Abstract
Background
Rheumatoid Arthritis (RA) is a chronic inflammatory disease that is primarily diagnosed and managed by rheumatologists; however, it is often primary care providers who first encounter RA-related symptoms. This study developed and validated a case definition for RA using national surveillance data in primary care settings.
Methods
This cross-sectional validation study used structured electronic medical record (EMR) data from the Canadian Primary Care Sentinel Surveillance Network (CPCSSN). Based on the reference set generated by EMR reviews by five experts, three machine learning steps: ‘bag-of-words’ approach to feature generation, feature reduction using a feature importance measure coupled with recursive feature elimination and clustering, and classification using tree-based methods (Decision Tree, Random Forest, and Extreme Gradient Boosting). The three tree-based algorithms were compared to identify the procedure that generated the optimal evaluation metrics. Nested cross-validation was used to allow evaluation and comparison and tuning of models simultaneously.
Results
Of 1.3 million patients from seven Canadian provinces, 5,600 people aged 19 + were randomly selected. The optimal algorithm for selecting RA cases was generated by the XGBoost classification method. Based on feature importance scores for features in the XGBoost output, a human-readable case definition was created, where RA cases are identified when there are at least 2 occurrences of text “rheumatoid” in any billing, encounter diagnosis, or health condition table of the patient chart. The final case definition had sensitivity of 81.6% (95% CI, 75.6–86.4), specificity of 98.0% (95% CI, 97.4–98.5), positive predicted value of 76.3% (95% CI, 70.1–81.5), and negative predicted value of 98.6% (95% CI, 98.0-98.6).
Conclusion
A case definition for RA in using primary care EMR data was developed based off the XGBoost algorithm. With high validity metrics, this case definition is expected to be a reliable tool for future epidemiological research and surveillance investigating the management of RA in CPCSSN dataset.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12911-024-02776-w.
Keywords: Rheumatoid arthritis, Case definition, EMR phenotyping, Electronic medical records, Machine learning
Introduction
Rheumatoid arthritis (RA) is an inflammatory condition, which affects approximately 1% of the general population [1]. Although an RA diagnosis requires specialist assessment, the involvement of family physicians (FPs) is key to its recognition and management. In general, FPs are a person’s first point of contact within the healthcare system, they may also provide long-term management of symptoms while the patient waits specialist consultation, and are central to the management of RA in the context of related comorbid conditions [2]. Although most evidence has examined the management of RA by specialists, there is a need to examine the services provided for RA in primary care [3].
The Canadian Primary Care Sentinel Surveillance Network (CPCSSN) database provides a unique opportunity to study RA in Canadian community patient populations. It extracts and standardizes electronic medical record (EMR) data from 12 primary care research networks across Canada and restructures it for health services research and epidemiology. CPCSSN provides an ideal data infrastructure to develop a case definition for RA relevant to the context of primary care [4]. Prior to the use of RA data from CPCSSN, a case definition is needed to identify cases. The aim of this paper is to use machine learning (ML) methods applied to many variables to develop a case definition to accurately distinguish RA cases and non-cases in the CPCSSN dataset. The cohort of people with RA managed in primary care settings identified by the case definition may then be used for future studies of the disease’s epidemiology and management.
Methods
Data source
The CPCSSN is a pan-Canadian collaboration of practice-based research and surveillance networks that collects, formats, and merges patient-level primary care EMR data [5]. As of December 31, 2020, CPCSSN contained data from two million de-identified primary care patients (including over 1.3 million adults aged 19+) and more than 1,300 primary care physicians’ practices across eight provinces. The database includes patient demographics, diagnoses (using the International Classification of Diseases, Ninth Revision, Clinical Modification (ICD-9-CM) for recording diagnoses), prescribed medications, physical measurements (e.g., blood pressure, height, weight, BMI), physician billing claims, behavioral risk factors (e.g., smoking, alcohol use, physical activity), laboratory test results, referrals to specialists, and medical procedures. However, imaging data and clinical notes are excluded for confidentiality reasons.
The data for this study was sourced from patients’ EMR, capturing all available health data from their first visit up until 2020. While most of the data spans from 2008 to 2020, it can include records dating back as far as 1990, depending on the patient’s history.
Prescribed medications in the CPCSSN database include those prescribed by both primary care physicians and specialists, such as rheumatologists, as long as the prescriptions were documented in the patient’s chart by the primary care provider.
To date, CPCSSN has developed and validated case definitions for 28 diseases and health conditions with validation metrics for sensitivity, specificity, positive predictive value and negative predictive values all greater than 70% [6]. Increasingly, ML is being used to develop case definitions.
Study sample
Given that RA is a low prevalence condition, we expect very few true cases in our sample, which most seriously affected our sample size. Hence, to identify the number of charts required for the validation set of our sample, we used the Wald 95% Confidence Interval for sensitivity [7].
where Sn is the expected sensitivity of the case definition, c is the full width of the confidence interval for the validation metrics, and p is the expected prevalence of the disease within the sample.
A preliminary search for RA in the dataset using an ICD-9-CM code for RA (714*) yielded a prevalence estimate of 0.8%. Using the formula above, to achieve validation metrics of at least 80% sensitivity while limiting the widest range of the 95% confidence interval to 10%, the minimum number of charts needed for the validation was estimated to be more than 300,000 charts, which was not practical. A practical option was to use a seeded sample with charts of patients judged highly likely to be RA cases (i.e., charts with at least two ICD-9 codes of 714) to create an artificial sample prevalence of approximately 10% [8]. With this adjustment, the minimum number of charts needed was reduced to a more feasible number − 2,459. The same number of charts was used for algorithm training and testing. In total 5,100 CPCSSN records were selected at random, supplemented by 500 ‘probably positive’ CPCSSN records. The total sample was representative of the entire CPCSSN dataset with regards to sex, age groups, and the ratio of rural-to-urban dwellers. There was no limitation was placed to exclude patients with ‘referral to specialists’ from the cohort.
Reference set development
An expert panel consisting of 5 reviewers with experience of CPCSSN data, including a physiotherapist, registered nurse, physician, epidemiologist, and health care researcher, reviewed the 5,600 CPCSSN records in order to create a reference standard of labelled RA cases and non-cases.
Data were uploaded to a secured platform, so that reviewers could review the records assigned to them securely and independently. A manual (Supplementary 1) to support the reviewers was developed by a practicing rheumatologist (CB) prior to the review activity with modifications and additional explanations based on questions raised during the training sessions. All reviewers attended three training sessions led by CB. The initial two training sessions each consisted of 30 records that were reviewed as a group. The third training session included 120 records that were reviewed independently. Reviewers did not know if a chart was a “probable RA” case or not, and they were instructed not to attempt to make a diagnosis but rather to recognize cases that had already been diagnosed and documented by the family physicians. Results from the third training session were used to calculate Cohen’s Kappa score to estimate interrater reliability and ensure that reviewers were consistent in their assessments of RA cases and non-cases [9].
Machine learning application
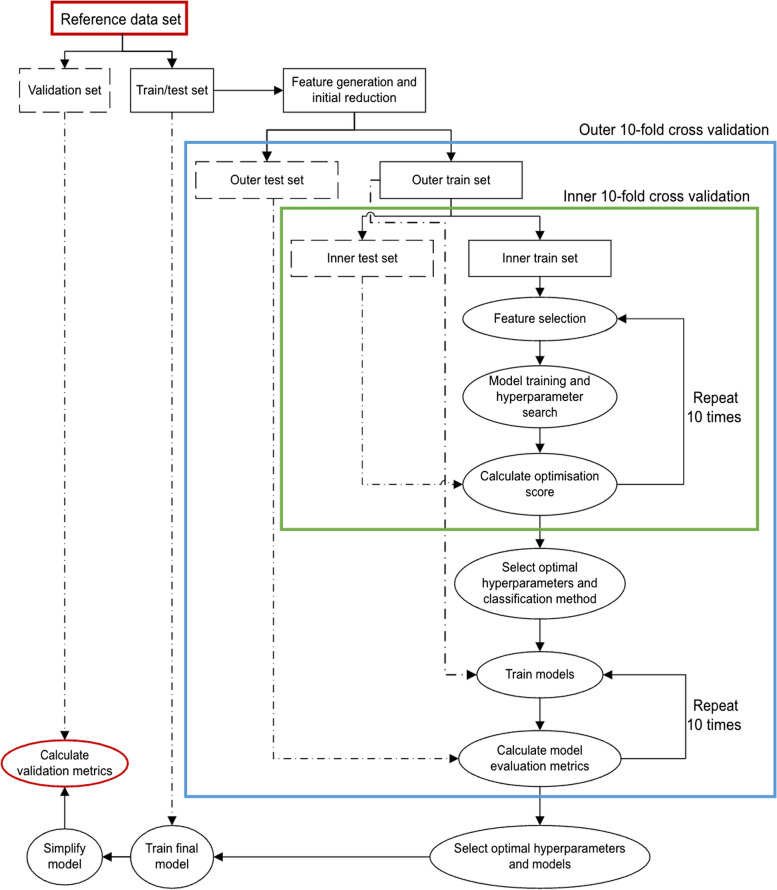
Case definition development and validation were completed using ML methods. The pipeline and nested cross-validation steps to train ML algorithms (Fig. 1) are described in more detail in our previous case definition development [10].
Fig. 1.
Steps used to train and optimize ML algorithms
The ML consisted of the following procedures.
Preparation for algorithm validation : To ensure that algorithms used information only from the training dataset and to avoid data leakage, we randomly divided the reference set into two independent data subsets before applying any data manipulation methods [11]. The first subset (n = 2,778) used labelled data to train and test algorithms to recognize RA cases. The accuracy of the final case definition was then validated by studying its classification using the second labelled subset (n = 2,777).
The final case definition was validated using the validation dataset that was set aside from the start of this analysis. To validate the final case definition, we compared the agreement between the reference set and the simplified case definition and reported its sensitivity, specificity, PPV, and NPV.
-
2.
Data pre-processing: We extracted many features for ML consideration including all possible single digit and combinations of texts and codes from different fields independently for the train/test set and the validation set. All features were derived from patient records within the training / testing set. Categorical features were selected from either the International Classification of Diseases, 9th Revision (ICD-9) for diagnoses and billing data, Anatomical Therapeutic Classification (ATC) for medications, Logical Observation Identifiers Names and Codes (LOINC) for laboratory test data, and Systematized Nomenclature of Medicine - Clinical Terms (SNOMED-CT) for referral codes. Free-text was extracted using a ‘bag-of-words’ model and presented each word or each pair of words (bi-gram) as binary variables (0 and 1 to indicate absence/presence of an attribute) as a feature to train ML algorithms.
To increase accuracy of the case definition, we permitted features that indicated one occurrence of the single feature at any time, two occurrences at any time, two occurrences within 12 months, and two occurrences within 24 months. In total, 183,476 features were generated.
To select the most relevant features from this large list, we calculated the difference between the number of true positive cases and false positive cases, divided by the total number of positive cases. This determined each feature’s ability to identify true positive cases while minimizing false positives (i.e., the greater this value, the more useful the feature). The features were ranked and the most common 200 features were used to train our models.
-
3.
Optimization of algorithms: We optimized our algorithms using the nested cross-validation method. It contained two layers, a 10-fold cross-validation in an inner layer to tune hyper-parameters and select a model with the best performance, and another 10-fold cross-validation in an outer layer to estimate the quality of models trained by the inner layer.
In the inner layer, we chose to maximize either F1-score or Matthews’ correlation coefficient (MCC) when tuning to balance the trade-off between sensitivity-specificity and positive predictive value (PPV) – negative predictive value (NPV) [12, 13]. The formula of F1-score and MCC were as follow [14]:
F1-score gave equal weight to sensitivity and PPV, hence optimizing the F1-score to maximize both sensitivity and PPV. Because true negatives were not included in F1-score, a risk of reducing specificity and NPV while maximizing sensitivity and PPV was possible. MCC, on the other hand, took into account all four metrics: sensitivity, specificity, PPV, and NPV; maximizing MCC could help to balance the trade-off between sensitivity-PPV and specificity-NPV but it could lead to lower sensitivity and PPV [15]. We selected both F1-score and MCC to compare algorithms’ performance.
Machine learning pipeline
To train and optimize performance of ML algorithms, the inner layer was a pipeline which contained a sequence of three steps – feature selection, model training and optimizing (supervised classification), and evaluation.
Step 1. Feature selection
Starting with the most common 200 features, we further reduced the number of features using k-best feature selection (kBF) method [16]. The kBF computed a statistical test (i.e., chi-square test) with the outcome (i.e., RA case or not) for each feature to find features with the highest statistical scores (i.e., closely dependent on the outcome) and included them in the ML algorithm. We also used Recursive Feature Elimination (RFE), a method of backward selection to limit the number of features included in ML training; like other backward selection methods, RFE started with all features in a fitting model to predict cases or non-cases with the least useful feature being eliminated at each calculation; a new model was then fitted and the process continued until the best subset of features was created [17].
Step 2. Model training and optimizing
After generating a list of the most relevant features using kBF and RFE approach in step 1, tree-based methods (i.e., Decision Tree, Random Forest, and XGBoost algorithms) were chosen to classify the outcome given their interpretability. The three methods shared the same logic of learning, in which a hierarchy of if/else questions are applied; a Decision Tree produced a straightforward tree with sets of rules to identify cases [18], a Random Forest produced multiple independent trees with parallel sets of rules [19], while Extreme Gradient Boosting (XGBoost) was the most complex method which produced one tree at a time and used that tree’s results to build the next tree [20].
Step 3. Internal evaluation
Using the training and testing dataset, we evaluated our models by comparing the agreement between classification from the chart reviewers and the classification identified by each model. The algorithm with the highest sensitivity, specificity, PPV, and NPV was selected to be the final case definition.
The outer layer included features and hyperparameters those were determined as the most effective from the inner layer. These values were used to train the models. Evaluation metrics (sensitivity, specificity, PPV, and NPV) obtained from the outer layer were used to identify the best algorithms.
Python 3.10.7 was used for all steps of this analysis. Ethics was secured from the University of Alberta Heath Research Board (PRO00107346).
Results
There were 5,600 CPCSSN patient charts that formed the reference set for the machine learning training, testing, and validating. Of these, 45 records that had no clinical data were excluded, leaving the remaining of 5,555 records. Among 120 charts used for the third reviewer training, 30 were ‘probably positive’ RA charts. A Cohen’s Kappa score of 0.75 was calculated, which indicated satisfactory agreement among reviewers [9]. The remaining 5,405 CPCSSN records were reviewed independently by the reviewers; however, 112 records did not have an outcome and were labelled ‘uncertain’. The rheumatologist (CB) re-reviewed these records. The team identified 414 RA cases (Fig. 2).
Fig. 2.
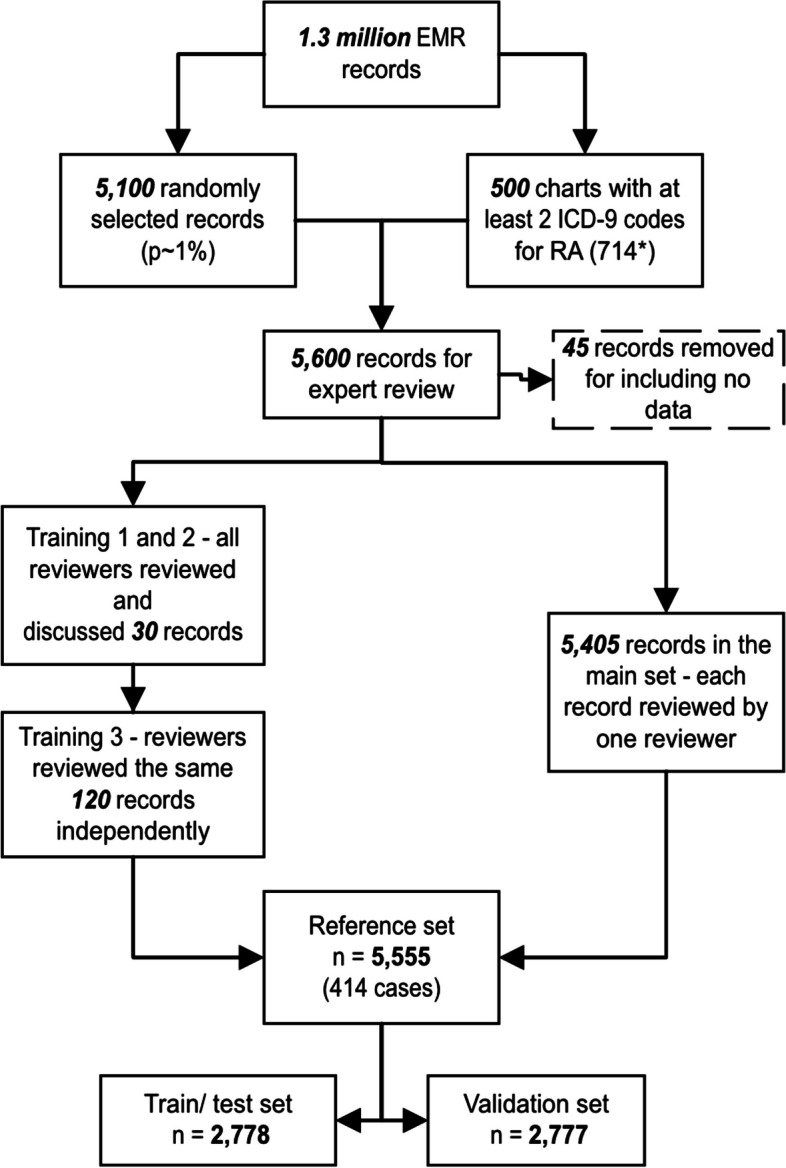
Flowchart for charts in the reference set
The reference set was based on 5,555 CPCSNN records, with 414 RA patients corresponding to a pre-test prevalence of 7.5%. The patient cohort, expectedly, was slightly older, included more females, more urban residents, and more people with multiple comorbidities than the full CPCSSN 2020 dataset (Table 1).
Table 1.
Baseline demographic characteristics of the sample
| Baseline characteristics | Record review cohort | CPCSSN 2020 dataset |
|---|---|---|
| N | 5,555 | 1,363,552 |
| Age, mean (SD) | 52.9 (19.5) | 51.9 (20) |
| Sex (Females), n (%) | 3,204 (57.8) | 759,635 (55.7) |
| Rural residence, n (%) | 882 (15.9) | 208,126 (16.3) |
| Number of chronic conditionsa, n (%) | ||
| 0 | 1,911 (34.4) | 586,853 (43.0) |
| 1 | 1,357 (24.4) | 330,288 (24.2) |
| 2 | 1,017 (18.3) | 204,673 (15.0) |
| 3+ | 1,270 (22.9) | 241,738 (17.8) |
aThe number of chronic conditions included those identified in CPCSSN 2020, including: Chronic obstructive pulmonary disease, Dementia, Depression, Diabetes Mellitus, Dyslipidemia, Epilepsy, Hypertension, Osteoarthritis, and Parkinsonism
We found that maximizing MCC produced good sensitivity and PPV with narrower 95%CI than maximizing F1-score. The XGBoost gave the highest PPV of 69.9% and a good sensitivity of 93% (Table 2). The rules that the XGBoost used for making decisions were complicated which was built on 130 trees and a maximum of 12 levels of branches for each tree. Hence, we extracted 10 features with highest importance scores from the tree to produce a human-readable case definition. The top 10 features are displayed in Table 3. We then created a total of 100 case definitions, including case definitions for each feature (10 total), and from each pair of features (45 combinations of two features using AND and 45 combinations of two features using OR).
Table 2.
Evaluation metrics for ML algorithms created by the pipelines
| Sensitivity % (95% CI) |
Specificity % (95% CI) |
PPVa % (95% CI) |
NPVa % (95% CI) |
|
|---|---|---|---|---|
| MCC | ||||
| Decision tree | 95.8 (90.6–100.0) | 96.3 (95.0–98.0) | 68.5 (60.1–79.3) | 99.6 (99.2–100.0) |
| Random forest | 95.8 (90.6–100.0) | 96.3 (95.0–98.3) | 68.8 (60.1–81.9) | 99.6 (99.2–100.0) |
| XGBoost | 94.8 (90.5–100.0) | 96.5 (95.3–98.0) | 69.4 (61.8–79.3) | 99.6 (99.2–100.0) |
| F1-score | ||||
| Decision tree | 93.0 (76.7–100.0) | 96.6 (95.3–98.3) | 69.9 (61.8–81.9) | 99.4 (8.0–100.0) |
| Random forest | 94.8 (90.5–100.0) | 96.4 (95.0–98.0) | 69.2 (60.3–79.3) | 99.6 (99.2–100.0) |
| XGBoost | 94.0 (76.7–100.0) | 96.4 (95.0–98.3) | 69.0 (60.1–81.9) | 99.5 (98.0–100.0) |
aMCC Matthew’s Correlation Coefficient, PPV Positive predictive values, NPV Negative predictive value, XGBoost Extreme Gradient Boosting
Table 3.
Top ten features with the highest important scores of the best performance XGBoost case definition
| Feature | Code Description | Data type | Importance score |
|---|---|---|---|
| One occurrence of text “rheumatoid arthritis” | Billing, encounter diagnosis or problem list | 0.2395 | |
| One occurrence of text “rheumatoid” | Billing, encounter diagnosis or problem list | 0.1461 | |
| Two occurrences of text “rheumatoid” | Billing, encounter diagnosis or problem list | 0.0562 | |
| Two occurrences of text “arthritis” | Billing, encounter diagnosis or problem list | 0.0297 | |
| Two occurrences of text “rheumatoid” in 24 months | Billing, encounter diagnosis or problem list | 0.0295 | |
| Two occurrences of text “other inflammatory” | Billing, encounter diagnosis or problem list | 0.0228 | |
| One ATC code P01BA02 | Hydroxychloroquine | Medication | 0.0225 |
| Two occurrences of text “arthritis” | Problem list | 0.0224 | |
| One ATC code B03BB01 | Folic acid | Medication | 0.0197 |
| Two occurrences of text “rheumatoid arthritis” in 24 months | Billing, encounter diagnosis or problem list | 0.0189 |
“Or any sub-codes” means all codes starting with the same characters (e.g., B03 (or any sub-codes) includes B03A, B03B, B03BB, etc.)
ATC Anatomical Therapeutic Chemical (ATC) Classification, ICD-9 International Classification of Diseases, Ninth Revision
Of these combinations, the RA case definition with the highest evaluation metrics is given by: At least 2 occurrences of text “rheumatoid” in any billing, encounter diagnosis, or health condition table. For validation purposes, we trained the XGBoost algorithm on the entire train/test data set, then validated it on the validation set. The simplified case definition was also validated using the validation set. The validation metrics are displayed in Table 4.
Table 4.
Validation metrics for the XG Boost and the human readable case definition derived from it
| Sensitivity % (95% CI) |
Specificity % (95% CI) |
PPV* % (95% CI) |
NPV* % (95% CI) |
|
|---|---|---|---|---|
| ML case definition |
88.7 (81.1–95.4) |
98.1 (97.0–98.8) |
80.1 (71.6–87.0) |
99.1 (98.4–99.6) |
| Human readable case definition |
81.6 (75.6–86.4) |
98.0 (97.4–98.5) |
76.3 (70.1–81.5) |
98.6 (98.0–98.6) |
Discussion
The diagnosis of RA is complex and typically requires a rheumatologist to diagnosis it; however, patients with RA are typically first seen by the FP. Identifying RA cases in primary care datasets can be challenging yet necessary to describe health services received in the community. Through rigorous approaches in the CPCSSN data, the optimal approach to identify RA cases in CPCSSN data was the XGBoost with maximized MCC, simplified by using importance scores. Although we expected to obtain a prevalence of 10% in the sample by selecting extra charts with at least two ICD-9 codes for RA (ICD 714 and its sub-codes), the prevalence of RA sample was only 7.5% (414/5,555). This supports our concern that using one single diagnosis code may lead to over-estimation of RA prevalence. The lower prevalence also led to wider 95% confidence intervals for sensitivity and PPV than the expected 10%; however, our developed case definition detected RA cases recorded with good validity with all metrics larger than 70% [21]. Using this case definition, a prevalence of 0.9% for RA was reported (12,083/1,365,121) in CPCSSN 2020 data for adults aged 19 and older, which aligns with Canadian national and provincial prevalence for RA [22, 23].
Previous attempts to develop an RA case definition in primary care by comparing cases identified by case criteria set by an expert panel have been published in the UK’s Clinical Practice Research Datalink [24], yet the validation metrics were not reported. Results from this UK study might be biased as the case definition mostly overlapped with the selection rules used to develop the reference set. Other approaches that used ML methods such as decision trees as the sole method [25] based on hospital diagnosis codes and medication codes yielded very good sensitivity (86.2%) and PPV (85.6%), with excellent specificity (94.6%).
In our study, the simplified case definition had lower sensitivity and PPV than metrics produced by using the ML algorithm. Transparency and readability of case definition rules produced by ML methods may not be ideal, because even ‘explainable’ rules from ML methods like logistic regression or simple tree-based models might still include so many variables that it makes answering the question ‘why is a case a case?’ not straightforward. Our hybrid method took advantage of ML methods to optimize a classification model, then extracted features that were deemed to most significantly contribute to the classification to generate a final case definition. This approach avoided expert bias for both the training / testing and validation sets and provides an algorithm that other users may apply to CPCSSN data.
In light of these findings, limitations that should be considered centred primarily on secondary uses of clinical data. Misclassification of RA may be related to a few possibilities. Firstly, the data used did not include all of the available data in the EMR such as clinical notes, referral letters, and imaging data, which might limit the ability of reviewers in recognizing RA cases. Secondly, the reference standard was created by an expert panel with most charts reviewed by only one reviewer. Although high inter-rater reliability was computed, the reference contained some false positives and false negatives. Lastly, the case definition was derived from standardized CPCSSN data, instead of the raw EMR data. This may have limited the accuracy of the chart review process and case definitions created, as the raw text may contain more details than the standardized text provides.
Conclusion
A validated case definition was derived for RA cases in CPCSSN electronic medical record data with very good validation metrics. RA cases are identified when there are at least two occurrences of “rheumatoid arthritis” in any diagnosis fields within 24 months or at least one occurrence of “rheumatoid arthritis” in the problem list of the patient chart. This case definition had sensitivity of 81.6% (95% CI, 75.6–86.4), specificity of 98.0% (95% CI, 97.4–98.5), PPV of 76.3% (95% CI, 70.1–81.5), and NPV of 98.6% (95% CI, 98.0–98.6). Future studies on people with RA identified by this case definition will inform understanding of the epidemiology, management, and burden of disease at a national level. There are other opportunities for ongoing community surveillance and practice quality improvement.
Supplementary Information
Acknowledgements
Appreciation is expressed to Matt Taylor (CPCSSN data manager) for creating and managing the Data Presentation Tool, which was used to review EMRs.
Appreciation is expressed to Michael Cummings (Southern Alberta Primary Care Research Network – CPCSSN data manager) for providing the Feature Generation tools, which was used to create a list of ML features, and to provide advice to the ML algorithm development.
Appreciation is expressed to Russell Pilling, Rebecca Miyagishima, Amanda Larocque, and Jack Fu for reviewing CPCSSN charts for the reference standard development.
Abbreviations
- ATC
Anatomical Therapeutic Classification
- CaRT
Classification and Regression Trees
- COPD
Chronic obstructive pulmonary disease
- CPCSSN
Canadian Primary Care Sentinel Surveillance Network
- EMR
Electronic medical record
- FN
False negative
- FP
False positive
- ICD-9
International Classification of Diseases – 9th version
- MCC
Matthews correlation coefficient
- ML
Machine learning
- NPV
Negative Predicted Value
- PPV
Positive Predicted Value
- RA
Rheumatoid Arthritis
- RFE
Recursive Feature Elimination
- TN
True negative
- TP
True positive
Authors’ contributions
ND, AJ, CB: conceptualization, methodology, manuscript reviewing and editing; AP: methodology, data analysis, original draft preparation; MC, TW: methodology, supervision; JW, CL, DK: conceptualization, manuscript reviewing and editing. All authors have read, revised, and approved of the final manuscript.
Funding
This study was funded by an Arthritis Society’s Strategic Operating Grant (Grant ID 20 − 000000018). AP received a Mitacs Accelerate postdoctoral fellowship (ID IT27834). The Arthritis Society and Mitacs had no influence on the study design, data analysis, findings interpretation, or any content of this manuscript.
Data availability
The data used in this analysis are available from the Canadian Primary Care Sentinel Surveillance Study (CPCSSN) upon reasonable request to ND, with permission from CPCSSN.
Declarations
Ethics approval and consent to participate
This study has received approval from the Health Research Ethics Board at the University of Alberta (Pro00107346) and the University of Calgary (REB21-0723) and adheres to all relevant guidelines and regulations for research involving de-identified health data (e.g. CHREB, Tri-Council Policy Statement on the Ethical Conduct for Research Involving Humans [TCPS2]).
The CPCSSN database has received ethics approval, including waivers of individual patient informed consent for their de-identified data to be used for surveillance and research, from each contributing network’s local Research Ethics Board (Queen’s University, Memorial University of Newfoundland, University of Ottawa, University of Calgary, Dalhouse University, University of Toronto, Western University, Bruyère Research Institute, University of British Columbia, University of Alberta, and University of Manitoba).
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cross M, Smith E, Hoy D, Carmona L, Wolfe F, Vos T, et al. The global burden of rheumatoid arthritis: estimates from the global burden of disease 2010 study. Ann Rheum Dis. 2014;73(7):1316–22. [DOI] [PubMed] [Google Scholar]
- 2.England BR, Roul P, Yang Y, Sayles H, Yu F, Michaud K, et al. Burden and trajectory of multimorbidity in rheumatoid arthritis: a matched cohort study from 2006 to 2015. Ann Rheum Dis. 2021;80(3):286–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Radu AF, Bungau SG. Management of rheumatoid arthritis: an overview. Cells. 2021;10(11):2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birtwhistle RV. Canadian Primary Care Sentinel Surveillance Network: a developing resource for family medicine and public health. Can Fam Physician Med Fam Can. 2011;57(10):1219–20. [PMC free article] [PubMed] [Google Scholar]
- 5.Garies S, Birtwhistle R, Drummond N, Queenan J, Williamson T. Data resource profile: national electronic medical record data from the Canadian Primary Care Sentinel Surveillance Network (CPCSSN. Int J Epidemiol. 2017;46(4):1091–2. [DOI] [PubMed] [Google Scholar]
- 6.CPCSSN. CPCSSN Case Definition Version 2. 2019. https://cpcssn.ca/wp-content/uploads/2023/03/CPCSSN-Case-Definitions-2022-Q4_v2.pdf. Cited 20 Jul 2023.
- 7.Vollset SE. Confidence intervals for a binomial proportion. Stat Med. 1993;12(9):809–24. [DOI] [PubMed] [Google Scholar]
- 8.Williamson T, Green ME, Birtwhistle R, Khan S, Garies S, Wong ST, et al. Validating the 8 CPCSSN case definitions for chronic disease surveillance in a primary care database of electronic health records. Ann Fam Med. 2014;12:367–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McHugh ML. Interrater reliability: the kappa statistic. Biochem Med. 2012;22(3):276–82. [PMC free article] [PubMed] [Google Scholar]
- 10.Pham ANQ, Cummings M, Yuksel N, Sydora B, Williamson T, Garies S et al. Development and Validation of a Machine Learning Algorithm for Problematic Menopause in the Canadian Primary Care Sentinel Surveillance Network (CPCSSN. 2023. 10.21203/rs.3.rs-2403081/v1. Cited 13 Mar 2023.
- 11.Hannun A, Guo C, van der Maaten L. Measuring Data Leakage in Machine-Learning Models with Fisher Information. arXiv; 2021. http://arxiv.org/abs/2102.11673. Cited 17 May 2023.
- 12.Trevethan R, Sensitivity. Specificity, and predictive values: foundations, pliabilities, and pitfalls in Research and Practice. Front Public Health. 2017;5:307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H, Wang B, Zhang X, Feng C. Relations among sensitivity, specificity and predictive values of medical tests based on biomarkers. Gen Psychiatry. 2021;34(2):e100453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seo S, Kim Y, Han HJ, Son WC, Hong ZY, Sohn I, et al. Predicting successes and failures of clinical trials with outer product–based convolutional neural network. Front Pharmacol. 2021;12:670670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chicco D, Jurman G. The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genomics. 2020;21(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011;12:2825–30. [Google Scholar]
- 17.Suresh S, Newton DT, Everett TH, Lin G, Duerstock BS. Feature selection techniques for a machine learning model to detect autonomic Dysreflexia. Front Neuroinformatics. 2022;16:901428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.TruicăCO Leordeanu C. Classication of an Imbalanced Data Set using Decision TreeAlgorithms. Univ Politeh Buchar Sci Bull Ser C - Electr Eng Comput Sci. 2017;79:69. [Google Scholar]
- 19.Breiman L. Random forests. Mach Learn. 2001;45(1):5–32. [Google Scholar]
- 20.Chen T, Guestrin C, XGBoost:. A Scalable Tree Boosting System. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining [Internet]. New York, NY, USA: Association for Computing Machinery; 2016. pp. 785–94. (KDD ’16). Available from: 10.1145/2939672.2939785. Cited 19 Sep 2023.
- 21.Williamson T, Green ME, Birtwhistle R, Khan S, Garies S, Wong ST, et al. Validating the 8 CPCSSN case definitions for chronic disease surveillance in a primary care database of electronic health records. Ann Fam Med. 2014;12(4):367–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Canadian Chronic Disease Surveillance System (CCDSS). https://health-infobase.canada.ca/ccdss/data-tool/. Cited 24 Apr 2023.
- 23.Widdifield J, Paterson JM, Bernatsky S, Tu K, Tomlinson G, Kuriya B, et al. The epidemiology of rheumatoid arthritis in Ontario, Canada. Arthritis Rheumatol Hoboken NJ. 2014;66(4):786–93. [DOI] [PubMed] [Google Scholar]
- 24.Muller S, Hider SL, Raza K, Stack RJ, Hayward RA, Mallen CD. An algorithm to identify rheumatoid arthritis in primary care: a clinical Practice Research Datalink study. BMJ Open. 2015;5(12):e009309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou SM, Fernandez-Gutierrez F, Kennedy J, Cooksey R, Atkinson M, Denaxas S, et al. Defining Disease phenotypes in Primary Care Electronic Health Records by a machine Learning Approach: a case study in identifying rheumatoid arthritis. PLoS ONE. 2016;11(5):e0154515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used in this analysis are available from the Canadian Primary Care Sentinel Surveillance Study (CPCSSN) upon reasonable request to ND, with permission from CPCSSN.