197–217. [DOI] [PubMed] [Google Scholar]
- 34.Gergondey R, Garcia C, Serre V, Camadro JM, Auchère F. The adaptive metabolic response involves specific protein glutathionylation during the filamentation process in the pathogen Candida albicans. Biochim Biophys Acta. 2016;1862(7):1309–23. [DOI] [PubMed] [Google Scholar]
- 35.Amador-Garcia A, Zapico I, Borrajo A, Malmstrom J, Monteoliva L, Gil C. Extending the Proteomic Characterization of Candida albicans Exposed to Stress and Apoptotic Inducers through Data-Independent Acquisition Mass Spectrometry. mSystems. 2021;6(5):e0094621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Groot PW, de Boer AD, Cunningham J, Dekker HL, de Jong L, Hellingwerf KJ, de Koster C, Klis FM. Proteomic analysis of Candida albicans cell walls reveals covalently bound carbohydrate-active enzymes and adhesins. Eukaryot Cell. 2004;3(4):955–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gil-Bona A, Amador-Garcia A, Gil C, Monteoliva L. The external face of Candida albicans: A proteomic view of the cell surface and the extracellular environment. J Proteomics. 2018;180:70–9. [DOI] [PubMed] [Google Scholar]
- 38.Gil-Bona A, Monteoliva L, Gil C. Global Proteomic Profiling of the Secretome of Candida albicans ecm33 Cell Wall Mutant Reveals the Involvement of Ecm33 in Sap2 Secretion. J Proteome Res. 2015;14(10):4270–81. [DOI] [PubMed] [Google Scholar]
- 39.Song N, Zhou X, Li D, Li X, Liu W. A Proteomic Landscape of Candida albicans in the Stepwise Evolution to Fluconazole Resistance. Antimicrob Agents Chemother. 2022;66(4): e0210521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vaz C, Pitarch A, Gomez-Molero E, Amador-Garcia A, Weig M, Bader O, Monteoliva L, Gil C: Mass Spectrometry-Based Proteomic and Immunoproteomic Analyses of the Candida albicans Hyphal Secretome Reveal Diagnostic Biomarker Candidates for Invasive Candidiasis. J Fungi (Basel). 2021;7(7). [DOI] [PMC free article] [PubMed]
- 41.Zore G, Abdulghani M, Kazi R, Shelar A, Patil R. Proteomic dataset of Candida albicans (ATCC 10231) Biofilm. BMC Res Notes. 2023;16(1):155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Egi M, Bellomo R, Stachowski E, French CJ, Hart GK, Hegarty C, Bailey M. Blood glucose concentration and outcome of critical illness: the impact of diabetes. Crit Care Med. 2008;36(8):2249–55. [DOI] [PubMed] [Google Scholar]
- 43.Owen DH, Katz DF. A vaginal fluid simulant. Contraception. 1999;59(2):91–5. [DOI] [PubMed] [Google Scholar]
- 44.Achcar F, Camadro J, Mestivier D. Autoclass@IJM: a powerful tool for Bayesian classification of heterogenous data in biology. Nucleic Acids Res. 2009;37(July):W63-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fan Y, He H, Dong Y, Pan H. Hyphae-specific genes HGC1, ALS3, HWP1, and ECE1 and relevant signaling pathways in Candida albicans. Mycopathologia. 2013;176(5–6):329–35. [DOI] [PubMed] [Google Scholar]
- 46.Alhameed RA, Semreen MH, Hamad M, Giddey AD, Sulaiman A, Al Bataineh MT, Al-Hroub HM, Bustanji Y, Alzoubi KH, Soares NC: Multi-Omics Profiling of Candida albicans Grown on Solid Versus Liquid Media. Microorganisms 2023;11(12). [DOI] [PMC free article] [PubMed]
- 47.Duvenage L, Walker LA, Bojarczuk A, Johnston SA, MacCallum DM, Munro CA, Gourlay CW: Inhibition of Classical and Alternative Modes of Respiration in Candida albicans Leads to Cell Wall Remodeling and Increased Macrophage Recognition. mBio 2019;10(1). [DOI] [PMC free article] [PubMed]
- 48.Knorre DA, Popadin KY, Sokolov SS, Severin FF. Roles of mitochondrial dynamics under stressful and normal conditions in yeast cells. Oxid Med Cell Longev. 2013;2013:139491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Naseem S, Gunasekera A, Araya E, Konopka JB. N-acetylglucosamine (GlcNAc) induction of hyphal morphogenesis and transcriptional responses in Candida albicans are not dependent on its metabolism. J Biol Chem. 2011;286(33):28671–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Naseem S, Min K, Spitzer D, Gardin J, Konopka JB. Regulation of Hyphal Growth and N-Acetylglucosamine Catabolism by Two Transcription Factors in Candida albicans. Genetics. 2017;206(1):299–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Du H, Ennis CL, Hernday AD, Nobile CJ, Huang G: N-Acetylglucosamine (GlcNAc) Sensing, Utilization, and Functions in Candida albicans. J Fungi (Basel) 2020, 6(3). [DOI] [PMC free article] [PubMed]
- 52.Williams RB, Lorenz MC: Multiple Alternative Carbon Pathways Combine To Promote Candida albicans Stress Resistance, Immune Interactions, and Virulence. mBio 2020; 11(1). [DOI] [PMC free article] [PubMed]
- 53.Gilmore SA, Naseem S, Konopka JB, Sil A. N-acetylglucosamine (GlcNAc) triggers a rapid, temperature-responsive morphogenetic program in thermally dimorphic fungi. PLoS Genet. 2013;9(9): e1003799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang G, Yi S, Sahni N, Daniels KJ, Srikantha T, Soll DR. N-acetylglucosamine induces white to opaque switching, a mating prerequisite in Candida albicans. PLoS Pathog. 2010;6(3):e1000806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rao HK, Paul S, Ghosh S. N-acetylglucosamine Signaling: Transcriptional Dynamics of a Novel Sugar Sensing Cascade in a Model Pathogenic Yeast, Candida albicans. J Fungi. 2021;7(1):65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alvarez FJ, Konopka JB. Identification of an N-acetylglucosamine transporter that mediates hyphal induction in Candida albicans. Mol Biol Cell. 2007;18(3):965–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gillum AM, Tsay EY, Kirsch DR. Kirsch DR: Isolation of the Candida albicans gene for orotidine-5’-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet. 1984;198(1):179–82. [DOI] [PubMed] [Google Scholar]
- 58.Vizcaino JA, Csordas A, Del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44(D1):D447-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been submitted to the ProteomeXchange Consortium via the PRIDE partner repository, with the dataset identifier PXD052475 [58].
The mass spectrometry proteomics data have been submitted to the ProteomeXchange Consortium via the PRIDE partner repository ProteomeXchange title: C. albicans cells exhibit specific proteomic profiles depending on the medium used to induce the filamentation process ProteomeXchange accession: PXD052475 Project Webpage: http://www.ebi.ac.uk/pride/archive/projects/PXD052475 FTP Download: https://ftp.pride.ebi.ac.uk/pride/data/archive/2024/10/PXD052475.
ProteomeXchange title: C. albicans cells exhibit specific proteomic profiles depending on the medium used to induce the filamentation process. ProteomeXchange accession: PXD052475.
Project Webpage: http://www.ebi.ac.uk/pride/archive/projects/PXD052475.
FTP Download: https://ftp.pride.ebi.ac.uk/pride/data/archive/2024/10/PXD052475.
The mass spectrometry proteomics data have been submitted to the ProteomeXchange Consortium via the PRIDE partner repository ProteomeXchange title: C. albicans cells exhibit specific proteomic profiles depending on the medium used to induce the filamentation process ProteomeXchange accession: PXD052475 Project Webpage: http://www.ebi.ac.uk/pride/archive/projects/PXD052475 FTP Download: https://ftp.pride.ebi.ac.uk/pride/data/archive/2024/10/PXD052475