Abstract
Motivation
Repeat elements, such as transposable elements (TE), are highly repetitive DNA sequences that compose around 50% of the genome. TEs such as Alu, SVA, HERV, and L1 elements can cause disease through disrupting genes, causing frameshift mutations or altering splicing patters. These are elements challenging to characterize using short-read genome sequencing, due to its read length and TEs repetitive nature. Long-read genome sequencing (lrGS) enables bridging of TEs, allowing increased resolution across repetitive DNA sequences. lrGS therefore present an opportunity for improved TE detection and analysis not only from a research perspective but also for future clinical detection. When choosing an lrGS TE caller, parameters such as runtime, CPU hours, sensitivity, precision, and compatibility with inclusion into pipelines are crucial for efficient detection.
Results
We therefore developed sTELLeR, (s) Transposable ELement in Long (e) Read, for accurate, fast, and effective TE detection. Particularly, sTELLeR exhibit higher precision and sensitivity for calling of Alu elements than similar tools. The caller is 5–48× as fast and uses <2% of the CPU hours compared to competitive callers. The caller is haplotype aware and output results in a variant call format (VCF) file, enabling compatibility with other variant callers and downstream analysis.
Availability and implementation
sTELLeR is a python-based tool and is available at https://github.com/kristinebilgrav/sTELLeR. Altogether, we show that sTELLeR is a fast, sensitive, and precise caller for detection of TE elements, and can easily be implemented into variant calling workflows.
1 Introduction
Transposable elements (TEs) are repetitive genomic sequences capable of changing their genomic location. There are two subtypes of TEs, DNA transposons and retrotransposons (RTs). DNA transposons move through a cut-and-paste mechanism, make up around 2% of the genome (Chenais 2022) and are not active in human genomes (Solyom and Kazazian 2012). RTs change their location through a copy–paste mechanism involving an RNA intermediate and make up around 50% of the genome. There are different families of RTs, where the most common ones are elements L1, Alu, SVA, and HERV. Some of these remain active, and the transposition rates for Alus range from 1:29–40 births to 1:63–117 births for L1 (Feusier et al. 2019, Borges-Monroy et al. 2021).
There are several examples where TEs have been disease causing, such as an SVA causing exon-trapping in MFSD8 (MIM# 610951) (Kim et al. 2019), and Alu insertions disrupting exons in NF1 and USH2A (Bilgrav Saether et al. 2023). Additionally, HERVs have been connected to cancer as well as autoimmunity (Alcazer et al. 2020). Detection of TEs is therefore clinically important, and understanding their mechanisms and characteristics is useful for determining their genomic consequences.
Long-read whole genome sequencing (lrGS) enables base-pair resolution of >10 kb stretches of continuous DNA. This facilitates characterization and resolution of complex and dynamic genomic regions (Logsdon et al. 2020). lrGS enabling full-length base-pair resolution of TEs is a significant improvement from short-read genome sequencing (srGS) where read lengths of 150 bp limit the discovery of genomic variation across repeat regions such as tandem repeats, segmental duplications, and TEs (Logsdon et al. 2020). Due to the previous complexity of analyzing these regions, they remain largely understudied.
Although the reference genome contains plenty of TEs, the majority of them differ across the population and are not represented in the reference genome (Ewing and Kazazian 2010, Sudmant et al. 2015, Bilgrav Saether et al. 2023). We have previously implemented an srGS nonreference TE insertion (TEI) detection workflow into our clinical analysis pipeline (Bilgrav Saether et al. 2023). However, as previously discussed, srGS provides limited resolution of TEs. With lrGS becoming more accessible and clinically applicable, an accurate, robust, and time-efficient lrGS TE caller is necessary in order to identify TEI. We here present a novel (s) Transposable ELement Long (e) Read (sTELLeR) caller, which is fast, sensitive, and precise at identifying nonreference TEI. We apply sTELLeR on simulated data as well as on a genome in a bottle (GIAB) trio and samples from the Human Pangenome Reference Consortium (HPRC).
2 Materials and methods
2.1 sTELLeR algorithm
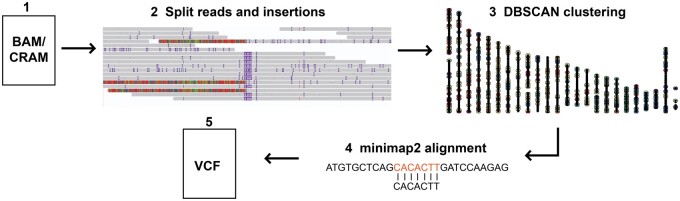
The sTELLeR algorithm entails five steps (Fig. 1). sTELLeR takes a bam or a cram file as input (1) and (2) extracts positions of split reads and insertions. The positions are (3) clustered using the density-based spatial clustering of applications with noise (DBSCAN) clustering algorithm. DBSCAN will cluster positions based on proximity to each other, where ε determines maximum distance between neighbors and a minimum number of positions is set to consider it a cluster. Once clusters are obtained, (4) the sequence of the insertions and split reads spanning the cluster is extracted and aligned to TEs which are provided in a fasta file using minimap2. Finally, (5) the aligned sequences are refined to a consensus nucleotide position and filtered to only include clusters where a minimum number of reads (user-defined, default 3) match a TE, and the length of the match needs to be at least 10% of the original insertion or split read. Lastly, results are provided as a VCF output.
Figure 1.
sTELLeR algorithm overview. (1) BAM or CRAM file can be provided as input. (2) sTELLeR identifies split reads and insertions. (3) The split reads and insertions are clustered using DBSCAN. (4) Sequences of clustered split reads and insertions are aligned to TE sequences provided. (5) Resulting matches are provided in a variant call format (VCF) file. Image adapted from (https://github.com/kristinebilgrav/sTELLeR).
2.2 sTELLeR benchmarking
sTELLeR was compared to multiple state-of-the-art lrGS TE callers, including xTEA, PALMER, TELR, and TLDR (Ewing et al. 2020, Shahid and Slotkin 2020, Chu et al. 2021, McDonald et al. 2021). The callers were compared based on sensitivity, precision, and runtime. Callers that did not complete within 48 h were excluded from the benchmark. Callers also need to be stable and compatible with inclusion into pipelines in, e.g. Snakemake (Mölder et al. 2021) or Nextflow (Di Tommaso et al. 2017). All analyses were run on the high-performance cluster Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX). Chosen callers were ran on data obtained from GIAB and the HPRC (Zook et al. 2016, Liao et al. 2023) as well as on simulated data.
The simulated data were created by inserting TE sequences from Alu, L1, HERV, and SVA at random in a masked reference file (GCF_000001405.26) (Supplementary File S1). The Alu, L1, and SVA sequences were obtained by extracting TE sequences from a GRCh38 reference file (GCF_000001405.26) using positions indicated by RepeatMasker. The HERV sequence was obtained from NCBI (AF020092) (Sayers et al. 2022). The exact positions were determined using the random positions. The fasta file containing TEs was used to create a simulated dataset using PBSIM3 (Ono et al. 2022). This resulted in a simulated BAM file with 20× coverage and where the reads have a similar error rate to those generated using the PacBio RS II. There were a total of 886 Alu, 888 L1, 444 SVA, and 452 HERV insertions in the dataset. Commands and scripts are given in Supplementary File S1.
An additional assembly-based callset was generated using the HPRC de novo assembly of the samples HG002 and HG01071. SVIM-asm (Heller and Vingron 2021) was utilized to call insertions on the assembly, and RepeatMasker (Smit et al. 2013) was used to determine the presence of Alu, L1, HERV, and SVA elements. Elements with a percent divergence >15% and under a certain consecutive length (SVA: 300, HERV: 1500, L1: 200, Alu: 100) were excluded. This resulted in 2193 Alu, 492 L1, 14 HERV, and 205 SVA elements in HG002 and 2262 Alu, 439 L1, 11 HERV, and 202 SVA elements in HG01071. sTELLeR, TLDR, and xTEA were subsequently ran on the PacBio samples downloaded from the same source and realigned to the GRCh38 reference (GCF_000001405.26) (Supplementary File S1).
For comparison of runtimes, PacBio BAM files for GIAB samples HG002, HG003, and HG004 were obtained from GIAB (Zook et al. 2016) and converted back to fastq files for alignment. ONT fastq files for the same samples were obtained from GIAB (Zook et al. 2016, Shafin et al. 2020). All fastq files were aligned to reference genome GRCh38 using minimap2 (Li 2018).
Commands for running the tools are listed in Supplementary File S1. The TE fasta sequences, used to run TLDR and sTELLeR, were downloaded from NCBI (Sayers et al. 2022) or extracted from the reference genome. These sequences are available through https://github.com/kristinebilgrav/sTELLeR_supplementary/, along with the scripts used to generate the assembly-based and simulated callset.
2.3 TE analysis in srGS
HG004 srGS bam file was downloaded from GIAB (Zook et al. 2016) and realigned to GRCh38 using bwa-mem (Li and Durbin 2009) and downsampled to 30× coverage. TE calling in the srGS data was performed using RetroSeq (Keane et al. 2013) and MELT2 (Gardner et al. 2017). The srGS TE caller RetroSeq is a caller shown to have high sensitivity in previous studies (Keane et al. 2013, Rishishwar et al. 2017, Vendrell-Mir et al. 2019). MELT2 is a popular TE caller which has been utilized to generate the GnomAD SV callset (Gardner et al. 2017, Collins et al. 2020). The commands used are given in Supplementary File S1.
3 Results
We downloaded and tested multiple callers such as xTEA (Chu et al. 2021), PALMER (McDonald et al. 2021), TELR (Han et al. 2022), and TLDR (Ewing et al. 2020). However, due to many of our mentioned demands from a caller, some fail to meet our requirements. PALMER was tested, but its runtime exceeded our runtime limits where it could not identify TEs on GS within 48 h. TELR (v1.1) was easy to install; however, a bug known to the developer, but not resolved, rendered it to be unstable on our data. A comprehensive table of runtimes for the excluded callers is found in Supplementary Table S1. The callers tested which were competitive to sTELLeR were TLDR and xTEA.
Runtime, CPU hours, and memory usage of sTELLeR, TLDR, and xTEA were assessed by running the tools on the GIAB Ashkenazim trio using the PacBio and ONT data (Table 1). Runtimes were faster for sTELLeR across all sample types and techniques.
Table 1.
sTELLeR, TLDR, and xTEA runtimes and memory usage for GIAB samples.
| Dataset | Coverage | Caller | Walltime (min) | CPU time (walltime×CPU) | Memory (GB) |
|---|---|---|---|---|---|
| HG002 PacBio | 52× | sTELLeR | 21 | 21 | 6 |
| TLDR | 111 | 1110 | 23 | ||
| xTEA | 1020 | 8161 | 12 | ||
| HG002 ONT | 43× | sTELLeR | 83 | 83 | 2 |
| TLDR | 231 | 2310 | 69 | ||
| xTEA | 2136 | 17 083 | 12 | ||
| HG003 PacBio | 61× | sTELLeR | 27 | 27 | 8 |
| TLDR | 127 | 1265 | 25 | ||
| xTEA | 1112 | 8893 | 13 | ||
| HG003 ONT | 78× | sTELLeR | 149 | 149 | 3 |
| TLDR | 468 | 7481 | 105 | ||
| xTEA | 2328 | 18 621 | 12 | ||
| HG004 PacBio | 60× | sTELLeR | 22 | 22 | 14 |
| TLDR | 135 | 1346 | 20 | ||
| xTEA | 1108 | 8866 | 12 | ||
| HG004 ONT | 78× | sTELLeR | 160 | 160 | 3 |
| TLDR | 471 | 7537 | 100 | ||
| xTEA | 2435 | 19 480 | 14 |
sTELLeR, TLDR, and xTEA were further tested on simulated data produced and run as described in Section 2. The simulated dataset contained 886 Alu, 888 L1, 452 HERV, and 444 SVA insertions. sTELLeR was able to identify 863 Alu, 859 L1, 443 HERV, and 435 SVA elements; TLDR 883 Alu, 52 L1, 186 HERV, and 14 SVA elements; and xTEA 866 Alu, no L1 or HERV, and 430 SVA elements (Table 2). This results in a sensitivity of 0.97 for Alu detection with sTELLeR, 0.99 using TLDR and 0.97 using xTEA. The precision was 1 for both sTELLeR and xTEA, and 0.97 for TLDR (Table 2). For L1 detection, the sensitivity was 0.96 for sTELLeR and 0.05 for TLDR and the precision 1 for sTELLeR and 0.82 for TLDR. For HERV, sTELLeR had a sensitivity of 0.98 and a precision of 1, while TLDR 0.41 and 1, respectively. For SVA, sTELLeR had a precision of 1, xTEA 0.99, while TLDR 0.82. The sensitivity was 0.97 for sTELLeR, 0.98 for xTEA, and 0.03 for TLDR.
Table 2.
Simulated metrics for the callers sTELLeR, TLDR, and xTEA ran on a simulated dataset (Alu = 886, L1 = 888, HERV = 452, SVA = 444).
| sTELLeR | TLDR | xTEA | ||
|---|---|---|---|---|
| True positives | Alu | 863 | 883 | 866 |
| L1 | 859 | 52 | NA | |
| HERV | 443 | 186 | NA | |
| SVA | 435 | 14 | 429 | |
| Sensitivity | Alu | 0.97 | 0.99 | 0.97 |
| L1 | 0.96 | 0.05 | NA | |
| HERV | 0.98 | 0.41 | NA | |
| SVA | 0.97 | 0.03 | 0.98 | |
| Precision | Alu | 1.0 | 0.97 | 1.0 |
| L1 | 1.0 | 0.82 | NA | |
| HERV | 1.0 | 1.0 | NA | |
| SVA | 1.0 | 0.82 | 0.99 | |
NA: Not applicable.
Furthermore, the callers were run on PacBio data from samples HG002 and HG01071. Resulting TEs were compared to SVIM-asm (Heller and Vingron 2021) insertion calls determined to be Alu, L1, HERV, or SVA elements by RepeatMasker (Smit et al. 2013). For HG002 (TEI = 2904), sTELLeR was able to call 1885 true positive TEI, TLDR 1741, and xTEA 1543. For HG01071 (TEI = 2914), sTELLeR identified 1914, TLDR 1808, and xTEA 1645 true positive TEI (Table 3).
Table 3.
Metrics for the callers sTELLeR, TLDR, and xTEA on samples HG002 (Alu = 2193, L1 = 492, HERV = 14, SVA = 205) and HG01071 (Alu = 2262, L1 = 439, HERV = 11, SVA = 202).
| sTELLeR |
TLDR |
xTEA |
|||||
|---|---|---|---|---|---|---|---|
| HG002 | HG01071 | HG002 | HG01071 | HG002 | HG01071 | ||
| True positives | Alu | 1572 | 1592 | 1494 | 1556 | 1357 | 1408 |
| L1 | 213 | 232 | 152 | 164 | 186 | 186 | |
| HERV | 4 | 5 | 4 | 5 | 0 | 0 | |
| SVA | 96 | 85 | 91 | 83 | 0 | 51 | |
| Sensitivity | Alu | 0.71 | 0.70 | 0.68 | 0.68 | 0.61 | 0.62 |
| L1 | 0.43 | 0.52 | 0.30 | 0.37 | 0.37 | 0.33 | |
| HERV | 0.28 | 0.45 | 0.03 | 0.45 | 0 | 0 | |
| SVA | 0.46 | 0.42 | 0.44 | 0.81 | 0 | 0.02 | |
| Precision | Alu | 0.87 | 0.92 | 0.86 | 0.88 | 0.86 | 0.87 |
| L1 | 0.54 | 0.36 | 0.63 | 0.61 | 0.84 | 0.73 | |
| HERV | 0.36 | 0.17 | 0.05 | 0.07 | 0 | 0 | |
| SVA | 0.75 | 0.77 | 0.45 | 0.94 | 0 | 0.94 | |
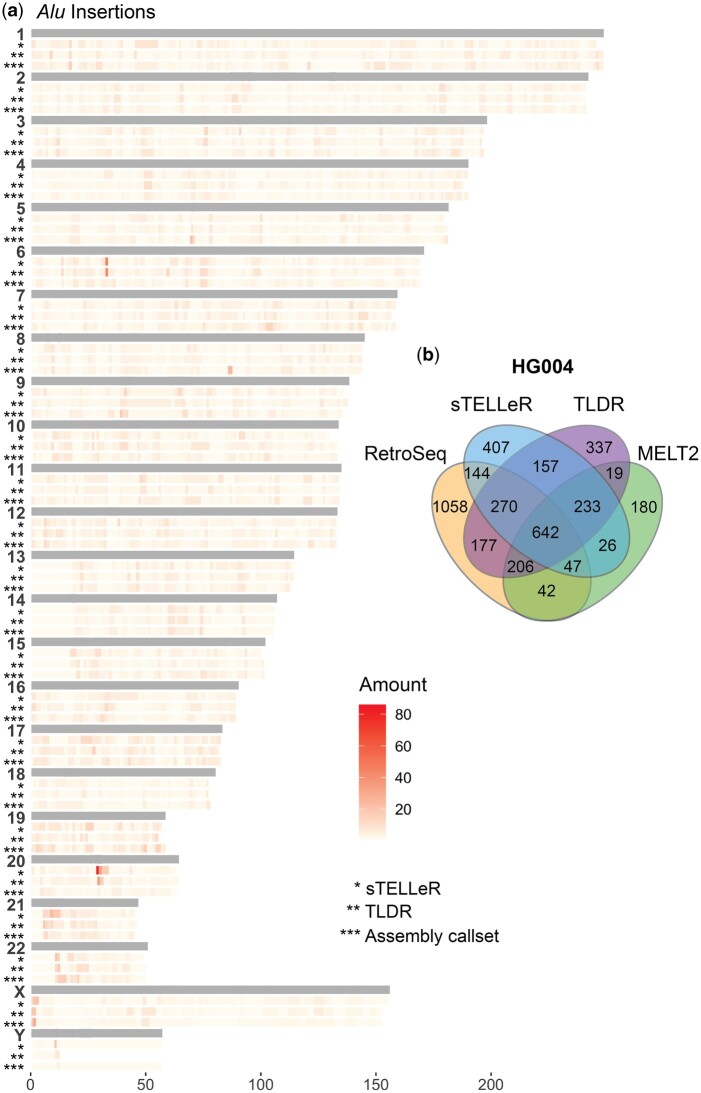
By binning the number of Alu elements across HG002 and HG01071 identified using the two most competitive callers (sTELLeR, TLDR) along with the assembly-based callset, one can observe clusters of Alu elements across the genome (Fig. 2a). The three methods have a similar distribution, where notably the acrocentric p-arms have low amount of Alu elements, while a region on the p-arm of X has high amounts. sTELLeR and TLDR share similar patterns of Alu detection and with a few exceptions is similar to the assembly-based callset. To further assess sTELLeR and TLDR, along with TE detection in lrGS, we called TEs Alu and L1 in sample HG004 using both srGS and lrGS. We ran sTELLeR and TLDR on the PacBio lrGS data and RetroSeq and MELT2 on the srGS data (Fig. 2b). We found the overlap of all callers to be 642. The total number of TEs undetected by the lrGS callers were 1280, while sTELLeR and TLDR agreed upon 1302 TEI.
Figure 2.
sTELLeR applied. (a) Heatmap of binned Alu insertions identified across HG002 and HG01071 with sTELLeR, TLDR, and the assembly-based callset. (b) VENN diagram of overlapping TEs called by RetroSeq, MELT2, sTELLeR, and TLDR in HG004.
4 Discussion
lrGS has come forth as a technique valuable for bridging repeats and regions previously challenging to resolve (Shahid and Slotkin 2020, Bilgrav Saether et al. 2023). Analysis of TEs have previously suffered from short-read lengths in srGS, as the length of repeats can be longer than a read. Thus, lrGS open for in-depth studies and characterization of TEs within the genome which was previously challenging. TEs implication in disease (Solyom and Kazazian 2012, Chenais 2022) makes them an important candidate in genome analysis, which have been underrepresented in clinical analysis workflows (Bilgrav Saether et al. 2023). In lrGS, a nonreference TE will be represented as insertions and split reads when aligned to a reference genome. The split reads or insertions will contain the sequence of the inserted element. This also applies to most other larger structural variants (SVs), and it is therefore necessary to be able to differentiate between TEIs and other SVs. With this, we wanted a TE caller for application on lrGS which can assist in both characterization and research of TEs as well as being compatible with future implementation of lrGS analysis workflows.
Implementation into research as well as diagnostic workflows require a fast, sensitive, and reproducible TE caller. A program should be easy to install and run, as well as be compatible with running in a pipeline environment. It should not be too computationally demanding. Ideally it should output a VCF in order to be compatible with outputs from other variant callers such as single-nucleotide variant, SV and copy-number variant callers. Many available callers today are often collected in a pipeline format, demanding extensive configuration. This makes further addition into in-house pipelines a complicated and inefficient step. Additionally, some callers explored had compilation errors or a runtime exceeding >48 h, which we deem too long for our analysis. The tested callers fit for our use were TLDR and xTEA. TLDR has been shown to achieve a sensitivity similar to srGS TE callers while assembling and annotating the insertions. However, the caller output results in a table format, and is not haplotype aware. xTEA display high sensitivity for Alu detection in both PacBio and ONT data (Chu et al. 2021); however, runtimes range from 22 to 46 h and output results in a text file format. Thus, in order to fulfill our requirements, we developed sTELLeR.sTELLeR is a python-based caller designed to identify nonreference insertions across the genome. Detection of reference TE polymorphisms, i.e. TEs present in the reference, but missing from the individuals, is not possible. The sTELLeR algorithm involves detecting split reads and insertions, upon which their positions are clustered using DBSCAN. DBSCAN clusters positions based on their distance to each other. The distance and number of positions needed to form a cluster can be altered (–sr) and optimized for intended use. This enables the sensitivity adaptable, providing flexibility and tailoring of the caller to the user’s needs. The user-input fasta file of TEs allows flexibility, making it possible to detect any type of nonreference insertion. sTELLeR is haplotype aware and can run on genome assemblies. Additionally, the output is provided in VCF output and any bam file can be submitted, meaning the tool can be applied to any species and nonreference insertion.
In research as well as clinical analysis, time, sensitivity, precision, and compatibility are important aspects. We show sTELLeR to be all the above, with easy installation through git or container, a runtime <30 min for a 60× PacBio genome (Table 2). Compared to similar tools such as TLDR and xTEA (Ewing et al. 2020, Chu et al. 2021), sTELLeR is 5–48× as fast and uses <2% of the CPU time.sTELLeR, TLDR, and xTEA were tested on a simulated dataset containing 886 Alu, 888 L1, 452 HERV, and 444 SVA insertions, as well as on samples HG002 and HG01071 from the HPRC (Wang et al. 2022). For the simulated dataset, sTELLeR has higher (TLDR) or similar (xTEA) precision and similar (xTEA) or slightly lower (TLDR) sensitivity for analysis of Alu elements than xTEA and TLDR (Table 2). For analysis of L1 elements, sTELLeR has higher sensitivity and precision than TLDR. Across both HERV and SVA elements, sTELLeR outperforms TLDR in both sensitivity and precision. xTEA is competitive at detecting SVA insertions. In the simulated dataset, xTEA was not able to identify any L1 or HERV elements, which could be due to the internal TE dataset not being compatible with the sequence of the elements used in the simulation, although retrieved from either the reference genome (GCF_000001405.26) or NCBI (AF020092).
In results from HG002 and HG01071, sTELLeR is more sensitive than both xTEA and TLDR for Alu and L1 element detection and more precise at Alu detection (Table 3). For SVA detection, sTELLeR has a precision >0.75 and sensitivity >0.42, while TLDRs sensitivity vary from 0.44 to 0.81 with a precision around 0.45. xTEA was not able to identify any SVA in sample HG01071 and for sample HG002 the sensitivity was very low (0.02), although the calls made were accurate (precision of 0.94). Furthermore, no HERV elements were detected by xTEA. HERV detection can be challenging as these elements are large, polymorphic and often not full length in the human population (Belshaw et al. 2005, Garcia-Montojo et al. 2018, Xue et al. 2020). This is reflected in the absence of true positive calls by xTEA and the low precision rate of both TLDR (<0.07) and sTELLeR (<0.36) on samples HG002 and HG01071 (Table 3). Overall, sTELLeR is able to identify a larger number of true positives across all TE types. Potential false positives can later be filtered using population databases. These results along with the low CPU usage show sTELLeR to be an efficient, precise, and sensitive TE caller.
In conclusion, we have developed a sensitive and precise caller which is fast and highly compatible with implementation into in-house workflows for research as well as clinical analysis.
Supplementary Material
Contributor Information
Kristine Bilgrav Saether, Department of Molecular Medicine and Surgery, Karolinska Institute, Stockholm 171 76, Sweden; Clinical Genomics Facility, Science for Life Laboratory, Stockholm 171 76, Sweden.
Jesper Eisfeldt, Department of Molecular Medicine and Surgery, Karolinska Institute, Stockholm 171 76, Sweden; Clinical Genomics Facility, Science for Life Laboratory, Stockholm 171 76, Sweden; Department of Clinical Genetics and Genomics, Karolinska University Hospital, Stockholm 171 77, Sweden.
Author contributions
K.B.S. and J.E. Conceptualization; Data curation; Formal analysis; Methodology; Project administration; Software; Resources; Validation; Writing–review & editing. K.B.S. Investigation; Visualization; Writing–original draft. J.E. Funding acquisition; Supervision.
Supplementary data
Supplementary data are available at Bioinformatics online.
Conflict of interest: None declared.
Funding
This work has been supported by the KID partial funding to doctoral students at Karolinska Institute (2020-01082) and KI Research Foundation Grants (2020–2021). The computations were enabled by resources provided by the National Academic Infrastructure for Supercomputing in Sweden, partially funded by the Swedish Research Council through grant agreement no. 2022-06725.
Data availability
sTELLeR can be found at https://github.com/kristinebilgrav/sTELLeR. A container is available as a docker image at https://hub.docker.com/r/kristinebilgrav/steller. Scripts used to generate the assembly-based and simulated callset as well as fasta sequences other scripts used in the project can be found at https://github.com/kristinebilgrav/sTELLeR_supplementary/. HPRC samples can be found at https://github.com/human-pangenomics/HPP_Year1_Data_Freeze_v1.0. GIAB samples can be found at https://github.com/genome-in-a-bottle/giab_data_indexes.
References
- Alcazer V, Bonaventura P, Depil S.. Human endogenous retroviruses (HERVs): shaping the innate immune response in cancers. Cancers (Basel) 2020;12:610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belshaw R, Dawson ALA, Woolven-Allen J. et al. Genomewide screening reveals high levels of insertional polymorphism in the human endogenous retrovirus family HERV-K(HML2): implications for present-day activity. J Virol 2005;79:12507–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilgrav Saether K, Nilsson D, Thonberg H. et al. Transposable element insertions in 1000 Swedish individuals. PLoS One 2023;18:e0289346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges-Monroy R, Chu C, Dias C. et al. Whole-genome analysis reveals the contribution of non-coding de novo transposon insertions to autism spectrum disorder. Mob DNA 2021;12:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenais B. Transposable elements and human diseases: mechanisms and implication in the response to environmental pollutants. Int J Mol Sci 2022;23:2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Borges-Monroy R, Viswanadham VV. et al. Comprehensive identification of transposable element insertions using multiple sequencing technologies. Nat Commun 2021;12:3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins RL, Brand H, Karczewski KJ. et al. ; Genome Aggregation Database Consortium. A structural variation reference for medical and population genetics. Nature 2020;581:444–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Tommaso P, Chatzou M, Floden EW. et al. Nextflow enables reproducible computational workflows. Nat Biotechnol 2017;35:316–9. [DOI] [PubMed] [Google Scholar]
- Ewing AD, Kazazian HH.. High-throughput sequencing reveals extensive variation in human-specific L1 content in individual human genomes. Genome Res 2010;20:1262–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing AD, Smits N, Sanchez-Luque FJ. et al. Nanopore sequencing enables comprehensive transposable element epigenomic profiling. Mol Cell 2020;80:915–28 e915. [DOI] [PubMed] [Google Scholar]
- Feusier J, Watkins WS, Thomas J. et al. Pedigree-based estimation of human mobile element retrotransposition rates. Genome Res 2019;29:1567–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Montojo M, Doucet-O’Hare T, Henderson L. et al. Human endogenous retrovirus-K (HML-2): a comprehensive review. Crit Rev Microbiol 2018;44:715–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner EJ, Lam VK, Harris DN. et al. ; 1000 Genomes Project Consortium. The mobile element locator tool (MELT): population-scale mobile element discovery and biology. Genome Res 2017;27:1916–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Dias GB, Basting PJ. et al. Local assembly of long reads enables phylogenomics of transposable elements in a polyploid cell line. Nucleic Acids Res 2022;50:e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller D, Vingron M.. SVIM-asm: structural variant detection from haploid and diploid genome assemblies. Bioinformatics 2021;36:5519–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane TM, Wong K, Adams DJ.. RetroSeq: transposable element discovery from next-generation sequencing data. Bioinformatics 2013;29:389–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Hu C, Moufawad El Achkar C. et al. Patient-customized oligonucleotide therapy for a rare genetic disease. N Engl J Med 2019;381:1644–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 2018;34:3094–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W-W, Asri M, Ebler J. et al. A draft human pangenome reference. Nature 2023;617:312–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon GA, Vollger MR, Eichler EE.. Long-read human genome sequencing and its applications. Nat Rev Genet 2020;21:597–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald TL, Zhou W, Castro CP. et al. Cas9 targeted enrichment of mobile elements using nanopore sequencing. Nat Commun 2021;12:3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mölder F, Jablonski KP, Letcher B. et al. Sustainable data analysis with Snakemake. F1000Res 2021;10:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono Y, Hamada M, Asai K.. PBSIM3: a simulator for all types of PacBio and ONT long reads. NAR Genom Bioinform 2022;4:lqac092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rishishwar L, Mariño-Ramírez L, Jordan IK.. Benchmarking computational tools for polymorphic transposable element detection. Brief Bioinform 2017;18:908–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers EW, Bolton EE, Brister JR. et al. Database resources of the national center for biotechnology information. Nucleic Acids Res 2022;50:D20–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafin K, Pesout T, Lorig-Roach R. et al. Nanopore sequencing and the Shasta toolkit enable efficient de novo assembly of eleven human genomes. Nat Biotechnol 2020;38:1044–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahid S, Slotkin RK.. The current revolution in transposable element biology enabled by long reads. Curr Opin Plant Biol 2020;54:49–56. [DOI] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P. RepeatMasker. Release Open-4.0. 2013. http://www.repeatmasker.org.
- Solyom S, Kazazian HH.. Mobile elements in the human genome: implications for disease. Genome Med 2012;4:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant PH, Rausch T, Gardner EJ. et al. ; 1000 Genomes Project Consortium. An integrated map of structural variation in 2,504 human genomes. Nature 2015;526:75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendrell-Mir P, Barteri F, Merenciano M. et al. A benchmark of transposon insertion detection tools using real data. Mob DNA 2019;10:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Antonacci-Fulton L, Howe K. et al. ; Human Pangenome Reference Consortium. The Human Pangenome Project: a global resource to map genomic diversity. Nature 2022;604:437–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue B, Sechi LA, Kelvin DJ.. Human endogenous retrovirus K (HML-2) in health and disease. Front Microbiol 2020;11:1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zook JM, Catoe D, McDaniel J. et al. Extensive sequencing of seven human genomes to characterize benchmark reference materials. Sci Data 2016;3:160025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
sTELLeR can be found at https://github.com/kristinebilgrav/sTELLeR. A container is available as a docker image at https://hub.docker.com/r/kristinebilgrav/steller. Scripts used to generate the assembly-based and simulated callset as well as fasta sequences other scripts used in the project can be found at https://github.com/kristinebilgrav/sTELLeR_supplementary/. HPRC samples can be found at https://github.com/human-pangenomics/HPP_Year1_Data_Freeze_v1.0. GIAB samples can be found at https://github.com/genome-in-a-bottle/giab_data_indexes.