Abstract
Introduction
Pseudomonas aeruginosa AUST‐03 (ST242) has been reported to cause epidemics in people with CF (pwCF) from Australia and has been associated with multidrug resistance and increased morbidity and mortality. Here, we report an epidemic P. aeruginosa (AUST‐03) strain in South African pwCF detected at a public hospital and characterize the genomic antibiotic resistance determinants.
Methods
The P. aeruginosa AUST‐03 (ST242) study isolates were analysed with whole genome sequencing using the Illumina NextSeq2000 platform. Raw sequencing reads were processed using the Jekesa pipeline and multilocus sequence typing and genomic antibiotic resistance characterization was performed using public databases. Genetic relatedness between the study isolates and global P. aeruginosa ST242 from public databases was determined using a maximum‐likelihood phylogenetic tree. Antibiotic susceptibility testing was performed using the disk diffusion and broth microdilution techniques.
Results
A total of 11 P. aeruginosa AUST‐03 isolates were isolated from two children with CF. The majority (8/11) of these isolates were multidrug‐resistant (MDR) or extensively drug resistant; and the multidrug efflux pumps MexAB‐OprM, MexCD‐OprJ, MexEF‐OprN, and MexXY‐OprM were the most clinically relevant antibiotic resistance determinants and were detected in all of the isolates. The study isolates were the most closely related to a 2020 P. aeruginosa AUST‐03 (ST242) CF isolate from Russia.
Conclusion
Epidemic MDR P. aeruginosa strains are present at South African public CF clinics and need to be considered when implementing segregation and infection control strategies to prevent possible spread and outbreaks.
Keywords: antibiotic resistance, cystic fibrosis, Pseudomonas aeruginosa AUST‐03, South Africa
1. INTRODUCTION
Cystic fibrosis (CF) is among the most common life‐limiting genetic diseases in South Africa and occurs at an incidence of 1:3000, 1:10,300, and 1:14,000 in White, mixed‐race and Black South African population groups, respectively. 1 , 2 Pseudomonas aeruginosa (P. aeruginosa) is recognized as one of the most significant lung pathogens among people with CF (pwCF) in South Africa 3 and persistent lung colonization with P. aeruginosa has been associated with poorer outcomes. 3 , 4 , 5
People with CF typically acquire P. aeruginosa independently from the environment and as such, each person with CF is often found to be infected or colonized with a single genetically distinct, non‐clonal strain of P. aeruginosa, that is unique to that person and not shared with other pwCF. 6 However, there have been multiple reports globally where pwCF that have shared environments such as CF clinics, or geographical locations, also shared genetically identical P. aeruginosa strains. 4 , 7 These shared P. aeruginosa strains, which have also been referred to as epidemic or transmissible strains have been associated with increased virulence, multidrug resistant (MDR) and higher mortality rates. 8 , 9 Australia has been reported to have some of the highest numbers of epidemic P. aeruginosa strains in pwCF, 10 , 11 with the most prevalent being P. aeruginosa AUST‐01 (ST649), AUST‐02 (ST775), and AUST‐03 (ST242). 4 , 11 The P. aeruginosa AUST‐03 (ST242) strain was first described in pwCF from Tasmania in 2003 and has caused outbreaks in Tasmania and other regions in Australia. 11 , 12
The use of advanced molecular techniques such as whole genome sequencing (WGS) that can aid in better understanding the underlying genetic mechanisms contributing to antibiotic resistance and also identify epidemic strains of P. aeruginosa in South Africa is limited. This was previously due to the high cost of WGS in South Africa, however, the recent decrease in WGS and global initiatives to make WGS more accessible in low to middle income countries (LMICs) has made the use of WGS more cost‐effective for researchers in South Africa. To our knowledge, epidemic strains of P. aeruginosa have not been reported in pwCF from South Africa. Here we report the presence of the epidemic P. aeruginosa AUST‐03 (ST242) strain detected with WGS in pwCF at a public hospital in Gauteng, South Africa. We further described the genomic characteristics conferring antibiotic resistance in the P. aeruginosa AUST‐03 (ST242) isolates from this study and made phylogenetic comparisons with global isolates of P. aeruginosa ST242 (AUST‐03) from pwCF.
2. MATERIALS AND METHODS
2.1. Ethics statement
The study was granted ethical approval by the University of the Witwatersrand Human Research Ethics Committee (Reference number: M1811104) and by the University of Pretoria, Faculty of Health Sciences, Research Ethics Committee (Reference number: 466/2018). Sputum specimens were obtained following the provision of written informed assent and consent from the study participants and their parents, respectively.
2.2. Study background and setting
The current study investigates the epidemic strain P. aeruginosa AUST‐03 (ST242) that was identified in two children with CF attending a clinic at a public hospital in Johannesburg, South Africa. This CF clinic was part of a larger parent study conducted over the period May 2019–February 2020 at this hospital and at a second hospital, that recruited a total of 10 pwCF that were known to be infected or colonized with P. aeruginosa. Both of these hospitals are among the largest public hospitals attending to pwCF in the Gauteng province of South Africa, however, no pwCF infected or colonized with P. aeruginosa AUST‐03 (ST242) were found at the second hospital. The hospital currently investigated in this study was the larger of the two with a population of about 56 pwCF and it exclusively attends to children with CF (≤18 years old). The majority of the children at this clinic are White (31/56), followed by children that were of Black (15/56), mixed race (6/56), and east Indian (4/56) ancestry. A total of seven of these children colonized with P. aeruginosa were recruited from this hospital for the parent study and an equal number of White (3 pwCF) and Black (3 pwCF) children were included in the parent study, with the seventh being a child of mixed‐race ancestry.
2.3. P. aeruginosa isolation and analysis
A single sputum specimen was collected from pwCF by means of spontaneous expectoration or after the administration of percussion exercises, with the assistance of the attending physiotherapist at the CF clinic. The sputum specimens were cultured on Pseudomonas CN agar (Oxoid) and incubated (Vacutec) at 37°C for up to 72 h. Multiple colonies were selected from each sputum specimen due to the possibility of pwCF being colonized with multiple or variant strains of P. aeruginosa. Up to 10 presumptive P. aeruginosa colonies of varying size and morphology were selected from the Pseudomonas CN agar (Oxoid) plate of each sputum specimen and sub‐cultured onto 5% sheep blood agar (Diagnostic Media Products). Genomic DNA was extracted using the Quick‐DNA Fungal/Bacterial Miniprep Kit (Zymo Research) and P. aeruginosa species confirmation was performed by conventional polymerase chain reaction (PCR) assays using previously described primers 13 and the following thermocycling conditions: 95°C for 5 min; 28 cycles of 95°C for 30 s, 57°C for 30 s, 72°C for 1 min; and 72°C for 10 min. P. aeruginosa ATCC 27853 was used as a positive control.
Antibiotic susceptibility testing (AST) was performed on PCR confirmed P. aeruginosa isolates using the disk diffusion technique for the following antibiotics: cefepime (30 µg), ceftazidime (30 µg), imipenem (10 µg), meropenem (10 µg), amikacin (30 µg), gentamicin (10 µg), tobramycin (10 µg), ciprofloxacin (5 µg), piperacillin/tazobactam (110 µg), and aztreonam (30 µg) and the broth microdilution technique for colistin according to the Clinical and Laboratory Standards Institute guidelines. 14 Multidrug resistant (MDR) P. aeruginosa isolates were defined as those displaying resistance to one or more anti‐pseudomonal antibiotics in at least three or more antibiotic classes, while extensively drug resistant (XDR) isolates were those displaying resistance to one or more anti‐pseudomonal antibiotics in all but two or less antibiotic classes. 15 , 16
2.4. P. aeruginosa whole genome sequencing (WGS) and analysis
Multiplexed, paired‐end libraries (2 × 150 bp) were prepared using the Illumina DNA Prep kit (Illumina), followed by WGS at 100x coverage on the PCR confirmed P. aeruginosa study isolates using the Illumina NextSeq. 2000 (Illumina Inc.) instrument. The raw paired‐end P. aeruginosa sequencing reads were processed using the Jekesa pipeline v1.0, using tools and methodology that included species identification and multi‐locus sequence typing (MLST) as previously outlined. 17 , 18 Genomic antibiotic resistance determinants in the study isolates were characterized using the Comprehensive Antibiotic Resistance Database (CARD) v3.2.2 19 and ResFinder 20 tools.
2.5. Phylogenetic comparison of global and study P. aeruginosa AUST‐03 (ST242) isolates
A total of 61 P. aeruginosa ST242 global genomes originating from human specimens were retrieved from the National Center for Biotechnology Information (NCBI) using the NCBI datasets tool v15.12.0 21 and were analyzed together with genome assemblies from our study isolates. Briefly, whole genome alignments were performed using scapper 22 and P. aeruginosa Zw92 strain (GenBank accession RXAY00000000.1) was used as a reference. IQ‐TREE v2.0.3 23 was used to generate a maximum‐likelihood phylogenetic tree using GTR + F + ASC + R4 with 1000 bootstrap approximations using UFBoot2 24 and the phylogenetic tree was visualized and annotated using Microreact (https://microreact.org/). 25 Global P. aeruginosa ST242 isolates that were the most closely related to the isolates from the study were also characterized for genomic antibiotic resistance determinants as previously discussed in Section 2.4.
3. RESULTS
3.1. P. aeruginosa AUST‐03 (ST242) colonized participants from the study
The P. aeruginosa AUST‐03 strain was found in a 16 year old male (P2) and an 8 year old female (P4), who both attended CF clinics at the hospital. Both children were determined to have been experiencing pulmonary exacerbations by attending clinicians and P4 who required oxygen, later passed away. Background details on the two study participants can be found in Table 1.
Table 1.
Demographics of cystic fibrosis (CF) study participants with Pseudomonas aeruginosa AUST‐03.
| Patient | P2 | P4 |
|---|---|---|
| Sex | Male | Female |
| Race | Black | White |
| Age | 16 | 8 |
| CFTR mutation | 3120 + 1 G > A x2 | ΔF508x1; 394delTTx1 |
| BMI | 19.3 | 14.1 |
| Comorbidities | Pancreatic insufficiency; asthma | Pancreatic insufficiency; cystic fibrosis related diabetes |
| Lung status | Exacerbation | Exacerbation |
| Sputum collection date | July 2019 | July 2019 |
3.2. Molecular identification and phenotypic characterization of P. aeruginosa AUST‐03 (ST242) isolates from the study
A total of 10 isolates of P. aeruginosa ST242 and one P. aeruginosa ST242 isolate as confirmed with PCR and MLST were isolated from the sputum of P2 and P4, respectively. In total, 11 isolates of the epidemic P. aeruginosa ST242 (AUST‐03) strain were recovered from the study participants and the AST results showed that the majority of these isolates from P2 [70% (7/10)] were MDR, while the one isolate from P4 was XDR. The following antibiotic resistance rates were recorded among the P. aeruginosa AUST‐03 isolates from the study: ciprofloxacin 100% (11/11), cefepime 73% (8/11), gentamicin 64% (7/11), amikacin 36% (4/11), tobramycin 18% (2/11), ceftazidime 18% (2/11), imipenem 18% (2/11), piperacillin‐tazobactam 18% (2/11), meropenem 9% (1/11), aztreonam 9% (1/11), and 9% colistin (1/11). Table 2 details the morphological characteristics and AST profiles of the study isolates.
Table 2.
Phenotypic morphological and antibiotic susceptible characteristics of the Pseudomonas aeruginosa AUST‐03 study isolates.
| Strain | Morphology | Cefepime | Ceftazidime | Imipenem | Meropenem | Amikacin | Gentamicin | Tobramycin | Ciprofloxacin | Piperacillin/tazobactam | Aztreonam | Colistin | MDR a |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P2 JHB I | Small round non‐mucoid | S | S | S | S | S | I | S | R | S | S | I | Non‐MDR |
| P2 JHB II | Small round non‐mucoid | R | R | I | S | R | R | I | R | R | I | I | MDR |
| P2 JHB III | Small flat mucoid | R | S | I | S | I | R | I | R | S | S | I | MDR |
| P2 JHB IV | Small round non‐mucoid | S | S | S | S | S | S | S | R | S | S | I | Non‐MDR |
| P2 JHB V | Small flat mucoid | R | I | I | S | R | R | R | R | I | S | I | MDR |
| P2 JHB VI | Medium round mucoid | R | S | S | S | I | R | I | R | S | S | I | MDR |
| P2 JHB VII | Small/medium mucoid | R | S | S | S | I | R | S | R | S | S | I | MDR |
| P2 JHB VIII | Small/medium mucoid | R | I | R | S | I | R | I | R | S | S | I | MDR |
| P2 JHB IX | Small/medium mucoid | S | S | S | S | I | I | S | R | S | S | I | Non‐MDR |
| P2 JHB X | Small flat mucoid | R | S | I | S | R | R | R | R | S | S | I | MDR |
| P4‐JHB‐III | Large non‐mucoid | R | R | R | R | R | I | S | R | R | R | R | XDR |
Abbreviations: I, intermediate; MDR, multidrug resistant; R, resistant; S, susceptible; XDR, extensively drug resistant.
MDR isolates were those displaying resistance to one or more anti‐pseudomonal antibiotics in at least three or more antibiotic classes; XDR isolates were those displaying resistance to one or more anti‐pseudomonal antibiotics in all but two or less antibiotic classes.
3.3. Phylogenetic and genomic antibiotic resistance characteristics of the P. aeruginosa AUST‐03 (ST242) isolates
Some genetic variation was observed in the 11 P. aeruginosa ST242 isolates from the study as the isolates formed multiple minor clusters and singletons (Figure 1), however the isolates were generally all very closely related. This variation may have been due to mutations caused by the varied microenvironment in the lungs. 26 Two isolates from each study participant (P2 JHB VIII and P4 JHB III) clustered very closely together which may have been indicative of strain sharing at this hospital, as they were unrelated and had no other common or shared environment. The P. aeruginosa ST242 study isolates were the most closely related to a P. aeruginosa ST242 isolate from Russia (SAMN23763320) that was isolated from a person with CF in 2020. The study isolates were also closely related to isolates from Australia, China, Spain and the USA.
Figure 1.
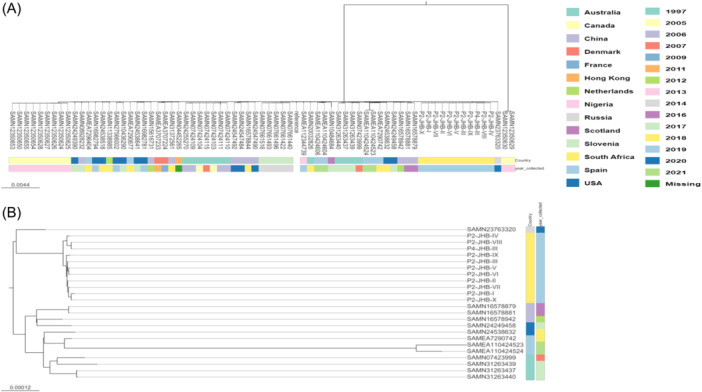
Maximum‐likelihood phylogenetic tree displaying the genetic relatedness of the Pseudomonas aeruginosa ST242 (AUST‐03) isolates from this study and from the global databases (A). Figure 1B displays a subtree from Figure 1A showing global isolates that were the most closely related to the isolates from this study.
The WGS results of the 11 P. aeruginosa AUST‐03 isolates from the study showed that the main genomic basis of antibiotic resistance in the P. aeruginosa isolates was efflux pump mediated. Genes conferring the MexAB‐OprM, MexCD‐OprJ, MexEF‐OprN, and MexXY‐OprM efflux pumps were detected in all of the study isolates and mutations in the mexR, mexT, nalC, and nfxB genes were detected in all of the isolates that conferred the upregulation of these efflux pumps. Additional efflux pumps emrE and pmpM genes were also detected in all of the study isolates that confer resistance to fluoroquinolones and aminoglycosides, respectively. The antibiotic resistance genes (ARGs): bla OXA‐50 , bla OXA‐1034 , bla PDC‐3 , bla PDC‐374 , and crpP were also found in all of the isolates, however, these genes are considered to play a minor role in P. aeruginosa antibiotic resistance. Acquired mutations in genes were found in the isolates that conferred resistance to fluoroquinolones (gyrA (3/11), gyrB (8/11) and parE (1/11)) and colistin (pmrAB (11/11)). Details on the major antibiotic resistance determinants of the P. aeruginosa AUST‐03 isolates from the study have been illustrated in Figure 2.
Figure 2.
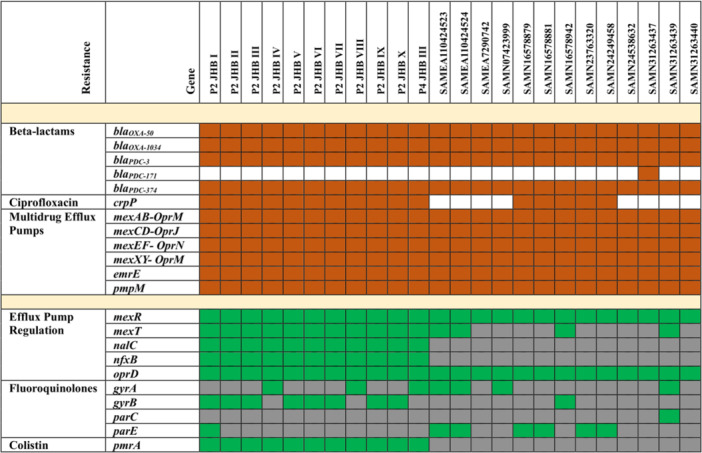
Genomic characterization of antibiotic resistant determinants and clinically relevant antibiotic resistance mutations in the Pseudomonas aeruginosa ST242 (AUST‐03) isolates from the study and the 13 most closely related global isolates. Brown squares denote genes that are present, green squares denote genes carrying nonsynonymous (missense, frameshift and nonsense) mutations, gray squares represent genes carrying wildtype/synonymous mutations with limited contribution to antibiotic resistance. White squares represent genes that were absent.
4. DISCUSSION
The P. aeruginosa AUST‐03 strain detected in the current study setting was found in two children with CF, a female aged 8 years old (P4) and a male aged 16 years old (P2). The children presented with pulmonary exacerbations and other co‐morbidities (Table 1). While limited data is available on pwCF infected or colonized with P. aeruginosa AUST‐03, available studies have reported that P. aeruginosa AUST‐03 has been associated with increased pulmonary exacerbations. 11 , 27 It is also difficult to determine whether P. aeruginosa AUST‐03 infects a specific age group of pwCF as only one other study was found in the literature that investigated P. aeruginosa AUST‐03 (then referred to as AES‐III). 27 In that study, Bradbury and colleagues found P. aeruginosa AUST‐03 in pwCF aged 19 to 34 years old, however, this study mostly recruited from outreach clinics attended by adults with CF. 27 The current study could also not establish a correlation between age and P. aeruginosa AUST‐03 infection as only two children had this strain. A larger study at the hospital involving adults with CF will be required to better establish the prevalence of this strain at the hospital and the age groups of pwCF affected. The P. aeruginosa AUST‐03 isolates infecting P2 were found to be monoclonal, while in P4, this strain occurred as a polyclonal P. aeruginosa infection together with a second novel strain (ST3820) that was described for the first time in this study. Polyclonal infection was also observed by Bradbury et al., 27 who found three different P. aeruginosa strains, including AUST‐03 infecting one study participant.
The majority (8/11) of the P. aeruginosa AUST‐03 isolates from the study were MDR or XDR, which is in accordance with global reports of P. aeruginosa AUST‐03 of this strain having an increased likelihood of displaying MDR antibiograms. 11 , 12 , 27 Ciprofloxacin resistance was common to all of the P. aeruginosa AUST‐03 isolates from the study, this antibiotic is recommended by the South African Cystic Fibrosis Consensus Guidelines (SACFCG) for the treatment of P. aeruginosa exacerbations. 5 The high frequency of gentamicin resistance (7/11) in the P. aeruginosa AUST‐03 study isolates is also noteworthy as inhaled gentamicin is recommended in pwCF from South Africa for the eradication and chronic suppressive treatment of P. aeruginosa. 5 In high income countries, tobramycin is used for this purpose, however, the high cost of this antibiotic in South Africa has limited its availability in public hospitals and as such, gentamicin is used as a more cost effective alternative in these settings. 28 This lack of access to tobramycin may also explain the low rates (18% (2/11)) of resistance to this antibiotic in the study isolates. While most P. aeruginosa AUST‐03 isolates from the study were MDR, resistance to colistin, which is an antibiotic of last resort 29 reserved for use in MDR Gram negative pathogens, was low. Only a single XDR P. aeruginosa AUST‐03 isolate (P4) was resistant to colistin and regrettably, this patient (P4) passed away.
The current study is among the few to characterize the antibiotic resistance determinants across the entire genomes of P. aeruginosa isolated from pwCF in South Africa. The basis of antibiotic resistance in the P. aeruginosa AUTS‐03 isolates was mediated by multidrug efflux pumps. The most clinically relevant of these were the MexAB‐OprM, MexCD‐OprJ, MexEF‐OprN, and MexXY‐OprM efflux pumps which were detected in all of the isolates. Furthermore, mutations in the regulatory genes: nalC (S209R and G71E), nfxB (Type A), and mexRT of the 11 study isolates were detected, that conferred the overexpression of the MexAB‐OprM, MexCD‐OprJ, and MexEF‐OprN efflux pumps, respectively. 30 , 31 The most clinically relevant acquired ARGs detected in the study isolates was crpP (11/11) which confers resistance to ciprofloxacin. 32 Additionally, acquired mutations in the DNA gyrase (gyrA_D87N (3/11) and gyrB (8/11)) and topoisomerase IV (parE (1/11)) genes were found that also confer resistance to fluoroquinolones such as ciprofloxacin. 33 While colistin resistance was low (1/11) in the study isolates, mutations were found in the lipid A regulatory genes (pmrAB) of all of the isolates that conferred enhanced colistin resistance, 33 creating a potential for the future development of colistin resistance in the isolates.
Australia, which is the country where P. aeruginosa AUST‐03 was first described is among the top 10 overseas countries with tourists visiting South Africa. 34 It could be plausible that the Australian P. aeruginosa AUST‐03 epidemic strain could have been introduced into South Africa through visitors or immigrants traveling between the two countries. While phylogenetic analysis found some genetic relatedness between the study isolates from South Africa and four isolates from Australia, a more in‐depth analysis using a larger and more representative number of isolates from both countries would be required to determine whether any direct introduction occurred from Australia to South Africa. Furthermore, it could not be determined whether the Australian isolates were associated with any outbreaks in pwCF. The study isolates were most closely related to a 2020 CF isolate from Russia, however, introduction of this isolate between these two countries could not be determined with the limited data set. The profiles of the antibiotic resistance genes carried in the genomes of the P. aeruginosa AUST‐03 isolates from this study were similar to those for the global isolates in the subtree shown in Figure 1B, however, the isolates from this study carried more genes related to crpP mediated ciprofloxacin resistance, colistin resistance mutations (pmrA) and the mutations involved upregulation of efflux pumps (nalC and nfxB) (Figure 2).
The current infection control strategies practised at the CF clinic investigated are based on the segregation of pwCF infected or colonized with P. aeruginosa from those that are not, by having the two groups attend CF clinics on different weeks. However, the study findings suggest that a revision of these practices may be required that isolates people with P. aeruginosa AUST‐03 during the weeks of the P. aeruginosa CF clinics. This would assist in preventing the spread of this strain among pwCF attending the clinics, as there is evidence of possible strain sharing between P2 and P4, as two isolates from these children clustered very closely together on the phylogenetic tree (Figure 1).
The detection in this study of the epidemic P. aeruginosa AUST‐03 strain in pwCF from South Africa highlights the importance of routine surveillance of CF lung pathogens. However, most CF clinics in public hospitals have limited financial resources and important CFTR modulator therapies such as Trikafta® are currently not available in South Africa or are too costly for pwCF to purchase internationally. Trikafta® could have been a life‐saving intervention for the 8 year old study participant, who carried the ΔF508 gene. 35 The main limitation from the study was the low numbers of pwCF that were investigated and that adults were not included in the study, as P. aeruginosa AUST‐03 was previously primarily reported in adults with CF. 27 Future studies investigating P. aeruginosa across larger sample sizes and geographic regions are warranted to establish the spread of the epidemic P. aeruginosa AUST‐03 strain across South Africa. Such studies may also be beneficial in detecting other epidemic strains of P. aeruginosa that can cause potential outbreaks in pwCF.
5. CONCLUSION
The current study presents to our knowledge, the first report of an epidemic strain of P. aeruginosa among pwCF from South Africa. The P. aeruginosa AUST‐03 from the study setting were MDR and similarly to strains from previous outbreaks, were found in pwCF experiencing pulmonary exacerbations. With the decrease in WGS costs in South Africa, more frequent genomic surveillance of CF pathogens is required. Epidemic strains of P. aeruginosa are present at South African CF clinics and it is essential to take them into account when implementing segregation and infection control strategies.
AUTHOR CONTRIBUTIONS
Thabo Hamiwe: Conceptualization; investigation; methodology; formal analysis; writing—original draft; writing—review and editing. Debbie A White: Writing—review and editing; resources; formal analysis. Stanford Kwenda: Writing—review and editing; software; formal analysis; resources. Arshad Ismail: Writing—review and editing; resources. Susan Klugman: Resources; writing—review and editing. Lore Van Bruwaene: Writing—review and editing. Ameena Goga: Writing—review and editing. Marleen M Kock: Resources; Writing—review and editing. Anthony M Smith: Funding acquisition; writing—review and editing; resources. Marthie M Ehlers: Conceptualization; funding acquisition; supervision; formal analysis; writing—review and editing.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
ACKNOWLEDGMENTS
The authors would like to thank the children, parents and members of staff who were involved or assisted with the study at the cystic fibrosis clinic conducted by the hospital. The authors would also like to acknowledge the National Research Foundation and the University of Pretoria for the provision of PhD scholarship funds. Sequencing of isolates in this study was made possible by support from the SEQAFRICA project which is funded by the Department of Health and Social Care's Fleming Fund using UK aid. The views expressed in this publication are those of the authors and not necessarily those of the UK Department of Health and Social Care or its Management Agent, Mott MacDonald. National Health Laboratory Service Trust, Grant No. 004_94726; SEQAFRICA project, Department of Health and Social Care's Fleming Fund using UK aid.
Hamiwe T, White DA, Kwenda S, et al. Detection of the epidemic Pseudomonas aeruginosa AUST‐03 (ST242) strain in people with cystic fibrosis in South Africa. Pediatr Pulmonol. 2024;59:3340‐3348. 10.1002/ppul.27202
Contributor Information
Thabo Hamiwe, Email: thabohamiwe@gmail.com.
Marthie M. Ehlers, Email: marthie.ehlers@up.ac.za.
DATA AVAILABILITY STATEMENT
Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the BioProject number PRJNA767316 and the following accession numbers: JBCARG000000000‐JBCARQ000000000.
REFERENCES
- 1. KZN Cystic Fibrosis Association . 2024. Accessed March 15, 2024. https://cysticfibrosis.co.za/
- 2. Zampoli M, Verstraete J, Frauendorf M, et al. Cystic fibrosis in South Africa: spectrum of disease and determinants of outcome. ERJ Open Res. 2021;7:00856‐2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vandenbroucke NJ, Zampoli M, Morrow B. Lung function determinants and mortality of children and adolescents with cystic fibrosis in South Africa 2007‐2016. Pediatr Pulmonol. 2020;55:1381‐1387. [DOI] [PubMed] [Google Scholar]
- 4. Fothergill JL, Walshaw MJ, Winstanley C. Transmissible strains of Pseudomonas aeruginosa in cystic fibrosis lung infections. Eur Respir J. 2012;40:227‐238. [DOI] [PubMed] [Google Scholar]
- 5. Zampoli M, Morrow B. The South African Cystic Fibrosis Consensus/Guidelines 5th ed. 2017. Accessed March 15, 2024. https://www.sacfa.org.za/wpcontent/uploads/20170914CFConsensusGuidelines2017.pdf
- 6. Duong J, Booth SC, McCartney NK, Rabin HR, Parkins MD, Storey DG. Phenotypic and genotypic comparison of epidemic and non‐epidemic strains of Pseudomonas aeruginosa from individuals with cystic fibrosis. PLoS One. 2015;10:e0143466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schmid J, Ling LJ, Leung JLS, et al. Pseudomonas aeruginosa transmission is infrequent in New Zealand cystic fibrosis clinics. Eur Respir J. 2008;32:1583‐1590. [DOI] [PubMed] [Google Scholar]
- 8. Workentine M, Poonja A, Waddell B, et al. Development and validation of a PCR assay to detect the prairie epidemic strain of Pseudomonas aeruginosa from patients with cystic fibrosis. J Clin Microbiol. 2016;54:489‐491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wee BA, Tai AS, Sherrard LJ, et al. Whole genome sequencing reveals the emergence of a Pseudomonas aeruginosa shared strain sub‐lineage among patients treated within a single cystic fibrosis centre. BMC Genomics. 2018;19:644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Aaron SD, Vandemheen KL, Ramotar K, et al. Infection with transmissible strains of Pseudomonas aeruginosa and clinical outcomes in adults with cystic fibrosis. JAMA. 2010;304:2145‐2153. [DOI] [PubMed] [Google Scholar]
- 11. Parkins MD, Somayaji R, Waters VJ. Epidemiology, biology, and impact of clonal Pseudomonas aeruginosa infections in cystic fibrosis. Clin Microbiol Rev. 2018;31:e00019‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bradbury RS, Champion AC, Reid DW. Epidemiology of Pseudomonas aeruginosa in a tertiary referral teaching hospital. J Hosp Infect. 2009;73:151‐156. [DOI] [PubMed] [Google Scholar]
- 13. Douraghi M, Ghasemi F, Dallal MM, Rahbar M, Rahimiforoushani A. Molecular identification of Pseudomonas aeruginosa recovered from cystic fibrosis patients. J Prev Med Hyg. 2014;55:50‐53. [PMC free article] [PubMed] [Google Scholar]
- 14. Clinical and Laboratory Standards Institute (CLSI) . Performance standards for antimicrobial susceptibility testing. 31st ed. CLSI Supplement M100. Clinical and Laboratory Standards Institute; 2021. [Google Scholar]
- 15. Botelho J, Grosso F, Peixe L. Antibiotic resistance in Pseudomonas aeruginosa‐mechanisms, epidemiology and evolution. Drug Resist Updates. 2019;44:100640. [DOI] [PubMed] [Google Scholar]
- 16. De Oliveira Santos IC, Pereira de Andrade NF, da Conceição Neto OC, et al. Epidemiology and antibiotic resistance trends in clinical isolates of Pseudomonas aeruginosa from Rio de Janeiro‐Brazil: importance of mutational mechanisms over the years (1995–2015). Infect Genet Evol. 2019;73:411‐415. [DOI] [PubMed] [Google Scholar]
- 17. Kwenda S, Khumalo ZTH, Mtshali S, Mnyameni F, Ismail A. Jekesa: an automated easy‐to‐use pipeline for bacterial whole genome typing. 2020. https://github.com/stanikae/jekesa
- 18. Smith AM, Erasmus LK, Tau NP, et al. Enteric fever cluster identification in South Africa using genomic surveillance of Salmonella enterica serovar Typhi. Microb Genom. 2023;9:001044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Alcock BP, Huynh W, Chalil R, et al. CARD 2023: expanded curation, support for machine learning, and resistome prediction at the comprehensive antibiotic resistance database. Nucleic Acids Res. 2023;51:D690‐D699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bortolaia V, Kaas RS, Ruppe E, et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother. 2020;75:3491‐3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sayers EW, Bolton EE, Brister JR, et al. Database resources of The National center for biotechnology information. Nucleic Acids Res. 2022;50:D20‐D26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Seemann T. Whole genome core alignments from multiple draft genomes. 2016. https://github.com/tseemann/scapper
- 23. Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ‐TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol Biol Evol. 2015;32:268‐274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 2018;35:518‐522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Argimón S, Abudahab K, Goater RJE, et al Microreact: visualizing and sharing data for genomic epidemiology and phylogeography. Microb Genom. 2016;30:e000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Van den Bossche S, De Broe E, Coenye T, Van Braeckel E, Crabbé A. The cystic fibrosis lung microenvironment alters antibiotic activity: causes and effects. Eur Respir Rev. 2021;30:210055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bradbury R, Champion A, Reid DW. Poor clinical outcomes associated with a multi‐drug resistant clonal strain of Pseudomonas aeruginosa in the Tasmanian cystic fibrosis population. Respirology. 2008;13:886‐892. [DOI] [PubMed] [Google Scholar]
- 28. Van Stormbroek B, Zampoli M, Morrow BM. Nebulized gentamicin in combination with systemic antibiotics for eradicating early Pseudomonas aeruginosa infection in children with cystic fibrosis. Pediatr Pulmonol. 2019;54:393‐398. [DOI] [PubMed] [Google Scholar]
- 29. Aghapour A, Gholizadeh P, Ganbarov K, et al. Molecular mechanisms related to colistin resistance in Enterobacteriaceae . Infect Drug Resist. 2019;1:965‐975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Aeschlimann JR. The role of multidrug efflux pumps in the antibiotic resistance of Pseudomonas aeruginosa and other Gram‐negative bacteria: insights from the society of infectious diseases pharmacists. Pharmacother: J Human Pharmacol Drug Ther. 2003;23:916‐924. [DOI] [PubMed] [Google Scholar]
- 31. Zahedi Bialvaei A, Rahbar M, Hamidi‐Farahani R, et al. Expression of RND efflux pumps mediated antibiotic resistance in Pseudomonas aeruginosa clinical strains. Microb Pathog. 2021;153:104789. [DOI] [PubMed] [Google Scholar]
- 32. Rehman A, Patrick WM, Lamont IL. Mechanisms of ciprofloxacin resistance in Pseudomonas aeruginosa: new approaches to an old problem. J Med Microbiol. 2019;68:1‐10. [DOI] [PubMed] [Google Scholar]
- 33. Pang Z, Raudonis R, Glick BR, Lin TJ, Cheng Z. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotech Adv. 2019;37:177‐192. [DOI] [PubMed] [Google Scholar]
- 34. Statistics South Africa tourism and migration statistical release P0351 . 2023. Accessed March 15, 2024. https://www.statssa.gov.za/publications/P0351/P0351October2023.pdf
- 35. Becq F, Mirval S, Carrez T, et al. The rescue of F508del‐CFTR by elexacaftor/tezacaftor/ivacaftor (Trikafta) in human airway epithelial cells is underestimated due to the presence of ivacaftor. Eur Respir J. 2022;59:2100671. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the BioProject number PRJNA767316 and the following accession numbers: JBCARG000000000‐JBCARQ000000000.


