Abstract
We describe a computer program, Assembly PCR Oligo Maker, created to automate the design of oligodeoxynucleotides for the PCR-based construction of long DNA molecules. This program is freely available at http://publish.yorku.ca/~pjohnson/AssemblyPCRoligomaker.html and has been specifically designed to aid in the construction of DNA molecules that are to be used for the production of RNA molecules by in vitro synthesis with T7 RNA polymerase. The input for Assembly PCR Oligo Maker is either the desired DNA sequence to be made or an RNA sequence. If RNA is the input, the program first determines the DNA sequence necessary to produce the desired RNA molecule. The program then determines the sequences of all the oligodeoxynucleotides necessary for a two-step assembly PCR-based synthesis of the desired DNA molecule. The oligodeoxynucleotide sequences outputted are designed to have a uniform melt temperature and are checked for regions of overlap outside of the desired priming regions necessary for the PCR reaction. The validity of the program was verified experimentally by synthesizing a 191-nt long DNA molecule using the DNA sequences suggested by the program.
INTRODUCTION
The construction of long DNA oligonucleotides, such as synthetic genes, is useful for a number of reasons. For protein expression, it is possible to improve the yield of a desired protein by optimizing codon usage to reflect the relative codon abundance in the host organism used for protein production. Long oligodeoxynucleotides can also be used to produce large amounts of RNA for structural or biochemical studies by in vitro transcription methods (1). Many RNA molecules studied to-date are under 100 nt in length and the DNA oligonucleotides needed for their production are readily synthesized and are commercially available (up to 100–150 nt long). As larger RNA molecules are studied, or if methods such as the use of cis-3′ and/or 5′ ribozymes to facilitate RNA production (2–4) or if RNA-based affinity tags to aid in RNA purification (5) are used, it is not possible to purchase DNA oligonucleotides of sufficient length to encode the desired RNA product. To produce these long DNA molecules (over 120–150 nt) it is necessary to use techniques originally developed for gene synthesis, such as assembly PCR.
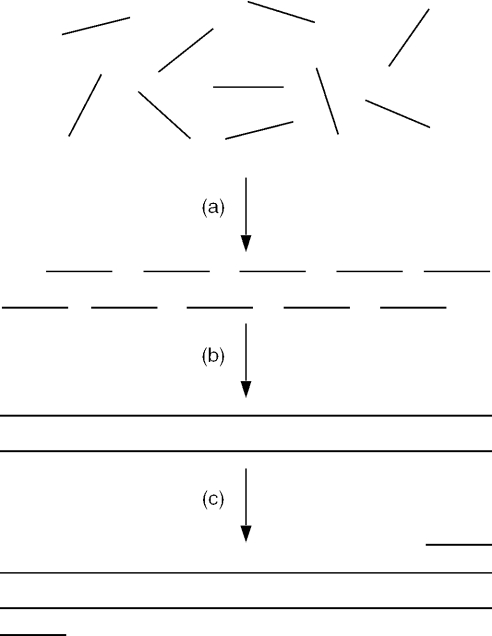
PCR is a commonly used method to amplify DNA sequences. PCR methods can be used to construct a synthetic gene through the use of a two-step PCR method referred to as assembly PCR (Figure 1) (6,7). In the first step of assembly PCR, multiple oligodeoxynucleotides that contain overlapping regions anneal, and the DNA polymerase extends the primers and fills in the regions between the primers. A range of products of different lengths results from the different possible combinations of annealing that involve less than all the oligodeoxynucleotides. In the second PCR step a pair of primers is introduced that is specific for the full-length oligodeoxynucleotide and the full-length product is selectively amplified from the mixture.
Figure 1.
The assembly PCR method for constructing long DNA molecules. (a) In the first PCR step a pool of oligodeoxynucleotides anneal and are (b) elongated to produce a full-length DNA molecule. In addition to the full-length product, a host of shorter molecules also results. (c) In the second PCR step the desired full-length molecule is selectively amplified from the mixture using primers specific for the desired full-length product.
MATERIALS AND METHODS
The code for Assembly PCR Oligo Maker is written in Java version 1.4.1 available from http://java.sun.com. Assembly PCR Oligo Maker is freely accessible on the web at http://publish.yorku.ca/~pjohnson/AssemblyPCRoligomaker.html. We have attempted to make this program compatible with most browsers, however, if there is any difficulty running the program we recommend downloading the latest Java Runtime Environment available from Sun Microsystems. The program was tested on the Windows (98, 2000, XP), Linux (Fedora Core 3), SGI (IRIX 6.5) and Mac (OSX 10.3) operating systems and is functional using virtually any web browser. The default method employed by the program for calculating DNA melting temperatures uses the nearest neighbor method employing the unified DNA–DNA hybridization thermodynamic parameters of Allawi and SantaLucia (8) with a monovalent cation correction to the entropy term as outlined by SantaLucia (9). The default sodium ion (monovalent cation) concentration is 50 mM, while the default DNA concentration is 0.5 μM. Alternately, the user can choose to calculate melt temperatures using the method of Wallace et al. (10) where melting temperature (Tm) = 2(A + T) + 4(G + C).
All oligodeoxynucleotides were purchased from Sigma-Aldrich Canada Ltd. Vent DNA polymerase was purchased from New England Biolabs (NEB). Upon receipt, the oligodeoxynucleotides for the first step of assembly PCR were diluted to 7 μM (0.125 μg/μl) with double distilled water, while the oligodeoxynucleotides for the second PCR step were diluted to 42 μM (0.25 μg/μl). For the first PCR reaction 4 μl of each oligo, 4 μl of 5 mM dNTPs, 10 μl of 10× thermopol buffer (NEB), 1.5 μl of Vent DNA polymerase (2000 U/ml) and 68.5 μl of double distilled water were combined. This mixture was then subjected to 8 cycles of amplification at 94°C (1.5 min), 54°C (2 min) and 72°C (3 min). During the first cycle the 94°C step was performed for 7 min. After the last cycle was completed an additional 5 min 72°C elongation step was performed. For the second PCR reaction, 1 μl of the crude mixture from the first PCR reaction was mixed with 4 μl of each primer, 4 μl of 5 mM dNTPs, 10 μl of 10× thermopol buffer (NEB), 1.5 μl of Vent DNA polymerase and 75.5 μl of double distilled water. This mixture was then subjected to 25 cycles of amplification. Each cycle consisted of a 30 s 94°C step, a 2 min 54°C step and a 1.5 min 72°C step. Before the first cycle a 5 min 94°C step was used. A 5 min 72°C elongation step was included following the final cycle. The PCR mixtures were analyzed by agarose gel electrophoresis.
The PCR amplified DNA and a sample of the pUC18 vector were then digested with the restriction enzymes HindIII and EcoRI (NEB). The PCR amplified DNA was then ligated into pUC18 using T4 DNA ligase (NEB) and transformed into DH5α Escherichia coli cells. DNA sequencing was performed in the Molecular Biology Core Facility at York University.
RESULTS
The Assembly PCR Oligo Maker program is designed to be flexible and easy to use. The user can input either an RNA sequence or a DNA sequence and can alter a number of settings used by the program during the determination of the oligodeoxynucleotide sequences. The user can input the target oligonucleotide sequence in either the 5′ to 3′ or 3′ to 5′ directions. For either RNA or DNA input, the user is queried as to whether the top17 sequence (1) for T7 RNA polymerase priming should be added or not. We will note that for highest RNA production the sequence following the T7 promoter should code for two or three guanosine nucleotides (1). If RNA is used as input, the corresponding DNA sequence to be produced is determined and provided as output. The simplest use of the program therefore is to determine the DNA sequence needed to produce a particular RNA molecule. At this point, the target DNA sequence to be produced is known and the program computes the necessary oligodeoxynucleotides for both steps of the two-step assembly PCR synthesis method. By default, the program only determines assembly oligodeoxynucleotide sequences for DNA targets >40 nt.
The program works by dividing the target DNA into an even number of segments with a user-defined maximum length. The user can define the maximum length of the calculated oligodeoxynucleotides to be 40, 50, 60 or 70 nt long. Flanking regions are added to these oligodeoxynucleotide sequences that overlap with a different oligodeoxynucleotide. The length of overlap between the different oligodeoxynucleotides is optimized so that the overlapping region matches a target Tm. This target Tm is constant for all of the overlapping regions. The target Tm value employed is a user defined variable with choices available between 50 and 60°C, the default value is 55°C. A consistent Tm value among the different oligodeoxynucleotides helps contributes to successful PCR reactions.
A key feature to ensure the success of assembly PCR is the necessity that the oligodeoxynucleotides only anneal where desired. If an oligodeoxynucleotides can anneal in multiple ways, in either an intramolecular or intermolecular fashion, the subsequent PCR reactions would produce a host of undesired products. The Assembly PCR Oligo Maker program tests the oligodeoxynucleotides calculated for both PCR reactions for regions of overlap outside of the desired location. If a stretch of nucleotides is found to overlap at a region outside of the desired site, the program calculates a Tm value for this region of overlap. If the Tm value of the overlapping region detected is greater than a user defined Tm threshold a warning appears notifying the user that an undesired overlapping region was detected. The region is also flagged in the results window. The user can then make changes to the sequence of the desired PCR product or ignore this warning and proceed to attempt the PCR reaction despite the presence of an unwanted overlap region. Alternately, upon detection of an undesired region of overlap the user can choose to calculate oligodeoxynucleotides with a smaller or greater maximum length in order to try and break up the region of unwanted overlap. This maximum oligodeoxynucleotide length option is controlled in the ‘customize’ window found under the ‘File’ menu. The default Tm value for triggering a warning for the presence of an undesired overlap is 40°C and is user variable between 30 and 45°C.
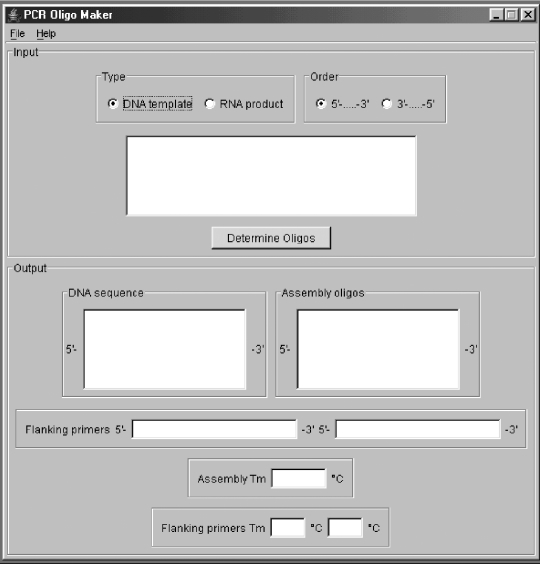
The output from the program is available in the main window (Figure 2) from which the user can cut and paste text in order to save the output. The output consists of all the necessary oligodeoxynucleotides to perform both of the PCR reactions. The calculated melt temperatures for the oligodeoxynucleotides are also reported. Alternately, the user can view a summary of the input and output in a single page available from ‘Show all results’ option under the ‘File’ menu. It is also possible to cut and paste text from this page to a file. Also present in the results window is a view of how the different oligodeoxynucleotides in the assembly PCR reaction align with each other.
Figure 2.
Interface of the Assembly PCR Oligo Maker program. The user enters the target sequence in the top window, presses the ‘Determine Oligos’ button and the output appears in the boxes below. The oligodeoxynucleotide sequences for the first PCR step are found in the ‘Assembly Oligo’ box, and the two oligodeoxynucleotide sequences for the second PCR step are found in the two ‘Flanking Primers’ boxes. User controlled settings and information about the program are accessible under the File and Help menus.
The output of the program was verified both in a theoretical and experimental fashion. Initially, a DNA oligonucleotide target was used as input, and the oligodeoxynucleotides determined by the program were analyzed to check that the correct DNA product would result from the two-step PCR DNA synthesis method. Next, the ability of the program to find regions of overlap outside of the desired region was confirmed. A target DNA oligonucleotide containing a repeat region was used as input. Depending on where the repeat region was in relation to where the target DNA product was divided, the program was able to detect it, and the calculated Tm of the unwanted overlap region was reported.
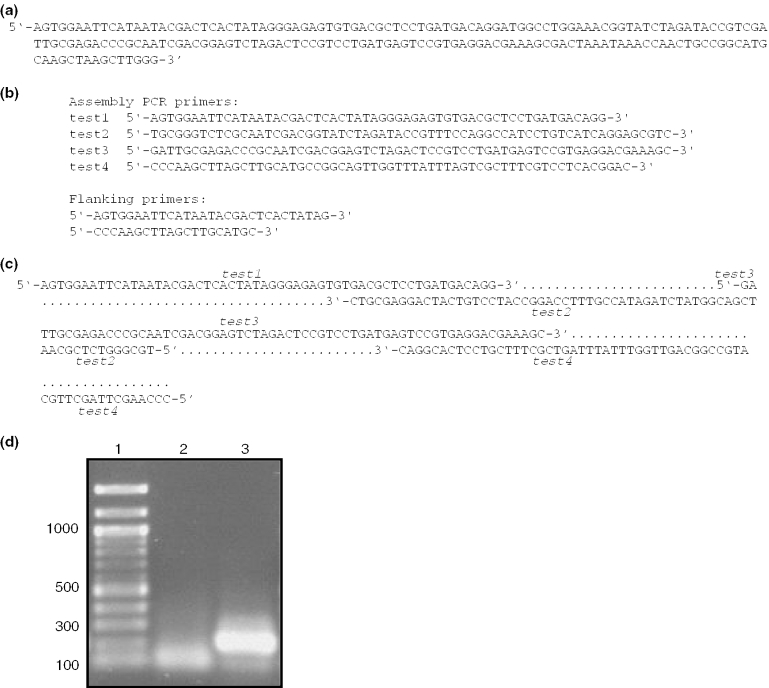
Finally, the program was experimentally verified by using the oligodeoxynucleotides determined by the program for the two-step assembly PCR construction of a DNA molecule that is to be used to produce an RNA molecule. The desired RNA product is a 191-nt molecule consisting of 5′ and 3′ cis hammerhead ribzoymes and a core 20-nt region that forms a hairpin structure and is having its structure studied in our lab by nuclear magnetic resonance methods. When using the default value of 70 for the maximum oligonucleotide length, the program broke this 191-nt DNA molecule into four segments for the first PCR reaction and produced the two oligodeoxynucleotide molecules for the second PCR reaction (Figure 3b). As shown by gel analyses (Figure 3d) the first PCR reaction produces a diffuse band or smear, while the desired full length product results from the second PCR reaction. This behavior is consistent with previous reports of assembly PCR gene construction (6,11,12). The product of the second PCR reaction was cloned into the pUC18 plasmid and the correctness of its sequence was verified by DNA sequencing.
Figure 3.
The construction of a 191-nt DNA molecule using the oligodeoxynucleotides determined by the Assembly PCR Oligo Maker program. (a) The sequence of the 191-nt DNA target to be produced. (b) The DNA sequences reported by Assembly PCR Oligo Maker program. Sequences for both steps of the assembly PCR reaction are reported. (c) Diagram showing how the four oligodeoxynucleotides anneal to produce the full-length dsDNA product (d) Agarose gel showing the results of the first (lane 2) and second (lane 3) PCR steps. The desired 191-nt molecule is visible after the second PCR step. DNA length markers are shown in lane 1.
DISCUSSION
The program we present here for determining the oligodeoxynucleotides needed for the assembly PCR construction of artificial genes has several advantages over similar applications that are currently available. These advantages are in addition to the ability of Assembly PCR Oligo Maker to handle RNA as an input and add the top17 sequence to determine the DNA sequence needed for RNA production. While this is a trivial process, the use of Assembly PCR Oligo Maker does remove the human element from this task that can inadvertently introduce sequence errors into the final product.
The other programs available that aid in oligodeoxynucleotide design for the assembly PCR construction of long DNA molecules include DNA Works (11), Gene2Oligo (12) and DNABuilder (http://cbi.swmed.edu/computation/cbu/DNABuilder.html). The major advantage of using Assembly PCR Oligo Maker is that the user has the option of determining the maximum length of oligodeoxynucleotides for assembly PCR that are created by the program. This option is not available in the other programs. The user thus has the option of choosing to create fewer but longer oligodeoxynucleotides than determined by other programs. Working with fewer oligodeoxynucleotides means that the process of dissolving and diluting them and setting up the assembly PCR reaction is simplified and that fewer opportunities for human error are available during the experiment. This function is shown in the example in Figure 3a where the 191-nt DNA target is broken into four oligodeoxynucleotides of length 56, 64, 64 and 59 nt for a total of 243 nt. For this calculation, a maximum length of 70 (the default value) was chosen as the maximum oligonucleotide length calculated. When the same target is used as input for DNABuilder the output was 6 oligodeoxynucleotides with a total of 306 nt; DNAWorks had an output of 10 oligodeoxynucleotides comprising a total of 371 nt; and using this sequence as input Gene2Oligo produced an output of 13 oligodeoxynucleotides made up of a total of 440 nt. This is almost twice the number of nucleotides as obtained from Assembly PCR Oligo Maker.
In Assembly PCR Oligo Maker the user has the option to choose to calculate more and shorter oligodeoxynucleotides if desired. Typically, the use of shorter oligodeoxynucleotides may be desired due to concerns that the user may have about using the longer oligodeoxynucleotides in a PCR reaction without performing additional purification steps, such as PAGE purification of the longer oligodeoxynucleotides. We will note that we have had no trouble using oligodeoxynucleotides 60–65 nt in length in the assembly PCR reaction using them as provided by the manufacturer, without employing any additional purification steps.
An additional advantage of our Assembly PCR Oligo Maker program is its flexibility in handling the input sequence. Assembly PCR Oligo Maker is able to correctly handle input containing spaces in the sequence, as you would include if working with the sequence in units of three nucleotides. The program also allows input in both the 3′ to 5′ direction as well as the 5′ to 3′ direction. This flexibility is not available in all of the other programs. Our program is readily accessible on the web and can be accessed with any web browser and does not depend on the computer operating system used. This is in contrast to being a downloadable program that is platform-specific. Finally, the output of Assembly PCR Oligo Maker is very clear, it lists the oligodeoxynucleotides needed for the assembly PCR reaction and the two oligodeoxynucleotides needed for the second PCR reaction as well as provide the predicted Tm values. All the oligodeoxynucleotides in the output are listed 5′ to 3′ which is in a form that is suitable for cutting and pasting the sequences into an email or web form for ordering oligodeoxynucleotides from vendors of users' choice.
In summary, we have created a program that is freely available on the internet to aid in the design of oligodeoxynucleotides for the construction of long DNA molecules. The program can assist in constructing any DNA molecule, but has a focus on creating DNA templates for RNA production. The sequences reported by Assembly PCR Oligo Maker were experimentally verified by the successful construction of a 191-nt DNA molecule.
Acknowledgments
This work was supported by funding from the Natural Sciences and Engineering Research Council of Canada (NSERC). We would like to thank the members of the Johnson lab for useful suggestions during the design of the program. Funding to pay the Open Access publication charges for this article was provided by NSERC.
Conflict of interest statement. None declared.
REFERENCES
- 1.Milligan J.F., Groebe D.R., Witherell G.W., Uhlenbeck O.C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Price S.R., Ito N., Oubridge C., Avis J.M., Nagai K. Crystallization of RNA–protein complexes. I. Methods for the large-scale preparation of RNA suitable for crystallographic studies. J. Mol. Biol. 1995;249:398–408. doi: 10.1006/jmbi.1995.0305. [DOI] [PubMed] [Google Scholar]
- 3.Ferré-D'Amaré A.R., Doudna J.A. Use of cis- and trans-ribozymes to remove 5′ and 3′ heterogeneities from milligrams of in vitro transcribed RNA. Nucleic Acids Res. 1996;24:977–978. doi: 10.1093/nar/24.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu L., Oost T.K., Schkeryantz J.M., Yang J., Janowick D., Fesik S.W. Discovery of aminoglycoside mimetics by NMR-based screening of Escherichia coli A-site RNA. J. Am. Chem. Soc. 2003;125:4444–4450. doi: 10.1021/ja021354o. [DOI] [PubMed] [Google Scholar]
- 5.Kieft J.S., Batey R.T. A general method for rapid and nonodenaturing purification of RNAs. RNA. 2004;10:988–995. doi: 10.1261/rna.7040604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dillon P.J., Rosen C.A. A rapid method for the construction of synthetic genes using the polymerase chain reaction. Biotechniques. 1990;9:298–300. [PubMed] [Google Scholar]
- 7.Stemmer W.P.C., Crameri A., Ha K.D., Brennan T.M., Heyneker H.L. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene. 1995;164:49–53. doi: 10.1016/0378-1119(95)00511-4. [DOI] [PubMed] [Google Scholar]
- 8.Allawi H.T., SantaLucia J., Jr Thermodynamics and NMR of internal G T mismatches in DNA. Biochemistry. 1997;36:10581–10594. doi: 10.1021/bi962590c. [DOI] [PubMed] [Google Scholar]
- 9.SantaLucia J., Jr A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl Acad. Sci. USA. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wallace R.B., Shaffer J., Murphy R.F., Bonner J., Hirose T., Itakura K. Hybridization of synthetic oligodeoxyribonucleotides to phi chi 174 DNA: the effect of single base pair mismatch. Nucleic Acids Res. 1979;6:3543–3557. doi: 10.1093/nar/6.11.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoover D.M., Lubkowski J. DNAWorks: an automated method for designing oligonucleotides for PCR-based gene synthesis. Nucleic Acids Res. 2002;30:e43. doi: 10.1093/nar/30.10.e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rouillard J.-M., Lee W., Truan G., Gao X., Zhou X., Gulari E. Gene2Oligo: oligonucleotide design for in vitro gene synthesis. Nucleic Acids Res. 2004;32:W176–W180. doi: 10.1093/nar/gkh401. [DOI] [PMC free article] [PubMed] [Google Scholar]