Abstract
Comparative genomics provides the means to demarcate functional regions in anonymous DNA sequences. The successful application of this method to identifying novel genes is currently shifting to deciphering the non-coding encryption of gene regulation across genomes. To facilitate the practical application of comparative sequence analysis to genetics and genomics, we have developed several analytical and visualization tools for the analysis of arbitrary sequences and whole genomes. These tools include two alignment tools, zPicture and Mulan; a phylogenetic shadowing tool, eShadow for identifying lineage- and species-specific functional elements; two evolutionary conserved transcription factor analysis tools, rVista and multiTF; a tool for extracting cis-regulatory modules governing the expression of co-regulated genes, Creme 2.0; and a dynamic portal to multiple vertebrate and invertebrate genome alignments, the ECR Browser. Here, we briefly describe each one of these tools and provide specific examples on their practical applications. All the tools are publicly available at the http://www.dcode.org/ website.
INTRODUCTION
To mine genomes for transcription regulatory elements (REs), a generic vertebrate genome can be viewed as containing three major categories of sequence elements: the exons of protein-coding genes, non-coding non-repetitive DNA and repetitive elements. While coding exons in general cover as little as 1–2%, non-coding and repetitive DNA elements are often equally represented in the remainder 98% of a vertebrate genome. The search for REs is usually performed in the non-coding non-repetitive part of the genome that generally contains promoters, introns, intergenic regions, non-coding RNAs, matrix attachment regions and other regulatory regions of unknown functions. Currently, it is still a major challenge to reliably identify REs and other functional non-coding elements in genomic sequences (1). In order to address this challenge, we have established a suite of computational resources available at www.dcode.org that can assist researchers in deciphering the information encrypted in the non-coding portion of genomes. These tools are primarily designed for addressing such questions as: What are the sequence signatures guiding proper spatial and temporal expression patterns? What is the structure of cis-regulatory modules (CRMs) of a particular function? What are the conservation parameters and the most adequate species used in genome comparisons that can highlight functional elements in the neutrally evolving genomic background? How do we distinguish transcriptionally active factor binding sites (TFBSs) from random TFBS patterns and identify complex TFBS modules with shared activity? How can we prioritize in silico predictions for in vivo biological testing? In this paper, we briefly review this collection of comparative tools and describe their applications.
Generating and visualizing DNA alignments: zPicture and Mulan
The genome encompasses biologically functional elements that have mutated at slower rates than the neutrally evolving genomic background. Therefore, comparative sequence analysis of different species that identifies evolutionary conserved regions (ECRs) facilitates the prediction of functional regions. The selection of species to be compared is of critical importance since the proper evolutionary distance has the power to highlight REs that would otherwise be undistinguished in alignments of closely related species (2). It is currently a widely employed technique to graphically represent sequence conservation profiles in reference to the base DNA sequence that is linear along the horizontal x-axis (Figure 1), while the vertical coordinate displays the percent identity ratio with the secondary sequence (3,4). Regions of conservation are graphically depicted as peaks and evolutionary thresholds can be defined to highlight (color) ECRs of user-defined minimal percent identity and length. Empirically, it was identified that 100 bp/70% identity threshold provides high sensitivity for analyzing human/mouse conservation profiles (5). A tighter threshold of 350 bp/77% identity identifies so-called coreECRs, increases selectivity of predictions and pinpoints to the most critical functional elements (6).
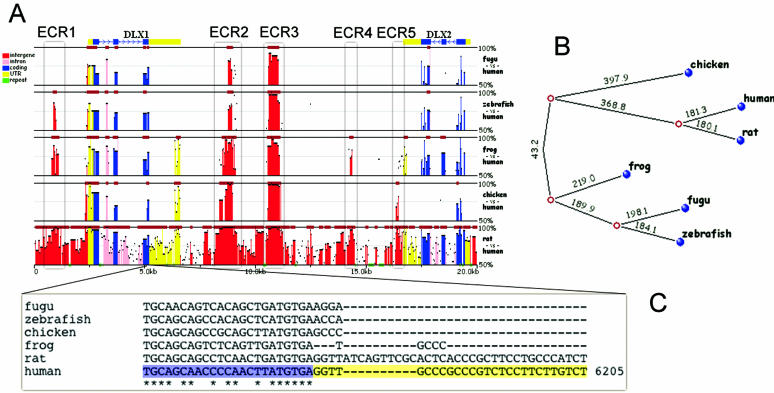
Figure 1.
Mulan conservation analysis for the human DLX1/DLX2 locus as compared with rat, chicken, frog, fugu and zebrafish species. Conservation profile (A) depicts the differential prediction of ncECRs in different species (legend on the left describes coloring of different type elements). Phylogenetic tree (B) represents the evolutionary history of the locus in different vertebrate lineages (with the numbers corresponding to the number of nucleotide mismatches per kb). DLX2 first exon alignment (C) displays sharp difference in coding (blue background) and UTR (yellow background) conservation.
Different alignment engines specialize in different utilities and applications for the compared sequences. ‘Local’ alignment tools are useful in comparing diverged sequences that have undergone several genomic inversion and/or rearrangement events. Several highly accurate and fast local aligners, blastz (7) or tba (8), are capable of recapitulating significant evolutionary modifications in pairwise and multiple-sequence alignments, respectively. We have utilized these aligners to create the zPicture (blastz-based, multiple-coverage) (http://zpicture.dcode.org/) (3) and Mulan (tba-based, single-coverage) (http://mulan.dcode.org/) (9) dynamic and interactive graphical conservation visualization tools. These tools can be used to align homologous nucleotide sequences of virtually any length and are equipped with several automation options that provide the users with the ability to automatically extract genomic sequences, repeat and gene annotations from the UCSC Genome browser (10), the GALA annotation database (11), the NCBI GenBank (12) and the ECR Browser (http://ecrbrowser.dcode.org/) (13).
Multi-species Mulan conservation analysis of the DLX1/DLX2 human locus highlights two non-coding ECRs (ncECRs) that are conserved in multiple vertebrate lineages, including fish, frog and chicken (ECR2 and ECR3, Figure 1A). These two ncECRs correlate to two known cis-REs that drive the expression of the DLX cluster of genes (14). The detection of these two elements is unfeasible from human/rodent alignments, because the majority of the locus (>76%) is highly conserved (100 bp/70%), and to achieve this aim, one requires the assistance of deep evolutionary analysis through the use of multiple species. While the majority of coding exons are conserved in all the species analyzed, this is not the case for non-coding elements. Non-coding conservation between the human sequence and either chicken, frog or the two fishes sharply disappears beyond the boundaries of coding exons (Figure 1C). This supports the hypothesis that the non-coding conservation of the human sequence in this locus with species more distantly related than rodents does not extend beyond the boundaries of functional elements. Therefore, the non-coding conservation with distant species should highlight functional elements in this locus. In addition to these two elements, Mulan presents an interesting conservation pattern for the DLX1/DLX2 locus with several other ncECRs differentially conserved across the phylogeny of the locus (Figure 1B). For example, if we exclude the human/rat conservation from the analysis, ECR5 is conserved only in chicken, ECR4 only in frog, and ECR1 only in frog and zebrafish. None of these ncECRs is conserved in fugu, which probably is the reason these elements were omitted in the original study of this locus (14), and are likely to play a role in species-specific differences.
Mulan performs a full local alignment (8), which is dynamically projected to one of the species at the graphical visualization step. Users have the option to interactively change the base species, thus changing the conservation profile visualization. It can be especially important for studies of loci like the current one that contains elements shared by only a select dataset of species. For example, one would not observe a sequence conservation peak for chicken/frog/fish specific element using human as the base species, but will immediately detect it by switching to having either chicken, frog or fish as the base species.
Phylogenetic shadowing of closely related species, eShadow and Mulan
Phylogenetic shadowing has emerged as a strategy for deciphering functional elements in comparisons of closely related species (such as different primates) (15). Standard pairwise comparisons between such sequences (that usually display 95% or higher level of sequence identity) fail to discriminate between slow- and fast-evolving regions due to a very low density of mutations. Phylogenetic shadowing overcomes this problem by comparing many closely related sequences simultaneously and combining mutations from all the sequences into a single conservation profile (15,16). If the mutations occur independently in different lineages, they would be differently distributed in different sequences. Therefore, combining sequence mismatches from N different closely related sequences increases the divergence rate by a factor of N, thus allowing the separation of slow- and fast-mutating regions (16).
Phylogenetic shadowing is implemented in two dcode.org tools, eShadow (16) and Mulan (9). Although performing similar functions, these two tools differ in their practical applications. Mulan provides easy sequence and annotation extraction through several venues combined with a fast and dynamic visualization interface. It is best used for the analysis of large sequence intervals with prior set up or known conservation detection parameters. eShadow is aimed at performing detailed analysis of sequence conservation and statistical optimization of conservation parameters for detecting functional elements in a given phylogeny. It is equipped with two methods of conservation analysis (HMM Islands, based on the hidden Markov model analysis of cumulative distribution of mismatches, and divergence threshold, that uses standard length/percent identity parameters in the analysis of multi-species conservation) and three methods of parameters optimization (Baum–Welch, Golden section search and maximum-likelihood estimations). We have previously shown that eShadow parameters optimization for the HMM Islands method provides a very sensitive method for predicting slow-mutating regions. It is also possible to apply this method to reliably recapitulate human/mouse conservation profiles using only human/baboon/chimp alignments (16).
The phylogenetic shadowing method is not limited to performing comparisons of closely related species. Recently, it was demonstrated that this method could be successfully applied to intra-species comparative analysis to identify functional coding and non-coding elements in the sea squirt, Ciona intestinalis (17). Here, we have tested the phylogenetic shadowing method in analyzing a different ‘conservation extreme’, the human HOXA gene cluster (Figure 2). This locus comprising four human HOXA genes is highly conserved in dog, mouse and rat genomes with inter-species ECRs completely covering the entire locus (Figure 2A, dark red bars on top of the layers) and, therefore, not permitting the discrimination between functional elements and the neutrally evolving background sequence. By combining human, dog, mouse and rat alignments into a single phylogenetic shadowing profile and increasing the ECR detection threshold to 260 bp/85% identity, we were able to filter out a substantial part of the original conservation plot to highlight only four non-coding (non-overlapping with exons) ECRs (Figure 2B). These are exactly the same ncECRs that would be identified by comparisons between the distantly related human and frog species (Figure 2C). This suggests that there are multiple areas where phylogenetic shadowing can be applied to, and may be a helpful strategy for cases of extremely high conservation and where sequences from several species are available.
Figure 2.
Phylogenetic shadowing of the HOXA4/HOXA5/HOXA6/HOXA7 locus visualized by the Mulan tool. Standard conservation profile of the human sequence as compared with the dog, mouse and rat homologs (100 bp/70%) (A). Phylogenetic shadowing profile of the human/dog/mouse/rat comparison (260 bp/85%) (B). Human/frog conservation profile (100 bp/70%) (C).
Detection of evolutionary conserved TFBS, rVista 2.0 and multiTF
The complexity of transcriptional regulation in vertebrates is achieved through the combinatorial and synchronized binding of different transcription factors to gene REs. These CRMs contain a specific footprint consisting of several TFBSs. CRMs are usually several hundred base pairs in length (2,5,6,14) and stand out of the neighboring genomic sequence as well-conserved regions. Unfortunately, there are no codon-like structures that would allow us to interpret the underlying sequence structure of CRMs. Their function can be inferred computationally only by functions that have been associated with known TFBS patterns present in CRM. There are several databases that document consensus sequences corresponding to known TFBS from different species (18,19). These databases are utilized to computationally identify TFBS in CRMs. Predicting functional TFBS is a very challenging process originating from the nature of binding sites that are very short in length (usually ranging from 6 to 12 bp). Therefore, TFBSs occur at a high frequency across a genome and result in an overabundance of false-positive predictions. For example, GATA-binding sites can be predicted every 30 bp or so in a random sequence using standard techniques (20). Two strategies have been developed that we successfully used to overcome the problem of false-positive predictions. First strategy is based on using sequence conservation to identify functional TFBS (3,20), while the second method utilizes independent optimization of TFBS similarity parameters to minimize the level of TFBS predictions in random genomic sequences (9). These two strategies are implemented into both rVista 2.0 (http://rvista.dcode.org/) (3) and multiTF (http://multitf.dcode.org/) (9) tools that overlay sequence alignments with TFBS predictions to identify evolutionary conserved TFBS.
rVista 2.0 operates with pairwise and multiTF with multiple-sequence alignments, respectively. This difference in the underlying conservation information provided for these two tools possesses different algorithmic requirements for the detection of conserved TFBS. The multiTF tool benefits from extensive sampling of the phylogeny and performs a search for TFBS that are represented in all the species by at least one fully conserved nucleotide (9). rVista 2.0, operates only with a pairwise alignment, searches for TFBS that are shared by the two species and are also located in areas of high local conservation (at least 80% sequence identity in a 20 bp window). Both multiTF and rVista 2.0 utilize TRANSFAC Professional database of TFBS (www.biobase.de). Prior to utilization, TRANSFAC position weight matrices (PWMs) representing TFBS consensus sequences undergo PWM thresholds optimization. This process decreases the number of false-positive predictions and ensures the probability of detecting a random TFBS pair in an average genomic element of 200 bp is <0.1% (9). rVista 2.0 and multiTF are interconnected with the ECR Browser, GALA, zPicture and Mulan tools. This allows the automatic forwarding of pre-calculated genome and user-generated alignments for TFBS analysis.
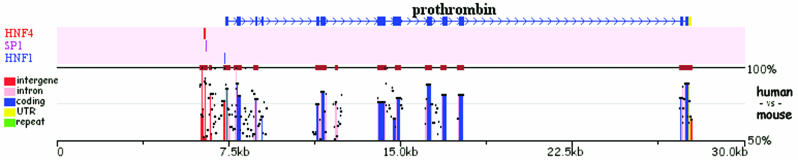
As an example for the application of rVista 2.0/multiTF tools to the annotation of conserved TFBS, we used rVista 2.0 to analyze the Prothrombin (F2) gene, which is the key component of blood coagulation associated with an increased risk of venous thrombosis. The human/mouse 30 kb alignment of the Prothrombin gene locus was automatically submitted from the ECR Browser for rVista 2.0 processing. We were specifically searching for HNF1/4 and SP1 TFBS known to enhance the expression of this gene (21). These three TFBS were identified as a single, well-defined cluster in the promoter region of the Prothrombin gene pinpointing exactly to the experimentally identified enhancer (Figure 3).
Figure 3.
rVista 2.0 identification of conserved TFBS from the Prothrombin gene locus. Identified TFBSs are depicted as colored tick marks above the conservation profile.
Identification of TFBS modules in promoters of co-expressed human genes, Creme 2.0
The rapidly emerging microarray datasets of co-regulated genes combined with genome scale information on genes involved in similar biological processes provide a valuable resource for studying genomic origins of gene expression. Assuming that the groups of co-expressed or co-functional genes consist of subgroups of genes that are transcriptionally co-regulated, large-scale genomic studies of promoters can be performed to identify specific gene regulatory signatures responsible for the co-regulated behavior of specific genes. We have created a tool, Creme 2.0 (http://creme.dcode.org/) that performs searches for genomic signatures specific to the promoters of a given set of genes. It does so by looking for clusters of TFBS that are enriched in the promoters of these genes compared with the random expectation across all the promoter sequences. All different combinations of up to four TFBSs per cluster are enumerated in the promoters of the studied genes as well as in all the conserved promoters of annotated genes in the human genome, thus establishing expectation values and identifying clusters specifically enriched in the set of studied genes. Structurally, Creme 2.0 consists of a web interface that accepts a list of LocusLink accession numbers describing a group of genes of interest, a database of human promoters TFBS that are conserved in mouse and rat, an enrichment detection statistical module, and the output interface that lists and visualizes enriched clusters of TFBS. One of the most direct applications for the Creme 2.0 tool is deciphering signatures of co-regulation from the information derived by human microarray studies (22,23).
The usefulness of Creme 2.0 was previously described using the microarray data on groups of genes co-expressed at different stages of the cell cycle (22,23). In this study, we used Creme 2.0 to detect TFBS modules specific to different Gene Ontology (GO) (22) (http://www.geneontology.org/) categories in an effort to identify co-regulation profiles and key transcriptional regulators in subgroups of genes that share a particular biological function. In order to do so, we selected two GO categories (muscle development and olfactory receptor activity) and extracted LocusLink accession numbers for the corresponding human genes that were subsequently inputted into the Creme 2.0 application. Thirteen TFBS clusters were identified in the analysis of 325 human genes from the ‘muscle development’ GO category. Eleven of them contained different combinations of TFBS consisting primarily of five TFBSs known to partake in muscle development, MEF2 (24,25), STAT1/4 (26,27), GATA6 (28,29) and IRF-1 (30). Creme 2.0 output for the analysis of the TFBS clusters corresponding to 467 human genes populating the ‘olfactory receptor activity’ category was very different. Only one cluster of the two TFBSs (TGIF and LEF1) was identified. Interestingly, there is evidence suggesting that TGIF (TGFbeta-induced factor) partakes in the transmission of nuclear signals in adults (31) and is associated with Holoprosencephaly (the most common severe brain anomaly in humans) (32) and myopia (or nearsightedness, a vision problem experienced by up to about one-third of the population) (33). Also, LEF1 is indirectly associated with the olfactory bulb in mice (34). These data support the link between in silico predictions of TFBS clusters and the biological processes they most likely are involved in. Creme 2.0 can be used to mine genomic data for initial clues on complex regulatory processes in groups of functionally interconnected genes. We are currently working on expanding Creme 2.0 applications to multiple genomes and on providing flexibility in conservation thresholds that could be applied to filtering of TFBS.
Genome alignments and genome-wide annotation of conserved TFBS, ECR Browser
The rapidly growing collection of sequenced vertebrate and invertebrate genomes provides a unique opportunity to expand the boundaries of comparative genomics to genome scale proportions. We have established an automated whole genome alignment strategy to explore the conservation information obtained from multiple genome comparisons. This strategy is based on the large-scale homology detection by blat and blast tools (35) followed by local blastz alignments (7) of homologous genomic segments. Finally, the ECR Browser tool (13) was created to provide an easy graphical interface to the generated alignments. Currently, the ECR Browser includes genome alignments of nine vertebrate species (human, mouse, rat, dog, chicken, frog, tetraodon, zebrafish and fugu) and six species of Drosophila. These alignments create multi-species graphical conservation profiles for different species and can be visualized for any particular region in the genome of choice. The ECR Browser multi-species conservation of the human genome highlights the change in the ratio of coding to non-coding ECRs throughout evolution with a rapid decrease in the density of ncECRs per gene as the phylogenetic distance to the reference species increases (6). Although, it is currently well established that the inter-genome comparisons of distant species, such as humans and fish, are very powerful in identifying critical distant gene REs (2,36,37), there are only 5% of the genes in the human genome that contain a human/fugu ncECR in their genomic neighborhood (6). Thus, the analysis with species more closely related than fish is required for many human genes to identify REs. Therefore, it is especially valuable that the ECR Browser provides flexibility in selecting the species to be used in comparative studies of any locus. Users can either select distant species for well-conserved loci, close species for fast-diverging loci or can simultaneously visualize comparisons with all the available species.
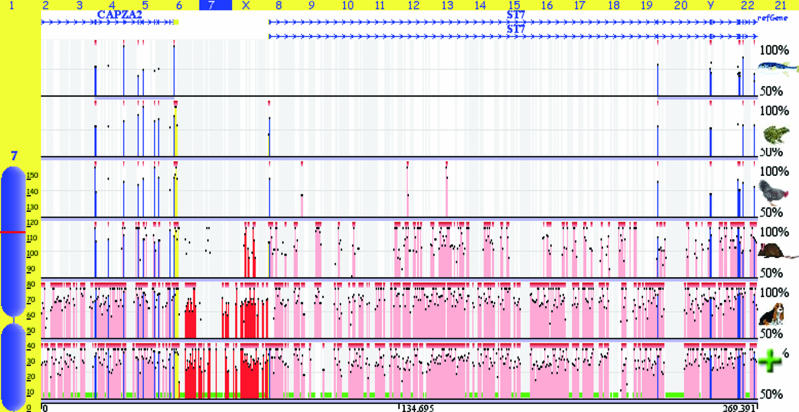
The Genome alignment feature of the ECR Browser tool allows users to arbitrary align novel sequences to several genomes of choice. The genome alignment is performed in a two-step manner. First, the sequence is mapped to the selected genome using the rapid blat utility and the homologous regions (up to five) are extracted from the genome. Subsequently, blastz alignments are performed for the submitted sequence with each homologous sequence. To name some of the features, upon generating the genome alignments, the tool creates graphical conservation plots, similarity dot-plots, extracts ECRs and detects conserved TFBS in the alignments. As a new added feature, the newly generated alignment can be inserted into the genome browser as an additional conservation track displayed alongside the pre-computed conservation profiles available in the ECR Browser. As an example, we input a randomly selected clouded leopard (Neofelis nebulosa) BAC sequence into the Genome alignment feature to align it with the human genome (Figure 4).
Figure 4.
Genome alignment of a randomly selected clouded leopard (N.nebulosa) BAC sequence (accession no. AC152898) sequenced by the ENCODE project (40) to the human genome is visualized as an additional conservation layer of the ECRs browser (it is marked by the green plus sign). It is overlaid with the standard pre-computed human conservation profiles with the dog, mouse, chicken, frog and fugu genomes.
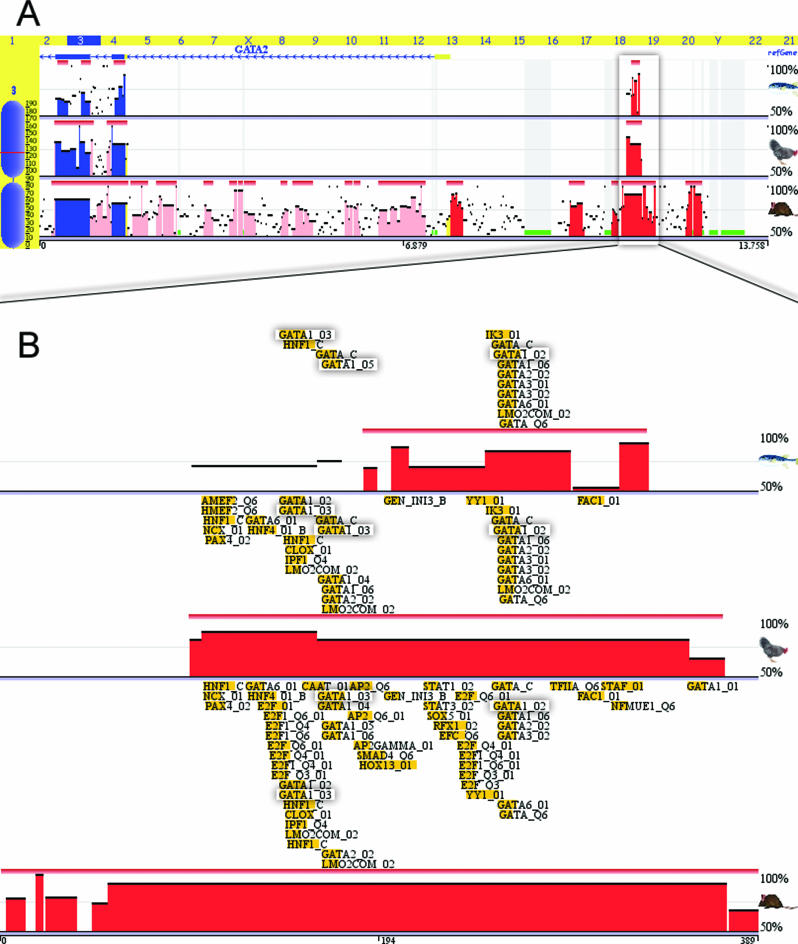
An additional new feature of the ECR Browser provides users with pre-computed annotation of conserved TFBS on a genome-wide scale, generated by the rVista 2.0 program. It is possible to dynamically overlay conserved TFBS for any genomic locus along with the annotation of genes and conservation profiles. Conserved and clustered TFBS may provide with valuable information on gene regulation for a locus of interest (38). As an example, we have applied this new feature to the analysis of the GATA2 locus. The GATA2 gene is a member of the GATA family of transcription factors, which contain zinc fingers in their DNA-binding domain, and are candidate regulators of gene expression in hematopoietic cells. GATA2 is expressed in hematopoietic progenitors and also in embryonic stem cells. Understanding the mechanisms of GATA2 regulation may provide clues for dissecting the core gene regulatory pathways. By analyzing the 105 kb GATA2 locus conservation, we have identified 33 human/mouse ncECRs that potentially represent REs for this gene. Interestingly, only one of these ECRs is conserved in human, mouse, chicken, frog and fugu species. It is a 389 bp long, 88% conserved human/mouse ECR located 3.2 kb upstream of the GATA2 gene (Figure 5A). The study of the human/mouse, human/chicken and human/fugu TFBS conservation profiles of this element performed through the ECR Browser identifies multiple conserved TFBS (Figure 5B). To refine the predictions and to potentially narrow the search down to the TFBS that have the highest probability of being functional, we performed a search for multiple copies of a conserved TFBS inside this element. This search identified a single cluster of three GATA1-binding sites, which is shared by all species. Despite the 400 MYs of evolution separating the two most distantly related species included, humans and fugu, and the sequence divergence inside this ECR, the sequence footprint of the three GATA1 sites was preserved intact. This observation suggests that the GATA1 protein regulates the expression of the GATA2 gene, and a multiple cluster of GATA1 TFBS is required for its function. Consistent with this hypothesis, it has been previously reported that in biochemical assays GATA1 controls the expression of GATA2 transcript (39). The current study identifies a putative GATA2 RE that providing a docking point for the cluster of three GATA1 regulatory molecules. Further biochemical studies of the identified element may confirm or disprove this hypothesis and provide some clues for more complex regulation of the GATA2 gene. Thus, the human/chicken and human/mouse TFBS conservation profiles suggest a putative role for other, HNF1/4 and E2F transcription factors in the regulation of this gene.
Figure 5.
ECR Browser conservation visualization of the GATA2 locus and the upstream ncECR shared by humans, mice, chicken and fugu (A). A zoomed in view of the conserved TFBS predicted for this ncECR identifies a cluster of three conserved GATA1 binding sites (shaded), which is present in all the species (B).
In the future, we plan to introduce two additional new features for the analysis of pre-computed ECR Browser conserved TFBS. First, we will provide users with the option to identify clustered TFBS and second will provide with an option to filter the sites conserved in multiple species. Finally, it is worth mentioning that the ECR Browser is dynamically interconnected with the rVista 2.0 and Mulan tools. A pairwise alignment for any genomic region or an ECR from the ECR Browser can be automatically forwarded to rVista 2.0 for the detection of conserved TFBS (through the ‘Synteny/Alignments’ and ‘Grab ECR’ links). Also sequences for any subset of comparison species from the active selection in the main window of the ECR Browser can be automatically forwarded to Mulan for the generation of a full local alignment. This effectively provides the option to generate phylogenetic trees, sequence similarity dot-plots, and allows subsequent forwarding of the generated alignment to the multiTF utility for the detection of cross-species conserved TFBS.
CONCLUSIONS
Comparative sequence analysis is a powerful approach for extracting functional information from DNA sequence. The overwhelming amount of sequence data generated from shot-gun sequencing of entire vertebrate, invertebrate and microbial genomes requires fast and reliable tools that can assist biologists analyzing the data on a whole-genome scale. Here, we have presented a collection of comparative genomic tools that are publicly available at www.dcode.org and are designed to help biologists generate and analyze pairwise (zPicture) and multiple (Mulan and eShadow) sequence alignments; to detect and decipher the function of transcriptional REs (rVista 2.0 and multiTF) and to predict DNA signature responsible for the shared behavior of co-regulated genes (Creme). In addition to these tools, we have created the ECR Browser that expands the boundaries of alignments to genome scale for multiple vertebrates and invertebrates and provides an access to the pre-computed annotation of conserved TFBS.
Acknowledgments
The work was performed under the auspices of the United States Department of Energy by the University of California, Lawrence Livermore National Laboratory Contract No. W-7405-Eng-48. G.G.L. was supported by LLNL LDRD-04-ERD-052 grant and I.O. was supported by DOE SCW0345 grant. Funding to pay the Open Access publication charges for this article was provided by the US Department of Energy SCW0345 grant.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hardison R.C. Comparative genomics. PLoS Biol. 2003;1:E58. doi: 10.1371/journal.pbio.0000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nobrega M.A., Ovcharenko I., Afzal V., Rubin E.M. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- 3.Loots G.G., Ovcharenko I. rVISTA 2.0: evolutionary analysis of transcription factor binding sites. Nucleic Acids Res. 2004;32:W217–W221. doi: 10.1093/nar/gkh383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwartz S., Zhang Z., Frazer K.A., Smit A., Riemer C., Bouck J., Gibbs R., Hardison R., Miller W. PipMaker—a web server for aligning two genomic DNA sequences. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loots G.G., Locksley R.M., Blankespoor C.M., Wang Z.E., Miller W., Rubin E.M., Frazer K.A. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science. 2000;288:136–140. doi: 10.1126/science.288.5463.136. [DOI] [PubMed] [Google Scholar]
- 6.Ovcharenko I., Stubbs L., Loots G.G. Interpreting mammalian evolution using Fugu genome comparisons. Genomics. 2004;84:890–895. doi: 10.1016/j.ygeno.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 7.Schwartz S., Kent W.J., Smit A., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W. Human–mouse alignments with BLASTZ. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanchette M., Kent W.J., Riemer C., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ovcharenko I., Loots G.G., Giardine B.M., Hou M., Ma J., Hardison R.C., Stubbs L., Miller W. Mulan: multiple-sequence local alignment and visualization for studying function and evolution. Genome Res. 2005;15:184–194. doi: 10.1101/gr.3007205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giardine B., Elnitski L., Riemer C., Makalowska I., Schwartz S., Miller W., Hardison R.C. GALA, a database for genomic sequence alignments and annotations. Genome Res. 2003;13:732–741. doi: 10.1101/gr.603103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wheeler D.L., Barrett T., Benson D.A., Bryant S.H., Canese K., Church D.M., DiCuccio M., Edgar R., Federhen S., Helmberg W., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2005;33:D39–D45. doi: 10.1093/nar/gki062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ovcharenko I., Nobrega M.A., Loots G.G., Stubbs L. ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 2004;32:W280–W286. doi: 10.1093/nar/gkh355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghanem N., Jarinova O., Amores A., Long Q., Hatch G., Park B.K., Rubenstein J.L., Ekker M. Regulatory roles of conserved intergenic domains in vertebrate Dlx bigene clusters. Genome Res. 2003;13:533–543. doi: 10.1101/gr.716103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boffelli D., McAuliffe J., Ovcharenko D., Lewis K.D., Ovcharenko I., Pachter L., Rubin E.M. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science. 2003;299:1391–1394. doi: 10.1126/science.1081331. [DOI] [PubMed] [Google Scholar]
- 16.Ovcharenko I., Boffelli D., Loots G.G. eShadow: a tool for comparing closely related sequences. Genome Res. 2004;14:1191–1198. doi: 10.1101/gr.1773104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boffelli D., Weer C.V., Weng L., Lewis K.D., Shoukry M.I., Pachter L., Keys D.N., Rubin E.M. Intraspecies sequence comparisons for annotating genomes. Genome Res. 2004;14:2406–2411. doi: 10.1101/gr.3199704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matys V., Fricke E., Geffers R., Gossling E., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V., et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sandelin A., Alkema W., Engstrom P., Wasserman W.W., Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loots G.G., Ovcharenko I., Pachter L., Dubchak I., Rubin E.M. rVista for comparative sequence-based discovery of functional transcription factor binding sites. Genome Res. 2002;12:832–839. doi: 10.1101/gr.225502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ceelie H., Spaargaren-Van Riel C.C., De Jong M., Bertina R.M., Vos H.L. Functional characterization of transcription factor binding sites for HNF1-alpha, HNF3-beta (FOXA2), HNF4-alpha, Sp1 and Sp3 in the human prothrombin gene enhancer. J. Thromb. Haemost. 2003;1:1688–1698. doi: 10.1046/j.1538-7836.2003.00393.x. [DOI] [PubMed] [Google Scholar]
- 22.Sharan R., Ben-Hur A., Loots G.G., Ovcharenko I. CREME: cis-regulatory module explorer for the human genome. Nucleic Acids Res. 2004;32:W253–W256. doi: 10.1093/nar/gkh385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharan R., Ovcharenko I., Ben-Hur A., Karp R.M. CREME: a framework for identifying cis-regulatory modules in human-mouse conserved segments. Bioinformatics. 2003;19(Suppl. 1):i283–i291. doi: 10.1093/bioinformatics/btg1039. [DOI] [PubMed] [Google Scholar]
- 24.Anderson J.P., Dodou E., Heidt A.B., De Val S.J., Jaehnig E.J., Greene S.B., Olson E.N., Black B.L. HRC is a direct transcriptional target of MEF2 during cardiac, skeletal, and arterial smooth muscle development in vivo. Mol. Cell. Biol. 2004;24:3757–3768. doi: 10.1128/MCB.24.9.3757-3768.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cripps R.M., Lovato T.L., Olson E.N. Positive autoregulation of the Myocyte enhancer factor-2 myogenic control gene during somatic muscle development in Drosophila. Dev. Biol. 2004;267:536–547. doi: 10.1016/j.ydbio.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 26.Stephanou A. Activated STAT-1 pathway in the myocardium as a novel therapeutic target in ischaemia/reperfusion injury. Eur. Cytokine Netw. 2002;13:401–403. [PubMed] [Google Scholar]
- 27.Wang I.M., Lin H., Goldman S.J., Kobayashi M. STAT-1 is activated by IL-4 and IL-13 in multiple cell types. Mol. Immunol. 2004;41:873–884. doi: 10.1016/j.molimm.2004.04.027. [DOI] [PubMed] [Google Scholar]
- 28.Lepore J.J., Cappola T.P., Mericko P.A., Morrisey E.E., Parmacek M.S. GATA-6 regulates genes promoting synthetic functions in vascular smooth muscle cells. Arterioscler. Thromb. Vasc. Biol. 2005;25:309–314. doi: 10.1161/01.ATV.0000152725.76020.3c. [DOI] [PubMed] [Google Scholar]
- 29.Yin F., Herring B.P. GATA-6 can act as a positive or negative regulator of smooth muscle specific gene expression. J. Biol. Chem. 2005;280:4745–4752. doi: 10.1074/jbc.M411585200. [DOI] [PubMed] [Google Scholar]
- 30.Lin Y., Zhu X., McLntee F.L., Xiao H., Zhang J., Fu M., Chen Y.E. Interferon regulatory factor-1 mediates PPARgamma-induced apoptosis in vascular smooth muscle cells. Arterioscler. Thromb. Vasc. Biol. 2004;24:257–263. doi: 10.1161/01.ATV.0000109170.43400.2f. [DOI] [PubMed] [Google Scholar]
- 31.Gripp K.W., Wotton D., Edwards M.C., Roessler E., Ades L., Meinecke P., Richieri-Costa A., Zackai E.H., Massague J., Muenke M., et al. Mutations in TGIF cause holoprosencephaly and link NODAL signalling to human neural axis determination. Nature Genet. 2000;25:205–208. doi: 10.1038/76074. [DOI] [PubMed] [Google Scholar]
- 32.Aguilella C., Dubourg C., Attia-Sobol J., Vigneron J., Blayau M., Pasquier L., Lazaro L., Odent S., David V. Molecular screening of the TGIF gene in holoprosencephaly: identification of two novel mutations. Hum. Genet. 2003;112:131–134. doi: 10.1007/s00439-002-0862-8. [DOI] [PubMed] [Google Scholar]
- 33.Lam D.S., Lee W.S., Leung Y.F., Tam P.O., Fan D.S., Fan B.J., Pang C.P. TGFbeta-induced factor: a candidate gene for high myopia. Invest Ophthalmol. Vis. Sci. 2003;44:1012–1015. doi: 10.1167/iovs.02-0058. [DOI] [PubMed] [Google Scholar]
- 34.Shimogori T., VanSant J., Paik E., Grove E.A. Members of the Wnt, Fz, and Frp gene families expressed in postnatal mouse cerebral cortex. J. Comp. Neurol. 2004;473:496–510. doi: 10.1002/cne.20135. [DOI] [PubMed] [Google Scholar]
- 35.Kent W.J. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woolfe A., Goodson M., Goode D.K., Snell P., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lettice L.A., Heaney S.J., Purdie L.A., Li L., de Beer P., Oostra B.A., Goode D., Elgar G., Hill R.E., de Graaff E. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- 38.Schroeder M.D., Pearce M., Fak J., Fan H., Unnerstall U., Emberly E., Rajewsky N., Siggia E.D., Gaul U. Transcriptional control in the segmentation gene network of Drosophila. PLoS Biol. 2004;2:E271. doi: 10.1371/journal.pbio.0020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohneda K., Yamamoto M. Roles of hematopoietic transcription factors GATA-1 and GATA-2 in the development of red blood cell lineage. Acta Haematol. 2002;108:237–245. doi: 10.1159/000065660. [DOI] [PubMed] [Google Scholar]
- 40.The ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]