Abstract
Key amino acid positions that are important for maintaining the 3D structure of a protein and/or its function(s), e.g. catalytic activity, binding to ligand, DNA or other proteins, are often under strong evolutionary constraints. Thus, the biological importance of a residue often correlates with its level of evolutionary conservation within the protein family. ConSurf (http://consurf.tau.ac.il/) is a web-based tool that automatically calculates evolutionary conservation scores and maps them on protein structures via a user-friendly interface. Structurally and functionally important regions in the protein typically appear as patches of evolutionarily conserved residues that are spatially close to each other. We present here version 3.0 of ConSurf. This new version includes an empirical Bayesian method for scoring conservation, which is more accurate than the maximum-likelihood method that was used in the earlier release. Various additional steps in the calculation can now be controlled by a number of advanced options, thus further improving the accuracy of the calculation. Moreover, ConSurf version 3.0 also includes a measure of confidence for the inferred amino acid conservation scores.
INTRODUCTION
The degree to which an amino acid position is recessive to substitutions is strongly dependent on its structural and functional importance. An amino acid that plays an essential role, e.g. in enzymatic catalysis, is likely to remain unaltered in spite of the random evolutionary drift. Hence, the level of evolutionary conservation is often indicative of the importance of the position in maintaining the protein's structure and/or function.
ConSurf is a web server for mapping the level of evolutionary conservation of each of the amino acid positions of a protein onto its 3D structure (1). The conservation scores are calculated based on the evolutionary relations among the protein and its homologs and the probability of residue replacement as reflected in amino acid substitution matrices (2,3). The scores are subsequently translated into a discrete coloring scale that is used to project them on a known 3D structure of one of the homologous proteins. The server is implemented in a user-friendly interface that enables scientists from the experimental biology as well as the bioinformatics communities to explore the evolutionary history of a protein of known 3D structure and to identify structurally and functionally important positions. We provide here a brief review of ConSurf with emphasis on the new features that were added recently.
METHODS
A short description of the methodology is provided here and a more detailed description is available at http://consurf.tau.ac.il/, under ‘OVERVIEW’, ‘QUICK HELP’ and ‘FAQ’.
ConSurf protocol
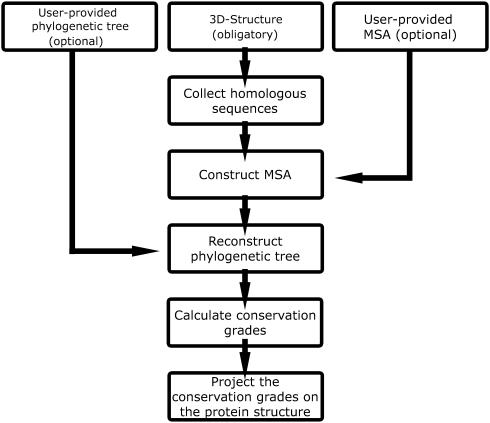
A flow chart, describing the ConSurf protocol, is presented in Figure 1. The minimal input requirement for ConSurf is a four-letter PDB (4) code and the relevant chain identifier of the query protein. Alternatively, a user-provided protein structure in the form of a PDB file can be uploaded. Using the 3D structure of the query protein as an input, the following steps are automatically carried out by ConSurf:
The amino acid sequence is extracted from the PDB file.
Homologous sequences in the SWISS-PROT database (5) are searched and collected using PSI-BLAST (6).
A multiple sequence alignment (MSA) of these sequences is computed using CLUSTAL W (7).
A phylogenetic tree is reconstructed based on the MSA, using the neighbor-joining algorithm (8) as implemented in the Rate4Site program (3).
Position-specific conservation scores are computed using the empirical Bayesian (2) or maximum-likelihood (3) algorithms.
The continuous conservation scores are divided into a discrete scale of 9 grades for visualization purpose. Grade 1 contains the most variable positions and is colored turquoise; grade 5 contains intermediately conserved positions and is colored white; and grade 9 contains the most conserved positions and is colored maroon.
The nine-color conservation grades are projected onto the 3D structure of the query protein.
Figure 1.
A flow chart of a ConSurf calculation.
The sensitivity and selectivity of the search for homologous proteins [step (ii) above] can be controlled by adjusting the number of PSI-BLAST iterations, the PSI-BLAST E-value cut-off and the maximum number of sequences extracted from PSI-BLAST (6). As an alternative to this automatic search, the server accepts a user-provided MSA. In such a case, steps (ii) and (iii) in the outline protocol are skipped.
ConSurf outputs
After the calculation begins, ConSurf produces a status page indicating the computation parameters along with the different stages of the server activity. The main result of a ConSurf calculation is under the link ‘View ConSurf Results with Protein Explorer’, which leads to the graphic visualization of the query protein, color coded by conservation scores, through the Protein Explorer interface (9). The continuous conservation scores of each of the amino acid positions are available under the link ‘Amino Acid Conservation Scores’, along with the color grades and additional data. The script command for viewing the 3D structure of the query protein, color coded by conservation scores, is available under the link ‘RasMol coloring script source’. This file can be downloaded and used locally with the RasMol program (10), thus producing the same color-coded scheme generated by the server. A PDB file, in which the conservation scores are specified in the temperature (B) factor field, can be downloaded through the link: ‘The PDB file updated with the conservation scores in the tempFactor field’. Thus, any 3D protein viewer, such as the RasMol program (10), which is capable of presenting the B factors, is suitable for mapping the conservation scores on the structure.
The ConSurf output also includes links to the PSI-BLAST results, the homologous sequences along with a link to their SWISS-PROT entry page, the MSA and the phylogenetic tree used in the calculation.
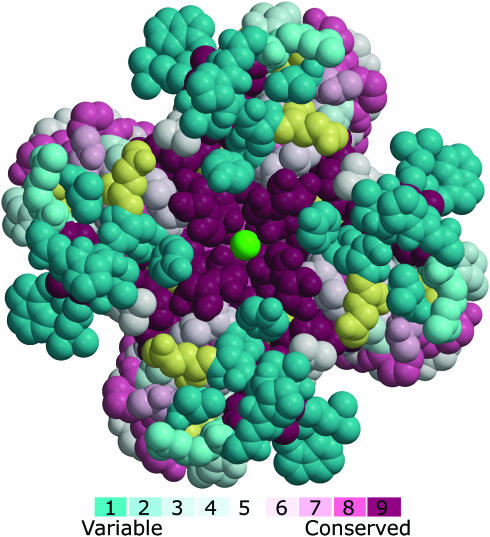
As an example, we provide in Figure 2 the main output of a ConSurf run of the Kcsa potassium-channel (11), a transmembrane protein from Streptomyces Lividans. Kcsa is a homotetramer with a 4-fold symmetry axis about its pore. The ConSurf calculations demonstrate the high level of conservation of the pore region as compared with the rest of the protein. The pore architecture provides the unique stereochemistry which is required for efficient and selective conduction of potassium ions (11). The biological importance of this stereochemistry is reflected by a strong evolutionary pressure to resist amino acid replacements in the pore. In contrast, the regions that surround the pore and face the extracellular matrix are highly variable.
Figure 2.
A ConSurf analysis of the Kcsa potassium channel. The tetrameric channel, which is viewed along the pore from the extracellular end, is presented using a space-filled model. The amino acids are colored by their conservation grades using the color-coding bar, with turquoise-through-maroon indicating variable-through-conserved. Amino acid positions, for which the inferred conservation level was assigned with low confidence, are marked with light yellow. The potassium ion at the channel pore is colored green. Conservation scores, which were calculated for one of the channel's subunits, were projected on the homotetrameric structure. The run was carried out using PDB code 1bl8 (11) and default ConSurf parameters. The picture was generated using MOLSCRIPT (21) and Raster3D (26).
NEW ADDITIONS AND IMPROVEMENTS IN ConSurf
An empirical Bayesian method to score conservation
The heart of the ConSurf server is the calculation of the conservation scores of each amino acid position. In the previous version of the server, the maximum-likelihood method (3) was used as the default option to this end. Recently, we showed that an empirical Bayesian method can significantly improve the accuracy of the estimated conservation scores (2). The empirical Bayesian method is particularly superior to the maximum-likelihood method when the number of homologous sequences analyzed is small (2). The new method is now integrated in ConSurf as the default option. The usage of the maximum-likelihood method is still available under the ‘Method’ pull-down menu.
Estimation of the reliability of the inferred conservation scores
An amino acid position that is conserved across all homologous sequences will always be assigned with the highest conservation grade. Yet, there is a difference if the conservation score is inferred based on a small MSA of, for example, 4 sequences, or based on a larger set of 30 sequences. Additionally, since the conservation calculation for positions with a lot of gaps is based on a fewer number of sequences, the conservation score for these positions will be less reliable than positions that have no gaps. The reliability of the conservation computation is not only determined by the number of sequences in each position but also by the evolutionary distances between the sequences, the phylogenetic tree topology and the evolutionary process.
One of the most important new features in ConSurf version 3.0 is the inclusion of a measure of the confidence of each of the inferred position-specific conservation scores. The measure is calculated using the empirical Bayesian method, as explained in (12) and at http://consurf.tau.ac.il/ under ‘OVERVIEW’. In short, it is based on a confidence interval that is defined by the lower and upper quartiles: the 25th and 75th percentiles of the inferred distribution of conservation scores, respectively. It gives the 50% confidence interval and also indicates the dispersion of each of the estimated scores. The confidence interval is usually large in positions with a small number of sequences, thus indicating a low level of support in the inferred conservation scores for these positions. When the number of sequences is large, the confidence interval is small, and the point score estimates are more assured. Amino acid positions, associated with confidence intervals that are too large to be trustworthy, are marked in the output files of the server and highlighted (in pale yellow) on the 3D structure of the protein (Figure 2).
Models of amino acid substitutions
The inference of the evolutionary conservation scores relies on a specified probabilistic model of amino acid replacements (3). The JTT matrix (13) was used to this end in the previous version of ConSurf. In version 3.0, we expanded the utility of ConSurf to support additional models of substitution for nuclear DNA-encoded as well as non-nuclear DNA-encoded proteins. The model of substitution can be chosen from the ‘Model of substitution for proteins’ pull-down menu, which is available in the ‘Advance Options’ section of the ConSurf main interface. The JTT (13), Dayhoff (14) and WAG (15) matrices are suitable for nuclear DNA-encoded proteins. The WAG matrix has been inferred from a large database of sequences comprising a broad range of protein families, and is thus suitable for distantly related amino acid sequences (15). The mtREV (16) and cpREV (17) matrices are suitable for mitochondrial and chloroplast DNA-encoded proteins, respectively. Examples that demonstrate the influence of using the different matrices on the calculations are available at http://consurf.tau.ac.il/ under ‘OVERVIEW’. The differences between ConSurf calculations using different matrices tend to be small but not negligible.
User-provided phylogenetic tree
A user-provided phylogenetic-tree (that should be consistent with the MSA) may be supplied as an additional input. In this case, steps (ii–iv) in the ‘ConSurf protocol’ (specified above) are skipped. We note that the accuracy of the conservation scores calculations relies on the correct reconstruction of the phylogeny (18). Default ConSurf runs are carried out using phylogenetic trees that are constructed with the neighbor-joining algorithm. The new feature enables the users to supply more accurate trees.
WORK UNDER DEVELOPMENT
We are currently integrating a few more enhancements to ConSurf. At present, ConSurf uses the neighbor-joining algorithm as a fast heuristic method to construct phylogenetic trees. Notwithstanding, the more exhaustive maximum-likelihood tree-reconstruction method is known to produce more accurate phylogenetic trees (19), which should increase the accuracy of the calculated conservation scores (18). We will integrate the maximum-likelihood-based SEMPHY program (20) into ConSurf. This program reconstructs phylogenetic trees dramatically faster than other maximum-likelihood tree-reconstruction methods (20), and can thus be used with little additional computational cost.
A computational tool will be developed, which will enable a simultaneous online view of the phylogenetic tree while analyzing the evolutionary profile of the protein. This interactive tool will allow the user to mark specific branches, which will be used for in-depth ConSurf analyses. For example, the selection of specific clades (sub-trees) may be used to define sub-families. The examination of ConSurf analysis of sub-families may reveal specific characters that are unique to each of them.
The main output of ConSurf is the projection of the conservation scores on the 3D structure of the query protein. In order to easily generate high-resolution color figures, we will provide a script command for the MOLSCRIPT program (21) as an additional output.
A planned enhancement to ConSurf will be the inclusion of all the visualization results in the header of the PDB file. The format that will be used to this end will allow an interactive offline view of the results using Protein Explorer on the user machine, exactly as they appear online.
CONCLUSIONS
ConSurf (1) is a web server that automatically calculates evolutionary conservation scores for each amino acid position and projects them onto the 3D structure of the protein. Evolutionary trace (ET) (22,23) is a related web server that may also be used to map conservation scores on the 3D structure. However, the ET method (23), which was developed for the identification of class-specific residues, is less accurate than ConSurf for scoring conservation (3). This may explain why biologically important regions that were detected using ConSurf were overlooked by the ET web server (1). (See http://consurf.tau.ac.il/, under ‘OVERVIEW’ for details). Other related web servers, such as MSA3D (9), ProtSkin (24) and COLORADO3D (25), may also be used to present conservation scores on protein structures. These web servers use a consensus approach to infer conservation, which is inferior to methods, such as the ET and ConSurf's maximum-likelihood and empirical Bayesian that explicitly take into account the phylogeny of the homologous sequences under study (2,3). Moreover, all the above servers are not fully automated as ConSurf and require a user-provided MSA.
The new version of ConSurf includes an improved algorithm for scoring evolutionary conservation and provides an index of confidence in the estimated scores. In addition, while ConSurf is still easy to use with default options, expert users can benefit from several advanced options that were added in order to provide more control over the calculations and so to increase the accuracy of the results.
Acknowledgments
The authors are grateful to the Bioinformatics Unit and the George S. Wise Faculty of Life Sciences at Tel Aviv University for providing technical assistance and computation facilities. This study was supported by a Research Career Development Award from the Israel Cancer Research Fund. T.P. was supported by a grant in Complexity Science from the Yeshaia Horvitz Association and from an ISF grant no. 1208/04. Development of Protein Explorer is supported by a grant to E.M. from the Division of Undergraduate Education of the US National Science Foundation. Funding to pay the Open Access publication charges for this article was provided by Tel Aviv University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Glaser F., Pupko T., Paz I., Bell R.E., Bechor-Shental D., Martz E., Ben-Tal N. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003;19:163–164. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- 2.Mayrose I., Graur D., Ben-Tal N., Pupko T. Comparison of site-specific rate-inference methods for protein sequences: empirical Bayesian methods are superior. Mol. Biol. Evol. 2004;21:1781–1791. doi: 10.1093/molbev/msh194. [DOI] [PubMed] [Google Scholar]
- 3.Pupko T., Bell R.E., Mayrose I., Glaser F., Ben-Tal N. Rate4Site: an algorithmic tool for the identification of functional regions in proteins by surface mapping of evolutionary determinants within their homologues. Bioinformatics. 2002;18:S71–S77. doi: 10.1093/bioinformatics/18.suppl_1.s71. [DOI] [PubMed] [Google Scholar]
- 4.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bairoch A., Apweiler R. The SWISS-PROT protein sequence data bank and its supplement TrEMBL in 1999. Nucleic Acids Res. 1999;27:49–54. doi: 10.1093/nar/27.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altschul S., Madden T., Schaffer A., Zhang J., Zhang Z., Miller W., Lipman D. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 9.Martz E. Protein Explorer: easy yet powerful macromolecular visualization. Trends Biochem. Sci. 2002;27:107–109. doi: 10.1016/s0968-0004(01)02008-4. [DOI] [PubMed] [Google Scholar]
- 10.Sayle R.A., Milner-White E.J. RASMOL: biomolecular graphics for all. Trends Biochem. Sci. 1995;20:374. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- 11.Doyle D.A., Morais Cabral J., Pfuetzner R.A., Kuo A., Gulbis J.M., Cohen S.L., Chait B.T., MacKinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 12.Susko E., Inagaki Y., Field C., Holder M.E., Roger A.J. Testing for differences in rates-across-sites distributions in phylogenetic subtrees. Mol. Biol. Evol. 2002;19:1514–1523. doi: 10.1093/oxfordjournals.molbev.a004214. [DOI] [PubMed] [Google Scholar]
- 13.Jones D.T., Taylor W.R., Thornton J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 14.Dayhoff M.O., Hunt L.T., Barker W.C., Schwartz R.M., Orcutt B.C. In: Atlas of Protein Sequence and Structure. Young C.L., editor. Washington, DC: National Biomedical Research Foundation; 1978. [Google Scholar]
- 15.Whelan S., Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 16.Adachi J., Hasegawa M. Model of amino acid substitution in proteins encoded by mitochondrial DNA. J. Mol. Evol. 1996;42:459–468. doi: 10.1007/BF02498640. [DOI] [PubMed] [Google Scholar]
- 17.Adachi J., Waddell P.J., Martin W., Hasegawa M. Plastid genome phylogeny and a model of amino acid substitution for proteins encoded by chloroplast DNA. J. Mol. Evol. 2000;50:348–358. doi: 10.1007/s002399910038. [DOI] [PubMed] [Google Scholar]
- 18.Mayrose I., Mitchell A., Pupko T. Site-specific evolutionary rate inference: taking phylogenetic uncertainty into account. J. Mol. Evol. 2005 doi: 10.1007/s00239-004-0183-8. in press. [DOI] [PubMed] [Google Scholar]
- 19.Felsenstein J. Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Methods Enzymol. 1996;266:418–427. doi: 10.1016/s0076-6879(96)66026-1. [DOI] [PubMed] [Google Scholar]
- 20.Friedman N., Ninio M., Pe'er I., Pupko T. A structural EM algorithm for phylogenetic inference. J. Comput. Biol. 2002;9:331–353. doi: 10.1089/10665270252935494. [DOI] [PubMed] [Google Scholar]
- 21.Kraulis P.J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Cryst. 1991;24:946–950. [Google Scholar]
- 22.Innis C.A., Shi J., Blundell T.L. Evolutionary trace analysis of TGF-beta and related growth factors: implications for site-directed mutagenesis. Protein Eng. 2000;13:839–847. doi: 10.1093/protein/13.12.839. [DOI] [PubMed] [Google Scholar]
- 23.Lichtarge O., Bourne H.R., Cohen F.E. An evolutionary trace method defines binding surfaces common to protein families. J. Mol. Biol. 1996;257:342–358. doi: 10.1006/jmbi.1996.0167. [DOI] [PubMed] [Google Scholar]
- 24.Deprez C., Lloubes R., Gavioli M., Marion D., Guerlesquin F., Blanchard L. Solution structure of the E.coli TolA C-terminal domain reveals conformational changes upon binding to the phage g3p N-terminal domain. J. Mol. Biol. 2005;346:1047–1057. doi: 10.1016/j.jmb.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 25.Sasin J.M., Bujnicki J.M. COLORADO3D, a web server for the visual analysis of protein structures. Nucleic Acids Res. 2004;32:W586–W589. doi: 10.1093/nar/gkh440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merritt E.A., Bacon D.J. Raster3D photorealistic molecular graphics. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]