Abstract
The WWW servers at http://www.icgeb.org/protein/ are dedicated to the analysis of protein 3D structures submitted by the users as the Protein Data Bank (PDB) files. CX computes an atomic protrusion index that makes it possible to highlight the protruding atoms within a protein 3D structure. DPX calculates a depth index for the buried atoms and makes it possible to analyze the distribution of buried residues. CX and DPX return PDB files containing the calculated indices that can then be visualized using standard programs, such as Swiss-PDBviewer and Rasmol. PRIDE compares 3D structures using a fast algorithm based on the distribution of inter-atomic distances. The options include pairwise as well as multiple comparisons, and fold recognition based on searching the CATH fold database.
INTRODUCTION
The advent of structural genomics initiatives has led to an increase in the number of protein 3D structures and hence there is a growing need for novel analysis tools (1–3). Maintenance of the various analysis programs on changing computer platforms is becoming a problem for many users. The Protein tools page at ICGEB is a collection of locally developed methods designed to assist users in the analysis of 3D structures. The underlying algorithms are designed to be simple and fast. Therefore, they are particularly suited for online use and for large-scale data management. All the three servers described here were written as standard C programs with PHP front end and run on a Beowulf type Linux cluster. The servers accept the Protein Data Bank (PDB) files (4), a description of the input/output options as well as the underlying theory is provided in the form of online help files.
The CX server is a visualization tool designed to highlight protruding atoms within a protein structure. Identification of protruding, or highly convex regions in proteins is relevant to the analysis of interfaces in protein–protein complexes, in the prediction of limited proteolysis cleavage sites and in the identification of possible antigenic determinant regions. CX (1–3,5) calculates the ratio between the external volume and the volume occupied by the protein within a sphere centered at every protein atom. Atoms in protruding regions will have a high ratio between the external and the internal volume, i.e. a high cx protrusion index. For protein structures, cx values can vary between 0 and 15. Only two independent parameters are used by CX: the average atomic volume and the sphere radius. The default value for the average atomic volume used by CX is set to 20.1 Å3. Given the approximate nature of the method and its purposes, slight variations in the average atomic volume do not affect the results in a remarkable way. The choice of the second parameter, the sphere radius, is rather empirical. Smaller values of R will make CX more sensitive to the local environment, whereas larger values will make it more sensitive to the global shape of the protein. The default radius used by CX (10 Å) is a good compromise to highlight both backbone and side chain protruding atoms in most applications (Figure 1A).
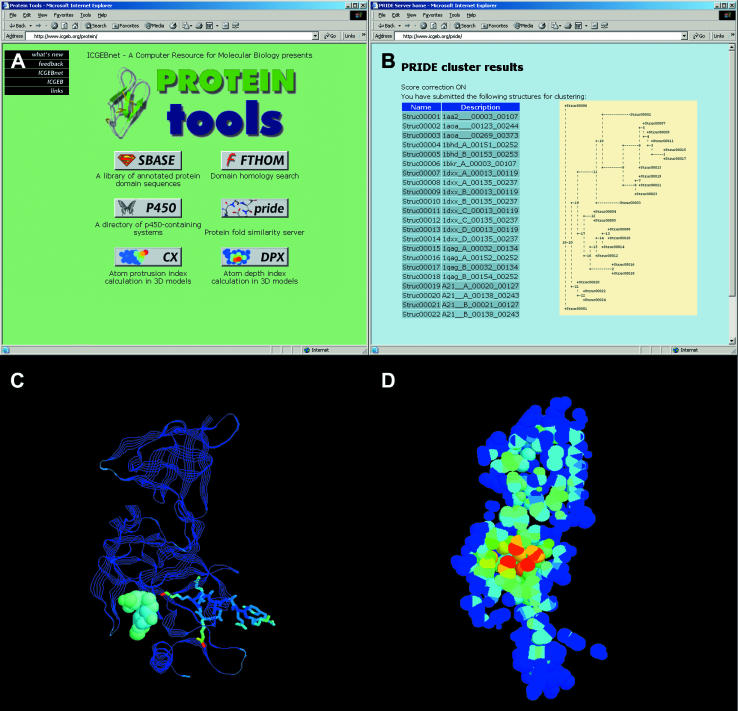
Figure 1.
The Protein tools server. (A) Title page [The SBASE (15), FTHOM (16) and P450 (17) services have been described elsewhere]. (B) Clustering of 24 CH domains by the PRIDE server. The tree is an ASCII rendering of the Newick file produced by the server. Bottom: structure of the human histone lysine N-methyltransferase SET7/9 complexed with a histone peptide and S-adenosyl-l-homocysteine (SAH), PDB: 1O9S. (C) Output of the CX server rendered with Rasmol (5). The enzyme is shown as ribbons, the peptide as sticks and SAH as a CPK model using; the structure is colored according to the cx values, calculated using a sphere radius of 8 Å. (D) Output of the DPX server rendered with Rasmol (5), The CPK model of the enzyme is shown in slab mode in the same orientation as in the left panel, and atoms colored according to their dpx values.
The DPX server is designed to facilitate the analysis of buried atoms within the protein interior. Parameters, such as the solvent accessible area (6) and the occluded surface, cannot distinguish buried residues that are close to the protein surface from those that are deep inside the protein core. Depth defined as the distance between a protein atom and the nearest water molecule surrounding the protein (7) was found to be a useful descriptor of the protein interior. Depth correlates better than solvent accessibility not only with amide H/D exchange rates for several proteins, but also with the difference in the thermodynamic stability of proteins containing cavity-creating mutations and with the change in the free energy formation of protein–protein complexes (8). We have developed the DPX index defined as the distance (Å) of a non-hydrogen buried atom from its closest solvent accessible protein neighbor (9,10) where buried and accessible atoms are identified using the rolling sphere algorithm. Although some information is lost for surface atoms (all solvent exposed atoms have dpx = 0 by default), the depth calculation is very fast because neither water molecules nor surface dots are explicitly considered. The only parameter that can be varied is the radius of the probe sphere, for which the default value is set to 1.4 Å (Figure 1B).
Both CX and DPX read ATOM lines from a PDB file submitted by the user. Non-standard residues, cofactors, metal ions and water molecules described in HETATM lines are not taken into account. Each chain in the PDB file is treated as an independent molecule but the results are written into a single output file in the PDB format, in which the cx or dpx values are written in place of the atomic displacement parameters (B-factors). The output file can thus be displayed using molecular graphics programs [e.g. Rasmol (11) and Swiss-PDBviewer (12)], and atoms colored according to their cx (or dpx) value. Mean residue cx (or dpx) values are also calculated.
The PRIDE server is designed to compare the fold (backbone conformation) of protein structures [for a review, see Carugo and Pongor (2) and the Database Issue 2005 of Nucleic Acids Research for current references]. PRIDE is based on comparing distributions of intramolecular Cα–Cα distances using a standard statistical process, contingency table analysis which gives a probability of identity or PRIDE score (13). For the calculation, the protein 3D structure is represented by 28 different Cα(i) − Cα(i + n) distance distributions (3 < n < 30) and the final PRIDE score for two protein structures is the average calculated from the results of the 28 comparisons (0 ≤ PRIDE ≤ 1). The calculation is extremely fast, so pairwise as well as multiple comparisons can be compared online. As PRIDE is a metric, it can be used to cluster and classify protein 3D structures through standard cluster analysis methods. As the calculation is based only on the Cα atoms, the input files may contain only the Cα lines. The PRIDE pair option compares two structures. Its output contains not only the final PRIDE score, but also the values it was derived from as well as a graphic representation of the underlying histograms. In case the PRIDE cluster option is used to analyze n protein 3D structures (presented as concatenated PDB files), the server provides three, easily downloadable output files: (i) the n × n square matrix where each i-th−j-th element is the distance, defined as 1-PRIDE, between the i-th and the j-th protein 3D structures; (ii) the dendrogram that summarizes a cluster analysis performed using the neighbor program of the PHYLIP software suite by applying the neighbor-joining criterion for cluster merging (Figure 1); and (iii) a Newick-format tree description that allows one to build its own dendrograms with the help of programs, such as njplot (http://pbil.univ-lyon1.fr/software/njplot.html) and TreeView (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html). In case the PRIDE/scan option is used, the database search option of the server makes it possible to compare a 3D structure with the folds of the CATH database (14). The search results are presented as a ranked list, and according to the statistical evaluation, in over 99.5 of the cases the most similar structure points to the correct topology group.
Acknowledgments
Funding to pay the Open Access publication charges for this article was provided by the International Centre for Genetic Engineering and Biotechnology, Trieste, Italy.
Conflict of interest statement. None declared.
REFERENCES
- 1.Domingues F.S., Koppensteiner W.A., Sippl M.J. The role of protein structure in genomics. FEBS Lett. 2000;476:98–102. doi: 10.1016/s0014-5793(00)01678-1. [DOI] [PubMed] [Google Scholar]
- 2.Carugo O., Pongor S. Recent progress in protein 3D structure comparison. Curr. Protein Pept. Sci. 2002;3:441–449. doi: 10.2174/1389203023380530. [DOI] [PubMed] [Google Scholar]
- 3.Carugo O., Pongor S. The evolution of structural databases. Trends Biotechnol. 2002;20:498–501. doi: 10.1016/s0167-7799(02)02082-6. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein F.C., Koetzle T.F., Williams G.J., Meyer E.F., Jr, Brice M.D., Rodgers J.R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J. Mol. Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 5.Pintar A., Carugo O., Pongor S. CX, an algorithm that identifies protruding atoms in proteins. Bioinformatics. 2002;18:980–984. doi: 10.1093/bioinformatics/18.7.980. [DOI] [PubMed] [Google Scholar]
- 6.Lee B., Richards F.M. The interpretation of protein structures. Estimation of static accessibility. J. Mol. Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 7.Pedersen T.G., Sigurskjold B.W., Andersen K.V., Kjaer M., Poulsen F.M., Dobson C.M., Redfield C. A nuclear magnetic resonance study of the hydrogen-exchange behaviour of lysozyme in crystals and solution. J. Mol. Biol. 1991;218:413–426. doi: 10.1016/0022-2836(91)90722-i. [DOI] [PubMed] [Google Scholar]
- 8.Chakravarty S., Varadarajan R. Residue depth: a novel parameter for the analysis of protein structure and stability. Struct. Fold Des. 1999;7:723–732. doi: 10.1016/s0969-2126(99)80097-5. [DOI] [PubMed] [Google Scholar]
- 9.Pintar A., Carugo O., Pongor S. DPX: for the analysis of the protein core. Bioinformatics. 2003;19:313–314. doi: 10.1093/bioinformatics/19.2.313. [DOI] [PubMed] [Google Scholar]
- 10.Pintar A., Carugo O., Pongor S. Atom depth as a descriptor of the protein interior. Biophys. J. 2003;84:2553–2561. doi: 10.1016/S0006-3495(03)75060-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sayle R.A., Milner-White E.J. RASMOL: biomolecular graphics for all. Trends Biochem. Sci. 1995;20:374. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- 12.Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 13.Carugo O., Pongor S. Protein fold similarity estimated by a probabilistic approach based on Cα–Cα distance comparison. J. Mol. Biol. 2002;315:887–898. doi: 10.1006/jmbi.2001.5250. [DOI] [PubMed] [Google Scholar]
- 14.Pearl F., Todd A., Sillitoe I., Dibley M., Redfern O., Lewis T., Bennett C., Marsden R., Grant A., Lee D., et al. The CATH Domain Structure Database and related resources Gene3D and DHS provide comprehensive domain family information for genome analysis. Nucleic Acids Res. 2005;33:D247–D251. doi: 10.1093/nar/gki024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vlahovicek K., Kajan L., Agoston V., Pongor S. The SBASE domain sequence resource, release 12: prediction of protein domain-architecture using support vector machines. Nucleic Acids Res. 2005;33:D223–D225. doi: 10.1093/nar/gki112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murvai J., Vlahovicek K., Barta E., Parthasarathy S., Hegyi H., Pfeiffer F., Pongor S. The domain-server: direct prediction of protein domain-homologies from BLAST search. Bioinformatics. 1999;15:343–344. doi: 10.1093/bioinformatics/15.4.343. [DOI] [PubMed] [Google Scholar]
- 17.Fabian P., Degtyarenko K.N. The directory of P450-containing systems in 1996. Nucleic Acids Res. 1997;25:274–277. doi: 10.1093/nar/25.1.274. [DOI] [PMC free article] [PubMed] [Google Scholar]