ABSTRACT
Association between ileal colonization by Adherent-Invasive Escherichia coli (AIEC) and Crohn’s disease (CD) has been widely described in high-incidence Western countries but remains unexplored in Asian countries with a fast increase in CD incidence. In the PACIFIC study, we compared the characteristics of AIEC pathobionts retrieved from ileal biopsies of CD patients enrolled in France (FR) and Hong Kong (HK). The prevalence of AIEC was similar in France (24.5%, 25/102) and Hong Kong (30.0%, 18/60) (p = 0.44). No difference was observed between the two populations of AIEC regarding adhesion and invasion levels. When tested for antibiotic resistance, the proportion of AIEC strains resistant to ampicillin, piperacillin, tobramycin, and gentamicin was significantly higher in HK AIEC strains compared to French strains. AIEC strains from FR or HK population were both able to persist in the mice intestine (DSS-treated CEABAC10 mice model). Moreover, genomic analysis of 25 FR and 17 hK AIEC strains using next-generation sequencing revealed the co-existence of several virulence factors associated with enteric E. coli pathotypes, although no single virulence factor was significantly associated with either country of origin or AIEC status. In vitro, all AIEC strains (FR and HK) were sensitive to the EcoActive™ phage cocktail, suggesting that it could be a promising option to target AIEC in CD across the world.
KEYWORDS: Crohn’s disease, adherent-invasive E. coli, trans-ethnic study, bacterial comparison
Introduction
Crohn’s disease (CD) is an inflammatory bowel disease (IBD) that can induce chronic inflammation of the gastrointestinal tract, leading to bowel damage and altered quality of life for patients.1 CD may affect different populations, including children, adults, and elderly people, regardless of geographic area or ethnicity.2,3 Epidemiology of CD varies according to the area of the world and can be summarized in distinguishing three distinct epidemiological stages: i) since 2020, Western countries (i.e., North America, Western Europe, and Oceania) display a relatively stable incidence but a sharp rise in the prevalence; ii) newly industrialized countries in Asia, Latin America, and the Middle East experience rising incidence but relatively low prevalence, and iii) in developing countries, we can observe the emergence of sporadic IBD cases.4–6 Epidemiological studies led in Eastern countries could be of great value to better understand the pathogenesis of CD and thus attempt to reduce its incidence. Although the exact etiology of CD remains unknown, experimental and observational data suggest that intestinal inflammation arises from an abnormal immune response to intestinal microbiota, favored by environmental factors and genetical susceptibility.5 Westernization of life habits could modify environmental exposure that can influence the microbiome and contribute to the genesis of CD.6,7 CD-associated dysbiosis is characterized by an increased number of mucosa-associated Enterobacterales, especially Escherichia coli, and a reduced overall biodiversity.8–11 Among these microbiota alterations, the association between ileal CD and a specific E. coli pathovar, adherent-invasive E. coli (AIEC), has now been widely demonstrated across the world.12 AIEC is able to adhere to and invade intestinal epithelial cells (IECs) as well as to replicate within macrophages, leading to increased levels of pro-inflammatory cytokines.12–14 However, owing to very different ecosystems according to the area of the world, assessing whether AIEC characteristics are similar or different across the world is paramount, while therapeutic strategies targeting AIEC (antibiotics, phages, fecal microbiota transplantation…) to treat patients with CD are currently being investigated. In this work, we aimed to compare the prevalence, the genomics, the phenotypic properties, the sensitivity to antibiotics, and bacteriophage cocktails between AIEC strains retrieved from the ileum of two different populations of patients with CD: Caucasians living in France and Chinese living in Hong Kong.
Methods
Ethical considerations
The study was performed in accordance with the Declaration of Helsinki, Good Clinical Practice, and applicable regulatory requirements. The French ethical committee, so-called “Comité de Protection des Personnes (CPP) Sud-Est 6” – France, approved the French study [AU 904]. The Chinese study was approved by the Ethics Committee of the Chinese University of Hong Kong [CRE 2014.026].
The in vivo experiments were carried out in strict accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals of Clermont Auvergne University (Clermont-Ferrand, France). The Committee for Research and Ethical Issues of the Department of Auvergne (CEMEA Auvergne) approved the animal protocol (Permit Number: CEMEAA 2,018,031,914,539,228). Mice were housed in an animal facility of Clermont Auvergne University (Clermont-Ferrand, France) in specific pathogen-free conditions, with access to food and water ad libitum.
Design of the study
Biopsies were taken from the ileum of 60 hong Kong Chinese (HK) CD patients undergoing colonoscopy at the Prince of Wales Hospital, the Chinese University of Hong Kong, and from the ileum of 102 French (FR) CD patients requiring ileo-colonoscopy at multiple IBD centers in France.15 Patients were included if they were 18 years or older with a diagnosis of ileal CD (ileal or ileocolonic); had active ileal disease at the time of endoscopy; and had stable CD-related medication. Patients with antibiotics, probiotics, prebiotics, or a history of enteric infection in the past 3 months were excluded. Tissues were immersed in the cell culture medium MEM supplemented with 15% sterile glycerol, snap frozen, and stored at −80°C until microbiological analysis.
Isolation and characterization of AIEC bacteria
Ileal biopsies were crushed in phosphate-buffered saline (PBS), and the lysate was plated on Drigalski agar to isolate Escherichia coli colonies after 24 hours of incubation at 37°C, as previously described.15,16 A random selection of 45 lactose-positive colonies per sample was validated for E. coli species using an automated mass spectrometry microbial identification system based on Matrix Assisted Laser Desorption Ionization Time-of-Flight (MALDI-TOF) technology, according to manufacturer’s recommendations (VitekII®, Biomérieux, France). They were then cultured in 96-well microplates in a Luria-Bertani medium at 37°C, supplemented with 15% glycerol, and stored at −80°C until AIEC characterization. Each French and Chinese partner has carried out the AIEC characterization by analyzing their abilities to adhere to and invade intestine-407 epithelial cells (ATCC, CCL-6) and to survive and replicate within THP-1 macrophages (ATCC, TIB-202) by conducting antibiotic protection assays, as previously described.14,15 Briefly, the 45 E. coli strains per biopsy were pre-screened for invasive abilities by mixing equitably and extemporaneously the overnight bacterial cultures and infecting I-407 cells at a multiplicity of infection of 100 bacteria per cell. Three hours later, infected I-407 cells were exposed for 1 h to 100 µg/ml of gentamicin or 50 µg/ml of meropenem (depending on strain resistance to antibiotics) to kill extracellular bacteria. Cells were lysed using Triton 1X, and the lysate was plated on Drigalski agar to isolate surviving bacteria. AIEC phenotype of these invasive strains was validated by conducting antibiotic protection assays to confirm their abilities to adhere to and invade I-407 cells and to survive and replicate within THP-1 macrophages.
Adhesion and invasion assays on AIEC bacteria
AIEC strains characterized from Hong Kong and from France were compared in the same experiment of adhesion and invasion to I-407 cells. As previously described,14 I-407 cells were seeded in 24-wells tissue culture plates at a density of 4.105 cells/well and incubated for 24 h. The cells were washed twice with PBS before being infected with a multiplicity of infection of 10 bacteria per cell in 1 mL of adequate cell culture medium. After a 3-h incubation period at 37°C with 5% CO2, the monolayers were washed 3 times with PBS. Invasion was assessed by adding a fresh cell culture medium containing 50 µg/ml of meropenem (gentamicin was substituted due to high resistance to gentamicin of HK strains) for 1 h to kill extracellular bacteria. For both adhesion and invasion assays, the cells were lysed using Triton 1X, and the samples were diluted and plated onto Luria Bertani (LB) agar plates to determine the number of colony-forming units. All of the assays were performed 3 times in separate experiments. The results were expressed as the number in CFU/cell of adherent or invasive bacteria compared to the number of bacteria present in the initial inoculum. For all phenotypical assays, the non-AIEC E. coli strain, K-12, was used as the negative control, and the AIEC reference strain LF82 was used as the positive control.
Bacterial strains and culture conditions
Four AIEC strains from each French (FR) or Hong Kong (HK) populations, selected for their belonging to B2 or D phylogroup, harboring or not S70/N78 mutations in the fimH adhesin gene17 and with the highest invasion rates in IECs among those that met the first two criteria, as well as AIEC LF82 strain,18 were used in the protocol of mouse infection (Supplemental Data Table S1). The strains were either naturally resistant to streptomycin or made resistant by insertion of the streptomycin-resistance encoding gene on the chromosome. Using a mobilizable mini-Tn7-based vector (pUC18R6KT-mini-Tn7T-Stp) delivered by E. coli MFDpir46 and E. coli MFDpir/pTNS3 strain encoding the tnsABCD genes necessary for the transposition of mini-Tn7 at the attTn7 insertion site, the AIEC recipient strain was modified as a streptomycin-resistant mutant.19,20 The AIEC strains and mutants were grown in Luria Bertani (LB) broth under static and aerobic conditions overnight at 37°C. On the day of infection, bacterial cultures were harvested by centrifugation at 4 500 × g for 10 min, and pellets were resuspended in PBS at a concentration of 5 × 109 bacteria/ml.
Mouse infection protocol
CEABAC10 FVB/N transgenic male mice (6–8 weeks old) were used in an in vivo model mimicking colitis and Crohn’s disease.21 They were orally pretreated for 3 days with 2.5 g/l of streptomycin and 0.5% (wt/vol) of dextran sulfate sodium (DSS; molecular mass = 36,000 –50,000 daltons; MP Biomedicals) in drinking water to promote the accessibility of bacteria to the intestine. Twenty-four hours later, mice were challenged with 0.2 ml of 109 AIEC bacteria (Figure 2a). Body weight was assessed over 10 days, and the severity of colitis was valued by the disease activity index (DAI) score ranging from 0 (healthy) to 12 (high colitis activity), as previously described.22 About 100 mg of fresh fecal samples were collected at different days postinfection (dpi) and homogenized in PBS, and serial dilutions were plated on LB agar medium containing 100 µg/ml streptomycin to select AIEC bacteria. After overnight incubation at 37°C, bacteria were numbered in CFU/ml. Ten days after oral infection, the mice were anesthetized with isoflurane and euthanized by cervical dislocation. Colon and ileum specimens were collected and washed. Serial dilutions in PBS were plated onto streptomycin-selective LB agar medium for overnight incubation at 37°C. AIEC bacteria associated with the tissue were counted in CFU/ml. Results reflecting the gain or loss of body weight between day 0 and day 10 are expressed as area under the body weight change curve (AUC) in percentage over 10 days. Results reflecting the score of DAI between day 0 and day 10 are expressed as area under the DAI score curve (AUC) in 10 days.
Figure 2.
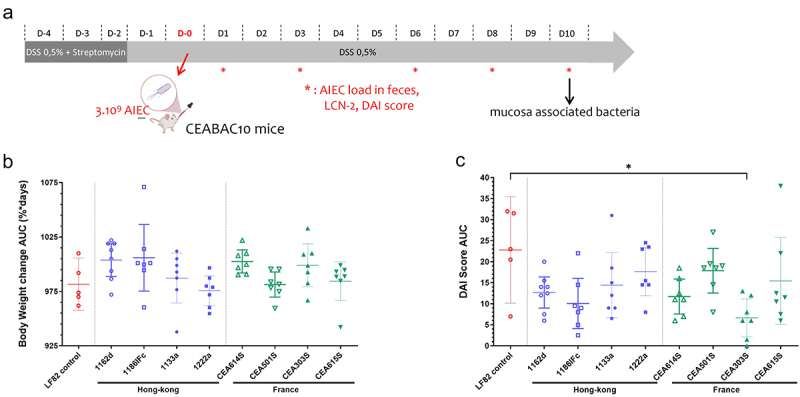
Clinical symptoms of colitis in CEABAC10 transgenic mice after an oral challenge with four FR strains (CEA614S, CEA501S, CEA303S, CEA615S), four HK strains (1162d, 1186IFc, 1133a, 1222a) and AIEC LF82 reference strain. (a) Schema of the dss-treated CEABAC10 transgenic mouse model infection protocol. AUC values represent the evolution of mice body weight (b) and DAI score (c) during 10 days after infection. n = 5-8 mice per group. Results are presented as mean with 95% confidence interval; Kruskal–Wallis test, *p < .05.
Whole-genome sequencing
Whole-genome sequencing (WGS) was conducted at the teaching hospital of Clermont-Ferrand, France, using a next-generation sequencing platform, with the analysis criteria previously described.23 Briefly, DNA extraction was carried out with the DNeasy UltraClean Microbial kit (Qiagen, Hilden, Germany), and libraries were prepared subsequently using the Nextera XT Kit (Illumina, San Diego, CA, USA). The reads generated on the Illumina MiSeq system were filtered for quality using Fastp software v0.19.10,24 and short reads were assembled with SPAdes.25
Molecular typing and virulence factor detection
E. coli phylogroups and multilocus sequence typing (MLST) were determined in silico according to the Clermont Typing method26 and Achtman’s MLST scheme.27 Core genome SNP-based typing (cgSNP) was performed with BactSNP v1.1.028 using the E. coli core genome downloaded from the Enterobase website (https://enterobase.warwick.ac.uk) as a reference, as previously described.29,30 After filtration of recombination zones detected by Gubbins,31 a phylogenetic tree was inferred from the resulting alignment by maximum likelihood using RAxML.32 The detection of virulence factors was performed with blastx DIAMOND33 using a threshold of 85% amino acid identity and the E. coli VFDB database (http://www.mgc.ac.cn/VFs/). After alignment with Muscle,34 minimum spanning trees were constructed from amino acid sequences with Grapetree.35
Antibiotics susceptibility testing
Identification of strains was confirmed by the Vitek MS MALDI TOF method (BioMérieux, La Balme, France). The production of extended-spectrum beta-lactamase (ESBL) was detected according to the recommendations of the Antimicrobial Committee of the French Society for Microbiology (CA- SFM, 2019 V2). Antibiotic susceptibilities were determined by the disk diffusion method according to the CA-SFM recommendations. ESBL production was confirmed by the double-disk synergy test. The following 29 antibiotics were tested: ampicillin, amoxicillin-clavulanic acid, ticarcillin, piperacillin, piperacillin-tazobactam, temocillin, cefalexin, cefuroxime, cefixime, mecillinam, temocillin, cefoxitin, ceftazidime, cefotaxime, aztreonam, cefepime, ertapenem, imipenem, amikacin, gentamicin, tobramycin, netilmicin, nalidixic acid, norfloxacin, ofloxacin, ciprofloxacin, fosfomycin, chloramphenicol, and trimethoprim (Biorad, Marnes-La-Coquette, France). These antibiotics were chosen according to the lists recommended by CA-SFM to identify most resistance mechanisms, especially those concerning beta-lactams (ESBL, carbapenemase especially).
Phages sensitivity testing
Seven individual phages composing the EcoActive™ cocktail were isolated by Intralytix or the Institut Pasteur. They were selected for their lysing capacity against AIEC strains during the spot test assay, as described by Titecat et al.36 The seven monophages were tested against 46 AIEC strains from FR and HK at ~2 × 104 and ~1 × 109 PFU/mL concentrations using the spot test assay.37 Next, the specificity of the monophages combined to produce the EcoActive™ phage cocktail was examined against the 46 AIEC strains at the same concentrations.
Statistical analysis
Values are expressed as percentage, mean with 95% confidence interval (CI) or as median with interquartile range (IQR). Statistical analyses were performed using GraphPad Prism 9 (version 9.3.1, GraphPad Software, San Diego, CA, USA). An unpaired Mann–Whitney test was performed for single comparisons, and the Kruskal–Wallis test with Dunn’s post hoc test was performed for multiple comparisons. A value of p < 0.05 was considered to be statistically significant.
Results
The prevalence and phenotype of AIEC associated to ileal mucosa are similar in France and Hong Kong Chinese populations
AIEC was detected in the ileal mucosa of 25 (24.5%) of 102 French patients with CD and 18 (30%) of 60 Chinese patients with CD (p = 0.44).
Phenotypical properties of AIEC strains from France and Hong Kong
Once identified with AIEC pathovar definition using adhesion, invasion in IECs, and survival within macrophage assays, the AIEC isolates from French patients were compared with isolates from Chinese patients for their abilities to adhere and to invade intestinal epithelial cells, the main phenotypic criteria. A total of 29 AIEC isolated from 24 French patients and 17 AIEC from 16 Chinese patients were submitted to the gentamicin protection assay, modified by using meropenem due to the presence of gentamicin-resistant strains in I-407 epithelial intestinal cells. The ability of these bacteria isolated in France or in Hong Kong to adhere to IECs was not different (p = 0.46), with a median of 6.7 [IQR, 0.543–12.74] CFU/cell for French AIEC isolates and a median of 1.245 [IQR, 0.755–9.417] CFU/cell for Chinese AIEC isolates (Figure 1a). The median level of invasion between FR AIEC isolates (0.027 [IQR, 0.010–0.111] CFU/cell) and HK AIEC isolates (0.043 [IQR, 0.023–0.102] CFU/cell) was no different (p = 0.57) (Figure 1b). These results indicate that AIEC strains interact similarly with intestinal epithelial cells regardless of their geographical origin.
Figure 1.
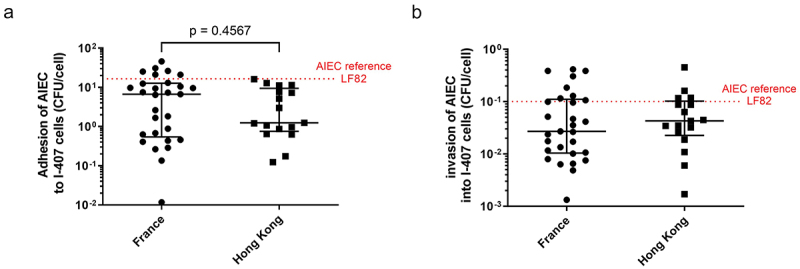
Adhesion and invasion of AIEC strains isolated from ileum of CD patients in France (n = 29) and Hong Kong (n = 17) into I-407 epithelial intestinal cell line. The number of adhered (a) or internalized (b) bacteria, to assess adhesion and invasion, respectively, was determined as described in section “materials and methods.” Results were expressed as numbers of CFU/cell and represented as median with interquartile range of triplicate experiments. Data were analyzed by Mann Whitney test, ns.
AIEC bacteria from France and Hong Kong colonize the intestine of mice with similar efficacy
The DSS-treated CEABAC10 transgenic mouse model expressing hCEACAM6 in epithelial cells represents a preclinical model allowing to decipher the role of AIEC bacteria in inflammatory bowel diseases. This model demonstrated the ability of LF82 bacteria, the reference for AIEC strains, to adhere to the intestinal epithelium and modulate intestinal permeability.18,21 To investigate and compare the gut persistence of AIEC bacteria isolated from FR and HK populations, seven mice per group were challenged with one of the four different FR strains (CEA614S, CEA501S, CEA303S, and CEA615S) or one of the four different HK strains (1162d, 1186IFc, 1133a, and 1222a), and the behavior was compared to mice infected with the AIEC LF82 reference strain.
To analyze the consequences of AIEC colonization, body weight loss of mice orally challenged with 109 bacteria was recorded. The disease activity index (DAI) score was determined at days 0, 1, 3, 6, 8, and 10 post-infection. There was a similar evolution between day 0 and day 10 in the body weight of the LF82 control group (AUC = 981.6%*days; 95% CI, 957.6–1006%*days) compared to other AIEC-infected groups (Figure 2b). No significant difference was observed in the DAI score AUC during this period of 10 days between the LF82 control group (AUC = 22.80; 95% CI, 10.15–35.45) and other AIEC-infected groups, except for the group of mice infected with CEA303S strain (AUC = 6.64; 95% CI, 2.14–11.15, p = .0157) (Figure 2c).
The levels of fecal AIEC bacteria, reflecting the persistence of the pathobiont within the gut lumen, were assessed for 10 days and revealed that there was no difference in bacterial counts between LF82 and each of the eight FR and HK strains at days 1, 3, 6, 8, and 10 post-infection (Figure 3a). On day 10 after infection, three-fourths of the CEABAC10 mice groups that were challenged with FR strains (i.e. CEA614S, CEA501S, and CEA615S) and two-fourths of the CEABAC10 mice groups that were challenged with HK strains (i.e. 1162d and 1186IFc) still presented more than 1 × 105 CFU per gram of feces, while mice challenged with LF82 strain harbored 1.1 × 108 [IQR, 5.3 × 107 − 2 × 108] CFU/g of feces. However, 10 days after infection, 1222a strain from HK and CEA303S strain from FR showed reduced colonization at the level of 3.8 × 102 [IQR, 1.0–1.5 × 103] and 1.1 × 103 [IQR, 1.7 × 102 − 1.9 × 107] CFU/g of feces, respectively.
Figure 3.
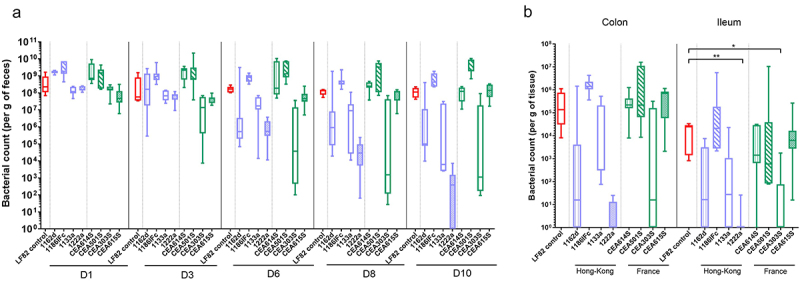
AIEC gut colonization after an oral challenge with four FR strains (CEA614S, CEA501S, CEA303S, CEA615S), four HK strains (1162d, 1186IFc, 1133a, 1222a) and AIEC LF82 reference strain in CEABAC10 transgenic mice. (a) Levels of fecal AIEC bacteria at days 1, 3, 6, 8 and 10 postinfection (Cfu/g of feces). (b) AIEC bacteria associated with the intestinal mucosa at day 10 postinfection (Cfu/g of tissue). n = 5-8 mice per group. Results are presented as box-plot with whiskers from minimum to maximum (red box = LF82 reference, purple boxes = hK AIEC strains, green boxes = FR AIEC strains); Kruskal-Wallis test, *p < .05, **p < .01.
To confirm AIEC colonization, we determined the number of mucosa-associated bacteria at euthanasia. Ten days after infection, the median colonic bacterial load of FR AIEC remained high for CEA614S, CEA501S, and CEA615S strains (respectively, 2.2 × 105 [IQR, 1.6 × 105 − 4.2 × 105], 2.2 × 105 [IQR, 6.4 × 104 − 1.1 × 107], and 7.0 × 105 [IQR, 5.8 × 104 − 8.4 × 105] CFU/g of tissue) and close to median LF82 load (1.4 × 105 [IQR, 6.2 × 104 − 8.1 × 105] CFU/g of tissue) (Figure 3b). 1186IFc HK strain was also maintained at a high level of colonization in the colon (1.4 × 106 [IQR, 1.1 × 106 − 2.3 × 106] CFU/g). Concerning 1162d, 1133a, 1222a, and CEA303S strains at day 10, a decreased bacterial load was noticed in the colon at the median level of 1.2 × 102 [IQR, 0–7.6 × 103], 3.3 × 102 [IQR, 3.0 × 102 − 2.1 × 105], 0.0 [IQR, 0–1.2 × 101], and 1.5 × 101 [IQR, 0–1.6 × 105] CFU/g of tissue, respectively.
The median ileal bacterial load of FR AIEC remained high for CEA614S, CEA501S, and CEA615S strains (respectively, 1.4 × 103 [IQR, 6.4 × 102 − 2.9 × 104], 5.9 × 102 [IQR, 8.1 × 101 − 1.0 × 107], and 6.3 × 103 [IQR, 2.7 × 103 − 1.6 × 104] CFU/g of tissue) and close to median LF82 load (2.5 × 104 [IQR, 1.6 × 103 − 2.9 × 104] CFU/g of tissue) (Figure 3b). Similar to colonic colonization, 1186IFc strain from HK was also maintained at a high level of colonization in the ileum (2.1 × 104 [IQR, 2.8 × 103 − 1.8 × 105] CFU/g of tissue). Consistent with colonic values, 1162d, 1133a, 1222a, and CEA303S strains displayed at day 10 in the ileum a decreased bacterial load at the median level of 1.3 × 102 [IQR, 0–3.3 × 103], 2.6 × 101 [IQR, 0–1.1 × 103], 0.0 [IQR, 0–0] and 0.0 [IQR, 0–7.5 101] CFU/g of tissue, respectively. This suggests that most AIEC strains could colonize mice ileum to similar levels as LF82, although a heterogeneity in the levels and persistence of colonization by certain AIECs over the course of days can be observed.
Genomic analysis revealed a majority of AIEC strains belonging to the B2 phylogroup but did not report a specific sequence typing (ST) dependent on AIEC origin
In silico MLST (Multi-Locus Sequence Typing) and phylogrouping were assessed from whole-genome sequencing data of 29 FR AIEC and 17 hK AIEC isolates. The core genome SNP-based phylogenetic tree representation showed that AIECs originated from the main phylogroups of E. coli (A, B1, B2, D, and F), belong to a wide diversity of ST lineages, and were not clustered according to their geographic origin (Figure 4). However, most AIEC strains belonged to the B2 phylogroup (19/46, 41.3%), followed by the D phylogroup (10/46, 21.7%). Within the frequent pandemic STs (for example, ST95, ST69, ST38, ST73, and ST131), AIEC samples from the two countries of origin can be found. These lineages were present among CD patients in isolated AIEC with two AIEC strains identified as ST131, two strains as ST95, five strains as ST73 belonging to B2 phylogroup, and six strains identified as ST69 and two AIEC as ST38 among D phylogroup. Of interest, the 1162d strain from the HK CD patient belonged to the same ST135 as the LF82 AIEC reference. Overall, the results show the genetic diversity within this pathovar and the absence of significant differences in the AIEC core genome from FR and HK.
Figure 4.
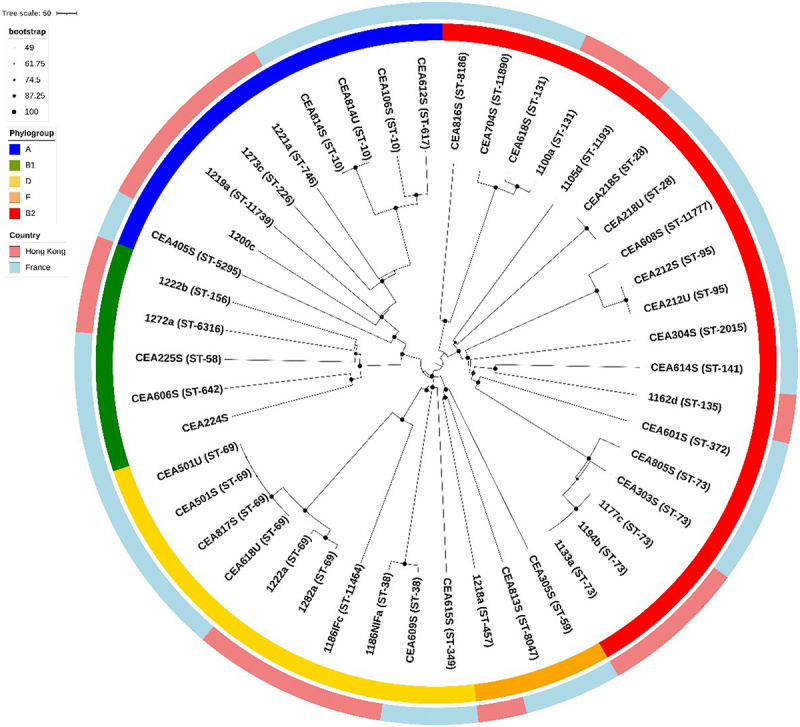
Phylogenetic analysis of AIEC isolated from French and Hong Kong Chinese CD patients. Concatenated core genome SNPs sequences were aligned and phylogenetic inferences obtained using Gubbins method and RAxML program. Sequence types (ST) correspond to Achtman’s MLST scheme. Countries where the sample collection originated are indicated by two different colors on the outer ring. All isolates were clustered into five phylogroups represented by five colors on the inner ring.
Genomic analysis revealed the co-existence of several virulence factors associated with enteric E. coli pathotypes across the sequenced AIEC strains
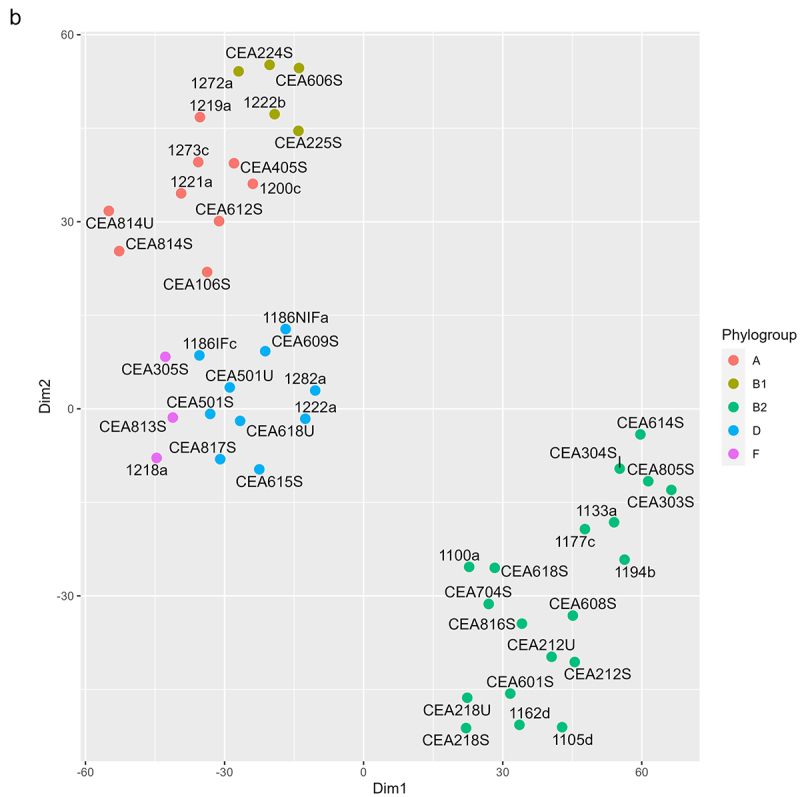
We examined the distribution of virulence factors in a gene sequencing dataset of the 46 AIEC strains of this study using a Blastx approach and the VFDB database. This procedure identified 128 virulence factors among the 46 strains (Figure 5a). Many of these virulence factors are involved in functions that are previously implicated in pathophysiology in IBD, including adhesion/invasion,38 flagella, iron-acquisition,39 serum resistance, secretion systems,40 and others like capsule synthesis.41 We observed that strains in the phylogroup B2 (44.8% from FR and 35.3% from HK collection) generally have more virulence factors than other strains and more virulence factors in common (Figure 5b). From the amino acid sequences of four virulence genes of interest (i. e. ompA, fimH, fliC, and chiA), minimum spanning trees were constructed to compare the phylogeny of the 46 AIEC strains of the study (Figure 6). We chose to focus on these specific virulence genes as they have been described as important AIEC virulence factors. In particular, previous studies showed the implication of ompA,42 fimH,17 fliC,43 and chiA44 in identifying variants in the pathogenicity of AIEC, leading to hypothesize that the presence of some AIEC-specific variants could promote the adhesion of AIEC to intestinal cells. In this study, we investigated whether AIEC isolated from HK exhibited the same variants as the strains from FR. The minimum spanning trees revealed that the majority of strains displayed the presence of ompA, fimH, fliC genes, and 50% of the strains, the presence of chiA gene. In general, the results showed the absence of one large cluster, with the AIEC isolates well-dispersed regardless of their origin. The various fimH variants identified were closely related as suggested by the short length of the edges (Figure 6). However, there was no observable relationship between the grouping of AIEC isolates and the country of origin.
Figure 5.
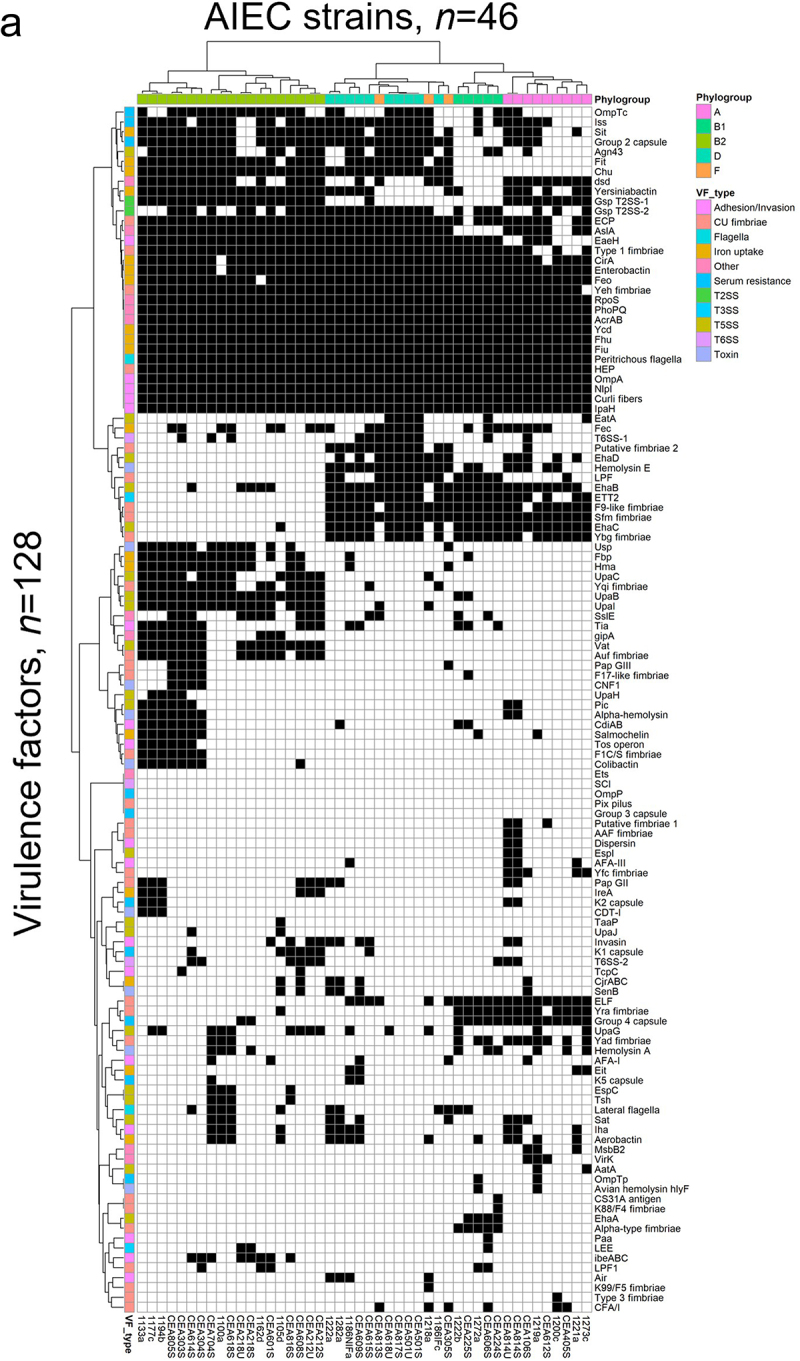
Analysis of virulence genes detected in the Illumina gene sequencing dataset of the 46 AIEC strains of this study. (a) Dendrogram analysis was performed for AIEC strains phylogenic relationships and virulence factors (VFs) relatedness using blastx DIAMOND. Black boxes indicate the presence of the gene listed on the right and white boxes, their absence. The scale on the right indicate the phylogroups and the family of the listed gene, identified by colors. (b) t-sne plot visualizing cluster assignments of AIEC strains. Strains are projected into t-sne space, with the first two t-sne components as the axes of the plot. Phylogroups denoted as distinct colors were assigned according to the Clermont method and cgSNP phylogeny.
Figure 5.
Continued.
Figure 6.
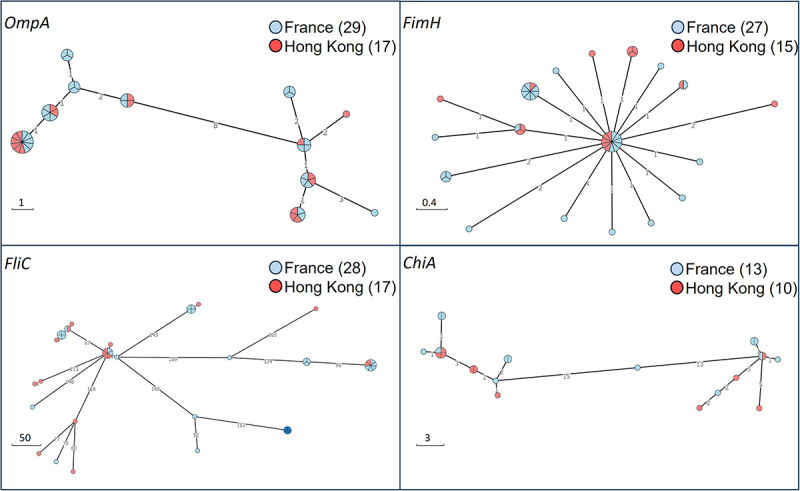
Minimum spanning trees of ompA, fimH, fliC and chiA genes variants deduced from the whole-genome sequences of the 46 AIEC strains of this study. The length of the edges is proportional to the number of amino acid substitutions (scale: substitution number), and the size of nodes to the number of strains (number of sectors). The geographic origin is indicated by the node color.
Antibiotics sensitivity of AIEC collection is higher in FR than in HK
The proportion of AIEC strains resistant to antibiotics was higher in HK AIEC strains compared to French strains regarding ampicillin (FR: 31%, n = 9; HK: 88%, n = 15; 2.9-fold higher, p = 0.0002), ticarcillin (FR: 31%, n = 9; HK: 88%, n = 15; 2.9-fold higher, p = 0.0002), piperacillin (FR: 31%, n = 9; HK: 82%, n = 14; 2.7-fold higher, p = 0.0018), tobramycin (FR: 3%, n = 1; HK: 70%, n = 12; 23-fold higher, p < 0.0001) and gentamicin (FR: 3%, n = 1; HK: 70%, n = 12; 23-fold higher, p < 0.0001) (Table 1). Over half of the total AIEC strains were resistant to ampicillin (52.3%), ticarcillin (52.3%), and piperacillin (50%) compared to sensitivity to other antibiotics tested in this study, independently of the AIEC origin. Furthermore, all isolates were susceptible to ertapenem, imipenem, amikacin, and fosfomycin. In conclusion, most FR AIEC strains were sensitive to beta-lactam antibiotics conventionally used in medical practice. In contrast, AIEC strains from HK displayed more resistance to this class of antibiotics.
Table 1.
Proportions of AIEC strains isolated from French (n = 29) and Hong Kong (n = 17) Chinese CD patients that are resistant to specific antibiotics.
| Antibiotic | France |
Hong Kong |
p-Value | ||
|---|---|---|---|---|---|
| n | (%) | n | (%) | ||
| Ampicillin | 9/29 | (31) | 15/17 | (88,2) | .0002 |
| Amoxicillin-clavulanate | 1/29 | (3,4) | 2/17 | (11,8) | .5453 |
| Ticarcillin | 9/29 | (31) | 15/17 | (88,2) | .0002 |
| Piperacillin | 9/29 | (31) | 14/17 | (82,4) | .0018 |
| Piperacillin-tazobactam | 0/29 | (0) | 1/17 | (5,9) | .3696 |
| Mecillinam | 1/29 | (3,4) | 1/17 | (5,9) | >.9999 |
| Temocillin | 4/29 | (13,8) | 4/17 | (23,5) | .4429 |
| Cefalexin | 0/29 | (0) | 5/17 | (29,4) | .0045 |
| Cefoxitin | 0/29 | (0) | 1/17 | (5,9) | .3696 |
| Cefixime | 0/29 | (0) | 4/17 | (23,5) | .0146 |
| Cefuroxime | 0/29 | (0) | 4/17 | (23,5) | .0146 |
| Ceftazidime | 0/29 | (0) | 3/17 | (17,6) | .0448 |
| Cefotaxime | 0/29 | (0) | 3/17 | (17,6) | .0448 |
| Aztreonam | 0/29 | (0) | 3/17 | (17,6) | .0448 |
| Cefepime | 0/29 | (0) | 2/17 | (11,8) | .1314 |
| Ertapenem | 0/29 | (0) | 0/17 | (0) | >.9999 |
| Imipenem | 0/29 | (0) | 0/17 | (0) | >.9999 |
| Tobramycin | 1/29 | (3,4) | 12/17 | (70,6) | <.0001 |
| Gentamicin | 1/29 | (3,4) | 12/17 | (70,6) | <.0001 |
| Amikacin | 0/29 | (0) | 0/17 | (0) | >.9999 |
| Nalidixic acid | 2/29 | (6,9) | 5/17 | (29,4) | .0832 |
| Norfloxacin | 2/29 | (6,9) | 3/17 | (17,6) | .3429 |
| Ofloxacin | 2/29 | (6,9) | 5/17 | (29,4) | .0832 |
| Ciprofloxacin | 2/29 | (6,9) | 3/17 | (17,6) | .3429 |
| Chloramphenicol | 1/29 | (3,4) | 3/17 | (17,6) | .1354 |
| Trimethoprim | 4/29 | (13,8) | 8/17 | (47,1) | .0187 |
| Sulfamethoxazol-trimethoprim | 3/29 | (10,3) | 8/17 | (47,1) | .0100 |
| Fosfomycin | 0/29 | (0) | 0/17 | (0) | >.9999 |
| Netilmycin | 0/29 | (0) | 6/17 | (35,3) | .0013 |
The antibiotics were chosen according to the lists recommended by the Antimicrobial Committee of the French Society for Microbiology (CA-SFM, 2019 V2).
AIEC collections from both sides of the globe are susceptible to the EcoActive™ phage cocktail, a potential therapy
The lytic range of each of the seven phages was assessed on 29 AIEC strains obtained from FR and 17 AIEC strains obtained from HK CD patients at two concentrations: 2 × 104 and 1 × 109 PFU/mL. As summarized in Table 2, 25 of the 29 FR AIEC (86.2%) and 15 of the 17 hK AIEC (88.2%) were susceptible at 2 × 104 PFU/mL to at least one of the monophages included in the EcoActiveTM cocktail. The efficacy reached 100% at the higher phage concentration [i.e., × 109 PFU/mL] more commonly used in the Spot Test assay, suggesting that the component phages included in the EcoActiveTM phage cocktail had a strong lytic potency against AIEC strains from both FR and HK.
Table 2.
Summary of the lytic activity of the AIEC targeting seven monophages and the EcoActiveTM cocktail (2 × 104 and 1 × 109 PFU/mL) against a panel of isolates from France (n = 29) and Hong Kong (n = 17).
| FRANCE |
HONG KONG |
|||
|---|---|---|---|---|
| Phage preparation | 2 × 104 | 1 × 109 | 2 × 104 | 1 × 109 |
| ECML-119 | 28% | 66% | 41% | 59% |
| ECML-1232 | 34% | 83% | 47% | 76% |
| ECML-359 | 38% | 72% | 35% | 76% |
| ECML-363 | 28% | 62% | 35% | 76% |
| CLBP2 | 45% | 76% | 65% | 82% |
| LF82P2 | 14% | 48% | 6% | 71% |
| LF82P8 | 24% | 66% | 6% | 47% |
| EcoActive | 86.2% | 100% | 88.2% | 100% |
Percentages represent the number of AIECs in the total collection from each country lysed by the phage or the cocktail at each concentration.
Discussion
In the PACIFIC study, we performed, for the first time, a direct comparison of AIEC strains retrieved from two very different geographic areas. We did not report any differences regarding adhesion and invasion properties, phylogenetic characteristics of the strains (mainly B2 phylogroup), or ability to colonize the gut in a mice model. While the antibiotics resistance profile was different between the two types of strains, we found a strong lytic potency of the EcoActiveTM phage cocktail against AIEC strains regardless of their origin (FR or HK), suggesting that phage therapy may offer a more promising therapeutic option to target AIEC across the world.
The PACIFIC study described for the first time the presence of AIEC among the Hong Kong Chinese population, a population of particular interest owing to a low but increasing CD incidence.6 The prevalence of CD patients colonized by AIEC in Hong Kong (30%)16 is comparable to the prevalence observed in French CD patients (24.5%).15 A meta-analysis from Kamali Dolatabadi et al.45 shows that the prevalence of CD patients colonized by AIEC in our study is comparable to the prevalence observed so far in different parts of the world (data published in 2004–2020). The prevalence observed in French CD patients is consistent with the prevalence of AIEC reported in CD patients from Europe and North America,46 suggesting that the presence of these bacteria does not depend on HK-specific factors such as different genetic backgrounds and lifestyles. Due to a significant resistance of HK AIEC strains to gentamicin, we compared in vitro HK and FR isolates for their invasion abilities in I-407 cells by replacing gentamicin with meropenem, a carbapenem antibiotic that does not enter eukaryotic cells47 in gentamicin protection assay. The absence of difference in adhesion and invasion levels between HK and FR AIEC isolates is a first indicator that AIEC properties seem comparable regardless of geographical and ethnic origin.
Thanks to our whole-genome sequencing of AIEC isolates from France and Hong Kong assessing the phylogenetic relationships between AIEC strains, we showed that major sequence types associated with AIEC phenotypes could be found in both countries. For the first time, based on the sequence typing method, we showed that AIEC strains from Asia can be related to AIEC from Europe. Moreover, a HK strain named 1162d shared the same ST as the LF82 AIEC reference isolated from French CD patients.48 These findings may imply that these strains have emerged from the same ancestral lineage, as is the case, for example, with diverse human Enterotoxigenic Escherichia coli (ETEC) isolates circulating in the human population today that have probably originated from globally widespread ETEC lineages.49 AIEC isolates from studies on E. coli isolated from patients with IBDs represent various serotypes with a large range of ST.39,40,50,51 Among them, the presence of pandemic STs has been reported in a few studies.52,53 On the other hand, numerous studies have reported the clonal dissemination of extraintestinal pathogenic E. coli (ExPEC) strains and five major pandemic clonal lineages of ExPEC detected in widespread infectionsin particular, urinary tract infections and bloodstream infections. These lineages include ST131, ST95, ST73, ST69, and ST38.54–58 In our study, these five clones were detected among AIEC strains. Martinez-Medina et al.53 reported that a subgroup of strains belonging to the AIEC pathovar is closely related to the ExPEC. The study demonstrated that 40% of the intestinal isolates belonged to ST131 and 21.7% to the ST73. Similarly, among some groups of patients with CRC, a high prevalence of AIEC ST131, ST95, and ST73 was observed.59 All these results suggest that some extraintestinal E. coli could cause intestinal inflammation or intestinal AIEC could lead to extraintestinal infections. These lineages are highly resistant to antibiotics,55,60 making them a major concern for public health. Focusing on phylogroups, we found that the most common phylogroup within the AIEC strains was B2. This is consistent with eight previous studies that reported the same most represented phylogroup among AIEC.45 In accordance with available scientific literature,51,61 we did not detect any virulence factor strictly associated with AIEC, but the co-existence of several virulence factors mostly associated with extraintestinal pathogenic E. coli. This could be explained by several hypotheses such as how AIEC emerged, the fact that the approaches used so far are not appropriate enough, or the lack of a standardized method for AIEC phenotypic characterization.62 Several genetic elements more frequently described in AIEC pathogenicity were also identified in our study. For example, little evidence exists on the putative role of OMPs in AIEC virulence, and Rolhion et al. in 201042 found ompA amino acid variants that could be responsible for the increased invasion ability. Nonetheless, in agreement with previous study63,64 that found that ompA gene variants were similar between AIEC, IPEC, ExPEC, and non-AIEC strains, we did not detect in our study a particular variant specific to AIEC or specific to AIEC about its origin. The same report was made with fliC, fimH, and chiA gene variants, which are heterogeneously distributed among AIEC strains regardless of their origin.
Nonetheless, results obtained on FimH, one of the most studied virulence factors in AIEC pathotype, showed that fimH gene sequence variants were not so distant from each other and a group of five FR strains and six HK strains shared the same variant. The identified sequence of this variant is consensus, meaning that it is the most common and evolutionarily primary fimH variant. On the other hand, some strains harbored a FimH that differed from the consensus sequence by different substitutions including N70S and S78N substitutions described in the AIEC collection by Dreux and colleagues.17 In accordance to Camprubi-Font et al. work,65 we reinforce the idea that no particular variants were associated with AIEC origin in the present work and that no exclusive pathoadaptive changes are associated with the AIEC phenotype but could be a mark of the transition from commensalism to pathobionts in E. coli.
The DSS-treated CEABAC10 transgenic mouse model expressing hCEACAM6 in epithelial cells represents a well-known preclinical model for assessing the capacity of AIEC bacteria to adhere to the intestinal epithelium,18 used to study the role of environmental factors such as a Western-style diet,66,67 and for highlighting the efficacy of various anti-AIEC strategies, i.e., bacteriophages,68 anti-virulence molecules or even yeast probiotics.69,70 In this work, we showed that a subset of four AIEC selected per country of origin confirms the persistence of these bacteria in the mice gut, regardless of their original affiliation. This finding reinforces the fact that not only the LF82 reference strain but a majority of AIEC strains strongly interact with the intestinal mucosa.15,71 Nonetheless, as we observed, some strains isolated from France or Hong Kong may struggle to colonize the gut of CEABAC10 mice. Bleich et al.72 recently demonstrated that the in vitro AIEC pathotype definition cannot truly predict colonization phenotype in vivo. Indeed, it would be rather the colonization-associated features and genomic features of E. coli strains that convey metabolic advantages (e.g., iron acquisition and carbohydrate consumption) that lead to efficient mucosal colonization than the high level of invasion assessed in vitro. So, we may assume that some AIEC strains chosen in our animal experiment do not satisfy all the conditions for effective colonization without challenging the AIEC gut colonizing potential, regardless of their origin. Kittana and colleagues73 challenged the theory of Bleich et al. and showed a strong positive association between E. coli survival and replication in macrophages and epithelial cells in vitro and strain pathogenicity in vivo. While they observed a heterogeneity regarding the behavior of different E. coli strains in vivo, our study found the same trend regarding AIEC bacteria. It is not surprising owing to the phenotypical definition of AIEC with huge variability of genomes.
Several arguments do not support antibiotic use in the treatment of CD. First, antibiotic use has been correlated with an increased risk of CD in several epidemiological studies of high-income countries.74 Then, although certain antibiotics are used in the treatment of CD, their effectiveness appears to be limited to certain manifestations of the disease.75 They are also known to cause changes within the microbiome that can lead to mild inflammation and altered nutrient availability within the intestine.76 In our study, most FR AIEC strains were sensitive to beta-lactam antibiotics conventionally used in medical practice. Some works in North America and Spain reported that ileal CD-associated E. coli manifest resistance to commonly used antimicrobials. Dogan et al. published that AIEC with resistance to one or more antimicrobials were present in 6/8 (75%) AIEC-colonized ileal CD patients and that 8/13 (62%) AIEC strains from these patients were resistant to one or more of the 17 tested antimicrobials.77 For example, resistance to ciprofloxacin and trimethoprim/sulfamethoxazole was found in 2/6 and 4/6 AIEC-colonized ileal CD patients. Camprubí-Font et al. showed that the presence of pic virulence gene and ampicillin resistance in E. coli strains have a probability of 82% to be AIEC.65 Cho et al.78 reported that higher levels of resistance to sulfamethoxazole/trimethoprim and ampicillin were detected in CD E. coli strains compared to control strains (17.5% vs 4.2% and 25% vs 20.8%, respectively). They determined that 20% of CD strains were carrying resistance to 2 or more antibiotics. In contrast, their work did not detect any gentamicin resistance among the 40 tested strains, while we found a low prevalence of resistance to gentamicin among AIEC strains from FR (1/29).
In contrast, we noticed that AIEC strains from HK exhibited more resistance to this panel of antibiotics, probably due to the economic status of the country and less-controlled access to medicines than European countries. Indeed, the global human antibiotic consumption was estimated by Browne et al. for 204 countries from 2000 to 2018 including children with lower respiratory tract infections.79 They identified large variations of antibiotic consumption in high-income and upper-middle income countries with the lowest levels estimated in sub-Saharan Africa and the highest in eastern Europe and central Asia. Both inappropriate antibiotic use and lack of access to antibiotics have been reported in low- and middle-income countries as previously highlighted,80,81 especially in south and southeast Asia, where self-medication and non-licensed antibiotic vendors are common place.82 Studies conducted in China between 2000 and 2012 described a high level of outpatient-prescribed antibiotics (50.3%), and excessive prescriptions are particularly effective in lower-level hospitals and in less developed western China.83 Interestingly, numerous available data state the heterogeneity in consumption rates according to antibiotic classes.79,84 While resistance to Beta-lactam antibiotics is more commonly acknowledged,60 HK AIEC strains in our study were more resistant to aminoglycoside antibiotics (tobramycin and gentamicin) than FR AIEC strains. This report and the work of Dogan et al.77 suggest that antibiotics will not be effective in the global fight against AIEC. Worse, they could risk exacerbating CD since it has been demonstrated that a wide range of antibiotic classes strongly potentiated initial AIEC infection and expanded AIEC in chronically infected mice.85
Unlike antibiotics, bacteriophages are highly specific, infecting only a limited number of strains within a given bacterial species, and have a much smaller impact on the microbiota composition.86,87 In this study, we demonstrated the high potential of the EcoActive™ phage cocktail to target and lyse the AIEC strains from both FR and HK. Part of the phages in this cocktail have already proved their efficacy in decreasing the intestinal colonization of LF82 AIEC strain in wild-type and CEACAM6-expressing mice.68 From that point, our results strongly support using this EcoActive™ phage cocktail as a new treatment option for targeting AIEC in CD patients worldwide. However, although phage therapy has been used in Eastern Europe for several decades, it is yet to be approved as an active antibacterial treatment for human use in the European Union or the United States, given that regulatory issues and quality standards need to be appropriately addressed before approval and widespread use.88,89 Scientific evidence from adequately powered, double-blind, placebo-controlled, randomized clinical trials is scarce. The results of a trial enrolling patients with Crohn’s disease at Mount Sinai Hospital in New York (NCT03808103) to evaluate the use of phages targeting this bacterial subpopulation are eagerly awaited. Thus, targeting microbiota is an attractive option to treat patients with CD.
In conclusion, the PACIFIC study showed that the prevalence and phenotype of AIEC bacteria are similar in FR and HK with close phylogenetic background. Unlike antibiotics resistance profile, phage therapy seems not to be impacted by the origin of the AIEC strains and could be a more promising therapeutic option to target AIEC in CD patients across the world.
Supplementary Material
Acknowledgments
Thanks to Digestive Disease Week® Chicago 2023, this work, whose title and summary were published, was the subject of a poster presentation by Chevarin et al. [90] We would like to thank Adeline Sivignon for her tips in mice experiments. We express our thanks to Joelle Woolston from Intralytix for logistics with the strain exchange. We also thank Microbiota I-Center members for their advice. We thank all financial support institutions.
Funding Statement
This work was supported by the “Ministère de l’Enseignement Supérieur, de la Recherche et de l’Innovation (MESRI), Inserm (Institut national de la santé et de la recherche médicale); [UMR1071], INRAE (Institut national de recherche en agriculture, alimentation et environnement); [USC 1382]; the PROCORE-France/Hong Kong Joint Research Scheme [F-CUHK402/15]; the Agence Nationale de la Recherche of the French government through the PACIFIC project (French National Research Agency (ANR)/Research Grants Council (RGC) Joint Research Scheme [A-CUHK402/17]; and the National Program “Microbiote” Inserm. N.B. The funders had no role in the study's design, data collection and analysis, decision to publish or preparation of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Author contributors
Study concept and design: CC, NB, AB, ZX, SCG, and JFC. Acquisition of data: CC, ZX, LM, FR, RB, AS, and RB. Analysis and interpretation of data: CC, ZX, AS, RB, and NB. Technical and platform support: LM and RB. Manuscript drafting: CC, NB, and AB. Critical revision of the manuscript for important intellectual content drafting: FR, ZX, SCG, RB, and JFC. Funding: NB, AB, and SCG. All authors approved the final version of the manuscript.
Data availability statement
The data that support the findings of this study are available in Recherche Data Gouv at https://doi.org/10.57745/AQUNJA.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/19490976.2024.2431645
References
- 1.Pariente B, Mary J-Y, Danese S, Chowers Y, De Cruz P, D’Haens G, Loftus EV, Louis E, Panés J, Schölmerich J, et al. Development of the Lemann index to assess digestive tract damage in patients with Crohn’s disease. Gastroenterology. 2015;148:52–63.e3. doi: 10.1053/j.gastro.2014.09.015. [DOI] [PubMed] [Google Scholar]
- 2.Molodecky NA, Soon IS, Rabi DM, Ghali WA, Ferris M, Chernoff G, Benchimol EI, Panaccione R, Ghosh S, Barkema HW, et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142(1):46–54.e42. quiz e30. doi: 10.1053/j.gastro.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Ng SC, Shi HY, Hamidi N, Underwood FE, Tang W, Benchimol EI, Panaccione R, Ghosh S, Wu JCY, Chan FKL, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st Century: a systematic review of population-based studies. Lancet Lond Engl. 2017;390(10114):2769–20. doi: 10.1016/S0140-6736(17)32448-0. [DOI] [PubMed] [Google Scholar]
- 4.Buie MJ, Quan BKin J, Windsor JW, Coward S, Hansen TM, King JA, Kotze PG, Gearry RB, Ng SC, Mak JW, et al. GLobal hospitalization trends for Crohn’s disease and ulcerative colitis: systematic review with temporal analyses. Clin Gastroenterol Hepatol. 2022;162(3):S45–S46. doi: 10.1053/j.gastro.2021.12.094. [DOI] [PubMed] [Google Scholar]
- 5.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et al. Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491(7422):119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaplan GG. The global burden of IBD: from 2015 to 2025. Nat Rev Gastroenterol Hepatol. 2015;12(12):720–727. doi: 10.1038/nrgastro.2015.150. [DOI] [PubMed] [Google Scholar]
- 7.Ng SC, Tang W, Leong RW, Chen M, Ko Y, Studd C, Niewiadomski O, Bell S, Kamm MA, de Silva HJ, et al. Environmental risk factors in inflammatory bowel disease: a population-based case-control study in Asia-Pacific. Gut. 2015;64(7):1063–1071. doi: 10.1136/gutjnl-2014-307410. [DOI] [PubMed] [Google Scholar]
- 8.Marteau P. Bacterial flora in inflammatory bowel disease. Dig Dis. 2009;27(Suppl 1):99–103. doi: 10.1159/000268128. [DOI] [PubMed] [Google Scholar]
- 9.Marteau P, Chaput U. Bacteria as trigger for chronic gastrointestinal disorders. Dig Dis. 2011;29(2):166–171. doi: 10.1159/000323879. [DOI] [PubMed] [Google Scholar]
- 10.Rhodes JM. The role of Escherichia Coli in inflammatory bowel disease. Gut. 2007;56(5):610–612. doi: 10.1136/gut.2006.111872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sasaki M, Sitaraman SV, Babbin BA, Gerner-Smidt P, Ribot EM, Garrett N, Alpern JA, Akyildiz A, Theiss AL, Nusrat A, et al. Invasive Escherichia Coli are a feature of Crohn’s disease. Lab Invest. 2007;87:1042–1054. doi: 10.1038/labinvest.3700661. [DOI] [PubMed] [Google Scholar]
- 12.Nadalian B, Yadegar A, Houri H, Olfatifar M, Shahrokh S, Asadzadeh Aghdaei H, Suzuki H, Zali MR. Prevalence of the pathobiont adherent-invasive Escherichia Coli and inflammatory bowel disease: a systematic review and meta-analysis. J Gastroenterol Hepatol. 2021;36(4):852–863. doi: 10.1111/jgh.15260. [DOI] [PubMed] [Google Scholar]
- 13.Boudeau J, Glasser AL, Masseret E, Joly B, Darfeuille-Michaud A, Orndorff PE. Invasive ability of an Escherichia Coli strain isolated from the ileal mucosa of a patient with Crohn’s disease. Infect Immun. 1999;67(9):4499–4509. doi: 10.1128/IAI.67.9.4499-4509.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser AL, Barnich N, Bringer MA, Swidsinski A, Beaugerie L, Colombel JF. High prevalence of adherent-invasive Escherichia Coli associated with ileal mucosa in Crohn’s disease. Gastroenterology. 2004;127(2):412–421. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- 15.Buisson A, Vazeille E, Fumery M, Pariente B, Nancey S, Seksik P, Peyrin-Biroulet L, Allez M, Ballet N, Filippi J, et al. Faster and less invasive tools to identify patients with ileal colonization by Adherent-invasive E. Coli in Crohn’s disease. U Eur Gastroenterol J. 2021;9:1007–1018. doi: 10.1002/ueg2.12161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhilu X, Xiangqian D, Keli Y, Caroline C, Jingwan Z, Yu L, Tao Z, Cheung CL, Yang S, Fengrui Z, et al. Association of adherent-invasive Escherichia Coli with severe gut mucosal dysbiosis in Hong Kong Chinese population with Crohn’s disease. Gut Microbes. 2021;13(1):1994833. doi: 10.1080/19490976.2021.1994833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dreux N, Denizot J, Martinez-Medina M, Mellmann A, Billig M, Kisiela D, Chattopadhyay S, Sokurenko E, Neut C, Gower-Rousseau C, et al. Point mutations in FimH adhesin of Crohn’s disease-associated adherent-invasive Escherichia Coli enhance intestinal inflammatory response. PLOS Pathog. 2013;9:e1003141. doi: 10.1371/journal.ppat.1003141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carvalho FA, Barnich N, Sivignon A, Darcha C, Chan CH, Stanners CP, Darfeuille-Michaud A. Crohn’s disease adherent-invasive Escherichia Coli colonize and induce strong gut inflammation in transgenic mice expressing human CEACAM. J Exp Med. 2009;206:2179–2189. doi: 10.1084/jem.20090741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferrières L, Hémery G, Nham T, Guérout A-M, Mazel D, Beloin C, Ghigo J-M. Silent mischief: bacteriophage mu insertions contaminate products of Escherichia Coli random mutagenesis performed using suicidal transposon delivery plasmids mobilized by broad-host-range RP4 conjugative machinery. J Bacteriol. 2010;192(24):6418–6427. doi: 10.1128/JB.00621-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sivignon A, Chervy M, Chevarin C, Ragot E, Billard E, Denizot J, Barnich N. An adherent-invasive Escherichia coli -colonized mouse model to evaluate microbiota-targeting strategies in Crohn’s disease. Dis Model Mech. 2022;15(10):dmm049707. doi: 10.1242/dmm.049707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Denizot J, Sivignon A, Barreau F, Darcha C, Chan HF, Stanners CP, Hofman P, Darfeuille-Michaud A, Barnich N. Adherent-invasive Escherichia coli induce Claudin-2 expression and barrier defect in CEABAC10 mice and crohnʼs disease patients§. Inflamm Bowel Dis. 2012;18(2):294–304. doi: 10.1002/ibd.21787. [DOI] [PubMed] [Google Scholar]
- 22.Sivignon A, Yan X, Alvarez Dorta D, Bonnet R, Bouckaert J, Fleury E, Bernard J, Gouin SG, Darfeuille-Michaud A, Barnich N. Development of heptylmannoside-based glycoconjugate antiadhesive compounds against adherent-invasive Escherichia Coli bacteria associated with Crohn’s disease. MBio. 2015;6:e01298–15. doi: 10.1128/mBio.01298-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beyrouthy R, Sabença C, Robin F, Poeta P, Igrejas G, Bonnet R. Successful dissemination of plasmid-mediated extended-spectrum β-lactamases in Enterobacterales over humans to Wild Fauna. Microorganisms. 2021;9(7):1471. doi: 10.3390/microorganisms9071471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen S, Zhou Y, Chen Y, Gu J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nurk S, Bankevich A, Antipov D, Gurevich AA, Korobeynikov A, Lapidus A, Prjibelski AD, Pyshkin A, Sirotkin A, Sirotkin Y, et al. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol. 2013;20(10):714–737. doi: 10.1089/cmb.2013.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beghain J, Bridier-Nahmias A, Le Nagard H, Denamur E, Clermont O. ClermonTyping: an easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb Genomics. 2018;4(7):e000192. doi: 10.1099/mgen.0.000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MCJ, Ochman H, et al. Sex and virulence in Escherichia Coli: an evolutionary perspective. Mol Microbiol. 2006;60(5):1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshimura D, Kajitani R, Gotoh Y, Katahira K, Okuno M, Ogura Y, Hayashi T, Itoh T. Evaluation of SNP calling methods for closely related bacterial isolates and a novel high-accuracy pipeline: BactSNP. Microb Genomics. 2019;5(5):e000261. doi: 10.1099/mgen.0.000261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phan M-D, Peters KM, Sarkar S, Lukowski SW, Allsopp LP, Moriel DG, Achard MES, Totsika M, Marshall VM, Upton M, et al. The serum resistome of a globally disseminated multidrug resistant uropathogenic Escherichia Coli clone. PLOS Genet. 2013;9(10):e1003834. doi: 10.1371/journal.pgen.1003834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ben Zakour NL, Alsheikh-Hussain AS, Ashcroft MM, Khanh Nhu NT, Roberts LW, Stanton-Cook M, Schembri MA, Beatson SA. Sequential acquisition of virulence and fluoroquinolone resistance has shaped the evolution of Escherichia Coli ST131. mBio. 2016;7(3):10 .1128/mbio.00347–16. doi: 10.1128/mBio.00958-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, Parkhill J, Harris SR. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43(3):e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12(1):59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 34.Edgar RC. High-accuracy alignment ensembles enable unbiased assessments of sequence homology and phylogeny. Nat Commun. 2022;13(1):6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou Z, Alikhan N-F, Sergeant MJ, Luhmann N, Vaz C, Francisco AP, Carriço JA, Achtman M. GrapeTree: visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 2018;28(9):1395–1404. doi: 10.1101/gr.232397.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Titécat M, Rousseaux C, Dubuquoy C, Foligné B, Rahmouni O, Mahieux S, Desreumaux P, Woolston J, Sulakvelidze A, Wannerberger K, et al. Safety and efficacy of an AIEC-Targeted bacteriophage cocktail in a mice colitis model. J Crohns Colitis. 2022;16(10):1617–1627. doi: 10.1093/ecco-jcc/jjac064. [DOI] [PubMed] [Google Scholar]
- 37.Carlson K. Working with bacteriophages: common techniques and methodological approaches. Bacteriophages: Biol Appl. 2005;1:437–494. [Google Scholar]
- 38.Barnich N, Darfeuille-Michaud A. Adherent-invasive Escherichia coli and crohnʼs disease. Curr Opin Gastroenterol. 2007;23(1):16–20. doi: 10.1097/MOG.0b013e3280105a38. [DOI] [PubMed] [Google Scholar]
- 39.Dogan B, Suzuki H, Herlekar D, Sartor RB, Campbell BJ, Roberts CL, Stewart K, Scherl EJ, Araz Y, Bitar PP, et al. Inflammation-associated adherent-invasive Escherichia Coli are enriched in pathways for use of propanediol and iron and M-Cell translocation. Inflamm Bowel Dis. 2014;20(11):1919–1932. doi: 10.1097/MIB.0000000000000183. [DOI] [PubMed] [Google Scholar]
- 40.Nash JH, Villegas A, Kropinski AM, Aguilar-Valenzuela R, Konczy P, Mascarenhas M, Ziebell K, Torres AG, Karmali MA, Coombes BK. Genome sequence of adherent-invasive Escherichia Coli and comparative genomic analysis with other E. Coli Pathotypes. BMC Genomics. 2010;11(1):667. doi: 10.1186/1471-2164-11-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinez-Medina M, Naves P, Blanco J, Aldeguer X, Blanco JE, Blanco M, Ponte C, Soriano F, Darfeuille-Michaud A, Garcia-Gil LJ. Biofilm formation as a novel phenotypic feature of adherent-invasive Escherichia Coli (AIEC). BMC Microbiol. 2009;9(1):202. doi: 10.1186/1471-2180-9-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rolhion N, Barnich N, Bringer MA, Glasser AL, Ranc J, Hebuterne X, Hofman P, Darfeuille-Michaud A. Abnormally expressed ER stress response chaperone Gp96 in CD favours adherent-invasive Escherichia Coli invasion. Gut. 2010;59(10):1355–1362. doi: 10.1136/gut.2010.207456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sevrin G, Massier S, Chassaing B, Agus A, Delmas J, Denizot J, Billard E, Barnich N. Adaptation of Adherent-Invasive E. Coli to gut environment: impact on flagellum expression and bacterial colonization ability. Gut Microbes. 2020;11(3):364–380. doi: 10.1080/19490976.2017.1421886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Low D, Tran HT, Lee IA, Dreux N, Kamba A, Reinecker HC, Darfeuille-Michaud A, Barnich N, Mizoguchi E. Chitin-binding domains of Escherichia Coli ChiA mediate interactions with intestinal epithelial cells in mice with colitis. Gastroenterology. 2013;145(3):602–12 e9. doi: 10.1053/j.gastro.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kamali Dolatabadi R, Feizi A, Halaji M, Fazeli H, Adibi P. The prevalence of adherent-invasive Escherichia Coli and its association with inflammatory bowel diseases: a systematic review and meta-analysis. Front Med. 2021;8:730243. doi: 10.3389/fmed.2021.730243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palmela C, Chevarin C, Xu Z, Torres J, Sevrin G, Hirten R, Barnich N, Ng SC, Colombel J-F. Adherent-invasive Escherichia Coli in inflammatory bowel disease. Gut. 2018;67(3):574–587. doi: 10.1136/gutjnl-2017-314903. [DOI] [PubMed] [Google Scholar]
- 47.Edwards JR. Meropenem: a microbiological overview. J Antimicrob Chemother. 1995;36(Suppl A):1–17. doi: 10.1093/jac/36.suppl_a.1. [DOI] [PubMed] [Google Scholar]
- 48.Darfeuille-Michaud A, Neut C, Barnich N, Lederman E, Di Martino P, Desreumaux P, Gambiez L, Joly B, Cortot A, Colombel JF. Presence of adherent Escherichia Coli strains in ileal mucosa of patients with Crohn’s disease. Gastroenterology. 1998;115:1405–1413. doi: 10.1016/S0016-5085(98)70019-8. [DOI] [PubMed] [Google Scholar]
- 49.Steinsland H, Lacher DW, Sommerfelt H, Whittam TS. Ancestral lineages of human enterotoxigenic Escherichia Coli. J Clin Microbiol. 2010;48(8):2916–2924. doi: 10.1128/JCM.02432-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Desilets M, Deng X, Rao C, Ensminger AW, Krause DO, Sherman PM, Gray-Owen SD. Genome-based definition of an inflammatory bowel disease-associated adherent-invasive Escherichia Coli pathovar. Inflamm Bowel Dis. 2016;22(1):1–12. doi: 10.1097/mib.0000000000000574. [DOI] [PubMed] [Google Scholar]
- 51.O’Brien CL, Bringer MA, Holt KE, Gordon DM, Dubois AL, Barnich N, Darfeuille-Michaud A, Pavli P. Comparative genomics of Crohn’s disease-associated adherent-invasive Escherichia Coli. Gut. 2016; doi: 10.1136/gutjnl-2015-311059. [DOI] [PubMed] [Google Scholar]
- 52.Petersen AM, Nielsen EM, Litrup E, Brynskov J, Mirsepasi H, Krogfelt KA. A phylogenetic group of Escherichia Coli associated with active left-sided inflammatory bowel disease. BMC Microbiol. 2009;9(1):171. doi: 10.1186/1471-2180-9-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martinez-Medina M, Mora A, Blanco M, Lopez C, Alonso MP, Bonacorsi S, Nicolas-Chanoine MH, Darfeuille-Michaud A, Garcia-Gil J, Blanco J. Similarity and divergence among adherent-invasive Escherichia Coli and extraintestinal pathogenic E. Coli strains. J Clin Microbiol. 2009;47(12):3968–3979. doi: 10.1128/jcm.01484-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kocsis B, Gulyás D, Szabó D. Emergence and dissemination of extraintestinal pathogenic high-risk international clones of Escherichia Coli. Life Basel Switz. Life. 2022;12(12):2077. doi: 10.3390/life12122077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Riley LW. Pandemic lineages of extraintestinal pathogenic Escherichia Coli. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis. 2014;20(5):380–390. doi: 10.1111/1469-0691.12646. [DOI] [PubMed] [Google Scholar]
- 56.Doumith M, Day M, Ciesielczuk H, Hope R, Underwood A, Reynolds R, Wain J, Livermore DM, Woodford N. Rapid identification of major Escherichia Coli sequence types causing urinary tract and bloodstream infections. J Clin Microbiol. 2015;53(1):160. doi: 10.1128/JCM.02562-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Palomino A, Gewurz D, DeVine L, Zajmi U, Moralez J, Abu-Rumman F, Smith RP, Lopatkin AJ. Metabolic genes on conjugative plasmids are highly prevalent in Escherichia Coli and can protect against antibiotic treatment. Isme J. 2023;17(1):151–162. doi: 10.1038/s41396-022-01329-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zong Z, Fenn S, Connor C, Feng Y, McNally A. Complete genomic characterization of two Escherichia Coli lineages responsible for a cluster of carbapenem-resistant infections in a Chinese hospital. J Antimicrob Chemother. 2018;73(9):2340–2346. doi: 10.1093/jac/dky210. [DOI] [PubMed] [Google Scholar]
- 59.Heidari A, Emami MH, Maghool F, Mohammadzadeh S, Kadkhodaei Elyaderani P, Safari T, Fahim A, Kamali Dolatabadi R. Molecular epidemiology, antibiotic resistance profile and frequency of integron 1 and 2 in adherent-invasive Escherichia Coli isolates of colorectal cancer patients. Front Microbiol. 2024;15:1366719. doi: 10.3389/fmicb.2024.1366719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nasrollahian S, Graham JP, Halaji M. A review of the mechanisms that confer antibiotic resistance in pathotypes of E. Coli. Front Cell Infect Microbiol. 2024;14. doi: 10.3389/fcimb.2024.1387497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barrios-Villa E, Martínez de la Peña CF, Lozano-Zaraín P, Cevallos MA, Torres C, Torres AG, Rocha-Gracia RDC. Comparative genomics of a subset of adherent/invasive Escherichia Coli strains isolated from individuals without inflammatory bowel disease. Genomics. 2020;112(2):1813–1820. doi: 10.1016/j.ygeno.2019.10.013. [DOI] [PubMed] [Google Scholar]
- 62.Camprubí-Font C, Martinez-Medina M. Why the discovery of adherent-invasive Escherichia Coli molecular markers is so challenging? World J Biol Chem. 2020;11(1):1–13. doi: 10.4331/wjbc.v11.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miquel S, Peyretaillade E, Claret L, de Vallee A, Dossat C, Vacherie B, Zineb el H, Segurens B, Barbe V, Sauvanet P, et al. Complete genome sequence of Crohn’s disease-associated adherent-invasive E. Coli strain LF82. PLOS ONE. 2010;5(9):e12714. doi: 10.1371/journal.pone.0012714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Camprubí-Font C, Ruiz Del Castillo B, Barrabés S, Martínez-Martínez L, Martinez-Medina M. Amino acid substitutions and differential gene expression of outer membrane proteins in adherent-invasive Escherichia Coli. Front Microbiol. 2019;10:1707. doi: 10.3389/fmicb.2019.01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Camprubí-Font C, Ewers C, Lopez-Siles M, Martinez-Medina M. Genetic and phenotypic features to screen for putative adherent-invasive Escherichia Coli. Front Microbiol. 2019;10:108. doi: 10.3389/fmicb.2019.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Martinez-Medina M, Denizot J, Dreux N, Robin F, Billard E, Bonnet R, Darfeuille-Michaud A, Barnich N. Western diet induces dysbiosis with increased E Coli in CEABAC10 mice, alters host barrier function favouring AIEC colonisation. Gut. 2014;63(1):116–124. doi: 10.1136/gutjnl-2012-304119. [DOI] [PubMed] [Google Scholar]
- 67.Agus A, Massier S, Darfeuille-Michaud A, Billard E, Barnich N. Understanding host-adherent-invasive Escherichia Coli interaction in Crohn’s disease: opening up new therapeutic strategies. Biomed Res Int. 2014;2014:1–16. doi: 10.1155/2014/567929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Galtier M, De Sordi L, Sivignon A, de Vallée A, Maura D, Neut C, Rahmouni O, Wannerberger K, Darfeuille-Michaud A, Desreumaux P, et al. Bacteriophages targeting adherent invasive Escherichia Coli strains as a promising new treatment for Crohn’s disease. J Crohns Colitis. 2017;11:840–847. doi: 10.1093/ecco-jcc/jjw224. [DOI] [PubMed] [Google Scholar]
- 69.Sivignon A, Bouckaert J, Bernard J, Gouin SG, Barnich N. The potential of FimH as a novel therapeutic target for the treatment of Crohn’s disease. Expert Opin Ther Targets. 2017;21(9):837–847. doi: 10.1080/14728222.2017.1363184. [DOI] [PubMed] [Google Scholar]
- 70.Sivignon A, de Vallee A, Barnich N, Denizot J, Darcha C, Pignede G, Vandekerckove P, Darfeuille-Michaud A. Saccharomyces cerevisiae CNCM I-3856 prevents colitis induced by AIEC bacteria in the transgenic mouse model mimicking crohnʼs disease. Inflamm Bowel Dis. 2015;21(2):276–286. doi: 10.1097/mib.0000000000000280. [DOI] [PubMed] [Google Scholar]
- 71.Barnich N, Carvalho FA, Glasser AL, Darcha C, Jantscheff P, Allez M, Peeters H, Bommelaer G, Desreumaux P, Colombel JF, et al. CEACAM6 acts as a receptor for adherent-invasive E. Coli, supporting ileal mucosa colonization in Crohn disease. J Clin Invest. 2007;117(6):1566–1574. doi: 10.1172/jci30504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bleich RM, Li C, Sun S, Barlogio CJ, Broberg CA, Franks AR, Bulik-Sullivan E, Dogan B, Simpson KW, Carroll IM, et al. A consortia of clinical E. Coli strains with distinct in-vitro adherent/invasive properties establish their own Co-colonization niche and shape the intestinal microbiota in inflammation-susceptible mice. Res Sq. 2023; rs.3.rs–2899665. doi: 10.21203/rs.3.rs-2899665/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kittana H, Gomes-Neto JC, Heck K, Juritsch AF, Sughroue J, Xian Y, Mantz S, Segura Muñoz RR, Cody LA, Schmaltz RJ, et al. Evidence for a causal role for Escherichia Coli strains identified as adherent-invasive (AIEC) in intestinal inflammation. mSphere. 2023;8(2):e00478–22. doi: 10.1128/msphere.00478-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Oberc A, Coombes BK. Convergence of external Crohn’s disease risk factors on intestinal bacteria. Front Immunol. 2015;6:6. doi: 10.3389/fimmu.2015.00558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nitzan O, Elias M, Peretz A, Saliba W. Role of antibiotics for treatment of inflammatory bowel disease. World J Gastroenterol. 2016;22(3):1078–1087. doi: 10.3748/wjg.v22.i3.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Spees AM, Wangdi T, Lopez CA, Kingsbury DD, Xavier MN, Winter SE, Tsolis RM, Bäumler AJ, Maloy S. Streptomycin-induced inflammation enhances Escherichia Coli gut colonization through nitrate respiration. mBio. 2013;4(4):e00430–13. doi: 10.1128/mBio.00430-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dogan B, Scherl E, Bosworth B, Yantiss R, Altier C, McDonough PL, Jiang ZD, Dupont HL, Garneau P, Harel J, et al. Multidrug resistance is common in Escherichia coli associated with ileal Crohnʼs disease. Inflamm Bowel Dis. 2013;19(1):141–150. doi: 10.1002/ibd.22971. [DOI] [PubMed] [Google Scholar]
- 78.Cho YH, Renouf MJ, Omotoso O, McPhee JB. Inflammatory bowel disease-associated adherent-invasive Escherichia Coli have elevated host-defense peptide resistance. FEMS Microbiol Lett. 2022;369(1):fnac098. doi: 10.1093/femsle/fnac098. [DOI] [PubMed] [Google Scholar]
- 79.Browne AJ, Chipeta MG, Haines-Woodhouse G, Kumaran EPA, Hamadani BHK, Zaraa S, Henry NJ, Deshpande A, Reiner RC, Day NPJ, et al. Global antibiotic consumption and usage in humans, 2000–18: a spatial modelling study. Lancet Planet Health. 2021;5(12):e893–e904. doi: 10.1016/S2542-5196(21)00280-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mendelson M, Røttingen J-A, Gopinathan U, Hamer DH, Wertheim H, Basnyat B, Butler C, Tomson G, Balasegaram M. Maximising access to achieve appropriate human antimicrobial use in low-income and middle-income countries. Lancet Lond Engl. 2016;387(10014):188–198. doi: 10.1016/S0140-6736(15)00547-4. [DOI] [PubMed] [Google Scholar]
- 81.Fink G, D’Acremont V, Leslie HH, Cohen J. Antibiotic exposure among children younger than 5 years in low-income and middle-income countries: a cross-sectional study of nationally representative facility-based and household-based surveys. Lancet Infect Dis. 2020;20(2):179–187. doi: 10.1016/S1473-3099(19)30572-9. [DOI] [PubMed] [Google Scholar]
- 82.Do NTT, Vu HTL, Nguyen CTK, Punpuing S, Khan WA, Gyapong M, Asante KP, Munguambe K, Gómez-Olivé FX, John-Langba J, et al. Community-based antibiotic access and use in six low-income and middle-income countries: a mixed-method approach. Lancet Glob Health. 2021;9(5):e610–e619. doi: 10.1016/S2214-109X(21)00024-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yin X, Song F, Gong Y, Tu X, Wang Y, Cao S, Liu J, Lu Z. A systematic review of antibiotic utilization in China. J Antimicrob Chemother. 2013;68(11):2445–2452. doi: 10.1093/jac/dkt223. [DOI] [PubMed] [Google Scholar]
- 84.Klein EY, Milkowska-Shibata M, Tseng KK, Sharland M, Gandra S, Pulcini C, Laxminarayan R. Assessment of WHO antibiotic consumption and access targets in 76 countries, 2000–15: an analysis of pharmaceutical sales data. Lancet Infect Dis. 2021;21(1):107–115. doi: 10.1016/S1473-3099(20)30332-7. [DOI] [PubMed] [Google Scholar]
- 85.Oberc AM, Fiebig-Comyn AA, Tsai CN, Elhenawy W, Coombes BK. Antibiotics potentiate adherent-invasive E. Coli infection and expansion. Inflamm Bowel Dis. 2019;25(4):711–721. doi: 10.1093/ibd/izy361. [DOI] [PubMed] [Google Scholar]
- 86.McCallin S, Alam Sarker S, Barretto C, Sultana S, Berger B, Huq S, Krause L, Bibiloni R, Schmitt B, Reuteler G, et al. Safety analysis of a Russian phage cocktail: from metagenomic analysis to oral application in healthy human subjects. Virology. 2013;443(2):187–196. doi: 10.1016/j.virol.2013.05.022. [DOI] [PubMed] [Google Scholar]
- 87.Galtier M, De Sordi L, Maura D, Arachchi H, Volant S, Dillies M-A, Debarbieux L. Bacteriophages to reduce gut carriage of antibiotic resistant uropathogens with low impact on microbiota composition. Environ Microbiol. 2016;18(7):2237–2245. doi: 10.1111/1462-2920.13284. [DOI] [PubMed] [Google Scholar]
- 88.Sybesma W, Pirnay JP, Alavidze Z, Aminov R, Betts A, Bardiau M, Bretaudeau L, Caplin J, Chanishvili N, Coffey A, et al. Expert round table on acceptance and re-implementation of bacteriophage therapy silk route to the acceptance and Re-implementation of bacteriophage therapy. Biotechnol J. 2016;11(5):595–600. doi: 10.1002/biot.201600023. [DOI] [PubMed] [Google Scholar]
- 89.Pelfrene E, Willebrand E, Cavaleiro Sanches A, Sebris Z, Cavaleri M. Bacteriophage therapy: a regulatory perspective. J Antimicrob Chemother. 2016;71(8):2071–2074. doi: 10.1093/jac/dkw083. [DOI] [PubMed] [Google Scholar]
- 90.Chevarin C, Xu Z, Martin L, Robin F, Beyrouthy R, Colombel JF, Sulakvelidze A, Bonnet R, Ng SC, Buisson A, et al. Mo1799 comparison of Crohn’s disease-associated adherent-invasive Escherichia coli (AIEC) from France and Hong-Kong: results from the Pacific study. Gastroenterology. 2023;164:S-915–S–916. doi: 10.1016/S0016-5085(23)03109-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available in Recherche Data Gouv at https://doi.org/10.57745/AQUNJA.


