Abstract
I-Mutant2.0 is a support vector machine (SVM)-based tool for the automatic prediction of protein stability changes upon single point mutations. I-Mutant2.0 predictions are performed starting either from the protein structure or, more importantly, from the protein sequence. This latter task, to the best of our knowledge, is exploited for the first time. The method was trained and tested on a data set derived from ProTherm, which is presently the most comprehensive available database of thermodynamic experimental data of free energy changes of protein stability upon mutation under different conditions. I-Mutant2.0 can be used both as a classifier for predicting the sign of the protein stability change upon mutation and as a regression estimator for predicting the related ΔΔG values. Acting as a classifier, I-Mutant2.0 correctly predicts (with a cross-validation procedure) 80% or 77% of the data set, depending on the usage of structural or sequence information, respectively. When predicting ΔΔG values associated with mutations, the correlation of predicted with expected/experimental values is 0.71 (with a standard error of 1.30 kcal/mol) and 0.62 (with a standard error of 1.45 kcal/mol) when structural or sequence information are respectively adopted. Our web interface allows the selection of a predictive mode that depends on the availability of the protein structure and/or sequence. In this latter case, the web server requires only pasting of a protein sequence in a raw format. We therefore introduce I-Mutant2.0 as a unique and valuable helper for protein design, even when the protein structure is not yet known with atomic resolution. Availability: http://gpcr.biocomp.unibo.it/cgi/predictors/I-Mutant2.0/I-Mutant2.0.cgi.
INTRODUCTION
When engineering proteins, an important problem to be considered is to which extent a mutation will affect the stability of the new protein with respect to the wild type. Different methods have been implemented in order to address this task. They are mainly based on the development of different energy functions, suited to compute the stability free energy changes (1–5).
Recently, an approach based on a neural network system was described (6). In this application, instead of directly estimating the relative stability changes upon protein mutation [the ΔΔG value, (1–5)], a neural network predicts the direction towards which the mutation shifts the stability of the protein (namely the ΔΔG sign).
All the methods developed so far, are however limited in that prediction can be carried out only when the protein 3D structure is available in the PDB database. In the post-genomic era, however, mutagenesis experiments may start from the proteome. Therefore, the development of predictors that help to design mutated proteins from the only sequence is urgent.
Here, we present a new server I-Mutant (2.0). I-Mutant2.0 can predict the stability change of the mutated protein structure, and, for the first time, it can predict to which extent a mutation in a protein sequence will or will not affect the stability of the folded protein. I-Mutant2.0 is based on support vector machines (SVMs) and it has been trained to predict both the direction (the ΔΔG sign) of the protein stability changes and the ΔΔG associated values. Thanks to the availability of a large database of thermodynamic data of mutated proteins (7), we show that for the specific task of predicting the ΔΔG sign, I-Mutant2.0 correctly predicts (with a cross-validation procedure) 80 or 77% of the data set, depending on the input of structural or sequence information, respectively. When predicting ΔΔG values associated with the mutation, the correlation of predicted with expected values, as taken from the experimental database, is 0.71 and 0.62, depending on the structure- and sequence-base prediction, respectively.
DESCRIPTION
I-Mutant2.0 was trained to accomplish four different tasks:
Prediction of the direction of the protein stability changes upon single point mutation from the protein tertiary structure (a classification task);
Prediction of the ΔΔG value of the protein stability changes upon single point mutation from the protein tertiary structure (a function approximation task);
Prediction of the direction of the protein stability changes upon single point mutation only from the protein sequence (a classification task);
Prediction of the ΔΔG value of the protein stability changes upon single point mutation only from the protein sequence (a function approximation task).
For each task, I-Mutant2.0 is based on SVMs. We tested several kernels and we found that the most convenient for the problems at hand is the one based on Radial Basis Functions . The results described here are therefore relative only to the RBF kernel.
For the classification task and for assigning the ΔΔG values, we basically adopt the same input code by identifying two labels: one represents the increased protein stability (ΔΔG > 0, label is +), the other is associated with the destabilizing mutation (ΔΔG < 0, label is −). The input vector consists of 42 values. The first two input values account, respectively, for the temperature and the pH at which the stability of the mutated protein was experimentally determined. The next 20 values (for 20 residue types) explicitly define the mutation (we set −1 to the element corresponding to the deleted residue and 1 to the new residue, all the remaining elements are kept equal to 0). Finally, the last 20 input values encode the residue environment that is a ‘spatial environment’ when the protein structure is available or the nearest sequence neighbors, when only the protein sequence is available. When the protein structure is known (and the prediction is performed on the protein structure) each of the 20 values is the number of the encoded residue type, to be found inside a sphere of a 0.9 nm radius and centered on the coordinates of the residue that undergoes mutation. Alternatively, when the prediction is performed starting from the protein sequence, each of the 20 input values is again the number of the encoded residue type, to be found inside a symmetrical window, centered at the mutated residue, that spans the sequence towards the left (N-terminus) and the right (C-terminus) for a total length of 19 residues.
The RSA value (Relative Solvent Accessible Area) can be calculated with the DSSP program (8) only when prediction is structure based, dividing the accessible surface area value of the mutated residue by the free residue surface. In this case, a further input value (for a total sum of 43 numbers) includes the relative solvent accessible area of the mutated residue only when the protein structure is considered. This strategy is similar to what was previously done with a neural network-based predictor for predicting stability changes upon mutation starting from the protein structure (6).
The RI value (Reliability Index) can be computed only when the sign of the stability change is predicted and is evaluated from the output of the SVM O as RI = 20*abs(O − 0.5).
I-Mutant2.0 accuracy
I-Mutant2.0 was trained and tested with a cross-validation procedure on a data set derived from the current release (December 2004) of the Thermodynamic Database for Proteins and Mutants [ProTherm, (7)]. The data set of proteins was extracted from ProTherm with the following constraints:
Only single point mutations were considered for each protein (no multiple mutations were taken into account).
The correspondent free energy change of protein stability (ΔΔG value) had been experimentally detected and experimental conditions (temperature and pH) were also listed in the database.
After this filtering procedure, our data set comprised 2087 different single mutations in 65 different proteins, 58 out of which were also known with atomic resolution. For each mutation, the corresponding free energy change was collected. The subset of structures known with atomic resolution (with a total of 1948 different single mutations) contains proteins uniformly distributed in the four predominant structural classes according to the SCOP classification (scop.mrc-lmb.cam.ac.uk). The final sets are available at http://gpcr.biocomp.unibo.it/~emidio/I-Mutant2.0/dbMut.html.
In Table 1, we list the accuracy obtained when I-Mutant2.0 is adopted as a classifier, and discriminates whether a given mutation increases (label +) or decreases (label −) the protein stability. The accuracy of the structure-based prediction is 0.80 and it is higher than that obtained when the prediction is sequence based. A similar value was previously obtained with a neural network-based predictor, trained/tested on a data set smaller than that adopted in this study (6). However, a remarkable result of this paper is that the overall accuracy is 0.77 even when the predictor is sequence based.
Table 1.
I-Mutant2.0 cross-validation accuracy
| I-Mutant2.0 input | Q2 | P(+) | Q(+) | P(−) | Q(−) | C |
|---|---|---|---|---|---|---|
| PDB | 0.80 | 0.73 | 0.56 | 0.83 | 0.91 | 0.51 |
| Sequence | 0.77 | 0.69 | 0.46 | 0.79 | 0.91 | 0.42 |
Q2 = Number of correct predictions/number of examples.
P(s) = Number of correct prediction for class s/all prediction made for s.
Q(s) = Number of correct prediction for class s/observed in class s.
C = Matthews's correlation coefficient [compare with (6)].
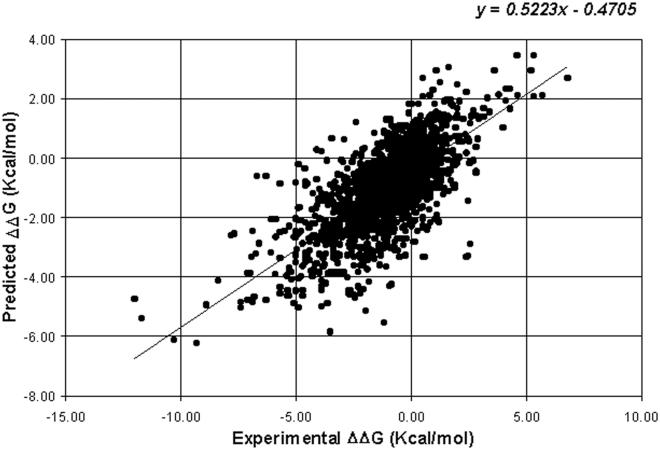
I-Mutant2.0 was also trained/tested to predict the value of the free energy stability change upon single point mutation, starting from the protein structure or sequence. In this case, the accuracy was evaluated by measuring the correlation between the predicted (adopting a cross-validation procedure) and the observed ΔΔG values. The correlation of the predicted and experimental data is 0.71, with a standard error of 1.3 kcal/mol, when the method is structure based (Figure 1). When it is sequence based, the correlation between the observed and the predicted data is 0.62 with a standard error of 1.45 kcal/mol.
Figure 1.
Correlation plot between the experimentally observed and the predicted ΔΔG values when the SVM method is structure based. The correlation is 0.71 and the corresponding standard error is 1.3 kcal/mol.
As a final observation, it must be considered that 89% of the proteins are contributing to the training/testing set adopted to implement both the sequence- and structure-based SVM method. With this in mind, the small scoring difference in accuracy that I-Mutant2.0 achieves starting from the protein structure or sequence (0.80 versus 0.77) suggests that to a large extent protein stability can also be correctly evaluated when specific interactions within the sequence neighbors are captured.
SERVER DETAILS
Inputs
On the I-Mutant2.0 web page, two alternatives are available: predicting the protein stability changes upon single point mutation starting from the protein structure or sequence (see Figure 2). In the first case, the PDB code of the protein is required; in the latter, the protein sequence needs to be pasted in the appropriate box. As discussed above and listed in Table 1, the accuracy is higher when the prediction is structure based. Nevertheless, even when it is sequence based the score is remarkable. Following the first option, two choices are available: (i) I-Mutant2.0 acts as a classifier, and the sign of the free energy stability change upon mutation is predicted; (ii) I-Mutant2.0 acts as a regression estimator and the value of free energy stability change (ΔΔG) upon mutation is predicted. In both cases, I-Mutant2.0 allows the selection of the prediction of protein stability changes at different ranges of temperature and pH (see Figure 2).
Figure 2.
Snapshot of the I-Mutant2.0 input pages for the prediction of the protein stability change upon mutation with the protein structure through its PDB code (top) or starting from the sequence (bottom).
Outputs
Four different outputs can be retrieved, depending on the selected mode. The user can start from the protein structure or sequence. In either case, the prediction of the value of free energy change or only its sign can be obtained. In all cases, a table of 19 rows (the 19 residues that are different from the one present in the sequence at a selected position) is returned. A further option, when activated, allows to highlight only the row corresponding to a given mutation.
The four different possible predictive options correspond to a different number of columns returned in the output table. The common number is six columns, listing respectively: the sequence position under consideration, the original residue name (one letter code), the mutated residue (one letter code), the predicted free energy change value (DDG) or the sign of the prediction (Increase/Decrease), the temperature and the pH at which the prediction has been carried out.
One more column may be present in the output table that lists in turn the reliability index value of the prediction (RI) or the solvent accessible surface area [RSA, computed with the DSSP program (8)]. This occurs when the sign of the stability change is predicted starting from the protein sequence or the DDG value of the free energy change is predicted starting from the protein structure, respectively.
Both RSA and RI are computed and present in the output table only when the sign of the protein stability change is predicted starting from the structure. In this case, the number of columns in the output table is 8.
From the above description, it is evident that both the structure-based and the sequence-based predictions of the sign of the protein stability change upon mutation are those endowed with the reliability scoring index (RI), and this allows the sorting out of the subset of more reliable predictions [see (6)].
When the DDG values are predicted from the protein structure or sequence, an estimate of the standard error can be evaluated from the linear regression between the predicted and the expected values and can be associated to the predicted value. Results obtained for the structure-based case are shown in Figure 1. With this procedure, the standard error value is 1.30 and 1.45 kcal/mol, when the prediction is structure or sequence based, respectively.
Acknowledgments
This work was supported by the following grants: ‘Hydrolases from Thermophiles: Structure, Function and Homologous and Heterologous Expression’ of the Ministero della Istruzione dell'Universita' e della Ricerca (MIUR) and a PNR 2001–2003 (FIRB art.8) project on Bioinformatics for Genomics and Proteomics, both delivered to R.C. E.C. is supported by a grant of the European Union's VI Framework Programme to the Bologna Node of the Biosapiens Network of Excellence project. P.F. acknowledges an MIUR grant on Proteases. Funding to pay the Open Access publication charges for this article was provided by local funding of the University of Bologna (ex 60%) delivered to R.C.
Conflict of interest statement. None declared.
REFERENCES
- 1.Casadio R., Compiani M., Fariselli P., Vivarelli F. Predicting free energy contributions to the conformational stability of folded proteins from the residue sequence with radial basis function networks. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1995;3:81–88. [PubMed] [Google Scholar]
- 2.Gilis D., Rooman M. Predicting protein stability changes upon mutation using database-derived potentials: solvent accessibility determines the importance of local versus non-local interactions along the sequence. J. Mol. Biol. 1997;272:276–290. doi: 10.1006/jmbi.1997.1237. [DOI] [PubMed] [Google Scholar]
- 3.Guerois R., Nielsen J.E., Serrano L. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J. Mol. Biol. 2002;320:369–387. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- 4.Khatun J., Khare S.D., Dokholyan N.V. Can contact potentials reliably predict stability of proteins? J. Mol. Biol. 2004;336:1223–1238. doi: 10.1016/j.jmb.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 5.Zhou H., Zhou Y. Distance-scaled, finite ideal-gas reference state improves structure-derived potentials of mean force for structure selection and stability prediction. Protein Sci. 2002;11:2714–2726. doi: 10.1110/ps.0217002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Capriotti E., Fariselli P., Casadio R. A neural network-based method for predicting protein stability changes upon single point mutations. Bioinformatics. 2004;20:I63–I68. doi: 10.1093/bioinformatics/bth928. [DOI] [PubMed] [Google Scholar]
- 7.Bava K.A., Gromiha M.M., Uedaira H., Kitajima K., Sarai A. ProTherm, version 4.0: thermodynamic database for proteins and mutants. Nucleic Acids Res. 2004;32:D120–D121. doi: 10.1093/nar/gkh082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]