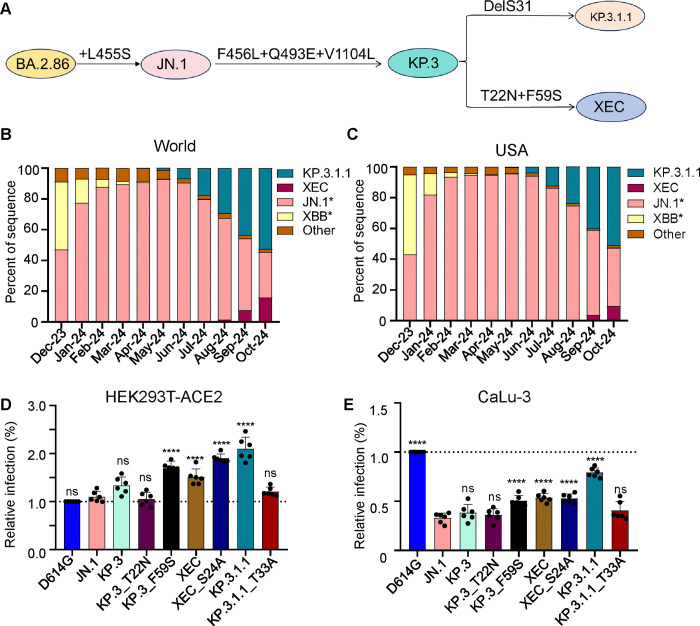
Figure 1: Mutations, circulation, and infectivity for JN.1-lineage variants XEC, KP.3.1.1 and KP.3.
(A) Schematic depiction of spike-defining mutations and relationships of JN.1, KP.3, KP.3.1.1, and XEC. (B-C) Frequency of sequences of KP.3.1.1, XEC, JN.1, and XBB worldwide (B) and the United States (C) represented by percentage. (D-E) Relative infectivity of lentivirus pseudotypes bearing spikes of interest as determined by secreted Gaussia luciferase, with D614G set to 1.0 for comparison, in HEK293T cells expressing human ACE2 (D) and CaLu-3 cells (E). Bars represent means with standard deviation of 3 biological replicate and 6 separate luciferase readings. Significance was determined by repeated measures one-way ANOVA in comparison to JN.1 and represented as ns p > 0.05 and ****p < 0.0001.