Abstract
PHYML Online is a web interface to PHYML, a software that implements a fast and accurate heuristic for estimating maximum likelihood phylogenies from DNA and protein sequences. This tool provides the user with a number of options, e.g. nonparametric bootstrap and estimation of various evolutionary parameters, in order to perform comprehensive phylogenetic analyses on large datasets in reasonable computing time. The server and its documentation are available at http://atgc.lirmm.fr/phyml.
INTRODUCTION
The ever-increasing size of homologous sequence datasets and complexity of substitution models stimulate the development of better methods for building phylogenetic trees. Likelihood-based approaches (including Bayesian) provided arguably the most successful advances in this area in the last decade. Unfortunately, these methods are hampered with computational difficulties. Different strategies have then been used to tackle this problem, mostly based on stochastic approaches. Markov chain Monte Carlo methods are probably the most valuable tools in this context as they provide computationally tractable solutions to Bayesian estimation of phylogenies (1,2).
Stochastic approaches have also been used to address optimization issues in the maximum likelihood framework. Hence, simulated annealing (3) and genetic algorithms (4,5) were proposed to estimate maximum likelihood phylogenies from large datasets. However, the hill climbing principle is usually considered faster than stochastic optimization and sufficient for numerous combinatorial optimization problems (6). Recently, Guindon and Gascuel (7) described a fast and simple heuristic based on this principle, for building maximum likelihood phylogenies. Several simulation studies (7,8) demonstrated that the tree topologies estimated with this approach are as accurate as those inferred using the best tree building methods currently available. These studies also showed that this new method is considerably faster than the other likelihood-based approaches. Using this heuristic, the analysis of large datasets is now achieved in reasonable computing time on any standard personal computer; e.g. only 12 min were required to analyse a dataset consisting of 500 rbcL sequences with 1428 bp from plant plastids.
This paper introduces PHYML Online, a web interface to the PHYML (PHYlogenetic inferences using Maximum Likelihood) software that implements the heuristic described by Guindon and Gascuel (7). PHYML Online provides a number of useful options (e.g. nonparametric bootstrap), and proposes quite recent models of sequence evolution [e.g. WAG (9) and DCMut (10)]. We first give an overview of the algorithm and present the web server thereafter.
ALGORITHM
The core of the heuristic is based on a well-known tree-swapping operation, namely ‘nearest neighbour interchange’, which defines three possible topological configurations around each internal branch (11). For each of these configurations, the length of the internal branch that maximizes the likelihood is estimated using numerical optimization. The difference of likelihood obtained under the best alternative topological configuration and the current one defines a score. A score with positive value indicates that the best alternative topological configuration yields an improvement of likelihood. A score with negative value indicates that the current topological configuration cannot be improved at this stage and only the length of the internal branch is adjusted. Each internal branch is examined in this manner and ranked according to its score. The optimal length of external branches is also computed. These calculations are performed independently for every branch and they define a set of (topological or numerical) modifications, each of which corresponds to an improvement of the current tree regarding the likelihood function.
The standard approach would only apply one of these modifications, typically that corresponding to the internal branch with best score. Here, a large proportion of all modifications computed previously is performed instead. This proportion is adjusted so as to increase the likelihood at each step, ensuring convergence of the algorithm. This way, the current tree is improved at each step, both in terms of topology and branch length, and only a few steps (usually a few dozen or less) are necessary to reach an optimum of the likelihood function. This explains the speed of this algorithm whose time complexity is O(pns), where p represents the number of refinement steps that have been performed and n is the number of sequences of length s.
PHYML ONLINE
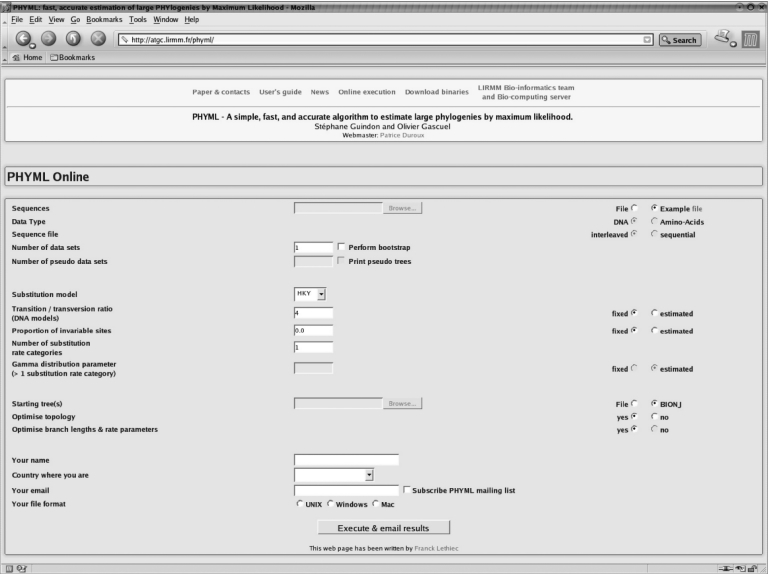
PHYML Online is a web interface to the PHYML algorithm (Figure 1). By default, the input data consists of a single text file containing one or more alignments of DNA or protein sequences in PHYLIP (12) interleaved or sequential format. Examples of sequence datasets in PHYLIP format are given in the ‘User's guide’ section of the web site.
Figure 1.
The PHYML Online interface.
Setting the parameters of a phylogenetic analysis through the interface is straightforward. The first step is the selection of the substitution model of interest. Alignments of homologous DNA and amino acid sequences can be examined under a wide range of models (JC69, K80, F81, F84, HKY85, TN93 and GTR for nucleotides, and Dayhoff, JTT, mtREV, WAG and DCMut for amino acids). Variability of substitution rates across sites and invariable sites can also be taken into account. The parameters that model the intensity of the variation of rates across sites and the proportion of invariables sites can be fixed by the user or estimated by maximum likelihood. Note that the parameters of the substitution model can be estimated under a fixed tree topology or not. The fixed topology option is useful when describing the evolutionary process is more important than estimating the history of sequences.
An option is available to assess the reliability of internal branches using nonparametric bootstrap (13) which is possible to achieve for even large datasets, thanks to the speed of PHYML optimization algorithm. The number of bootstrap replicates is fixed by the user. The bootstrap values are displayed on the maximum likelihood phylogeny estimated from the original dataset. Trees estimated from each bootstrap replicate, as well as the corresponding substitution parameters, can also be saved in separate files for further analysis (e.g. computation of confidence intervals for the substitution parameters or estimation of a consensus bootstrap tree, as performed by PHYLIP's CONSENSE).
Several datasets can be analysed in a single run. This option is especially useful in multiple gene studies. Multiple trees can also be used as input and further optimized by the algorithm described above. This might prevent the tree searching heuristic to be trapped in local maxima. When combined with the fixed tree option, the multiple input trees approach also facilitates the comparison of the fit of different phylogenies estimated from a single dataset. The ‘User's guide’ section gives details on the format of multiple sequence and tree files.
Sequences [and starting tree(s) if provided] are uploaded on our server, a 16-processor IBM computer running Linux 2.6.8-1.521custom SMP, and a maximum likelihood analysis is performed using the PHYML algorithm. Results are then sent to the user by electronic mail. The first file presents a summary of the options selected by the user, maximum likelihood estimates of the parameters of the substitution model that were adjusted, and the log likelihood of the model given the data. The second file shows the maximum likelihood phylogeny(ies) in NEWICK format. Trees can be viewed through an applet available on the PHYML Online server. This applet runs the program ATV (14) that provides numerous options to display and manipulate large phylogenetic trees.
AVAILABILITY
The PHYML Online server is located at ‘Laboratoire d'Informatique, de Robotique et de Microélectronique de Montpellier’: http://atgc.lirmm.fr/phyml.
PHYML can also be downloaded for local installation at http://atgc.lirmm.fr/phyml/binaries.html. The PHYML software has been implemented in C ANSI and is available under GNU general public licence. Sources are available upon request. Binaries, example datasets, sources and documentation are distributed free of charge for academic purpose only.
Acknowledgments
Thanks to Emmanuel Douzery and Stephanie Plön for carefully reading this article. This work was funded by ACI IMPBIO (French Ministry of Research) and Réseau des Génopoles. S.G. is supported by a postdoctoral fellowship from the Allan Wilson Centre for Molecular Ecology and Evolution, New Zealand. Funding to pay the Open Access publication charges for this article was provided by CNRS-STIC.
Conflict of interest statement. None declared.
REFERENCES
- 1.Rannala B., Yang Z. Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. J. Mol. Evol. 1996;43:304–311. doi: 10.1007/BF02338839. [DOI] [PubMed] [Google Scholar]
- 2.Huelsenbeck J.P., Ronquist F. MrBayes: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 3.Salter L., Pearl D. Stochastic search strategy for estimation of maximum likelihood phylogenetic trees. Syst. Biol. 2001;50:7–17. [PubMed] [Google Scholar]
- 4.Lewis P. A genetic algorithm for maximum likelihood phylogeny inference using nucleotide sequence data. Mol. Biol. Evol. 1998;15:277–283. doi: 10.1093/oxfordjournals.molbev.a025924. [DOI] [PubMed] [Google Scholar]
- 5.Lemmon A., Milinkovitch M. The metapopulation genetic algorithm: an efficient solution for the problem of large phylogeny estimation. Proc. Natl Acad. Sci. USA. 2002;99:10516–10521. doi: 10.1073/pnas.162224399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aarts E., Lenstra J.K., editors. Local Search in Combinatorial Optimization. Chichester, UK: Wiley; 1997. [Google Scholar]
- 7.Guindon S., Gascuel O. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 8.Vinh L.S., von Haeseler A. IQPNNI: moving fast through tree space and stopping in time. Mol. Biol. Evol. 2004;21:1565–1571. doi: 10.1093/molbev/msh176. [DOI] [PubMed] [Google Scholar]
- 9.Whelan S., Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 10.Kosiol C., Goldman N. Different versions of the Dayhoff rate matrix. Mol. Biol. Evol. 2005;22:193–199. doi: 10.1093/molbev/msi005. [DOI] [PubMed] [Google Scholar]
- 11.Swofford D., Olsen G., Waddel P., Hillis D. Phylogenetic inference. In: Hillis D., Moritz C., Mable B., editors. Molecular Systematics. 1996. chapter 11 Sinauer Sunderland, MA. [Google Scholar]
- 12.Felsenstein J. 1993. PHYLIP (PHYLogeny Inference Package) version 3.6a2, Distributed by the author, Department of Genetics, University of Washington, Seattle, WA.
- 13.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 14.Zmasek C., Eddy S. ATV: display and manipulation of annotated phylogenetic trees. Bioinformatics. 2001;17:383–384. doi: 10.1093/bioinformatics/17.4.383. [DOI] [PubMed] [Google Scholar]