Abstract
Complete genomic sequences of several oral pathogens have been deciphered and multiple sources of independently annotated data are available for the same genomes. Different gene identification schemes and functional annotation methods used in these databases present a challenge for cross-referencing and the efficient use of the data. The Bioinformatics Resource for Oral Pathogens (BROP) aims to integrate bioinformatics data from multiple sources for easy comparison, analysis and data-mining through specially designed software interfaces. Currently, databases and tools provided by BROP include: (i) a graphical genome viewer (Genome Viewer) that allows side-by-side visual comparison of independently annotated datasets for the same genome; (ii) a pipeline of automatic data-mining algorithms to keep the genome annotation always up-to-date; (iii) comparative genomic tools such as Genome-wide ORF Alignment (GOAL); and (iv) the Oral Pathogen Microarray Database. BROP can also handle unfinished genomic sequences and provides secure yet flexible control over data access. The concept of providing an integrated source of genomic data, as well as the data-mining model used in BROP can be applied to other organisms. BROP can be publicly accessed at http://www.brop.org.
INTRODUCTION
Over the past few years, several important oral pathogens have been completely or partially sequenced [for a review see (1)]. Table 1 provides an updated list of the current genomics tools and databases available for oral pathogens. While many of these databases and tools provide useful and unique information regarding the same genomes, difficulties are often encountered when users try to compare or combine information available for the same genes. For example, at least two independently annotated databases are currently available for the genomes of Porphyromonas gingivalis, Streptococcus mutans and Treponema denticola, i.e. in The Comprehensive Microbial Resource of The Institute of Genomic Research (TIGR CMR) and the Oral Pathogen Sequence Databases of Los Alamos National Laboratory (LANL), respectively (also refer to Table 1). Discrepancies occur when different criteria were used in either gene identification, naming or functional annotation. Table 2 shows the different numbers of genes present in various annotation sources for the genome of P.gingivalis. While these independently maintained databases provide useful and unique information and tools, they also present to users a great challenge for comparing and integrating the information for the same genes from these multiple sources. The Bioinformatics Resource For Oral Pathogens (BROP) is a web-base resource center providing bioinformatics tools and databases for oral pathogens with the primary goal of presenting integrated information for the same genome from multiple sources of data.
Table 1.
Genomics databases and tools available for oral pathogensa
| Organism | Strain | Genome size (Mb) | Collaborating Institute | Funding | Statusb | Databases and tools |
|---|---|---|---|---|---|---|
| Actinobacillus actinomycetemcomitans | HK1651 | 2.90 | University of Oklahoma | NIDCR | 1 | Downloadc: OU |
| BLASTd: OU1, NCBI, BROP, LANL | ||||||
| Databasee: BROP, LANL | ||||||
| Softwaref: BROP, LANL | ||||||
| Actinomyces naeslundii | MG1 | 3 | TIGR | NIDCR | NA | NA |
| Tannerella forsythensis (Bacteroides forsythus) | FDC 92A2 | 3.40 | TIGR | NIDCR | 1 | Download: TIGR1 |
| BLAST: TIGR | ||||||
| Database: TIGR | ||||||
| Software: TIGR | ||||||
| Candida albicans | SC5314 | NA | Stanford Genome Technology Center | NIDCR/Burroughs Wellcome Fund | NA | Download: Stanford |
| BLAST: Stanford | ||||||
| Candida albicans | 1161 | 15 | The Sanger Institute | Beowulf Genomics | NA | Download: Sanger |
| BLAST: Sanger | ||||||
| Fusobacterium nucleatum | ATCC 10953 | 2.4 | Baylor College of Medicine | NIDCR | 101 | Download: BCM |
| BLAST: BCM | ||||||
| Fusobacterium nucleatum | ATCC 25586 | 2.17 | Integrated Genomics | NIH | 1 | Download: IG, NCBI |
| BLAST: NCBI, BROP | ||||||
| Database: LANL BROP | ||||||
| Software: LANL BROP | ||||||
| Fusobacterium nucleatum vincentii | ATCC 49256 | NA | Integrated Genomics | NIH | 302 | Download: IG |
| Porphyromonas gingivalis | W83 | 2.34 | The Forsyth Institute/TIGR | NIDCR | 1 | Download: TIGR2, NCBI |
| BLAST: TIGR, NCBI, BROP, LANL | ||||||
| Database: TIGR, BROP, LANL | ||||||
| Software: TIGR, BROP, LANL | ||||||
| Microarrayg: OPMD | ||||||
| Prevotella intermedia | 17 | 3.8 | TIGR | NIDCR | 1 | Download: TIGR2 |
| BLAST: TIGR | ||||||
| Database: TIGR | ||||||
| Software: TIGR | ||||||
| Streptococcus gordonii (Challis) | NCTC 7868 | NA | TIGR | NIDCR | 273 | Download: TIGR1 |
| BLAST: TIGR | ||||||
| Database: TIGR | ||||||
| Software: TIGR | ||||||
| Streptococcus mitis | NCTC 12261 | 2.2 | TIGR | NIDCR | 1 | Download: TIGR2 |
| BLAST: TIGR | ||||||
| Database: TIGR | ||||||
| Software: TIGR | ||||||
| Streptococcus mutans | UA159 (ATCC 700610) | 2.03 | University of Oklahoma | NIDCR | 1 | Download: OU, NCBI |
| BLAST: OU2, NCBI, BROP, LANL | ||||||
| Database: BROP LANL | ||||||
| Software: BROP LANL | ||||||
| Streptococcus sanguis | SK36 | NA | Virginia Commonwealth University | NIDCR | NA | Download: VCU |
| BLAST: VCU | ||||||
| Streptococcus sobrinus | 6715 | NA | TIGR | NIDCR | NA | Download: TIGR1 |
| Treponema denticola | ATCC 35405 | 2.8 | Baylor College of Medicine/TIGR | NIDCR | 1 | Download: BCM, NCBI |
| BLAST: TIGR, BCM, NCBI, BROP | ||||||
| Database: BROP, LANL | ||||||
| Software: BROP, LANL TIGR | ||||||
| Treponema lecithinolyticum | OMZ 684T | 2.3 | The Forsyth Institute | NIDCR | 1001 | BLAST: BROP |
| Database: BROP | ||||||
| Software: BROP |
aAn up-to-date list is maintained at http://www.brop.org.
bStatus of sequencing is indicated by the number of assembled contigs.
cURLs for sequence download: University of Oklahoma (OU), ftp://ftp.genome.ou.edu/pub; The Institute for Genomic Research (TIGR1), http://www.tigr.org/tigr-scripts/ufmg/ReleaseDate.cgi; The Institute for Genomic Research (TIGR2), ftp://ftp.tigr.org/pub/data/Microbial_Genomes/; Stanford Genome Technology Center (Stanford), http://www-sequence.stanford.edu/group/candida/download.html; The Sanger Institute (Sanger), ftp://ftp.sanger.ac.uk/pub/yeast/sequences/candida; Baylor College of Medicine (BCM), ftp://ftp.hgsc.bcm.tmc.edu/pub/data; Integrated Genomics (IG), http://www.integratedgenomics.com/genomereleases.html and Virginia Commonwealth University (VCU), http://www.sanguis.mic.vcu.edu/.
dURLs for BLAST search tools: OU1, http://www.genome.ou.edu/act_blast.html; OU2, http://www.genome.ou.edu/smutans_blast.html; National Center for Biotechnology Information (NCBI), http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi; Bioinformatics Resource for Oral Pathogens (BROP), http://www.brop.org/modules.php?op=modload&name=News&file=article&sid=10; TIGR, http://tigrblast.tigr.org/ufmg/; Stanford http://www.sequence.stanford.edu/group/candida/search.html; Sanger, http://www.sanger.ac.uk/cgi-bin/blast/submitblast/c_albicans; BCM, http://hgsc.bcm.tmc.edu/microbial/microbialblast.cgi?organism=Fnucleatum and VCU, http://www.sanguis.mic.vcu.edu/.
eURLs for annotation database: BROP, http://genome.brop.org; Los Alamos National Laboratory (LANL), http://www.brop.lanl.gov/ and TIGR, http://www.tigr.org/tigr-scripts/CMR2/CMRHomePage.spl.
fURLs for analysis tools: BROP, http://genome.brop.org; LANL and http://www.brop.lanl.gov.
gURL for microarray database: Oral Pathogen Microarray Database (OPMD), http://array.brop.org.
Table 2.
Numbers of P.gingivalisgenes predicted by various sources of annotation databases and used in the microarrays designed by TIGR
bData source: ftp://bpublic.lanl.gov/compbio/data/oralgen.
dOriginal array information was obtained from TIGR, available at http://array.brop.org.
eRefers to the first version of TIGR P.gingivalis arrays, on which DNA samples based on PCR amplicons were spotted. Detail information about this array is available at: http://array.brop.org.
The second goal of BROP is to provide up-to-date genomic annotation. To date, homologous sequence matching remains the most useful way of functional inference for newly identified genes in a genome. The number of sequences in the public databases, against which most new sequences are searched, continues to increase exponentially. Many public resources provide daily updates and exchange of their databases (2–4). However most genomes, once annotated, published and deposited to the public databases, are not updated or reannotated. Thus, more frequent and repeated homologous search on these rapidly updated sequence databases can provide new or updated functional annotation of previously unknown genes in a timely manner. BROP employs a cluster of computers to continuously update the annotations for the genomic sequences of oral pathogens. A pipeline of automatic data-mining algorithms against several frequently updated sequence databases was implemented to repeatedly cycle the annotation through all target genomes. The annotation results can be viewed and searched through a centralized interface that provides interlinks to additional internal and external information. Thus the information provided in BROP will be always up-to-date; new tools and data-mining schemes or algorithms can always be added.
The third goal of BROP is to provide tools and databases for post-genomic research data such as those from microarray and proteomics experiments. For example, at the time of writing (September, 2004), DNA microarrays for two oral pathogens—P.gingivalis and S.mutans have been made available to the research community (http://www.nidcr.nih.gov/Research/Extramural/NIDCR_TIGR_Facility.htm) and studies have been performed based on these arrays (5). BROP provides microarray database and statistics analysis tools specially designed for the oral pathogen microarray data. Furthermore the data in the microarray database are linked to the genomic tools provided in BROP, making it very convenient for researchers to process, store, analyze and interpret their microarray data.
TOOLS
Genome Viewer
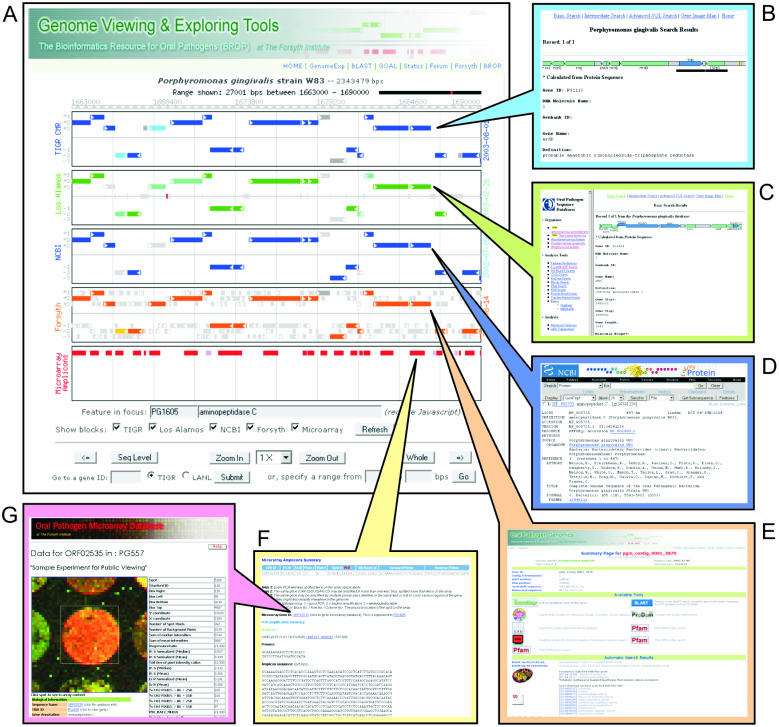
To alleviate the inconvenience encountered when comparing two different sets of annotations for the same genome, Genome Viewer provides a graphical, six-frame transnational view of the same region of the genome with individual panels showing different sets of annotations. It has easy navigating features including zooming, centering and searching by gene ID. The zooming ranges from 100 bp, which shows the actual nucleotide sequence, to as large as the entire genome. Figure 1 is a screen shot of the Genome Viewer showing a range of genome of P.gingivalis with four individual panels of information that present various information annotated by TIGR CMR, LANL ORALGEN, the NCBI GenBank record and BROP (see below for BROP annotation method). When the browser pointer is placed over (i.e. mouse over) a feature (e.g. an open reading frame or ORF) in a panel, the ID and definition of the element is shown in a separate JavaScript window located below the panels. This leaves the panels with a less crowded appearance since they are not cluttered by the annotation text, thus they are easier to view and navigate. Clicking on any of the features in the panels leads to the original and detailed information (illustrated in Figure 1, callout boxes B–D). Currently, for P.gingivalis, Genome Viewer also provides an additional panel showing PCR amplicons used in the microarrays manufactured by TIGR, which have been made available to the research community (5). Detail amplicon information is available by clicking the amplicons in the panel, which is linked to the Oral Pathogen Microarray Database (OPMD, described below; illustrated in Figure 1E and F). For other oral pathogen genomes, once the microarray information is available, additional panel can be added to the Genome Viewer. Currently, Genome Viewer also provides viewing of all the microbial genome sequences that are available at the National Center for Biotechnology Information (NCBI).
Figure 1.
Screen shot of the Genome Viewer software (A) showing side-by-side comparison of annotations from multiple sources for the same region of the genome P. gingivalis W83. The clickable links in the viewer are depicted with the rectangular callout boxes: (B) TIGR CMR; (C) LANL Oralgen; (D) NCBI; (E) BROP Gene Summary; (F) Array Amplicon Information; and (G) OPMD.
Genome-wide ORF Alignment (GOAL)
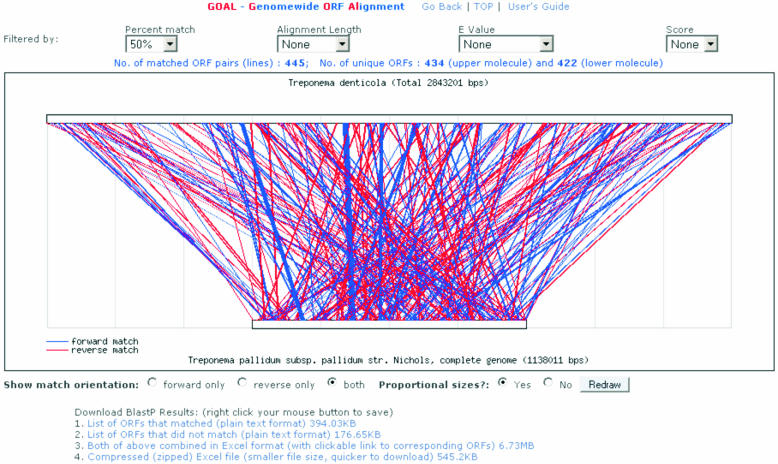
Similar to Bugspray provided by LANL (http://biosphere.lanl.gov/bugspray_std/cgi-bin/wc.cgi) GOAL is a comparative genomic tool that provides graphical view of whole-genome alignments between any two chosen genomes/molecules, based on the protein sequence homology of the ORFs between them. Each of the ORFs of the first selected genome is searched against every ORF in the second genome using the NCBI BLASTP program (ftp://ftp.ncbi.nih.gov/blast). Homologous regions are then plotted out between two genomes, based on the BLASTP matches and filtering criteria selected by users (e.g. percent align, alignment length, statistical E-values and scores of the matched ORFs). Detailed BLASTP results are made available for downloading in either plain text, tab-delimited or Microsoft Excel Spreadsheet formats. The Excel result file also provides convenient web links to the corresponding annotation databases for all ORFs. Currently GOAL allows the alignment of any two chosen genomes that are being curated or maintained in the database, including both finished and unfinished oral pathogen genomes, as well as all current microbial genomes available at NCBI. Figure 2 shows a sample alignment between two genomes using GOAL.
Figure 2.
Visualization of whole-genome ORF alignment between the genomes of P.gingivalis W83 and Bacteroides thetaiotaomicron VPI-5482 using the GOAL tool. Actual display of the forward and reverse matches was shown in blue and red colors, respectively.
Genome Explorer
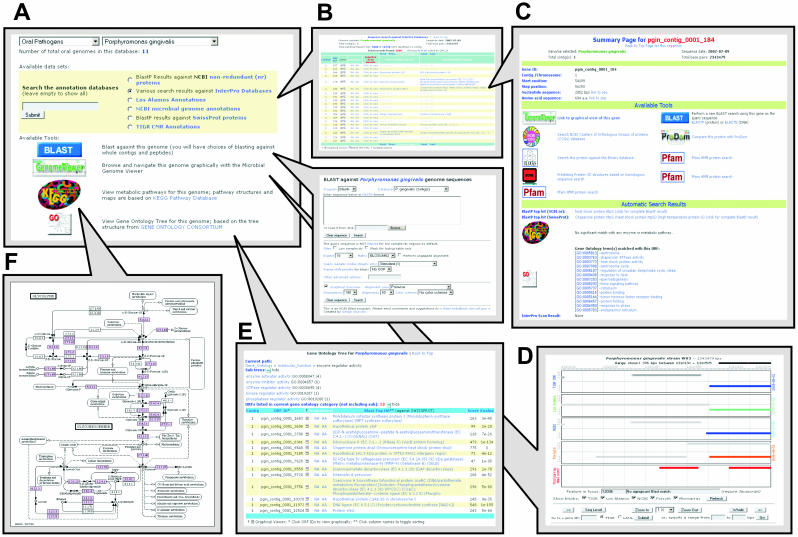
Genome Explorer is a centralized web interface that inter-connects all the oral pathogen genomics resources. The front-end of Genome Explorer is a user-friendly interface that allows investigators to easily navigate among all the genomics information provided in its database. Once a target genome is chosen, the interface dynamically presents all the databases and tools available for the selected genome, such as the data-mining results against frequently updated sequence databases (described below). Other options include links to the Genome Viewer, KEGG pathways (6), Gene Ontology (GO) Tree (7), BLAST and InterProScan search results (8) for the selected genome. The back-end of Genome Explorer is a searchable annotation database that integrates all the results generated from the data-mining pipeline described below. The search result is presented in a paginated and sortable table that also provides web links to (i) a summary page for individual ORF, (ii) Genome Viewer to show the exact location of the target ORF in the genome and (iii) the original BLAST or InteProScan results. The summary page provides all the information and tools available for a specific ORF, including all the data-mining results mentioned above, as well as convenient links to other web tools for performing fresh search and analysis. In short, Genome Explorer is a one-stop site for all the genomic information available for each target genome or gene. A sample screen shot of the Genome Explorer is shown in Figure 3.
Figure 3.
Screen shot of the Genome Explorer software (A) showing a plethora of tools and information available for a particular gene selected. The interface contains links to (B) a text-based annotation searching result; (C) a summary page for individual ORF; (D) the Genome Viewers; (E) Gene Ontology information; and (F) KEGG metabolic pathway.
PIPELINE OF CONTINUOUS GENOME ANNOTATION AND DATABASE UPDATE
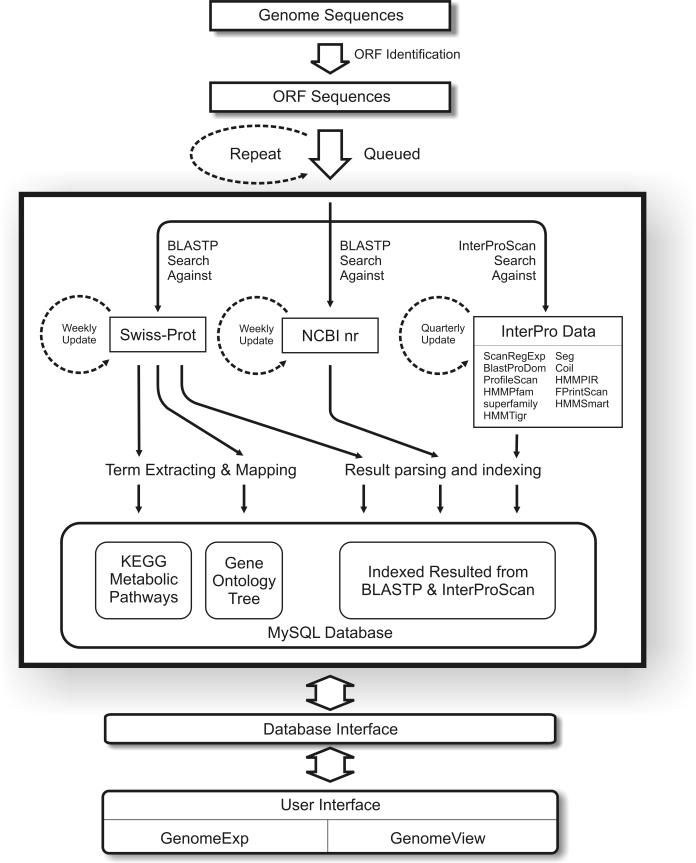
Although the amount of sequence data has been rapidly growing, so does computing power available for analysis. Oral pathogens are a group of organisms that are of interest to scientists who study infectious oral or dental diseases. So far <20 genomes from this group have been sequenced. The relative small number and size of genomes makes it feasible to keep the annotation data up-to-date almost on a weekly basis with the increasing computing power provided by modern hardware technology. BROP employs a cluster of dedicated computer servers to continuously mine the information from genomes of oral pathogens. Figure 4 depicts the pipeline of several inter-connected data-mining schemes that constantly fill in and update the BROP databases. Current BROP data-mining algorithms include: (i) BLASTP (http://www.ncbi.nih.gov/BLAST/) (8) search against weekly updated NCBI non-redundant protein data (ftp://ftp.ncbi.nih.gov/blast/db/nr.tar.gz); (ii) BLASTP search against Swiss-Prot protein data (http://us.expasy.org/sprot/) (9); and (iii) InterProScan search (http://www.ebi.ac.uk/InterProScan/) (10) against ScanRegExp, BlastProDom, ProfileScan, HMMPfam, Superfamily, HMMTigr, Seg, Coil, HMMPIR, FPrintScan and HMMSmart databases (http://www.ebi.ac.uk/interpro/databases.html). Swiss-Prot is a set of well-annotated protein sequences which contains interlinks to the ENZYME (11) and Gene Ontology (12) data and thus the BLASTP search result against Swiss-Prot can be further processed for the construction of KEGG metabolic pathway and GO trees. BROP is dedicated to the annotation of oral pathogen genomes. Currently it is constantly mining the data for 11 genomes, of which 8 have been completed (assembled to a single sequence contig) and 3 are unfinished. BROP also provides a live statistics and status web page for monitoring all the data-mining work so that users are aware of the date of the data they are exploring.
Figure 4.
Pipeline of automatic annotation of oral pathogen genomes.
ORAL PATHOGEN MICROARRAY DATABASE (OPMD)
The National Institute of Dental and Craniofacial Diseases (NIDCR) has been providing no-cost, oligonucleotide genomic DNA microarray slides for oral bacteria to the research community (http://www.nidcr.nih.gov/Research/Extramural/NIDCR_TIGR_Facility.htm). To use these arrays, the investigators have to agree to the release of data in a timely manner to a public database. The data should also be documented in adherence to standards for the recording and reporting of microarray-based gene expression data.
OPMD serves as a public repository for microarray experiments on oral pathogens. It was constructed based on the Longhorn Array Database (LAD) (13)—an open source version of the Stanford Microarray Database (SMD) (14). OPMD stores two-color raw and normalized microarray data as well as their corresponding image files, which can be viewed online. The data are compliant with the ‘minimum information about a microarray experiment’ (MIAME) standard (15). OPMD also provides interfaces for data retrieval, analysis and visualization. Analysis tools in OPMD are specifically designed to process the oral pathogen microarray data. For example, the Significance Analysis of Oral Pathogen Microarray Data (SAOPMD) can accept microarray data from two versions of P.gingivalis microarrays that have been manufactured and distributed to the research community by TIGR. Users can upload multiple array data files (currently accepts two-color data file in GenePix format) for a two-condition experiment. Data are first normalized within each slide, then between slides, and repeated data for genes of the same ID from multiple slides are grouped together for statistics significance analysis. Results are presented to the users in plain and hyper-linked text, as well as in Excel format for downloading.
THE BROP WEBSITE
In addition to tools and data described above, BROP also provides links and information relevant to bioinformatics researches on oral pathogens. The BROP web site was constructed based on a content management system (CMS)—PostNuke (http://www.postnuke.org), and provides additional features such as discussion forum for the research community. The versatile users and groups management system of PostNuke provide ideal usage monitoring as well as controlling the accessibility of any subset of data or information. This is helpful when certain data are not yet ready to be accessed by general public (1). For the data that are still in private domain (e.g. unfinished sequences and their annotations), users need to apply for an account at BROP and obtain permission to access the data. The universal resource locator (URL) for BROP website is http://www.brop.org.
CONCLUSIONS
Genomic sequences have provided a plethora of information to the scientific community and have profoundly advanced our understanding of biology. As genome sequencing technologies have become more efficient and affordable, more and more genomes have been or are being sequenced by many institutes (http://www.genomesonline.org). While this is all very encouraging, this information avalanche often proves daunting to biologists for there are great difficulties encountered in searching, retrieving, interpreting or managing the data. The multiple sources of the data representing the same genomic entity, as described in this report, make the task even tougher. BROP is a suite of software tools and databases that originated from the daily and practical needs of a group of biologists at our institute who study the oral pathogens. Quite frequently genomic data are available, but at scattered locations and without proper tools for analyzing data from different sources or in different formats. BROP provides integrated and updated genomic information which will help biologists access and understand the genomic data. Although the focus of BROP is on oral pathogens, these concepts can be readily applied to bioinformatics software design for other organisms.
Acknowledgments
We thank Drs Floyd Dewhirst, Margaret Duncan, Jacques Izard and Mark Maiden of The Forsyth Institute for valuable comments and suggestions, which often were turned into new or improved features in BROP. We also thank Mr Ronald Sutherland and Dr Douglas B. Hanson of the Office of Computing and Network Technology at The Forsyth Institute for their assistance. This work was supported by the NIDCR grant K22 DE14742. Funding to pay the Open Access publication charges for this article was provided by NIDCR.
Conflict of interest statement. None declared.
REFERENCES
- 1.Duncan M.J. Genomics of oral bacteria. Crit. Rev. Oral Biol. Med. 2003;14:175–187. doi: 10.1177/154411130301400303. [DOI] [PubMed] [Google Scholar]
- 2.Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L. GenBank: update. Nucleic Acids Res. 2004;32:D23–D26. doi: 10.1093/nar/gkh045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kulikova T., Aldebert P., Althorpe N., Baker W., Bates K., Browne P., van den Broek A., Cochrane G., Duggan K., Eberhardt R., et al. The EMBL Nucleotide Sequence Database. Nucleic Acids Res. 2004;32:D27–D30. doi: 10.1093/nar/gkh120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miyazaki S., Sugawara H., Ikeo K., Gojobori T., Tateno Y. DDBJ in the stream of various biological data. Nucleic Acids Res. 2004;32:D31–D34. doi: 10.1093/nar/gkh127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen T., Hosogi Y., Nishikawa K., Abbey K., Fleischmann R.D., Walling J., Duncan M.J. Comparative whole-genome analysis of virulent and avirulent strains of Porphyromonas gingivalis. J. Bacteriol. 2004;186:5473–5479. doi: 10.1128/JB.186.16.5473-5479.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanehisa M. The KEGG database. Novartis Found Symp. 2002;247:91–101. Discussion 101–103, 119–128, 244–152. [PubMed] [Google Scholar]
- 7.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boeckmann B., Bairoch A., Apweiler R., Blatter M.C., Estreicher A., Gasteiger E., Martin M.J., Michoud K., O'Donovan C., Phan I., et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zdobnov E.M., Apweiler R. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 11.Bairoch A. The ENZYME database in 2000. Nucleic Acids Res. 2000;28:304–305. doi: 10.1093/nar/28.1.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Camon E., Magrane M., Barrell D., Binns D., Fleischmann W., Kersey P., Mulder N., Oinn T., Maslen J., Cox A., et al. The Gene Ontology Annotation (GOA) project: implementation of GO in SWISS-PROT, TrEMBL, and InterPro. Genome Res. 2003;13:662–672. doi: 10.1101/gr.461403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Killion P.J., Sherlock G., Iyer V.R. The Longhorn Array Database (LAD): an open-source, MIAME compliant implementation of the Stanford Microarray Database (SMD) BMC Bioinformatics. 2003;4:32. doi: 10.1186/1471-2105-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sherlock G., Hernandez-Boussard T., Kasarskis A., Binkley G., Matese J.C., Dwight S.S., Kaloper M., Weng S., Jin H., Ball C.A., et al. The Stanford Microarray Database. Nucleic Acids Res. 2001;29:152–155. doi: 10.1093/nar/29.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brazma A., Hingamp P., Quackenbush J., Sherlock G., Spellman P., Stoeckert C., Aach J., Ansorge W., Ball C.A., Causton H.C., et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nature Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]