Abstract
We present the second and improved release of the TOUCAN workbench for cis-regulatory sequence analysis. TOUCAN implements and integrates fast state-of-the-art methods and strategies in gene regulation bioinformatics, including algorithms for comparative genomics and for the detection of cis-regulatory modules. This second release of TOUCAN has become open source and thereby carries the potential to evolve rapidly. The main goal of TOUCAN is to allow a user to come to testable hypotheses regarding the regulation of a gene or of a set of co-regulated genes. TOUCAN can be launched from this location: http://www.esat.kuleuven.ac.be/~saerts/software/toucan.php.
INTRODUCTION
The fundamental drive to characterize the largely unknown functional non-coding part of a genome and the rapid evolution of systems biology approaches to decipher gene regulatory networks, currently depend on the understanding of the regulatory regions (promoters, enhancers, silencers, insulators, etc.) that govern transcriptional regulation. This implies the localization of such regions, the determination of the transcription factors that bind to it, and ultimately the deciphering of the logic behind the combinatorial control and the details on how information is processed to confer a specific expression pattern of the controlled gene. It is generally accepted that in Metazoa, a gene's regulatory system is built up in a modular fashion, consisting of one or more cis-regulatory modules (CRMs), each being responsible for a particular aspect of the parent's expression. Two important characteristics of CRMs have emerged and both appear to be crucial for their computational detection: (i) a CRM is a cluster of transcription factor binding sites (TFBSs) (1), and (ii) the sequence that is covered by a CRM is often conserved between related species, provided that the evolutionary distance between the species is neither too small nor too large [e.g. human–mouse (2), human–fish (3), Drosophila melanogaster–Drosophila pseudoobscura (4)], depending on the regulatory system under study. During the past few years, a number of research papers have been published in specialized journals describing new algorithms for comparative genomics (5,6) and for the detection of CRMs (7–12). There have also appeared a number of publications describing studies on gene regulation where regulatory regions are first predicted using such algorithms, and are then validated further using experiments in the laboratory (13–16). Such studies, however, require extensive bioinformatics expertise to use, parametrize and combine several methods (for example, construct a pipeline that consists of sequence retrieval, pairwise or multiple alignments, motif discovery, motif detection, module discovery, genome-wide module scanning and in silico validation of the predictions). It is, therefore, becoming more and more difficult for a bench biologist and even for a bioinformaticist who is focused on another domain (e.g. microarray data analysis) to perform a thorough regulatory sequence analysis.
TOUCAN (17) was developed to integrate several data and algorithmic resources and to implement new analysis strategies on top of the data and the algorithm layers. The most important feature of the first release was the ability to find over-represented TFBSs in the proximal promoters or the distal CNSs of a set of co-regulated or co-expressed genes. Here, we present a second release of TOUCAN with several new services that are mainly focused on comparative genomics and on the detection of CRMs. We have conducted several example analyses with TOUCAN that are summarized in online tutorials.
GENERAL SOFTWARE SETUP
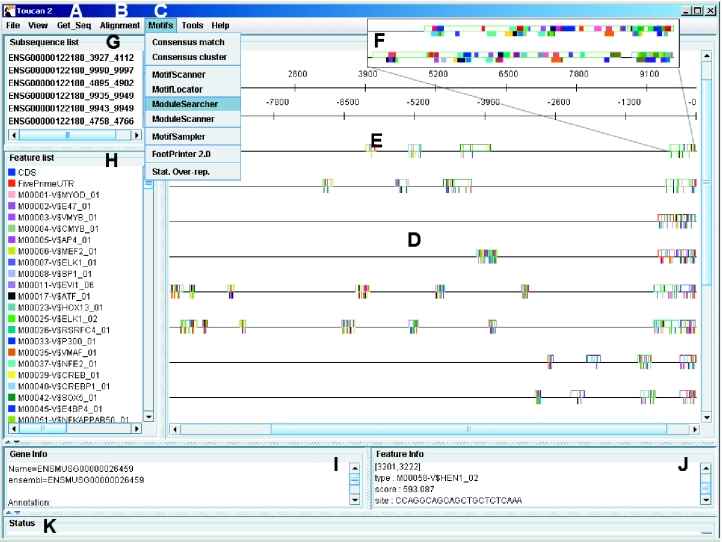
TOUCAN is a client–server application. The client is a Java Graphical User Interface (GUI) that can be launched automatically with Java Web Start from this URL: http://www.esat.kuleuven.ac.be/~saerts/software/toucan.php, provided that Java 2 is installed on the client machine. A screenshot of the GUI is shown in Figure 1. Most of the algorithmic tasks (described below) that can be accessed within this GUI are not executed at the client side, but the tasks are sent as extensible markup language (XML) messages to one of the TOUCAN servers (e.g. the default server at our department ESAT), using SOAP (Simple Object Access Protocol). After completion, the results of such a web service are sent back as XML messages and annotated on the respective sequences. This setup makes it possible to include new algorithmic or data access services easily and independent of the used programming language.
Figure 1.
Screenshot of the TOUCAN software. (A) The Get_Seq menu allows for automated sequence retrieval from the EMBL nucleotide database or from the Ensembl genomic databases. Whole gene sequences or upstream sequences can be retrieved from the latter, together with the corresponding sequences of orthologous genes. (B) All pairs of orthologous sequences of a set of genes can be aligned automatically using AVID, LAGAN or Blastz. (C) Motifs and CRMs can be predicted by various algorithms (see text). (D) The sequence-feature map is sensitive to mouse clicks and allows manipulations, such as cutting, reverse complementing, etc. (E) Features are annotated as colored boxes. The large rectangle shown under (E) is a region that is conserved with the region in the mouse sequence that has the same color, right below it (i.e. a CNS). The small vertical lines within the CNSs are predicted TFBSs. (F) By zooming in, the individual binding site predictions become visible. (G) Sequence sublist where regions like CNSs are collected so that certain analyses can be focused only on the sublist. The sublist can also be saved or be opened in a new window. (H) Feature list where features can be removed, or given another visualization characteristics (color, fill, etc), or where certain combinations of features can be selected to be visualized. (I) Information about a sequence. (J) Information about a feature that is left-clicked on. (K) Status window.
SEQUENCE RETRIEVAL
The sequence retrieval within TOUCAN uses the Java API of Ensembl (i.e. the ensj-core library), combined with direct MySQL queries on the Ensembl database. Because of the link with Ensembl and the rapid advances in genome sequencing and genome annotation, the new release of TOUCAN allows for the sequence retrieval of many more Metazoan species and supports the automatic retrieval of all available orthologous sequences of a given gene. A second improvement in sequence retrieval, again because of improvements in Ensembl, is the automatic mapping of diverse gene identifiers, such as cDNA microarray and Affymetrix chip clone identifiers. Thereby, it is straightforward to retrieve all the upstream regions and their orthologous sequences of a gene cluster obtained by microarray data analysis.
COMPARATIVE GENOMICS
The use of phylogenetic footprinting (PF) was discussed in recent reviews (18–20). One can distinguish two types of PF: (i) detect evolutionary conserved short sequence motifs in a set of orthologous promoters, taking the phylogenetic relationships among the orthologs into account [e.g. FootPrinter (21)]; (ii) use specialized alignment algorithms [e.g. AVID (5), LAGAN (6), BLASTZ (22)] to align large genomic regions around orthologous genes and to select the conserved non-coding sequence (CNS) as putative regulatory regions. As compared with the first version of TOUCAN, where only AVID was available, we have added web services for both types of PF: FootPrinter for the first, and LAGAN and BLASTZ for the second. The comparison of the results from more than one alignment algorithm on a sequence pair can be useful, especially between global (AVID and LAGAN) and local (BLASTZ) alignments (23). For the analysis of co-regulated gene sets, we automated the pairwise alignments so that all available pairs can be aligned with a single instruction. The resulting CNSs can be selected or extracted automatically from all sequences, to be used in the motif detection and module detection steps.
MOTIF DETECTION
The motif detection services are the same as in the first release: (i) a regular expression matcher for consensus sequences; (ii) MotifSampler for the discovery of new motifs by Gibbs sampling (24); (iii) the MotifLocator algorithm to score sequences with position weight matrices (PWMs) on a score cutoff basis; and (iv) the MotifScanner algorithm to score sequences with PWMs on a probabilistic basis (17). Currently, the available PWM libraries are TRANSFAC (25), JASPAR (26) and several smaller libraries that we compiled from the literature. The statistical analysis uses the binomial formula to select sites that are significantly over-represented in a set of sequences (27,17). Files with expected frequencies of motif instances in several (sub-)genomes, to be used in the binomial analysis, can either be downloaded from our website or created from local sequence sets within TOUCAN.
MODULE DETECTION
Genome-wide detection of CRMs by searching for clusters of a given combination of transcription factors (represented by known PWMs) was recently shown to result in high success rates. Schroeder et al. (16) and Berman et al. (15) tested putative enhancers that control transcription during segmentation in Drosophila, predicted with the Ahab (7) and eCIS-ANALYST (28,15) algorithms, respectively. A large fraction of the predicted modules was shown to be functional. Genomic scanning for module instances in TOUCAN is performed using the ModuleScanner algorithm (11) on a database of CNSs. We have prepared a number of CNS databases (currently human–mouse, human–fugu, human–zebrafish and human–chicken CNS databases are available) by aligning the 10 kb orthologous upstream sequences using AVID (75% identity in 100 bp).
When the user has no information on which combination of transcription factors is involved in a particular process, the combination itself can be predicted in TOUCAN using either of the two versions of the ModuleSearcher algorithm [A* (11) and Genetic Algorithm (12)] implementations. The ModuleSearcher searches for the optimal combination of PWMs (out of a library of available PWMs) in a set of sequences, e.g. all CNSs of a set of co-regulated or co-expressed genes (e.g. obtained from microarray data clustering). The newly found combination of PWMs can be validated by using it as a query in a genome-wide scan with the ModuleScanner. In case a significant part of the original co-regulated set is recovered within the top N (e.g. top 100) scoring genes, then the new module is more likely to be specific. Another way of validating a new module is to compare the function of the top N scoring genes with the function of the original gene set, e.g. using Gene Ontology annotation (GO summarization tools can be found at http://www.geneontology.org) or textual annotation [e.g. TxtGate (29)], as performed in (11).
OPEN SOURCE
The second release of TOUCAN has become open source software. Developers can either contribute to the TOUCAN client tool (e.g. visualization aspects, project management, etc.) or to TOUCAN web services (algorithmic services or data services). Information on how to obtain the source code and how to make a web service out of an algorithm can be found on the TOUCAN website.
EXAMPLE PROTOCOLS
Recently, Mayer et al. (30) published new findings of the regulatory mechanisms governing inflammatory gene expression that were found with TOUCAN. We have compiled all the performed steps of the TOUCAN analysis into an online tutorial that is available on the TOUCAN website (http://www.esat.kuleuven.ac.be/~saerts/software/toucan.php). The tutorial starts at the bench with a microarray experiment, then leads to a testable hypothesis about TFBSs involved in the process under study, and ends again at the bench with the validation of the newly predicted regulatory motifs and the respective DNA–protein interactions.
Next to this inspiring tutorial, we have made several other tutorials available, so that most of the functionalities of TOUCAN are explained within one or more worked-out cases.
Acknowledgments
We wish to thank all groups and consortia that made the following data and algorithms freely available: Ensembl, EMBL, JASPAR, public release of TRANSFAC, SCPD, PlantCARE, AVID, VISTA, LAGAN, MLAGAN, BLASTZ, PIPMAKER and FootPrinter. We also thank Wout Van Delm for testing. S.A. is a postdoctoral researcher of the K.U.Leuven. P.V.L. is sponsored by the Fund for Scientific Research Flanders (FWO). This work is partially supported by Instituut voor de aanmoediging van Innovatie door Wetenschap en Technologie in Vlaanderen (IWT) (STWW-00162, STWW-Genprom, GBOU-SQUAD-20160), Research Council KULeuven (GOA Mefisto-666, GOA-Ambiorics, IDO genetic networks), FWO (G.0115.01 and G.0413.03) and IUAP V-22. Funding to pay the Open Access publication charges for this article was provided by DWTC IUAP P5O22.
Conflict of interest statement. None declared.
REFERENCES
- 1.Davidson E.H. Genomic Regulatory Systems. San Diego, CA: Academic Press; 2001. [Google Scholar]
- 2.Wasserman W.W., Palumbo M., Thompson W., Fickett J.W., Lawrence C.E. Human–mouse genome comparisons to locate regulatory sites. Nature Genet. 2000;26:225–228. doi: 10.1038/79965. [DOI] [PubMed] [Google Scholar]
- 3.Ahituv N., Rubin E., Nobrega M. Exploiting human–fish genome comparisons for deciphering gene regulation. Hum. Mol. Genet. 2004;13(Suppl. 2):R261–R266. doi: 10.1093/hmg/ddh229. [DOI] [PubMed] [Google Scholar]
- 4.Sinha S., Schroeder M., Unnerstall U., Gaul U., Siggia E. Cross-species comparison significantly improves genome-wide prediction of cis-regulatory modules in Drosophila. BMC Bioinformatics. 2004;5:129. doi: 10.1186/1471-2105-5-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bray N., Dubchak I., Pachter L. AVID: a global alignment program. Genome Res. 2003;13:97–102. doi: 10.1101/gr.789803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brudno M., Do C., Cooper G., Kim M., Davydov E., Green E., Sidow A., Batzoglou S. LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 2003;13:721–731. doi: 10.1101/gr.926603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rajewsky N., Vergassola M., Gaul U., Siggia E. Computational detection of genomic cis-regulatory modules applied to body patterning in the early Drosophila embryo. BMC Bioinformatics. 2002;3:30. doi: 10.1186/1471-2105-3-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johansson O., Alkema W., Wasserman W., Lagergren J. Identification of functional clusters of transcription factor binding motifs in genome sequences: the MSCAN algorithm. Bioinformatics. 2003;19(Suppl. 1):I169–I176. doi: 10.1093/bioinformatics/btg1021. [DOI] [PubMed] [Google Scholar]
- 9.Sharan R., Ovcharenko I., Ben-Hur A., Karp R. CREME: a framework for identifying cis-regulatory modules in human–mouse conserved segments. Bioinformatics. 2003;19(Suppl. 1):I283–I291. doi: 10.1093/bioinformatics/btg1039. [DOI] [PubMed] [Google Scholar]
- 10.Grad Y., Roth F., Halfon M., Church G. Prediction of similarly-acting cis-regulatory modules by subsequence profiling and comparative genomics in D.melanogaster and D.pseudoobscura. Bioinformatics. 2004;20:2738–2750. doi: 10.1093/bioinformatics/bth320. [DOI] [PubMed] [Google Scholar]
- 11.Aerts S., Van Loo P., Thijs G., Moreau Y., De Moor B. Computational detection of cis-regulatory modules. Bioinformatics. 2003;19(Suppl. 2):II5–II14. doi: 10.1093/bioinformatics/btg1052. [DOI] [PubMed] [Google Scholar]
- 12.Aerts S., Van Loo P., Moreau Y., De Moor B. A genetic algorithm for the detection of new cis-regulatory modules in sets of coregulated genes. Bioinformatics. 2004;20:1974–1976. doi: 10.1093/bioinformatics/bth179. [DOI] [PubMed] [Google Scholar]
- 13.Yuh C., Brown C., Livi C., Rowen L., Clarke P., Davidson E. Patchy interspecific sequence similarities efficiently identify positive cis-regulatory elements in the sea urchin. Dev. Biol. 2002;246:148–161. doi: 10.1006/dbio.2002.0618. [DOI] [PubMed] [Google Scholar]
- 14.Nobrega M., Ovcharenko I., Afzal V., Rubin E. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- 15.Berman B., Pfeiffer B., Laverty T., Salzberg S., Rubin G., Eisen M., Celniker S. Computational identification of developmental enhancers: conservation and function of transcription factor binding-site clusters in Drosophila. Genome Biol. 2004;5:R61. doi: 10.1186/gb-2004-5-9-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schroeder M., Pearce M., Fak J., Fan H., Unnerstall U., Emberly E., Rajewsky N., Siggia E., Gaul U. Transcriptional control in the segmentation gene network of Drosophila. PLoS Biol. 2004;2:E271. doi: 10.1371/journal.pbio.0020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aerts S., Thijs G., Coessens B., Staes M., Moreau Y., De Moor B. TOUCAN: deciphering the cis-regulatory logic of coregulated genes. Nucleic Acids Res. 2003;31:1753–1764. doi: 10.1093/nar/gkg268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ureta-Vidal A., Ettwiller L., Birney E. Comparative genomics: genome-wide analysis in metazoan eukaryotes. Nature Rev. Genet. 2003;4:251–62. doi: 10.1038/nrg1043. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Z., Gerstein M. Of mice and men: phylogenetic footprinting aids the discovery of regulatory elements. J. Biol. 2003;2:11. doi: 10.1186/1475-4924-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wasserman W., Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nature Rev. Genet. 2004;5:276–287. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- 21.Blanchette M., Tompa M. FootPrinter: a program designed for phylogenetic footprinting. Nucleic Acids Res. 2003;31:3840–3842. doi: 10.1093/nar/gkg606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwartz S., Kent W.J., Smit A., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W. Human–mouse alignments with BLASTZ. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nardone J., Lee D., Ansel K., Rao A. Bioinformatics for the ‘bench biologist’: how to find regulatory regions in genomic DNA. Nature Immunol. 2004;5:768–774. doi: 10.1038/ni0804-768. [DOI] [PubMed] [Google Scholar]
- 24.Thijs G., Marchal K., Lescot M., Rombouts S., De Moor B., Rouze P., Moreau Y. A Gibbs sampling method to detect overrepresented motifs in the upstream regions of coexpressed genes. J. Comput. Biol. 2002;9:447–464. doi: 10.1089/10665270252935566. [DOI] [PubMed] [Google Scholar]
- 25.Wingender E., Chen X., Hehl R., Karas H., Liebich I., Matys V., Meinhardt T., Pruss M., Reuter I., Schacherer F. TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res. 2000;28:316–319. doi: 10.1093/nar/28.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sandelin A., Alkema W., Engstrom P., Wasserman W., Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:91–94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Helden J., Andre B., Collado-Vides J. Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies. J. Mol. Biol. 1998;281:827–842. doi: 10.1006/jmbi.1998.1947. [DOI] [PubMed] [Google Scholar]
- 28.Berman B.P., Nibu Y., Pfeiffer B.D., Tomancak P., Celniker S.E., Levine M., Rubin G.M., Eisen M.B. Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc. Natl Acad. Sci. USA. 2002;99:757–762. doi: 10.1073/pnas.231608898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glenisson P., Coessens B., Van Vooren S., Mathys J., Moreau Y., De Moor B. TXTGate: profiling gene groups with text-based information. Genome Biol. 2004;5:R43. doi: 10.1186/gb-2004-5-6-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mayer H., Bilban M., Kurtev V., Gruber F., Wagner O., Binder B., de Martin R. Deciphering regulatory patterns of inflammatory gene expression from interleukin-1-stimulated human endothelial cells. Arterioscler. Thromb. Vasc. Biol. 2004;24:1192–1198. doi: 10.1161/01.ATV.0000131263.06296.77. [DOI] [PubMed] [Google Scholar]