Abstract
Regulation of post-transcriptional gene expression by microRNAs (miRNA) has so far been validated for only a few mRNA targets. Based on the large number of miRNA genes and the possibility that one miRNA might influence gene expression of several targets simultaneously, the quantity of ribo-regulated genes is expected to be much higher. Here, we describe the web tool MicroInspector that will analyse a user-defined RNA sequence, which is typically an mRNA or a part of an mRNA, for the occurrence of binding sites for known and registered miRNAs. The program allows variation of temperature, the setting of energy values as well as the selection of different miRNA databases to identify miRNA-binding sites of different strength. MicroInspector could spot the correct sites for miRNA-interaction in known target mRNAs. Using other mRNAs, for which such an interaction has not yet been described, we discovered frequently potential miRNA binding sites of similar quality, which can now be analysed experimentally. The MicroInspector program is easy to use and does not require specific computer skills. The service can be accessed via the MicroInspector web server at http://www.imbb.forth.gr/microinspector.
INTRODUCTION
Micro RNAs (miRNA) are a class of genome-encoded small, single-stranded RNAs of ∼20 nt that are negative regulators of gene expression. Discovered three years ago (1–3), miRNAs have attracted a lot of attention and a large number of recent reviews summarize the biogenesis, phylogenetic relation and function of miRNAs, which can be found in animals and plants (4–11). MiRNAs operate by base-pairing interactions with an mRNA target. However, perfect sequence complementarity to an miRNA is observed only for some plant mRNAs (12), but in the majority of residual cases, including the first identified miRNA target pairs (13), the base-pairing interaction between the mRNA target and the riboregulator is imperfect. There seems to be a preference for a strong interaction at the 5′ side of the miRNA (14) and a symmetrical interaction is preferred (15), and most likely, the RNA–RNA interaction requires assistance of protein factors. Collectively, >1500 miRNAs have been identified so far for plants, nematodes, insects and mammals. This large number of recognized miRNAs contrasts with only a few dozen of target RNAs, for which a regulatory miRNA binding has been experimentally verified. Some miRNAs are expected to form regulatory networks controlling several mRNA targets. Lai (16) has found that some short sequence elements (boxes) that had been previously recognized as negative modulators of translational gene expression are actually binding sites for certain classes of miRNAs. For example, the K box is negatively regulating gene expression in several gene families, which are involved in early developmental processes in Drosophila melanogaster and at least four miRNAs (miR2, miR6, miR11 and miR13) are at their 5′ end complementary to the K box. However, not every miRNA of the K-box family will bind to each K box containing mRNA, suggesting that at least some subsets of miRNAs are composed of at least two modular elements, which we had termed ‘first name’ and ‘family’ motif (17). Several attempts have been made to identify miRNA targets by bioinformatics (18–22). In Arabidopsis thaliana, this approach was quite successful, since plant miRNAs seem to base-pair with higher stringency (23,24). For animal miRNAs and especially for mammalian miRNAs, this computational strategy will only identify those mRNA targets that have a high degree of sequence complementarity. However, some of the genetically verified miRNA/mRNA interactions (13,25) are not particularly strong in terms of RNA–RNA interaction. On the other hand, if one allows weak interactions, the number of false positive hits will raise in computational screens. Brennecke and Cohen (26) have addressed these difficulties by incorporation of phylogenetic parameters into the computer algorithm, which improves target identification.
Here, we describe a different computational approach to identify miRNA/mRNA interactions. Whereas most programs available start with a specific miRNA and attempt the identification of as many mRNA targets as possible, we ask a different and more modest question by analysing whether, in a given mRNA sequence a binding site can be found for any miRNA that originates from this organism and that is available in the database. The MicroInspector program will generate a list of possible target sites, sorted by free energy values. Adaptation of temperature and free energy settings, followed by visual inspection of secondary structures allows a detailed analysis. This approach allows more detailed examination of an mRNA sequence, identifying also weaker interactions, which can then be subjected to experimental tests. Several mRNAs that contain validated miRNA binding sites were subjected to analysis by the MicroInspector software, and all these interactions could be identified. However, in many other cases, we identified so far non-described interactions with lower energy values than those of the validated targets, suggesting that many more miRNAs/mRNA interactions are likely to exist. Their biological relevance requires subsequent experimental validation.
Usage of the program
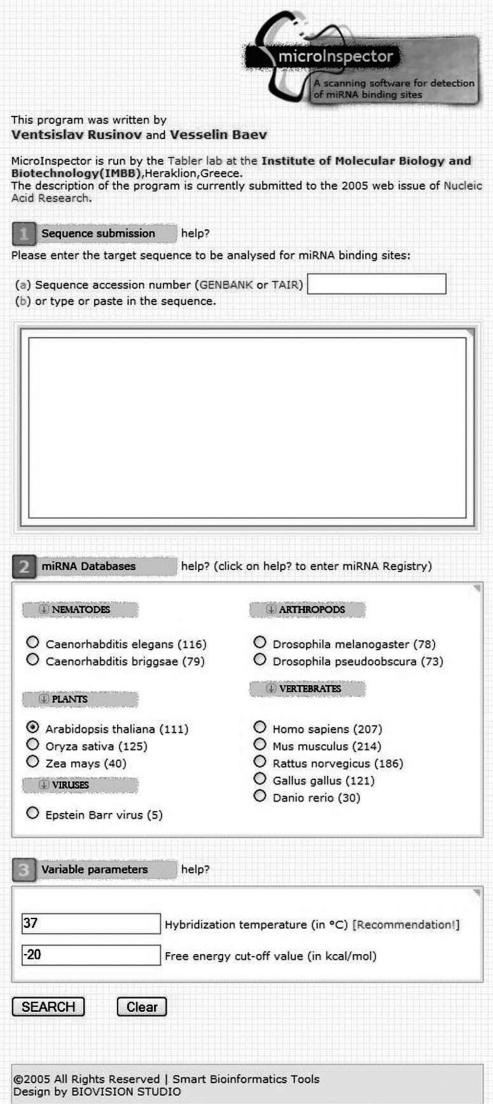
MicroInspector is a web-based tool for searching miRNA binding sites in a target RNA sequence, potentially regulated by such a small RNA. The interface of the program is given in Figure 1. The user needs to follow a few simple steps to perform a quest for potential miRNA binding sites. The first step is ‘entering the sequence’ to be analysed, which is typically an mRNA (the program treats DNA sequences as RNA). This can be done in two ways, either by providing the GenBank or TAIR accession number or by simply typing or pasting in the sequence (the program is designed that all gaps, numbers and non-defined characters will be ignored), which is useful for the analysis of unknown sequences or for detailed analysis of certain mRNA domains, e.g. 3′-untranslated regions (3′-UTRs).
Figure 1.
The MicroInspector interface. The user has to enter three categories of input parameters for scanning a target RNA for miRNA binding sites. There is a help pop-up window with brief explanations for each of the data fields.
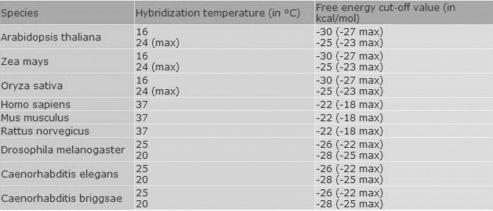
As a next step, the user needs to set a ‘hybridization temperature’: the default is 37°C, but evidently this value is not relevant for plants and insects, for which we recommend the values in Figure 2. Further, a value for the ‘free energy’ cut-off needs to be entered (default −20 kcal/mol), which characterizes the stability of the miRNA/mRNA interaction. Only results with lower energy than the cut-off value will be displayed, so that this parameter will influence the number of hits. The energy value should be varied in accordance with the temperature according to Figure 2. As an indication, it might be helpful to add that the free energies of validated miRNA/mRNA interactions range from −17 kcal/mol (bantam/hid5 at 25°C—Drosophila melanogaster) to −41 kcal/mol (CUC/miR164 at 25°C—Arabidopsis thaliana).
Figure 2.
Recommended settings for hybridization temperatures (°C) and corresponding free energy cut-off values in kcal/mol for different species.
Finally, the user needs to select an ‘miRNA database’, matching the biological origin of the target sequence. These local miRNA databases (in multifasta format) are based on entries of ‘the miRNA registry’ (http://www.sanger.ac.uk/Software/Rfam/mirna/index.shtml). Unless automatic retrieving of new miRNA entries will be possible, we will update the databases manually in regular intervals.
Principle of the program
Initial scanning and filtering
The user-defined target sequence is analysed for every miRNA sequence of the chosen database in a consecutive manner. The target sequence is scanned simultaneously and independently with two windows of 6 nt. The first 6-nt window represents nucleotides 1–6 (from the 5′ of the miRNA), and the second window nucleotides 2–7. They are slid through the target sequence (by steps of 1 nt) and the program performs analysis of complementarity. It is known that pairing to the 5′ portion of the miRNA, particularly nucleotides 2–7, appears to be most important for target recognition by vertebrate miRNAs. The most 5′-terminal miRNA nucleotide may or may not participate in binding.
A complementarity pre-filter seeks for each of the two 6-nt windows for domains having 5 Watson–Crick base pairs or 4 Watson–Crick base pairs with at least one additional G:U pair. If neither of the two windows fulfil this requirement, the data are ignored and the 6-nt windows are moved by 1 nt towards the 5′ end of the mRNA. When the sequence analysis identifies at least one 6-nt window as described above, the program will initiate a detailed analysis of this site. It extracts a 32-nt sequence of the mRNA terminating at the nucleotide that matches the 5′ end of the miRNA, i.e. the 5′-terminal nucleotide of the first 6-nt window. Subsequently, the miRNA sequence and the 32-nt potential target sequence domain are subjected to a pair-wise hybridization folding algorithm.
Dynamic hybridization and folding algorithm
MicroInspector uses a dynamic algorithm for the primary window alignment that is based on the complementarity of nucleotides—it allows Watson–Crick and G:U wobble basepairs. For calculation of thermodynamic properties of a predicted duplex in the algorithm, we integrated some folding routines from the Vienna RNA secondary structure programming library (RNAlib) from the Vienna RNA 1.5 version package (27,28) (see http://www.tbi.univie.ac.at/~ivo/RNA/RNAlib.html), which itself makes use of the RNA energy parameters of the Turner laboratory (29) (http://rna.chem.rochester.edu/).
This folding analysis will reveal the free energy, as well as the secondary structure of this RNA–RNA interaction. We chose a limit of 32 nt, because most miRNA–mRNA interactions will cover a smaller region than this. Therefore, only few significant hits are likely to be missed, in cases where longer binding domains are present. Hits below the selected threshold value for the free energy will be saved and subjected to a post-filter analysis.
Post-filter—2D analysis
The second filter of the program can discard binding sites that do not fit known features of miRNA–mRNA duplexes. This filter inspects the RNA–RNA structure after folding, and eliminates any hit characterized by two unpaired nucleotides on either the 5′ or the 3′ side of the miRNA sequence. The filter will also exclude structures with low folding energy values that are the result of self-complementarity in one of the two RNA strands. For example, this applies when the target domain forms an intramolecular hairpin. Further, entries will be eliminated if too large interior or bulge loops are predicted, or if large loops are located too close to the end of the secondary structure (>10 unpaired nucleotides). Central interior loops will be tolerated even if the loop size is large.
Output of the program
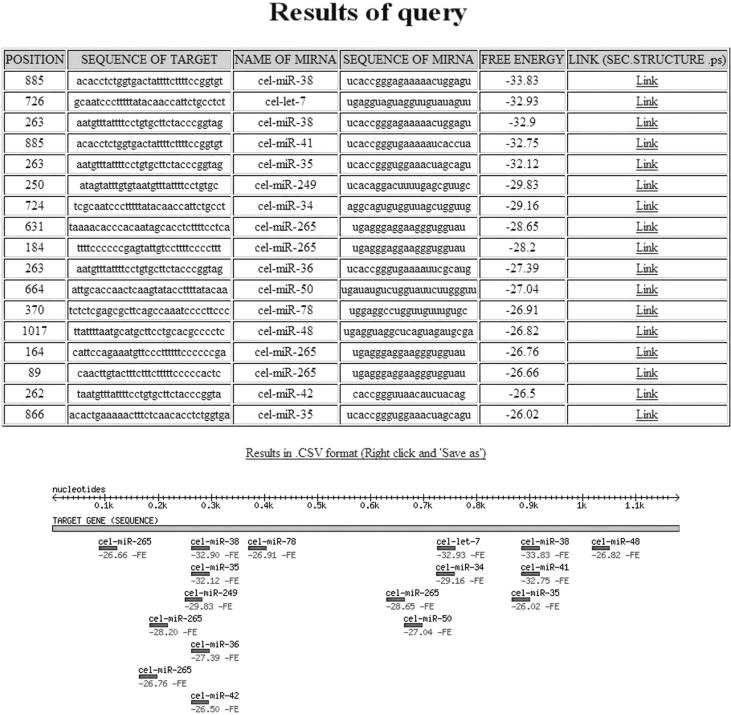
To illustrate the output given by MicroInspector, we present as an example an analysis of the miRNA binding sites for the 3′-UTR sequence of the Caenorhabditis elegans gene lin-41, which is known to interact with miRNA let7 (Entry name 3CEL000914 3′-UTR in Caenorhabditis elegans LIN41A (lin41A) mRNA, complete cds, from LION SRS database).
The main results of this MicroInspector query are represented as a table (see example in Figure 3). The first column of the table lists the ‘position’ of the 5′ end of the binding-site in the target RNA. The second column indicates the ‘target RNA name’ (accession number) which can be used as a link to access the sequence entry of the GenBank database. This column will be empty if the sequence has been entered by typing or pasting in. The third column indicates the ‘target sequence’ (capital letters) of the domain potentially interacting with the miRNA, followed by the ‘miRNA name’ (according to ‘the miRNA registry’) and the ‘miRNA sequence’ (lowercase letters) of the matching miRNA in columns four and five. Both sequences are given 5′ to 3′.
Figure 3.
Example of a data output of a MicroInspector analysis seeking for miRNA binding sites in the 3′-UTR sequence of the Caenorhabditis elegans gene lin-41. Please note that the verified interaction of miRNA let-7 is identified at position 726. In addition, the program identifies other interactions, including the interaction with miR-38 (top result on table), which is stronger than the interaction with let-7. The significance of each identified interaction can be analysed by activating the link that will display the secondary structure of the specific interaction as demonstrated in Figure 4; for further details see text.
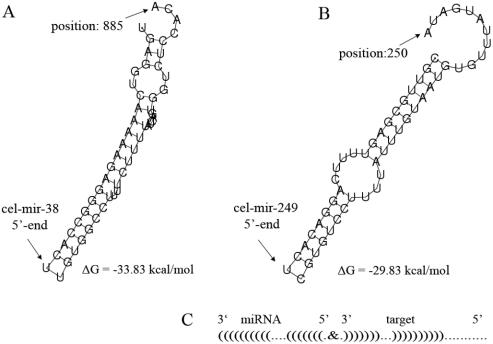
In the ‘free energy’ column the Gibbs free energy (ΔG) of the duplex structure is indicated in kcal/mol. Entries are sorted by free energy (lowest values on top). However, the ΔG value is not the only characteristic feature of a good binding site. For example, a longer miRNA, or a miRNA that is rich in GC, is more likely to yield predicted low energy binding sites. Also the symmetry of binding is an important factor, as is the stability of the base-pairing at the 5′ end of the miRNA. These restrictions require a detailed manual inspection of a particular binding site. For this reason, the rightmost column contains a link to the graphics (PostScript format) displaying the secondary structure of the actual RNA–RNA interaction as exemplified in Figure 4. Inspection of the individual structures revealed that the binding site of miR-38 (top of the list in Figure 3) might not be functional despite its low free energy (Figure 4A), while the interaction with miR-249 (number 6 on the list of Figure 3) results a in symmetrical RNA–RNA interaction (Figure 4B) that is likely to be biologically relevant.
Figure 4.
Representation of pair-wise interaction between miRNA and mRNA target. Examples of secondary structure graphics that can be displayed when the link of the right column in the result table (see Figure 3) is activated. (A) This specific example displays the predicted interaction of miR-38 with lin-41 (top result in table of Figure 3), which shows that the interaction of the miRNA and the target mRNA is restricted to the 5′ side of the miRNA; at the 3′ side the interaction is rather weak. Despite the low ΔG value, this interaction might not be functional. (B) The interaction of miR-249 with lin-41 is symmetrical and more likely to be relevant. (C) The same interaction as in (B) in a schematic representation; this simplified illustration of an RNA–RNA interaction is used in the downloadable results file ‘Results in .CSV format’ (see Figure 3).
The MicroInspector program also offers the download of the results as a single file for off-line analysis. A link to the result file is located at the bottom of the table—‘Results in .CSV format’. The file format ‘Comma separated value’ can be imported into Excel tables. The result file contains additional helpful information such as the date of analysis, the filename of the secondary structure graph and a schematic representation of the secondary structure of the duplex as shown in Figure 4C.
At the very bottom of the result page, the positions of the binding sites of the miRNAs with respect to the mRNA target are shown as an overview. Every potential interaction lists the name of the miRNA and the binding strength (ΔG value). If binding sites overlap, the potential interactions will be sorted so that those with the lowest free energy are on top.
Implementation (computer data)
The program is implemented as a Perl CGI-script, taking advantage of the modular design, allowing the use of specialized packages such as BioPerl (modules for developers of Perl-based software for life science research). The program was tested on a PC with an Intel Pentium IV processor 2.8 GHz and 1 GB RAM memory. The operation system is Fedora Core 2.0 by Red Hat Linux. The versions used are 5.8.5 for Perl (www.perl.com) and version 1.4 for BioPerl (www.bioperl.org). The access to the multi-fasta format sequence files and to the online databases is accomplished by the BioPerl modules. The results and all additional pieces of information are saved in a mySQL database for each session. The tables and files with the secondary structures will remain available for 3 days after the researcher's query. Every target analysis is loaded in an individual table in the corresponding mySQL database.
Acknowledgments
We thank Viktor Ivanov (University of Plovdiv) for the graphic design of the site. V.R. and V.B. have been supported by the European Union (EU) via Marie Curie training fellowships (contract HPMT-CT-2000-00175) and are currently supported in the same program under contract EST-7295-FAMED. Further, this work was supported in parts by grants to I.M. by the projects G3-02 and K1202/02 of the Bulgarian National Science Council and to M.T. by the General Secretariat for Research and Technology of the Hellenic Ministry of Development via the Bulgarian-Greek cooperation program (PN18/3-1-2003) and by the European Union FP6-2003-LIFESCIHEALTH-I program, within project FOSRAK (contract LSH-CT-2004-005120). The Open Access publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 2.Lau N.C., Lim L.P., Weinstein E.G., Bartel D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 3.Lee R.C., Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 4.Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- 5.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–676. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 6.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 7.He L., Hannon G.J. MicroRNAs: small RNAs with a big role in gene regulation. Nature Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 8.Nelson P., Kiriakidou M., Sharma A., Maniataki E., Mourelatos Z. The microRNA world: small is mighty. Trends Biochem. Sci. 2003;28:534–540. doi: 10.1016/j.tibs.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 9.Murchison E.P., Hannon G.J. miRNAs on the move: miRNA biogenesis and the RNAi machinery. Curr. Opin. Cell. Biol. 2004;16:223–229. doi: 10.1016/j.ceb.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 10.He Z., Sontheimer E.J. ‘siRNAs and miRNAs’: a meeting report on RNA silencing. RNA. 2004;10:1165–1173. doi: 10.1261/rna.7900204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lai E.C. microRNAs: runts of the genome assert themselves. Curr. Biol. 2003;13:R925–R936. doi: 10.1016/j.cub.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 12.Reinhart B.J., Weinstein E.G., Rhoades M.W., Bartel B., Bartel D.P. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 14.Doench J.G., Sharp P.A. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiriakidou M., Nelson P.T., Kouranov A., Fitziev P., Bouyioukos C., Mourelatos Z., Hatzigeorgiou A. A combined computational-experimental approach predicts human microRNA targets. Genes Dev. 2004;18:1165–1178. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lai E.C. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nature Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 17.Boutla A., Delidakis C., Tabler M. Developmental defects by antisense-mediated inactivation of micro-RNAs 2 and 13 in Drosophila and the identification of putative target genes. Nucleic Acids Res. 2003;31:4973–4980. doi: 10.1093/nar/gkg707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adai A., Johnson C., Mlotshwa S., Archer-Evans S., Manocha V., Vance V., Sundaresan V. Computational prediction of miRNAs in Arabidopsis thaliana. Genome Res. 2005;15:78–91. doi: 10.1101/gr.2908205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai E.C., Tomancak P., Williams R.W., Rubin G.M. Computational identification of Drosophila microRNA genes. Genome Biol. 2003;4:R42. doi: 10.1186/gb-2003-4-7-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim L.P., Lau N.C., Weinstein E.G., Abdelhakim A., Yekta S., Rhoades M.W., Burge C.B., Bartel D.P. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Enright A.J., John B., Gaul U., Tuschl T., Sander C., Marks D.S. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.John B., Enright A.J., Aravin A., Tuschl T., Sander C., Marks D.S. Human MicroRNA Targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bartel D.P., Chen C.Z. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nature Rev. Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 24.Carrington J.C., Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- 25.Brennecke J., Hipfner D.R., Stark A., Russell R.B., Cohen S.M. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 26.Brennecke J., Cohen S.M. Towards a complete description of the microRNA complement of animal genomes. Genome Biol. 2003;4:228. doi: 10.1186/gb-2003-4-9-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hofacker I.L., Fontana W., Stadler P.F., Bonhoeffer S., Tacker M., Schuster P. Fast folding and comparison of RNA secondary structures. Monatshefte f. Chemie. 1994;125:167–188. [Google Scholar]
- 28.Hofacker I.L. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Serra M.J., Turner D.H. Predicting thermodynamic properties of RNA. Methods Enzymol. 1995;259:242–261. doi: 10.1016/0076-6879(95)59047-1. [DOI] [PubMed] [Google Scholar]