Abstract
Using fragment profiling, PCR, and Southern hybridization, we found that Salmonella enterica serovar Choleraesuis harbored virulence plasmids of various sizes, whereas serovars Typhimurium, Enteritidis, and Dublin carried a plasmid of a unique size. Also, the virulence plasmid of Typhimurium contained genes in the same order detected in the other three plasmids, all of which contained deletions.
The virulence plasmids of several Salmonella enterica serovars (1, 2, 4, 5, 8, 12, 16, 17, 19, 22, 26) invariably carry the spv operon (7), which plays a role in the virulence of the host strain. The size of these plasmids varies with each serovar, ranging from 50 to 285 kb (21), and thus the plasmids can be classified into at least two incompatibility groups (21). Some virulence plasmids can express the virulence in a heterologous host (2, 10). Their close relationship is shown in a heteroduplex analysis, which indicates that the level of closeness runs, in descending order, from the virulence plasmid of Typhimurium (pSTV) to that of Enteritidis (pSEV), then to that of Choleraesuis (pSCV), and finally to that of Dublin (pSDV) (18). Earlier, we had shown that pSEV, pSTV, and the virulence plasmid of Gallinarum-Pullorum contain the F-like oriT region, whereas pSCV and pSDV do not (23). Recently, during the mapping of some genes and determination of the nucleotide sequences of repA of FIB and FIIA of pSEV, a conclusion was reached that the genetic organizations of pSTV and pSEV are identical (25). A number of virulence plasmid operons and genes (3, 6, 9, 13–20, 24, 25, 27, 30), some of which are listed in Table 1 and Fig. 1, have lately been identified, and their nucleotide sequences have been determined. We determined and compared the sizes and physical and genetic maps of pSCV, pSDV, pSEV, and pSTV. Choleraesuis harbored pSCV plasmids of various sizes, whereas each of the other serovars harbored a virulence plasmid of a unique size. Furthermore, compared with pSTV, the other three contained deletions, and except for the missing genes due to the deletions, the three plasmids’ genes and their order could be detected in pSTV.
TABLE 1.
Characteristics of the primers used in this study
| Name | Gene regiona | Strand | Nucleotide sequence | Source or reference |
|---|---|---|---|---|
| Par1 | parBS | + | ggcgtcaatggttgagatgact | 3 |
| Par2 | parBS | − | gtccagttcatcctgaaccact | 3 |
| IncR1 | incR, parA | + | agcacgtttgacagggtaacg | 3 |
| IncR2 | incR, parA | − | gtggcgactttccgtaactgct | 3 |
| SpvR | spvRA | + | aacaccatgattagtaagaactaatcagt | 15 |
| SpvA | spvRA | − | cctgaacaatgacgtcgctcagat | 15 |
| SpvC1 | spvC | + | cttgcacaaccaaatgcggaagat | 15 |
| SpvC2 | spvC | − | ctctgcatttcaccaccatcacg | 15 |
| Rsk1 | rsk | + | ccctacccaggtgttgaagtcat | 30 |
| Rsk2 | rsk | − | ccttctccctctcagcagcttcat | 30 |
| RepA1 | repA (RepFIB) | − | gaaccggcaaggaagcgcaatgt | 25 |
| RepA2 | repA (RepFIB) | + | ccctacccaggtcttgaaatcgt | 25 |
| PefA | pefAC | + | ccgaaggtgacttcaagtctgt | 6 |
| PefC | pefAC | − | cggcatttgcataggcactggt | 6 |
| PefD1 | pefD, orf5 | + | gcagcagtacggtgtatatggt | 6 |
| PefD2 | pefD, orf5 | − | cctccggtgaattttgccggaat | 6 |
| Rck1 | rck | + | tcgttctgtcctcactgctgct | 9 |
| Rck2 | rck | − | accggtaaccgacaccaacgtt | 9 |
| RepB1 | repA (RepFIIA) | + | ccctgccgttctgtcgtaagct | 25 |
| RepB2 | repA (RepFIIA) | − | tggtaggtaatcagccccagct | 25 |
| TraX-1 | traX, finO | + | aaccgtggcgctgctgctgat | This study |
| TraX-F | traX, finO | − | cttccacttcgggggcgtggt | This study |
| TraT-1 | traT | + | ggttacactggtcagttccactct | 24 |
| TraT-2 | traT | − | gccagttgttcttccagaactggt | 24 |
| Spt5 | oriT | + | ggttacgggattccttccatgaaat | This study |
| TraM-F2 | oriT | − | atatctttatctctcgccccttcct | This study |
| Sam1 | samA | + | gaggaactggatctgaatgcct | 20 |
| Sam2 | samA | − | gatttcctccaccggttgcagt | 20 |
parBS, parB-parS; pefAC, pefA-pefC; spvRA, spvR-spvA.
FIG. 1.
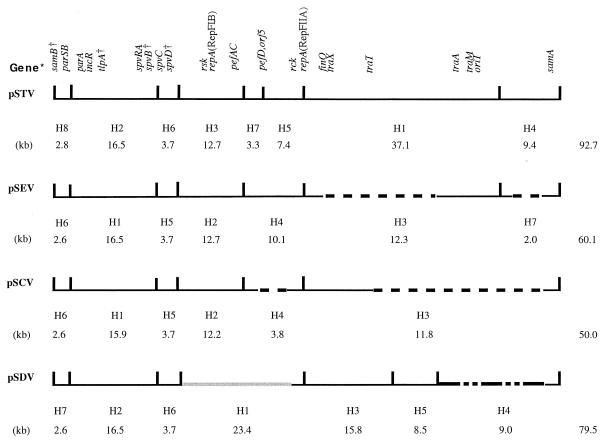
Physical and genetic maps of the virulence plasmids of four Salmonella serovars aligned to pSTV. The vertical line indicates the recognition site of HindIII;the number below the fragment designation is the molecular size (in kilobases) of the fragment. The number listed on the right is the molecular size (in kilobases [the sum of the size of each fragment]) of the virulence plasmid. *, parSB, parS-parB; spvRA, spvR-spvA; pefAC, pefA-pefC. †, the presence of these genes was not checked. 

 , deletion;
, deletion;  , heterologous region;
, heterologous region;  █ █
█ █  , undetermined deletion region. The size and location of the deletion in the nucleotide sequence are as follows (the arrow indicates the site of deletion): pSEV, in H3, about 22 kb (exact size on H1 of pSTV unknown [25]), and in H7, 7.4 kb (TGGCG↓GTCGCC [this work]); pSCV in H4, 6.8 kb (TGGC↓GGCCGGG; in pefD coordinate, C = 6814 and G = 13627 [6]), and in H3, 35 kb (CATCC↓GGCGG; for their sequences, see reference 24 for traT and reference 20 for sam [this work]). Note that for pSDV, in H4 the site was undetermined.
, undetermined deletion region. The size and location of the deletion in the nucleotide sequence are as follows (the arrow indicates the site of deletion): pSEV, in H3, about 22 kb (exact size on H1 of pSTV unknown [25]), and in H7, 7.4 kb (TGGCG↓GTCGCC [this work]); pSCV in H4, 6.8 kb (TGGC↓GGCCGGG; in pefD coordinate, C = 6814 and G = 13627 [6]), and in H3, 35 kb (CATCC↓GGCGG; for their sequences, see reference 24 for traT and reference 20 for sam [this work]). Note that for pSDV, in H4 the site was undetermined.
For this study, both laboratory and clinical strains (the latter obtained from Chang Gung Memorial Hospital, Linkou, Taiwan) were used. Bacteria were routinely grown in Penassay broth and Luria-Bertani agar medium. The presence of plasmids was checked by the method used earlier (11). The plasmid DNA was extracted and purified by the CsCl gradient method described elsewhere (22). Restriction fragment profiles were generated with restriction endonucleases BamHI, BglII, and HindIII, which were used according to the procedure recommended by the manufacturer (Bethesda Research Laboratories, Inc., Gaithersburg, Md.), and the fragments were electrophoresed in a 0.8% agarose slab gel. DND-DNA hybridization was performed according to the method of Southern (29), and the stringency condition employed was that described by the supplier (Bio-Rad) of the material (Zeta-probe membrane). The probes were prepared by PCR amplification of the 14 gene (or operon [Table 1]) fragments from OU5045 (Typhimurium strain C5, a laboratory strain) with the primers listed in Table 1. The probes were labeled with [32P]dCTP (specific activity of 3,000 Ci/mmol; Amersham) by the random primer method described by the supplier (Bethesda Research Laboratories), and the hybridized DNA was detected with an X-ray film with an intensifying screen. Note that sometimes two or more different segments in an operon were amplified. The PCR (solution supplied by Epicentre Technologies), with a 50-μl reaction mixture, was performed essentially under the conditions previously described (4). For cloning and sequence determination, the fragment in the gel was eluted and purified with an agarose gel DNA extraction kit (Boehringer Mannheim). The nucleotide sequence was determined by the dideoxy method (28).
The plasmids found in clinical isolates were examined for the presence of the spv genes. A virulence plasmid was defined here as a plasmid that carried an spv gene. Of the strains that were found to contain a virulence plasmid, all 203 clinical and laboratory strains of Typhimurium harbored a 95-kb virulence plasmid; all 27 Enteritidis strains harbored a 60-kb plasmid, and all 7 Dublin strains harbored an 80-kb plasmid. Only Choleraesuis strains harbored virulence plasmids of various sizes: 10 strains harbored 50-kb plasmids, 2 harbored 100-kb plasmids, and 5 harbored 110-kb plasmids.
Whether or not the four serovar plasmids carried the nine known operons or genes (Table 1) was then determined. All nine operons or genes were detected on pSTV, and the other three virulence plasmids all carried the spv operon (Table 2), as expected, as well as samA, operon par, and repA of RepFIIA. However, oriT and rck were absent in pSCV, while pSEV lacked traT and traX-finO regions. Furthermore, pSDV carried no oriT, repA of FIB, pefD, orf5, and rsk. It appears, therefore, that the lack of oriT is the reason for the inability of pSCV and pSDV to be mobilized by an F plasmid (23). Also, the presence of repA of RepFIIA, operon par, and incR, the genes involved in incompatibility, in all plasmids confirms the earlier observation that these plasmids are all incompatible with pSTV (21).
TABLE 2.
Genes detected by PCR and DNA-DNA hybridization
| Gene | Result for virulence plasmida:
|
|||
|---|---|---|---|---|
| pSTV | pSEV | pSCV | pSDV | |
| parBS | + | + | + | + |
| incR, parA | + | + | + | + |
| spvRA | + | + | + | + |
| spvC | + | + | + | + |
| rsk | + | + | + | − |
| repA (RepFIB) | + | + | + | − |
| pefAC | + | + | + | −/+ |
| pefD, orf5 | + | + | −/+ | − |
| rck | + | + | − | + |
| repA (RepFIIA) | + | + | + | + |
| traX, finO | + | − | + | + |
| traT | + | − | + | + |
| oriT | + | + | − | − |
| samA | + | + | + | + |
+, positive reaction; −, negative reaction; −/+, PCR negative and Southern blot positive.
The physical (HindIII restriction fragment) and genetic maps of the four plasmids examined are shown in Fig. 1. All pSEV and pSTV plasmids, respectively, generated identical fragment profiles. For pSCV, all 50-kb plasmids produced identical fragment profiles (Fig. 1), whereas the larger plasmids produced profiles that differed from one another. Also, two fragment profiles were produced from pSDV: one seen in the pSDV (Fig. 1) of the two laboratory strains and the other (not shown) derived from strain Lane (supplied by J. Fierer [15]) and all clinical isolates. The fragment profile of the former, being more closely related to the other three than the Lane type, was presented here.
In Fig. 1, pSEV, pSCV, and pSDV are aligned with pSTV. Fragment H3 of pSEV contained a large deletion (22 kb) (25). This is consistent with the observation of Montenegro et al. (18). In addition, a smaller deletion was found in H7. The location of these deletions has been determined in terms of the nucleotide sequence of the area and the counterparts in pSTV. Each of the H3 and H4 fragments of pSCV (50 kb) also contained a deletion (Fig. 1). These deletions apparently were the reasons for the smaller molecular sizes of pSEV and pSCV (50 kb). With regard to pSDV (pOU1100 and pOU1113 of the laboratory strains), H4 contained some deletion, the location of which had not been precisely determined. All four virulence plasmids contained a region of parB-parS (in the 2.8-kb H6 of pSEV and 2.6-kb fragments of the others)-parA-incR-(tlpA)-spvR-spvA-(spvB)-spvC. This region corresponds presumably to the homologous region shown in the heteroduplexes (18). The region from repA of RepFIIA to samA, which contained finO-traX-traT-traA-traM-oriT, was carried in the 46.5-kb (H1+H4) fragment of pSTV. The corresponding fragments in the other three plasmids (H3+H7 of pSEV, H3 of pSCV, and H3+H5+H4 of pSDV) were all shorter, because these regions contained deletions of various sizes as described above. Consequently, some gene regions would naturally be missing in these fragments. The region from rsk-repA (RepFIB) to the pef operon to part of repA (RepFIIA) was contained in the 23.4-kb fragment (H3+H7+H5) of pSTV, 22.4-kb fragment (H2+H4) of pSEV, and 16.8-kb fragment (H2+H4), which contained a deletion in H4, of pSCV. Interestingly, the corresponding region, the 23.4-kb fragment (H1), of pSDV produced positive results for rck and repA (RepFIIA) but not for rsk, repA (RepFIB), and operon pef, although positive DNA-DNA hybridization was observed for pefA to pefC (Table 2). This indicated that this region was quite different from the corresponding region of the other three plasmids. Thus, excluding the genes absent due to the deletions, all genes (or operons) and their order detected are generally identical in all four plasmids.
The above observations suggest, as others have suggested (25), that the four plasmids may share a common ancestor and that pSEV, pSCV, and pSDV might all have been derived from pSTV. The genes present are presumably evolutionarily advantageous to these plasmids. Serovars Typhimurium and Enteritidis carry two groups of SOS genes: umuC-umuD on the chromosome and samA-samB on the plasmid (20). These two serovars can thus repair the DNA damage caused by UV light and chemicals, increasing their chances of survival. Whether Choleraesuis and Dublin carry umuC-umuD is unknown. The evolutionary process, however, may have helped each of these four plasmids to evolve into a plasmid uniquely adapted to its respective host. The plasmid size unique to each serovar may be a manifestation of this adaptation.
Acknowledgments
This work was supported in part by grants DOH87-HR-606, from the National Health Research Institute, Department of Health; NSC87-2314-B-182-067, from the National Research Council, Executive Yuan; and CMR697, from the Chang Gung Research Fund, Chang Gung University, Taoyuan, Taiwan.
REFERENCES
- 1.Barrow P A, Lovell M A. Functional homology of virulence plasmids in Salmonella gallinarum, S. pullorum, and S. typhimurium. Infect Immun. 1989;57:3136–3141. doi: 10.1128/iai.57.10.3136-3141.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beninger P R, Chikami G, Tanabe K, Roudier C, Fierer J, Guiney D G. Physical and genetic mapping of the Salmonella dublin virulence plasmid pSDL2. J Clin Investig. 1988;81:1341–1347. doi: 10.1172/JCI113461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cerin H, Hackett J. The parVP region of the Salmonella typhimurium virulence plasmid pSLT contains four loci required for incompatibility and partition. Plasmid. 1993;30:30–38. doi: 10.1006/plas.1993.1031. [DOI] [PubMed] [Google Scholar]
- 4.Chiu C-H, Ou J T. Rapid identification of Salmonella serovars in feces by specific detection of virulence genes, invA and spvC, by enrichment broth culture-multiplex PCR combination assay. J Clin Microbiol. 1996;34:2619–2622. doi: 10.1128/jcm.34.10.2619-2622.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colombo M M, Leoeri G, Rubino S, Barbato A, Cappuccinelli P. Phenotypic features and molecular characterization of plasmids in Salmonella abortusovis. J Gen Microbiol. 1992;138:725–731. [Google Scholar]
- 6.Friedrich M J, Kinsey N E, Vila J, Kander R J. Nucleotide sequence of a 13.9 kb segment of the 90 kb virulence plasmid of Salmonella typhimurium: the presence of fimbrial biosynthetic genes. Mol Microbiol. 1993;8:543–558. doi: 10.1111/j.1365-2958.1993.tb01599.x. [DOI] [PubMed] [Google Scholar]
- 7.Gulig P A, Danbara H, Guiney D G, Lax A J, Norel F, Rhen M. Molecular analysis of spv virulence genes of the salmonella virulence plasmids. Mol Microbiol. 1993;7:825–830. doi: 10.1111/j.1365-2958.1993.tb01172.x. [DOI] [PubMed] [Google Scholar]
- 8.Gulig P A, Curtiss R., III Cloning and transposon insertion mutagenesis of virulence genes of the 100-kilobase plasmid of Salmonella typhimurium. Infect Immun. 1988;56:3262–3271. doi: 10.1128/iai.56.12.3262-3271.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heffernan E J, Harwood J, Fierer J, Guiney D. The Salmonella typhimurium virulence plasmid complement resistance gene rck is homologous to a family of virulence-related outer membrane protein genes, including pagC and ail. J Bacteriol. 1992;174:84–91. doi: 10.1128/jb.174.1.84-91.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hovi M, Sukupolvi S, Edwards M F, Rhen M. Plasmid-associated virulence of Salmonella enteritidis. Microb Pathog. 1988;4:385–391. doi: 10.1016/0882-4010(88)90066-6. [DOI] [PubMed] [Google Scholar]
- 11.Kado C, Liu S-T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawahara K, Haraguchi Y, Tsuchimoto M, Terakado N, Danbara H. Evidence of correlation between 50-kilobase plasmid of Salmonella choleraesuis and its virulence. Microb Pathog. 1988;4:155–163. doi: 10.1016/0882-4010(88)90057-5. [DOI] [PubMed] [Google Scholar]
- 13.Koski P, Saarilahti H, Sukupolvi S, Taira S, Riikonen P, Osterlund K, Hurme R, Rhen M. A new α-helical coiled protein encoded by the Salmonella typhimurium virulence plasmid. J Biol Chem. 1992;267:12258–12265. [PubMed] [Google Scholar]
- 14.Krause M, Guiney D G. Identification of a multimer resolution system involved in stabilization of the Salmonella dublin virulence plasmid pSDL2. J Bacteriol. 1991;173:5754–5762. doi: 10.1128/jb.173.18.5754-5762.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krause M, Roudier C, Fierer J, Haewood J, Guiney D. Molecular analysis of the virulence locus of the Salmonella dublin plasmid pSDL2. Mol Microbiol. 1991;5:307–316. doi: 10.1111/j.1365-2958.1991.tb02111.x. [DOI] [PubMed] [Google Scholar]
- 16.Lax A J, Pullinger G D, Baird G D, Williamson C M. The virulence plasmid of Salmonella dublin: detailed restriction map and analysis by transposon mutagenesis. J Gen Microbiol. 1990;136:1117–1123. doi: 10.1099/00221287-136-6-1117. [DOI] [PubMed] [Google Scholar]
- 17.Martinetti G, Altwegg M. rRNA gene restriction patterns and plasmid analysis as a tool for typing Salmonella enteritidis. Res Microbiol. 1990;141:1151–1162. doi: 10.1016/0923-2508(90)90088-8. [DOI] [PubMed] [Google Scholar]
- 18.Montenegro M A, Monelli G, Helmuth R. Heteroduplex analysis of Salmonella plasmids and their prevalence in isolates of defined sources. Microb Pathog. 1991;11:391–397. doi: 10.1016/0882-4010(91)90035-9. [DOI] [PubMed] [Google Scholar]
- 19.Nakamura M, Sato S, Ohya T, Suzuki S, Ikeda S. Possible relationship of a 36-megadalton Salmonella enteritidis plasmid to virulence in mice. Infect Immun. 1985;47:831–833. doi: 10.1128/iai.47.3.831-833.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nohmi T, Hakura A, Nakai Y, Watanabe M, Murayama S Y, Sofuni T. Salmonella typhimurium has two homologous but different umuDC operons: cloning of a new umuDC-like operon (samAB) present in a 60-megadalton cryptic plasmid of S. typhimurium. J Bacteriol. 1991;173:1051–1063. doi: 10.1128/jb.173.3.1051-1063.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ou J T, Baron L S, Dai X, Life C A. The virulence plasmids of Salmonella serovars typhimurium, choleraesuis, dublin, and enteritidis, and the cryptic plasmids of Salmonella serovars copenhagen and sendai belong to the same incompatibility group, but not those of Salmonella serovars durban, gallinarum, give, infantis and pullorum. Microb Pathog. 1990;8:101–107. doi: 10.1016/0882-4010(90)90074-z. [DOI] [PubMed] [Google Scholar]
- 22.Ou J T, Baron L S. Strain differences in expression of virulence by the 90-kilobase pair virulence plasmid of Salmonella serovar Typhimurium. Microb Pathog. 1991;10:247–251. doi: 10.1016/0882-4010(91)90058-i. [DOI] [PubMed] [Google Scholar]
- 23.Ou J T, Lin M-Y, Chao H-L. Presence of F-like oriT base-pair sequence on the virulence plasmids of Salmonella serovars Gallinarum, Enteritidis, and Typhimurium, but absent in those of Choleraesuis and Dublin. Microb Pathog. 1994;17:13–21. doi: 10.1006/mpat.1994.1048. [DOI] [PubMed] [Google Scholar]
- 24.Qi S Y, Mafune K I, O’Connor D, Rhen M. Characterization of the traT gene and mutants that increase outer membrane permeability from the Salmonella typhimurium virulence plasmid. Mol Microbiol. 1990;4:49–57. doi: 10.1111/j.1365-2958.1990.tb02014.x. [DOI] [PubMed] [Google Scholar]
- 25.Rodriguez-Pena J M, Buisan M, Ibanez M, Rotger R. Genetic map of the virulence plasmid of Salmonella enteritidis and nucleotide sequence of its replicons. Gene. 1997;188:53–61. doi: 10.1016/s0378-1119(96)00776-7. [DOI] [PubMed] [Google Scholar]
- 26.Rourier C, Krause M, Fierer J, Guiney D G. Correlation between the presence of sequences homologous to the vir region of the Salmonella dublin plasmid pSDL2 and the virulence of twenty-two Salmonella serotypes in mice. Infect Immun. 1990;58:1180–1185. doi: 10.1128/iai.58.5.1180-1185.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rychlik I, Loell M A, Barrow P A. The presence of genes to the K88 genes faeH and faeI on the virulence plasmid of Salmonella gallinarum. FEMS Microbiol Lett. 1998;159:255–260. doi: 10.1111/j.1574-6968.1998.tb12869.x. [DOI] [PubMed] [Google Scholar]
- 28.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Southern E M. Detection of specific sequence among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 30.Vandenbosch J L, Rabert D K, Urdangaray R, Jones G W. Sequence analysis of rsk, a portion of the 95-kilobase plasmid of Salmonella typhimurium associated with resistance to the bactericidal activity of serum. Infect Immun. 1989;57:850–857. doi: 10.1128/iai.57.3.850-857.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]