Abstract
The Polymerase chain reaction–restriction fragment length polymorphism (PCR–RFLP) is a relatively simple and inexpensive method for genotyping single nucleotide polymorphisms (SNPs). It requires minimal investment in instrumentation. Here, we describe a web application, ‘SNP Cutter,’ which designs PCR–RFLP assays on a batch of SNPs from the human genome. NCBI dbSNP rs IDs or formatted SNPs are submitted into the SNP Cutter which then uses restriction enzymes from a pre-selected list to perform enzyme selection. The program is capable of designing primers for either natural PCR–RFLP or mismatch PCR–RFLP, depending on the SNP sequence data. SNP Cutter generates the information needed to evaluate and perform genotyping experiments, including a PCR primers list, sizes of original amplicons and different allelic fragment after enzyme digestion. Some output data is tab-delimited, therefore suitable for database archiving. The SNP Cut-ter is available at http://bioinfo.bsd.uchicago.edu/SNP_cutter.htm.
INTRODUCTION
Single nucleotide polymorphism (SNP) plays an important role in the study of complex genetic diseases (1), in pharmacogenetic analysis (2) and, in population genetics and evolutionary studies (3). Many methods are available for SNP genotyping, including hybridization, allele-specific PCR, primer extension, oligonucleotide ligation and endonuclease cleavage. However, each of these methods has its specific advantages and disadvantages (4–6). Polymerase chain reaction–restriction fragment length polymorphism (PCR–RFLP) is a classic and relatively inexpensive method of genotyping that is based on endonuclease cleavage. An SNP that alters a restriction sequence can be genotyped by ‘natural PCR–RFLP’. SNPs that do not affect any restriction sequences can be applied to a so-called ‘mismatch (or mismatched) PCR–RFLP’. Mismatch PCR–RFLP uses a primer containing additional mismatch base(s) adjacent to the SNP site (7,8). This method, however, requires the selection of appropriate restriction enzymes, the design of appropriate PCR primers, as well as the introduction of mismatched primers if mismatch PCR–RFLP is used. This process could be time-consuming and error-prone, especially if many SNPs need to be genotyped.
A comprehensive web-based application, SNP Cutter, was created to simplify the PCR–RFLP assay design. Starting from SNP sequence data preparation, SNP Cutter performs batch and automated assay design for PCR–RFLP, using a pre-selected or customizable list of restriction enzymes. Important assay parameters are calculated and provided.
SYSTEM
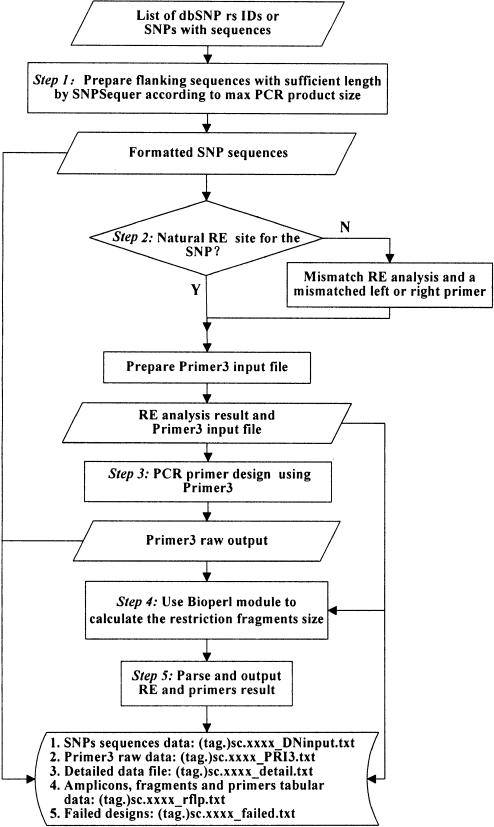
A Perl code CGI-driven web interface is provided for SNP Cutter. Primer3 (9) is used for PCR primer design. SNPSequer (http://bioinfo.bsd.uchicago.edu/SNPSequer.htm) is used to prepare SNP sequence inputs. The workflow of the SNP Cutter is illustrated in Figure 1.
Figure 1.
SNP Cutter workflow. Rectangles and rhombi show the task that the program performs. It includes five steps, shown as Step 1 through 5 in the figure. Parallelograms show the data that the user supplies or every step produces. The program produces five files as shown. ‘(tag.)sc.xxxx_DNinput.txt’ is the output of Step 1 SNPSequer; ‘(tag.)sc.xxxx_PRI3.txt’ is the output of Step 3 of Primer3 analysis; ‘(tag.)sc.xxxx_detail.txt’ and ‘(tag.)sc.xxxx_rflp.txt’ are the outputs of Step 5. ‘(tag.)sc.xxxx_failed.txt’ logs all SNPs failed at assay design. ‘RE’ is the abbreviation for ‘Restriction Enzyme.’ ‘(tag.)’ is an optional string user provides as identifier in the filename. ‘xxxx’ is a random number that the system produces.
PROGRAM INPUTS
Inputs of SNP Cutter are entered through its web interface. The inputs include SNPs to be genotyped, a customizable list of restriction enzymes and other parameters for primer design.
SNPs to be genotyped. SNP Cutter accepts two alternative formats of SNPs as input. The first format choice is a list of NCBI dbSNP rs IDs which simplify the preparation of sequence data (from dbSNP). The second format choice is the user's own list of formatted SNPs with flanking sequences (tab or space delimited text with SNP IDs, allelic nucleotides, 5′ and 3′ flanking sequences), which facilitates users who want to genotype SNPs absent in dbSNP as many investigators are discovering novel SNPs in their own labs. For both choices, SNPSequer extracts SNP flanking sequences with sufficient length from the latest version of the human genomic DNA sequence assembly for the primer design.
Customizable list of restriction enzymes. SNP Cutter provides four flavors of the restriction enzyme lists. The first choice list contains enzymes from REBASE (version 501, December 2004, http://rebase.neb.com/rebase/rebase.html) which are commercially available. The second list contains the comparatively less expensive restriction enzymes (<$0.13/U, according to the retail price in the US and Chinese market). The third list contains commonly used and relatively reliable enzymes that had already been successfully used for previous genotyping procedures. This list contains the consolidated data from 34 published papers. Users can also submit their preferred enzymes to the author of the SNP Cutter, so that this list will be accumulated. With the fourth choice, users can also define specifically his or her list of preferred restriction enzymes. Enzymes recognizing ambiguous restriction sequences were removed from all of the enzyme lists provided by SNP Cutter. The enzyme lists are updated periodically according to the updates from REBASE, the price in the market and the data collected by author or submitted by users.
Other parameters for primer design. Some other adjustable parameters include parameters for Primer3 primer design and option settings for output files. Primer3 parameters are separated into two parts, one for natural PCR–RFLP and another for mismatch PCR–RFLP. For natural PCR–RFLP, the ‘PCR product size range’ is defined in the same way as in the original Primer3. For mismatch PCR–RFLP, the default PCR product size range is ‘100–200’ bp as SNP site is adjacent to 3′ end of the mismatch primer. The preferred mismatch PCR product size is 100–200 bp. This ensures that the digested allelic fragments can be easily resolved on the electrophoresis gel. Users can decide how many nucleotides at the 3′ end of a primer should be free of mismatch. Putting the mismatch on the last two nucleotides of the primer is discouraged. Also, introducing multiple mismatches in SNP Cutter is not recommended because multiple mismatches and 3′ end mismatch in PCR primer could potentially create problems for PCR optimization.
PROGRAM OUTPUTS
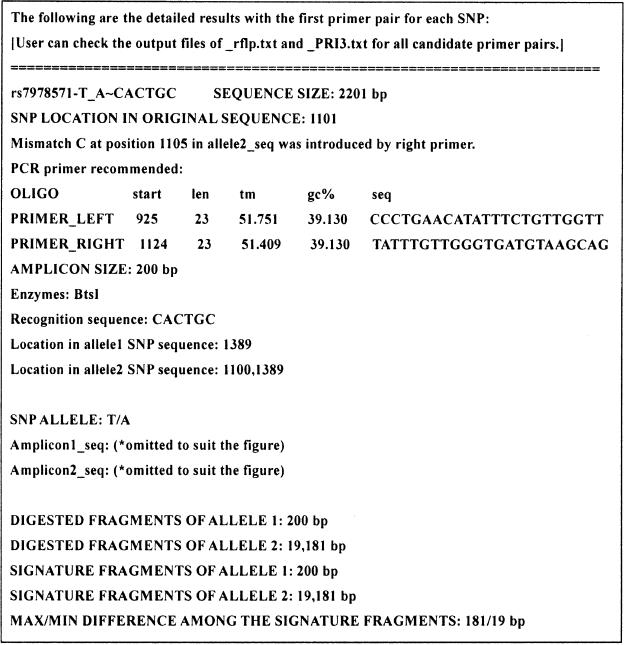
SNP Cutter provides five output files including two data files for databasing. While allowing user to add a string as identifier at the beginning of the filenames, all output files are named with an ‘sc’ prefix while different endings of file names differentiate them. (i) A detailed data file with ‘detail.txt’ as the end of the filename contains all data that users need to design and carry out PCR–RFLP genotyping experiments. This file is composed of three sections. The first section presents PCR primers and enzymes data. The second section presents sequences of the two allelic amplicons if primers were designed successfully. If no primer was designed, this section will show the original SNP sequences of the two alleles. The third section presents allelic fragments data, which includes the size of the digested fragments, ‘signature fragments’ in two alleles, and maximum and minimum size differences among signature fragments. Signature fragments of one allele are the unique after-digestion fragments whose sizes are different from all the fragments of another allele. The data of sizes of signature fragments and differences among these fragments are important for the assay evaluation, since they are directly correlated with the resolution of electrophoresis bands on a gel. It is much easier to resolve and correctly called for genotype on allelic bands of a 100 bp versus 200 bp than of a 90 bp versus 100 bp. The data also supplies log information, such as SNPs that failed to find a proper restriction enzymes or primers. Figure 2 shows one example of part of a detailed data file. (ii) The data file names ending in ‘PRI3.txt’ contains the original Primer3 output. All the primer design parameters are presented in this file so that PCR primer designs can be viewed. (iii) The SNP Cutter supplies a tab-delimited text format file ending with ‘rflp.txt’ if both the PCR primer design and restriction enzyme selection are successfully completed. This file contains the assorted information about primers, enzymes, allelic fragments and signature fragments. In contrast to detailed data file in which only the first set of candidate primers are presented, all candidate primers are presented here in ‘rflp.txt’ file. Tab-delimited text data can be easily imported and managed in a spreadsheet or a database. (iv) Data file names ending in ‘DNinput.txt’ designates a tab-delimited data file containing SNP sequences prepared by SNPSequer. It can be used to save run-time in case users need to redesign assay for some SNPs with modified conditions. (v) A file ending with ‘failed.txt’ will present all the assay design failure information to help the users to identify SNPs which need a redesign.
Figure 2.
Illustration of an example of detail data file. All the information about the primers, amplicons, enzymes and fragments are shown in this file. The real sequence data of ‘Amplicon1_seq’ and ‘Amplicon2_seq’ were omitted to save space.
The outputs can be either delivered to the user by email or downloaded from URLs showed at web page. The user also has the options of obtaining results by email attachments or by an email notice containing URLs to access the results.
DISCUSSION AND CONCLUSION
Comparisons were made between the SNP Cutter and the other existing PCR–RFLP assay design tools (see Supplementary Material): PIRA PCR (10,11), SNPKit (12), dCAPS Finder 2.0 (13) and SNP2CAPS(14). The results indicated that SNP Cutter is more efficient and informative than the other tools, especially for input data preparation, enzyme selection, and output format and contents.
All the existing tools only take inputs as formatted sequences, leaving the burden of sequence preparation to users. In contrast, SNP Cutter processes batches of SNPs which simplifies the data preparation process. For a given chromosomal region, users may retrieve the list of SNPs rs IDs from NCBI Map Viewer (http://www.ncbi.nlm.nih.gov/mapview/) or UCSC Genome Browser (http://genome.ucsc.edu/cgi-bin/hgGateway?org=Human). These rs IDs can be directly used for PCR–RFLP assay design. Alternatively, the formatted SNP sequence data as the second choice of input gives users an opportunity to design assays for SNPs that have not been submitted to dbSNP. Additionally, the embedding SNPSequer program in SNP Cutter makes data preparation even simpler, as >30% of the 5′ or 3′ flanking sequence of the reference SNPs in dbSNP are <200 bp (C. Liu and H. Zhu, unpublished data).
SNP Cutter compiled a list of pre-selected inexpensive and reliable enzymes including EcoRI, PstI, SmaI, HindIII, HaeIII, MspI, RsaI and TaqI. This is one of the first efforts to compile a list of preferred enzymes for PCR–RFLP assay. This curated enzyme list is believed to be able to help producing more robust genotyping assays.
The success rate of assay design in SNP Cutter varies from 45 to 85% depending on the parameter settings. Using a curated enzyme list with limited number of enzymes and stringent PCR conditions could lead to lower success rate of design, but the designed assays are more robust.
We found it convenient to manage experimental data in spreadsheets or databases and, SNP Cutter outputs two tab-delimited text files that simplify database management. This feature is not provided by the other tools.
SNP Cutter presents sizes data of amplicons, digested allelic fragments and signature fragments. This data is important as a guide for evaluating genotyping results. The SNP Cutter might supply multiple alternative choices of restriction enzymes for genotyping of one SNP, and the users can select one of them according to these data and the price of the enzymes.
Although several methods have been developed to make PCR–RFLP technology work for large-scale SNP genotyping such as Microplate Array Diagonal Gel Electrophoresis (MADGE) (15), Terminal RFLP (T-RFLP) (16) and fluorescent RFLP (Frflp) (17). PCR–RFLP per se is not generally recognized as a high-throughput SNP genotyping method comparing with many other methods such as TaqMan and Illumina. But PCR–RFLP does have its advantages and still plays important role in many small labs. SNP Cutter was developed to assist those investigators who are using PCR–RFLP to perform SNP genotyping.
In summary, the SNP Cutter provides batch rs IDs inputs, multiple choices of pre-selected enzyme list, tabular format output, experimental data, the integrated pipeline of SNP sequences extraction, restriction enzymes searching and primer design which makes PCR–RFLP genotyping assay design more efficient.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
This work was supported by a Young Investigator Grant to C.L. from NARSAD (National Alliance for Research in Schizophrenia and Affective Disorders) and NIH R01 MH65560-01 and R01 MH59535 to Elliot S. Gershon and by the Chinese State ‘863’ program (2002BA711A07-08, 2002BA711A07-03), ‘973’ program (2001CB510302, 2004CB518601) and National Natural Science Foundation of China (30070410, 30123006, 30371530 and 30340078). Support from the Geraldi Norton Memorial Corporation, Mr and Mrs Peterson, the Eklund Family and Anita Kaskel Roe are also gratefully acknowledged. Funding to pay the Open Access publication charges for this article was provided by the National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1.Marnellos G. High-throughput SNP analysis for genetic association studies. Curr. Opin. Drug Discov. Devel. 2003;6:317–321. [PubMed] [Google Scholar]
- 2.Mooser V., Waterworth D.M., Isenhour T., Middleton L. Cardiovascular pharmacogenetics in the SNP era. J. Thromb. Haemost. 2003;1:1398–1402. doi: 10.1046/j.1538-7836.2003.00272.x. [DOI] [PubMed] [Google Scholar]
- 3.Hacia J.G., Fan J.B., Ryder O., Jin L., Edgemon K., Ghandour G., Mayer R.A., Sun B., Hsie L., Robbins C.M., et al. Determination of ancestral alleles for human single-nucleotide polymorphisms using high-density oligonucleotide arrays. Nature Genet. 1999;22:164–167. doi: 10.1038/9674. [DOI] [PubMed] [Google Scholar]
- 4.Syvanen A.C. Accessing genetic variation: genotyping single nucleotide polymorphisms. Nature Rev. Genet. 2001;2:930–942. doi: 10.1038/35103535. [DOI] [PubMed] [Google Scholar]
- 5.Tsuchihashi Z., Dracopoli N.C. Progress in high throughput SNP genotyping methods. Pharmacogenomics. J. 2002;2:103–110. doi: 10.1038/sj.tpj.6500094. [DOI] [PubMed] [Google Scholar]
- 6.Kwok P.Y., Chen X. Detection of single nucleotide polymorphisms. Curr. Issues Mol. Biol. 2003;5:43–60. [PubMed] [Google Scholar]
- 7.Haliassos A., Chomel J.C., Tesson L., Baudis M., Kruh J., Kaplan J.C., Kitzis A. Modification of enzymatically amplified DNA for the detection of point mutations. Nucleic Acids Res. 1989;17:3606. doi: 10.1093/nar/17.9.3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haliassos A., Chomel J.C., Grandjouan S., Kruh J., Kaplan J.C., Kitzis A. Detection of minority point mutations by modified PCR technique: a new approach for a sensitive diagnosis of tumor-progression markers. Nucleic Acids Res. 1989;17:8093–8099. doi: 10.1093/nar/17.20.8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rozen S., Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 10.Ke X., Collins A., Ye S. PIRA PCR designer for restriction analysis of single nucleotide polymorphisms. Bioinformatics. 2001;17:838–839. doi: 10.1093/bioinformatics/17.9.838. [DOI] [PubMed] [Google Scholar]
- 11.Ke X., Collins A., Ye S. PCR designer for restriction analysis of various types of sequence mutation. Bioinformatics. 2002;18:1688–1689. doi: 10.1093/bioinformatics/18.12.1688. [DOI] [PubMed] [Google Scholar]
- 12.Hao K., Niu T., Sangokoya C., Li J., Xu X. SNPkit: an efficient approach to systematic evaluation of candidate single nucleotide polymorphisms in public databases. Biotechniques. 2002;33:822. doi: 10.2144/02334st06. 824–826, 828. [DOI] [PubMed] [Google Scholar]
- 13.Neff M.M., Turk E., Kalishman M. Web-based primer design for single nucleotide polymorphism analysis. Trends Genet. 2002;18:613–615. doi: 10.1016/s0168-9525(02)02820-2. [DOI] [PubMed] [Google Scholar]
- 14.Thiel T., Kota R., Grosse I., Stein N., Graner A. SNP2CAPS: a SNP and INDEL analysis tool for CAPS marker development. Nucleic Acids Res. 2004;32:e5. doi: 10.1093/nar/gnh006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaunt T.R., Hinks L.J., Rassoulian H., Day I.N. Manual 768 or 384 well microplate gel ‘dry’ electrophoresis for PCR checking and SNP genotyping. Nucleic Acids Res. 2003;31:e48. doi: 10.1093/nar/gng048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bruce K.D., Hughes M.R. Terminal restriction fragment length polymorphism monitoring of genes amplified directly from bacterial communities in soils and sediments. Mol. Biotechnol. 2000;16:261–269. doi: 10.1385/MB:16:3:261. [DOI] [PubMed] [Google Scholar]
- 17.Lazzaro B.P., Sceurman B.K., Carney S.L., Clark A.G. fRFLP and fAFLP: medium-throughput genotyping by fluorescently post-labeling restriction digestion. Biotechniques. 2002;33:539–546. doi: 10.2144/02333st04. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.